ABSTRACT
The centrosome-associated proteins Ninein (Nin) and Ninein-like protein (Nlp) play significant roles in microtubule stability, nucleation and anchoring at the centrosome in mammalian cells. Here, we investigate Blastoderm specific gene 25D (Bsg25D), which encodes the only Drosophila protein that is closely related to Nin and Nlp. In early embryos, we find that Bsg25D mRNA and Bsg25D protein are closely associated with centrosomes and astral microtubules. We show that sequences within the coding region and 3′UTR of Bsg25D mRNAs are important for proper localization of this transcript in oogenesis and embryogenesis. Ectopic expression of eGFP-Bsg25D from an unlocalized mRNA disrupts microtubule polarity in mid-oogenesis and compromises the distribution of the axis polarity determinant Gurken. Using total internal reflection fluorescence microscopy, we show that an N-terminal fragment of Bsg25D can bind microtubules in vitro and can move along them, predominantly toward minus-ends. While flies homozygous for a Bsg25D null mutation are viable and fertile, 70% of embryos lacking maternal and zygotic Bsg25D do not hatch and exhibit chromosome segregation defects, as well as detachment of centrosomes from mitotic spindles. We conclude that Bsg25D is a centrosomal protein that, while dispensable for viability, nevertheless helps ensure the integrity of mitotic divisions in Drosophila.
KEY WORDS: RNA localization, Centrosome, Dynein, Pole plasm
Summary: In humans, mutations in Ninein or Ninein-like protein result in microcephaly and other severe diseases. We show that while flies lacking the Ninein orthologue can survive, many die as embryos with defects in mitosis.
INTRODUCTION
Establishment of embryonic patterning in Drosophila melanogaster requires localized translation of numerous maternally deposited mRNAs in specific regions of the embryo during the initial nuclear divisions in the syncytial stage of embryogenesis (Lasko, 2012). Drosophila primordial germ cells, often called pole cells, are specified by localized posterior determinants, many of which are translated from mRNAs that accumulate in the posterior pole plasm of the oocyte and early embryo. At least 11 mRNAs known to be involved in pole cell development and/or embryonic patterning transiently accumulate in a perinuclear pattern around the pole cell nuclei during nuclear division 9 in embryogenesis, namely germ cell-less (Jongens et al., 1992), polar granule component (Hanyu-Nakamura et al., 2008), nanos (nos) (Wang and Lehmann, 1991), spire (Dahlgaard et al., 2007), Tao (Sato et al., 2007), arrest (Parisi et al., 2001), exuperantia (Winslow et al., 1988), oo18 RNA-Binding Protein (Lantz et al., 1994), tramtrack (Read et al., 1992), cyclin B (Kadyrova et al., 2007), and pumilio (Asaoka-Taguchi et al., 1999; Lécuyer et al., 2007). One of these mRNAs, nos, is first anchored to the posterior actin cytoskeleton, and then transported to the migrating posterior nuclei by the motor protein Dynein along astral microtubules (Lerit and Gavis, 2011). Vasa protein (Vas), a DEAD-box helicase essential for germ cell specification, localizes in the same pattern (Lerit and Gavis, 2011). It is assumed that other mRNAs with the same distribution pattern localize through a similar mechanism, although this has not been directly investigated.
In the early Drosophila embryo the first ten rounds of nuclear divisions are synchronous and are not coupled to cytokinesis (Foe and Alberts, 1983). During this period, nuclei migrate toward the periphery of the embryo. The next three rounds of nuclear division remain synchronous and uncoupled to cell divisions, except at the posterior pole of the embryo, where nuclei migrate through the germ plasm, slow their divisions and become incorporated within pole cells, the first distinctive cells to form in the embryo. Centrosomes that associate with the nuclei that migrate to the posterior trigger the release of germ plasm components, such as nos and Vas, from the embryonic posterior cortex, enabling Dynein-dependent transport into the pole cells as they form (Lerit and Gavis, 2011). This suggests that germ cell specification might be particularly sensitive to the activities of centrosome-associated proteins. Consistent with this, a role for Neurl4, a centrosome-associated protein, in germ cell specification and integrity has recently been revealed (Jones and Macdonald, 2015).
A centrosome typically consists of a pair of centrioles surrounded by pericentriolar material, and it is from this structure that spindle and astral microtubules emanate (Azimzadeh and Bornens, 2007). Centrioles contain nine triplets of microtubules with proximal and distal ends (Vorobjev and Nadezhdina, 1987). The pericentriolar material is a highly organized structure (Fu and Glover, 2012; Lawo et al., 2012). Stringent control of the centrosome and centrioles is vital since abnormalities in spindle pole function can lead to genomic instability. Centrosomes in syncytial Drosophila embryos differ in composition from those of other animals; they are considered immature because they are shorter and have no clear difference between their proximal and distal ends (Gonzalez et al., 1998). In addition, centriole duplication occurs after centrosome separation (Callaini and Riparbelli, 1990). This is unlike in mammalian cells where duplication takes place prior to centrosome division (Callaini et al., 1997). Despite these differences, many proteins involved in centrosomal structure and function are conserved between mammals and flies (Woodruff et al., 2014).
In the context of the link between centrosomes and targeting of pole plasm components to the presumptive pole cells, we decided to investigate Blastoderm specific gene 25D (Bsg25D), because it produces an mRNA that localizes to the posterior pole in a similar pattern to nos and the other RNAs mentioned above, and because it encodes a protein related to mammalian Ninein (Nin) and Ninein-like Protein (Nlp), centrosomal proteins involved in microtubule organization (Casenghi et al., 2003; Stillwell et al., 2004). Bsg25D was among the first genes to be molecularly characterized in Drosophila, and was initially reported to be transcribed only during the blastoderm stage of embryogenesis (Boyer et al., 1987; Roark et al., 1985). However, recent results with more sensitive techniques have shown that, while Bsg25D is most abundantly expressed in early embryos, it is also expressed during many developmental stages (Roy et al., 2010; Wasbrough et al., 2010; BDGP in situ homepage: http://insitu.fruitfly.org/cgi-bin/ex/insitu.pl). Bsg25D mRNA is both maternally contributed during oogenesis and zygotically expressed in syncytial blastoderm stage embryos (Lécuyer et al., 2007; Tomancak et al., 2007). This transcript is localized to the posterior pole plasm in early embryogenesis and exhibits a prominent perinuclear pattern around the pole cell nuclei during nuclear division 9, similar to nos and the other mRNAs discussed above, as well as peri-centrosomal localization in the somatic region of the embryo (Iampietro et al., 2014).
To explore the cellular and developmental roles of Bsg25D, we produced loss-of-function mutants and used them to investigate its functions during oogenesis and early embryogenesis. We show that Bsg25D protein and mRNA co-localize to centrosomes and microtubules in vivo and that a purified form of Bsg25D protein can bind to microtubules in vitro. Furthermore, the localization of Bsg25D in oogenesis and embryogenesis is dictated by separable localization elements within the coding region and 3′ UTR, while mislocalization of Bsg25D in oocytes affects microtubule polarity and subsequent embryonic patterning. Finally, we find that maternal and zygotic expression of Bsg25D is important for full embryonic viability and that mutant embryos frequently exhibit mitotic divisions prior to the midblastula transition (MBT) in embryogenesis.
RESULTS
Localization of Bsg25D mRNA and protein is dynamic in oogenesis
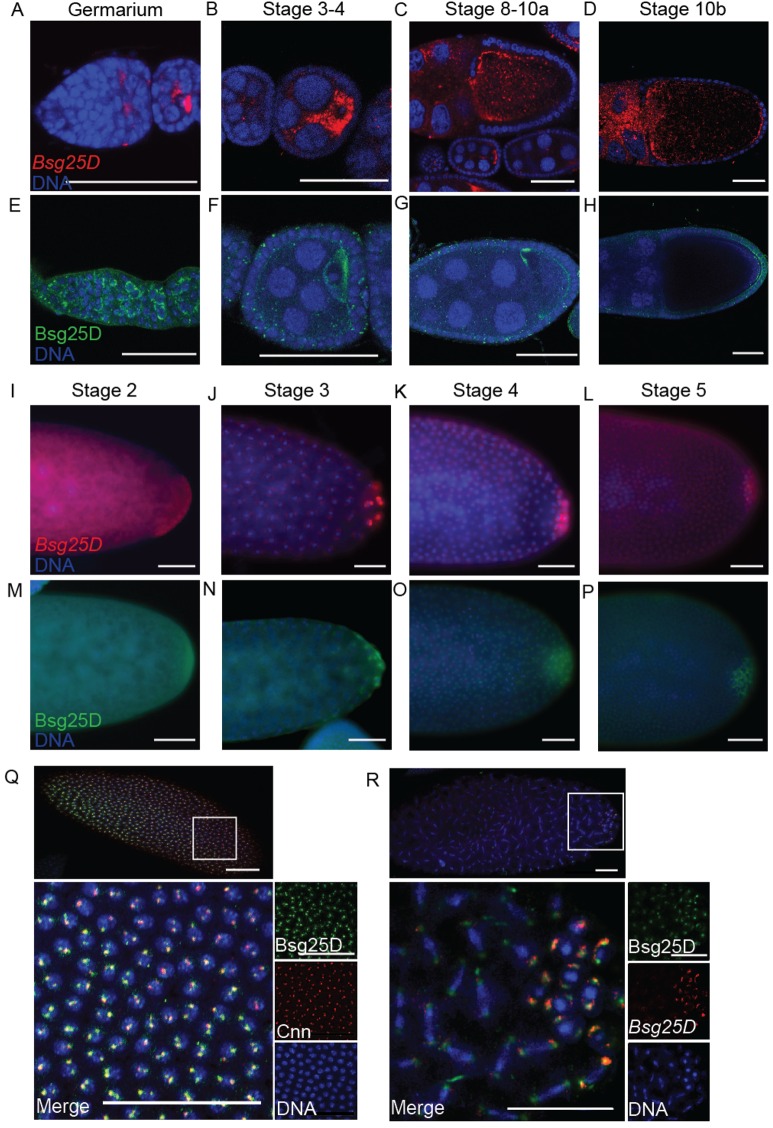
We first sought to characterize the distribution of Bsg25D mRNA and protein in oogenesis and early embryogenesis as a means of identifying possible sites where its function is required. In situ hybridization experiments indicated that Bsg25D mRNA is expressed throughout oogenesis. Like many other mRNAs that are ultimately destined for the posterior pole plasm, Bsg25D accumulates in the oocyte of stage 2-7 egg chambers. Unlike these others, however, Bsg25D mRNA becomes most concentrated at both the anterior and posterior poles of the oocyte from stage 10 onward (Fig. 1A-D). Immunohistochemical experiments using an antiserum that recognizes Bsg25D (Iampietro et al., 2014) revealed that its protein expression largely mirrored that of its mRNA until stage 10, when anterior accumulation of the protein is not as apparent as for the mRNA (Fig. 1E-H).
Fig. 1.
Localization of Bsg25D RNA and protein in oogenesis and early embryogenesis. (A-D) Bsg25D RNA (red) and (E-H) Bsg25D protein (green) share a similar localization pattern in oogenesis. (I-L) Localization of Bsg25D RNA (red) and (M-P) Bsg25D protein (green) in stage 2-5 of embryos. Photographs show only the posterior halves of embryos to emphasize the accumulation of Bsg25D mRNA and protein in the posterior pole plasm. (Q) High-magnification image showing Bsg25D (green) and Cnn (red) accumulation near the somatic nuclei (blue) of a syncytial blastoderm stage embryo. (R) High-magnification image showing the distribution of Bsg25D RNA (red) and Bsg25D protein (green) in posterior the pole cells and mitotic somatic nuclei of a syncytial blastoderm stage embryo. For all images scale bar=50 µm.
In situ hybridization experiments confirmed previous evidence indicating that Bsg25D mRNA is enriched at the posterior of the embryo in its earliest stages of development, and then accumulates in the pole buds and pole cells as they form (Fig. 1I-L). Localization of Bsg25D mRNA to the posterior pole plasm is not absolute and some is apparent in somatic regions of the embryo as well, where it accumulates in two puncta on opposite sides of each nucleus (Iampietro et al., 2014; Lécuyer et al., 2007) (Fig. 1J). Immunohistochemical staining showed that Bsg25D protein is distributed in a similar pattern (Fig. 1M-P). To determine the relationship between Bsg25D puncta and centrosomes, we carried out double labeling experiments with antisera recognizing Bsg25D and the expression of centrosomal component Centrosomin fused to GFP (Cnn-GFP). We observed that Bsg25D and Cnn-GFP foci were closely associated, and that the foci of Bsg25D staining are generally larger than those of Cnn-GFP (Fig. 1Q). In many cases the Cnn signal was completely enveloped by the Bsg25D signal. Using a similar approach we also observed colocalization between Bsg25D and γ-tubulin throughout mitosis, but not with α-tubulin (Fig. S1). These results indicate that Bsg25D is a component of Drosophila centrosomes, or associates closely with them. Remarkably, Bsg25D mRNA also co-localized with centrosomes, both in posterior pole cells and in the somatic region of the embryo (Fig. 1Q,R). The accumulation of both Bsg25D mRNA and Bsg25D protein at centrosomes suggests that Bsg25D mRNA is translated locally there.
Generation of Bsg25D mutant alleles
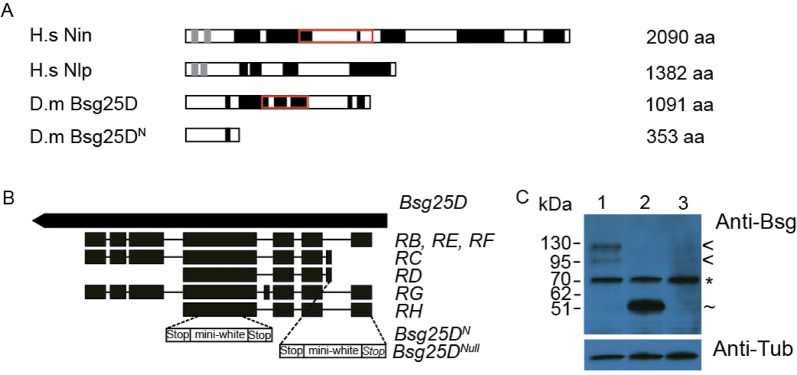
To investigate Bsg25D function, we next used the ends-out gene targeting method (Maggert et al., 2008) to produce premature-termination (Bsg25D N, first 353 amino acids) and null (Bsg25D Null) alleles of Bsg25D (Fig. 2B). Bsg25D, like Nin and Nlp, has numerous coiled-coil domains (Fig. 2A). It also contains a predicted Smc chromosome segregation ATPase domain (Marchler-Bauer et al., 2015) that is also found in Nin, but not in Nlp (Fig. 2A). Nin and Nlp also contain predicted EF-hand domain pairs in their N-terminal regions (Fig. 2A). While substantial sequence similarity between Bsg25D and these proteins is present in their N-termini, some of the conserved residues of EF-hands are absent in Bsg25D. Based on these relationships we consider Bsg25D to be a closer orthologue to Nin than to Nlp. Bsg25D N lacks the Smc domain and all but one of the coiled domains (Fig. 2A-C). However, a similar N-terminal fragment of mouse Nin was found to co-purify, in co-immunoprecipitation and pull-down experiments, with γ-tubulin containing complexes (Delgehyr et al., 2005). The Bsg25D Null allele does not detectably express Bsg25D protein at all (Fig. 2B,C).
Fig. 2.
Mutant alleles of Bsg25D generated through gene targeting. (A) Schematic of Nin, Nlp, Bsg25D and BsgN proteins. Nin isoform 1 (UniProt identifier Q8N4C6-1) and Nlp isoform 1 (UniProt identifier Q9Y2I6-1), both considered the canonical isoform, and Bsg25D-PB are shown here. EF-hand domains (light grey), coiled-coil domains (black), and the Smc domain (red box) are shown (Dinkel et al., 2016; Hong et al., 2000; Letunic et al., 2006; Sigrist et al., 2012; Wang and Zhan, 2007). (B) Schematic diagram of Bsg25D, showing the ORFs of seven different predicted Bsg25D transcripts and the sites at which the gene targeting vector pw25.5 was inserted. (C) Immunoblot of ovary lysates from Oregon-R wild-type controls (lane 1), Bsg25D N/Bsg25D N (lane 2) and Bsg25D Null/Bsg25D Null (lane 3). The bands observed correspond to Bsg25D full-length isoforms from different alternatively spliced transcripts (<), a truncated isoform corresponding to the size predicted for Bsg25DN/Bsg25DN (∼) and a non-specific band present in all lysates (*). α-tubulin was used for a loading control.
Bsg25D mRNA localization involves elements within both its coding region and its 3′ UTR
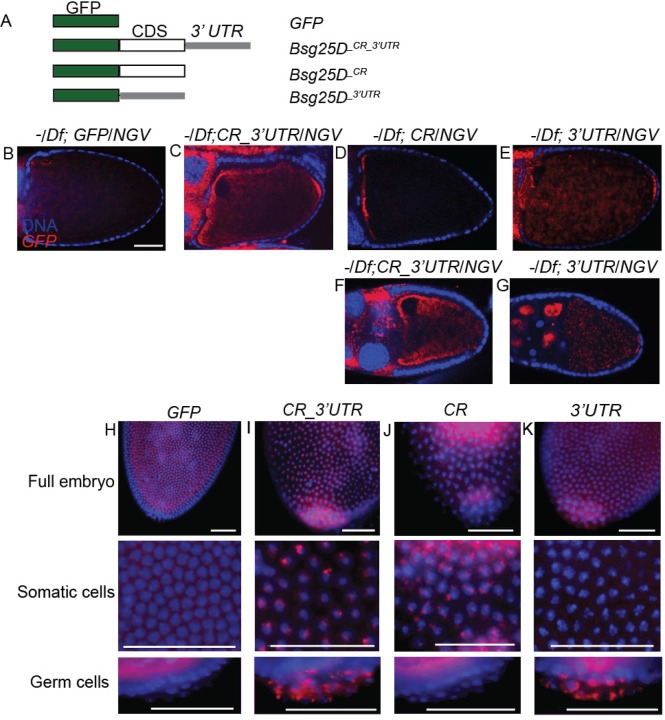
We next aimed to identify potential localization elements in Bsg25D mRNA and to investigate whether localization of the mRNA is relevant to its in vivo function, we generated a series of transgenic fly lines (Fig. 3A). The genotypes of these flies were confirmed through PCR, northern blot, and immunostaining (Fig. S2A-D). UASp-eGFP-Bsg25D _CR_3′UTR (CR_3′UTR) flies expressed the full-length coding region of Bsg25D (isoform RB) along with the 3′ UTR, UASp-eGFP-Bsg25D _CR (CR) expressed the full-length coding region but lacked the 3′ UTR, and UASp-eGFP-Bsg25D _3′UTR (3′UTR) expressed only the 3′ UTR. UASp-eGFP (GFP) flies, expressing eGFP alone, served as a negative control.
Fig. 3.
Bsg25D RNA contains localization elements within the coding region and 3′ UTR. (A) Schematic diagram of the transgenic constructs used, eGFP (green), Bsg25D-PB coding region (white), Bsg25D 3′ UTR (grey). (B-E) Distribution of transgenically-expressed mRNAs in stage 10 oocytes as shown by in situ hybridization, using a probe recognizing GFP, in the Bsg25D Null/Df(2L)6011 (−/Df) genetic background. The coding region alone promotes localization to the anterior pole, while the 3′ UTR promotes posterior localization. (F,G) Posterior localization is not apparent in early stage 10 oocytes expressing full-length GFP-Bsg25D (F) but is evident in similar stage oocytes expressing only GFP fused to the Bsg25D 3′ UTR (G). (H-K) Distribution of GFP-reporter mRNAs in early embryos expressing the transgenes as shown by in situ hybridization, using a probe recognizing GFP, in a wild-type genetic background. The 3′ UTR is essential for accumulation of these RNAs into the pole plasm and pole cells. All images scale bar=50 µm.
Using these transgenic lines, we conducted in situ hybridization experiments with an eGFP probe to examine the distribution of transgenic eGFP-Bsg25D mRNAs in oocytes lacking endogenous Bsg25D. As expected, eGFP was uniformly distributed in the oocytes of Bsg25D Null/Df; GFP/NGV (expressed with nanos-GAL4-VP16) females (Fig. 3B). When the full-length eGFP-Bsg25D transgene containing the 3′ UTR was expressed (Bsg25D Null/Df; CR_3′UTR/NGV), eGFP distribution faithfully reproduced the pattern of endogenous Bsg25D (Fig. 3C, compare with Fig. 1D). In contrast, eGFP-Bsg25D mRNA containing the coding region alone (Bsg25D Null/Df; CR/NGV) was concentrated at the anterior of the oocyte and did not accumulate at the oocyte posterior at stage 10a (Fig. 3D). Conversely, transgenic flies expressing only the eGFP-Bsg25D 3′ UTR (Bsg25D Null/Df; 3′UTR/NGV) precociously localized eGFP-Bsg25D RNA to the posterior at early stage 10 (Fig. 3E-G). Similar localization patterns are observed when these transgenes are expressed in a wild-type background (Fig. S2E-H).
Next, we examined the distribution of these transgenic mRNAs in otherwise wild-type embryos. In embryos from NGV; GFP mothers, eGFP is found generally in the cytoplasm (Fig. 3H). However, in embryos from flies expressing the Bsg25D coding region and 3′ UTR (NGV; CR_3′UTR), the chimeric mRNA localized to centrosomes in both the germline and the somatic region of the embryo (Fig. 3I). By contrast, eGFP- Bsg25D _CR (NGV; CR) mRNA specifically associated with somatic centrosomes and was largely absent from pole cells (Fig. 3J), while eGFP- Bsg25D _3′UTR (NGV; 3′UTR) mRNA was mostly enriched in pole cells (Fig. 3K). This suggests the presence of separable localization elements, one residing in the Bsg25D coding region mediating anterior/somatic targeting, and the other within the 3′ UTR of Bsg25D directing posterior localization, a pattern established during oogenesis.
Mislocalization of Bsg25D affects microtubule polarity and Gurken deployment in the developing oocyte
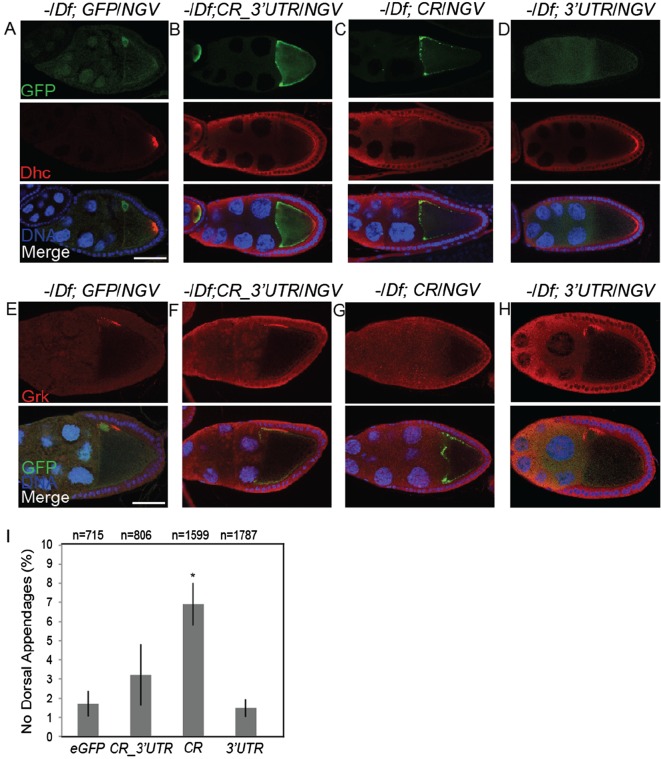
Ninein family members are involved in microtubule anchoring and nucleation. Therefore, we investigated whether mislocalizing Bsg25D would have an effect on microtubule arrangement in oogenesis, since microtubules are dynamic throughout oogenesis (Parton et al., 2011). We investigated microtubule organization in oocytes expressing the various forms of eGFP-Bsg25D used to study localization of its mRNA. First, we compared the distribution of eGFP signal to the distribution of the transgenic chimeric mRNAs. When eGFP protein was expressed on its own (Bsg25D Null/Df; GFP/NGV; Fig. 4A), we detected weak accumulation near the oocyte nucleus, most likely as a consequence of the K10 terminator element present in the vector. Much more robust targeting of GFP to the oocyte resulted from expression of eGFP-Bsg25D with or without its 3′ UTR, and the distribution of eGFP reflected that of the corresponding mRNA (Fig. 4B,C, compare with Fig. 3C,D). Asymmetric localization of eGFP requires the presence of the Bsg25D coding region, since eGFP alone expressed with the Bsg25D 3′ UTR does not accumulate at the posterior of the oocyte (Bsg25D Null/Df; 3′UTR/NGV; Fig. 4D). Immunostaining for Dhc was then used as an indirect means of assessing microtubule polarization; in wild-type oocytes Dhc accumulates at the posterior pole during mid-oogenesis in a microtubule-dependent manner (Palacios and St Johnston, 2002). In a Bsg25D Null genetic background, transgenic lines expressing eGFP, eGFP-Bsg25D _CR_3′UTR, or eGFP-Bsg25D _3′UTR, showed correct posterior localization of Dhc (Fig. 4A,B,D). However, when only the coding region of Bsg25D was expressed in fusion with eGFP, (Bsg25D Null/Df; CR/NGV), posterior accumulation of Dhc was reduced (Fig. 4C). The NGV driver produced variable levels of expression of eGFP-Bsg25D_CR in different individual egg chambers, and we observed that expression of eGFP-Bsg25D _CR correlated inversely with posterior Dhc localization. With very low expression of eGFP-Bsg25D_CR, Dhc localization is unaffected (Fig. S3A-C). This suggests that, while loss of Bsg25D does not affect microtubule polarity as measured in this assay, ectopic expression and mislocalization of Bsg25D may disrupt the polarization of microtubule minus-ends to the posterior pole of the oocyte.
Fig. 4.
Ectopic expression of GFP-Bsg25D affects microtubule polarity and Grk deployment. (A-D) GFP (A) or GFP-Bsg25D (B,C) distribution in the transgenic lines also examined in Fig. 5, in a Bsg25D Null/Df(2L)6011 (−/Df) genetic background. Note that the anterior accumulation of GFP-Bsg25D protein is more pronounced when the 3′ UTR is absent from the mRNA (compare C with B). (D) GFP protein alone lacking Bsg25D sequences does not stably accumulate at the posterior of the oocyte (top panel). Posterior localization of endogenous Dhc (red) is reduced in oocytes expressing GFP-Bsg25D lacking the 3′UTR as compared to other constructs (middle panels, compare C with the others). (E-H) Distribution of Gurken in oocytes expressing transgenic GFP-Bsg25D constructs. Expression of GFP-Bsg25D lacking the 3′UTR correlates with reduced accumulation of Grk near the oocyte nucleus (compare G with the others). Scale bar=50 µm. (I) Graph showing the proportion of embryos lacking dorsal appendages produced from females expressing transgenic GFP-Bsg25D constructs. Asterisks indicate statistical significance (P<0.05), and graph displays mean±s.e.m.
Next, we examined Gurken (Grk) localization in these oocytes, since proper targeting of this protein is dependent on microtubule polarization and on Dynein (MacDougall et al., 2003). In wild-type oogenesis Grk localizes to the antero-dorsal cortex at stage 8, forming a crescent around the oocyte nucleus (Neuman-Silberberg and Schüpbach, 1996). This localization pattern was observed in ovaries from Bsg25D Null flies expressing eGFP, eGFP-Bsg25D _CR_3′UTR, or eGFP-Bsg25D _3′UTR (Fig. 4E,F,H,J). Grk localization is reduced in stage 8 egg chambers expressing eGFP-Bsg25D _CR (Fig. 4G), although like Dynein, the intensity of localized Grk inversely correlates with the level of eGFP-Bsg25D _CR expression (Fig. S3D). There also appeared to be posterior extension of the Grk domain in some oocytes expressing eGFP-Bsg25D _CR (Fig. 4G), although variable expression of the transgene, as well as variability in the Grk domain even in wild-type egg chambers (Caceras and Nilson, 2005), makes it difficult to draw firm conclusions about the robustness of this phenotype. We also observed that more embryos produced by females expressing eGFP-Bsg25D _CR lacked dorsal appendages, as compared to those produced from wild-type females or from females expressing any of the other transgenic constructs (Fig. 4I). This phenotype could be rescued by endogenous Bsg25D (Fig. S3E-F). This suggests that the persistence of Bsg25D at the oocyte anterior may result in a failure to properly target Grk during mid-oogenesis, affecting dorsal appendage formation.
Bsg25D can bind microtubules, and with Dynein can move along microtubules toward their minus-ends, in vitro
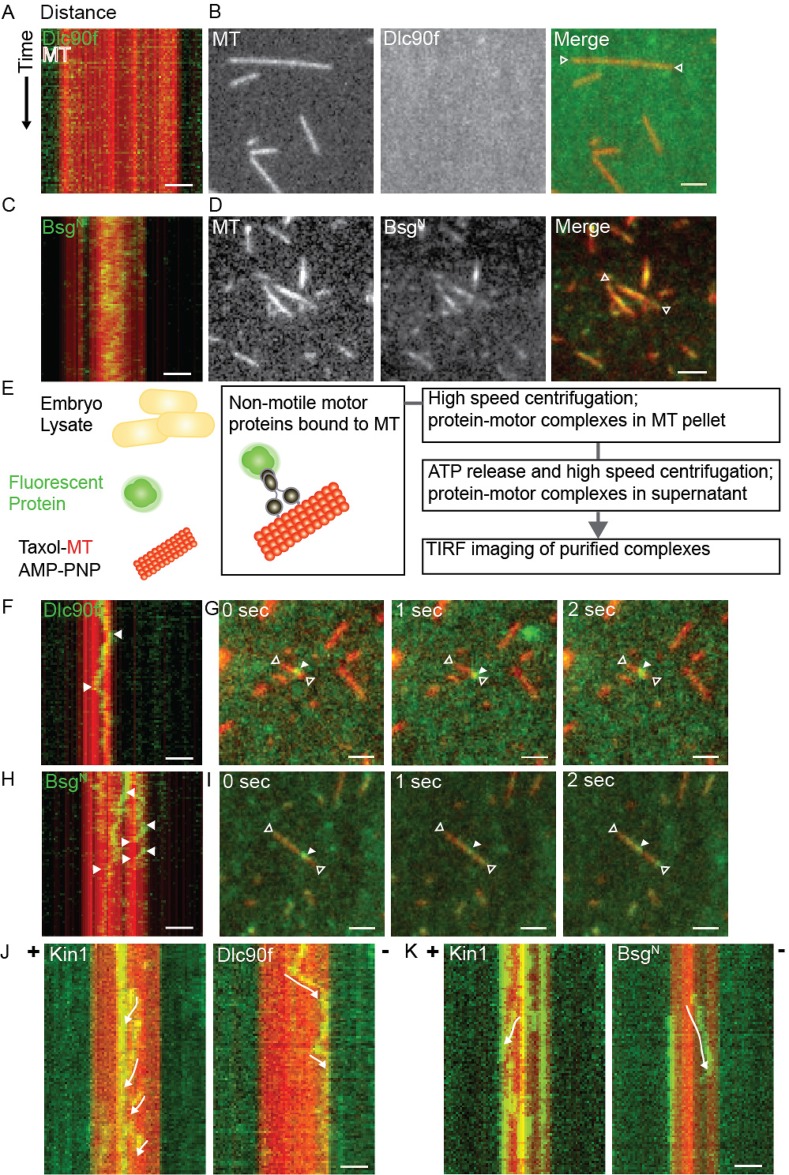
Next, we explored the dynamics of Bsg25D association with microtubules in vitro. To determine whether Bsg25D can associate with microtubules, we used total internal reflection fluorescence microscopy (TIRF) to conduct live imaging of purified Bsg25D and microtubules (Gell et al., 2010). For these experiments, because we were unable to express full-length Bsg25D in bacteria despite repeated efforts, we used BsgN, the truncated form of Bsg25D containing only the N-terminal 353 amino acids (Fig. S4). We found that purified BsgN alone in BRB80 buffer bound efficiently to microtubules, unlike the control protein Dynein light chain 90f (Dlc90f), which requires Dynein intermediate chain (Dic) and Dynein heavy chain (Dhc) to bind to microtubules (Fig. 5A-D; Movies 1, 2) (Song et al., 2007). BsgN bound diffusely to microtubules, with no specific preference for minus- or plus-ends (Fig. 5C,D), and no binding events were ever observed for Dlc90f (Fig. 5A,B). Next, we used a microtubule pull-down assay (modified from Amrute-Nayak and Bullock, 2012; Lindesmith et al., 2001) to purify motor proteins from Drosophila embryo lysates (Fig. 5E). These isolated motor protein complexes were then imaged by TIRF microscopy to observe potential transport of Dlc90f and BsgN protein molecules. In this manner, we detected transport of both Dlc90f (Fig. 5F,G; Movie 3) and BsgN (Fig. 5H,I; Movie 4). Movement events were observed more frequently for BsgN than for the control protein Dlc90f (Fig. 5D). Furthermore, purified motor protein Kinesin-1 (Kin1) was used to indicate microtubule plus-ends after Bsg25DN imaging. This was done by flowing buffer into the channel to wash away Bsg25DN protein-motor protein complexes, and subsequently flowing into the same channel Kin1 and 1 mM ATP. In this way we determined that the movement of these proteins is in the minus-end direction, suggesting this movement is Dynein-dependent for Dlc90f (Fig. 5J; Movies 5, 6), and BsgN (Fig. 5K; Movies 7, 8).
Fig. 5.
Bsg25D is a microtubule binding protein. (A,B) TIRF imaging of purified Alexa Fluor 488-labelled GST-Dlc90f (green) and tetramethylrhodamine-labelled bovine microtubules (red) in BRB80 buffer. No binding is observed either in the kymograph (A) or in direct images (B). (C,D) TIRF imaging of purified Alexa Fluor 488-labelled GST-BsgN (green) and tetramethylrhodamine-labelled bovine microtubules (red) in BRB80 buffer. Binding is evident both in the kymograph (C) and in direct images (D). (E) Schematic diagram of the microtubule pull-down assay used to purify motor proteins from Drosophila embryo lysate, a detailed description is provided in Materials and Methods. (F,G) With addition of purified motor proteins, movement events along microtubules were recorded for GST-Dlc90f in (F) a kymograph (arrows point to the beginning and end of a movement) and in (G) a series of still images from a movie. In (G) the open arrows point to the ends of a microtubule, while the closed arrow points to a GST-Dlc90f molecule. (H,I) With addition of purified motor proteins, movement events along microtubules were also recorded for GST-BsgN in (H) a kymograph and in (I) a series of still images from a movie. Arrows are as in F,G. (J,K) Kymographs comparing movement events of (J) GST-Dlc90f and (K) His-BsgN with those of Kinesin-1 imaging to indicate direction of movement. Movements are indicated with white arrows, GST-Dlc90f and GST-BsgN move in the opposite direction to Kinesin-1. In all images, scale bar=2 µm.
We measured the speed of Kin1 movement as 0.864±0.047 μm s−1 (means±s.e.m.; n=34), which is consistent with earlier measurements of its in vitro velocity (Howard et al., 1989). We also found that Dlc90f and BsgN moved at nearly identical speeds of 0.983±0.109 μm s−1 (n=23) and 0.978±0.074 μm s−1 (n=30), respectively, and these measurements are consistent with earlier analyses of Dynein movement in 1 mM ATP (Paschal et al., 1987; Ross et al., 2006).
Bsg25D functions in vivo to ensure accurate chromosome segregation during early embryonic nuclear divisions
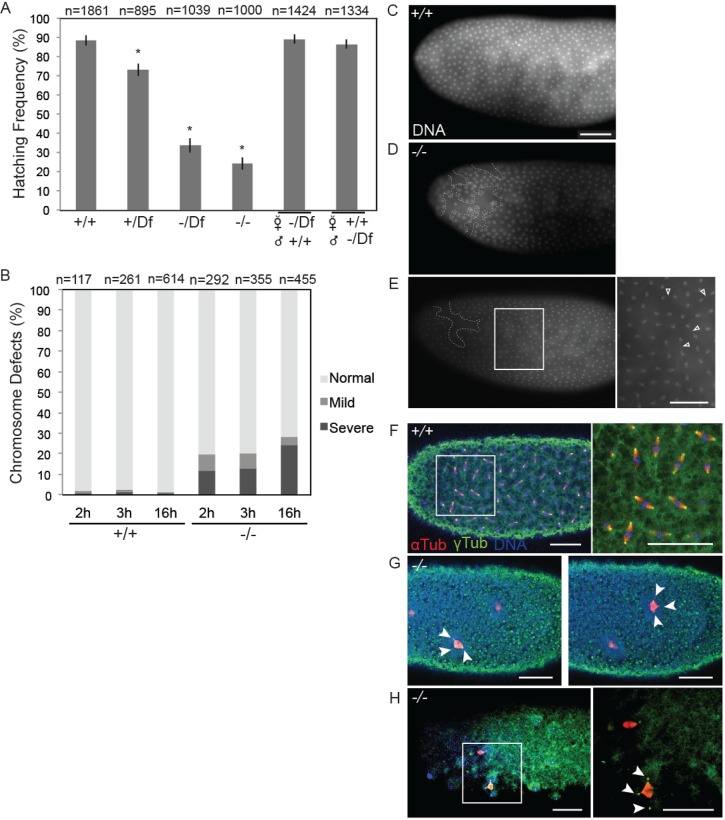
Our analysis of hemizygous flies, in which the Bsg25D Null allele was combined with a deficiency chromosome, Df(2L)Exel6011, which deletes the Bsg25D locus, revealed that Bsg25D Null/Df(2L)Exel6011 flies are viable and sufficiently fertile to be maintained as a stock. To evaluate more clearly whether Bsg25D loss-of-function impacts embryonic development, we next performed quantitative embryo viability assays following different crossing schemes. We first examined embryos from Bsg25D Null/Df(2L)Exel6011 females crossed to Bsg25D Null/Df(2L)Exel6011 males, which lack both maternally- and zygotically-expressed Bsg25D, and found however that approximately 70% failed to complete embryogenesis and did not hatch (Fig. 6A). We obtained similar results from embryos produced by Bsg25D Null/Bsg25D Null females crossed to Bsg25D Null/Bsg25D Null males, and from Bsg25D N/Bsg25D N females crossed to Bsg25D N/Bsg25D N males. Complete viability was recovered when Bsg25D Null/Df(2L)Exel6011 females were crossed to wild-type males or when wild-type virgin females were crossed to Bsg25D Null/Df(2L)Exel6011 males (Fig. 6A). We conclude that Bsg25D function is required for embryonic development and that either the maternal contribution of Bsg25D, or its early zygotic expression, is sufficient for its function in embryogenesis.
Fig. 6.
Bsg25D mutant embryos have a reduced hatching rate and exhibit mitotic defects. (A) Graph comparing frequency of hatching for embryos produced by females of the following genotypes crossed to males of the same genotype: Oregon-R (+/+), Df(2L)6011 heterozygotes (+/Df), Bsg25D Null/Df(2L)6011 (−/Df), and Bsg25D Null/Bsg25D Null (−/−). Asterisks indicate statistical significance (P<0.05) and data is displayed as mean±s.e.m. Embryonic viability is recovered when either Bsg25D Null/Df(2L)Exel6011 females were crossed to wild-type males, or wild-type females were crossed to Bsg25D Null/Df(2L)Exel6011 males. (B) Graph showing the frequency of Oregon-R (+/+) and Bsg25D Null (−/−) embryos exhibiting mitotic errors at three developmental time points. (C) DAPI staining of a wild-type embryo illustrating a typical regular arrangement of syncytial nuclei. (D,E) Examples of embryos graded as having mild mitotic defects. (D) An embryo with a region of nuclear fallout (white dashed line) and (E) an embryo with numerous anaphase bridges (arrows). (F) Anti-α-tubulin, anti-γ-tubulin immunostaining and DAPI staining of a wild-type embryo showing mitotic spindles. (G,H) Examples of embryos graded as having severe mitotic defects. (G) Two images of the same embryo showing two tripolar mitotic divisions (arrows) with γ-tubulin attached to mitotic spindle and (H) an embryo showing a tripolar spindle with γ-tubulin delocalized from mitotic spindle. Inset, with DAPI channel removed for better visualization of α-tubulin and γ-tubulin. In all images scale bar=50 µm.
We next examined the initial nuclear divisions in Bsg25D Null/Bsg25D Null embryos by staining their chromosomes with DAPI. This revealed that many such embryos exhibit excessive nuclear clearance from the embryo cortex (nuclear fallout) compared to wild-type controls (Fig. 6C,D). Moreover, Bsg25D Null/Bsg25D Null embryos showed abnormal nuclear aggregates, both large and small, and bridges between chromosomes, indicating failed chromatid separation (Fig. 6B,E). These defects in nuclear division ranged from mild (Fig. 6B-E), where the normal uniform pattern of nuclear divisions continues through the usual 13 rounds despite the phenotypes described above, to severe, where nuclear divisions fail resulting in embryonic lethality (Fig. 6B,G,H). Severely affected embryos do not cellularize and often display monopolar (not shown) and tripolar spindles (Fig. 6G,H). Many embryos were observed to have chromosome segregation defects in their initial nuclear cycles; however, in these early stage embryos the centrosomal marker γ-tubulin appeared to be localized normally, exhibiting a tight association with the mitotic spindle, as seen in wild-type specimens (Fig. 6F,G; Movie 9). In slightly older Bsg25D Null embryos, which had undergone more nuclear cycles, delocalization of the centrosome from the mitotic spindle can be observed (Fig. 6H; Movie 10). This suggests that a subset of Bsg25D Null embryos fail to properly anchor centrosomes to the mitotic spindle, which may contribute to embryonic lethality.
Finally, to determine whether pole cell specification is particularly sensitive to Bsg25D function, we counted pole cells from embryos lacking Bsg25D that developed as far as the cellular blastoderm stage and from control embryos. We observed a modest decrease in pole cell number in progeny embryos from crosses among Bsg25D Null/Df(2L)Exel6011 male and female flies that was statistically significant with respect to wild-type controls (29±0.81 vs 34±1.97, P=0.010). When we made other similar comparisons, we also observed small decreases consequent to loss of Bsg25D activity that were however not statistically significant (Bsg25D Null/Df(2L)Exel6011; GFP/NGV, 29±1.87 pole cells, P=0.056 when compared with wild-type, and Bsg25D N/Bsg25D N, 30±1.58 pole cells, P=0.102 when compared with wild-type). We therefore cannot conclude that Bsg25D has a particular function in pole cell specification. However, since a significant number of embryos lacking Bsg25D activity are unable to complete more than nine rounds of nuclear divisions, and consequently do not reach the stage of development when pole cells would form, we could not include such embryos in our analyses.
DISCUSSION
In this work, we observed that a purified fragment of Bsg25D can bind to microtubules in vitro, and in the presence of purified motor proteins, move along them primarily in the minus-end direction at a velocity similar to that of Dynein. An association between Bsg25D and Dlc90f was previously identified in a high-throughput protein-protein interaction study (Giot et al., 2003). Our results are also consistent with results obtained with both Nin and Nlp, which bind to cytoplasmic Dynein through their N-termini (Casenghi et al., 2005). Targeting Nin and Nlp to the centrosome occurs through Dynein-mediated transport and is dependent on the microtubule cytoskeleton (Casenghi et al., 2005). While the overall direction of Bsg25DN movement was minus-end directed, we also documented instances of movement toward the plus-end of a microtubule. This is consistent with imaging data from live mammalian epithelial cells demonstrating bidirectional microtubule-directed movement for Nin (Moss et al., 2007). Bidirectional movement has been observed in other single molecule assays and is a well-established characteristic of Dynein (Amrute-Nayak and Bullock, 2012; Reck-Peterson et al., 2006). One potential difference between Bsg25D and its mammalian counterparts is that our in vitro experiments indicate that Bsg25D binds to microtubules in the absence of Dynein, which has not been demonstrated for Nin or Nlp. More targeted experiments could in the future establish the mechanistic relationship between Bsg25D and Dynein. These could include depleting for Dynein the motor protein complexes used for the TIRF imaging and determining whether there are effects on BsgN mobility, or mapping and mutating the site on BsgN necessary for Dynein binding, and then determining whether such a mutated protein can move along microtubules.
While Bsg25D has extensive sequence similarity to Nin and Nlp, it appears to be more closely related to Nin. Like both Nin and Nlp, Bsg25D contains numerous coiled-coil domains. In addition, one of three cAMP-dependent protein kinase (PKA) phosphorylation sites in Nin is conserved in Bsg25D (amino acids 124-130). This may be important for Bsg25D function, as Nin phosphorylation has been linked for centrosomal localization of certain Nin isoforms, and phosphorylation by PKA has been found to play a critical role in mitotic progression (Chen et al., 2003; Hong et al., 2000; Kotani et al., 1998; Lin et al., 2006). Bsg25D shares a conserved D-box domain with Nlp that is not present in Nin (Bsg25D amino acids 268-276), but it does not have the D-box or Ken-box motifs that for Nlp have been experimentally shown to be important for cell cycle dependent degradation (Nlp amino acids 633-641 and 495-497) (Wang and Zhan, 2007). Nlp has phosphorylation sites for Aurora B or Cdc2/cyclin B1 kinases that do not appear to be conserved in Bsg25D, however independent mass spectrometry analyses of Nlp and Bsg25D reveals the presence of many phosphoserine and phosphothreonine residues at similar locations within both proteins (Casenghi et al., 2003; Zhai et al., 2008). Nlp phosphorylation by Plk is required for Nlp dissociation from the Dynein-Dynactin complex allowing for cell cycle progression in human cell lines (Casenghi et al., 2005).
While Bsg25D mutants can survive to become fertile adults, we observed that a majority of embryos that lack maternal and zygotic Bsg25D fail to hatch, and exhibit mitotic defects ranging from mild to very severe. Our experiments do not allow us to distinguish whether the mitotic defects are a cause or a consequence of the failure of many such embryos to develop. However, since Bsg25D associates with centrosomes, we can hypothesize that it contributes to their functions in microtubule nucleation and/or anchoring, and that its loss may cause abnormal mitotic spindle formation. While this manuscript was under review, we became aware of another study of Drosophila Bsg25D (Zheng et al., 2016). That paper reports similar results to ours with respect to localization of Bsg25D to centrosomes in early embryos. While both studies found Bsg25D null mutants to be viable, in contrast to our results their nin1 allele did not produce a significant decrease in embryonic viability. This quantitative difference in our results could be a consequence of differences in culture conditions or genetic backgrounds.
A role for mammalian Nin in connecting microtubules to the centrosome has been proposed (Shinohara et al., 2013). As well, siRNA-mediated knockdown of Nin in human immortal cell lines resulted in mitotic catastrophe, cell cycle arrest in G2/M phase and apoptosis (Kimura et al., 2013). Given this severe phenotype, it is surprising that Bsg25D function is not required for viability in Drosophila under laboratory conditions, especially since only one Ninein-related protein is present in flies as opposed to two in mammals. In humans the rare disease Seckel syndrome-7 (SCKL7) is caused by missense mutations in the NIN gene (Dauber et al., 2012). SCKL7 results in a growth phenotype called microcephalic primordial dwarfism, which is a severe form of growth failure wherein growth restriction occurs in utero and continues after birth (Bober et al., 2010; Dauber et al., 2012). These patients, however, often survive until adulthood. Furthermore, Nlp has been linked to ciliopathies, Usher syndrome and Leber congenital amaurosis (van Wijk et al., 2009). Knockout mice for both Nin and Ninl (which encodes Nlp) have been prepared (Brown and Moore, 2012) but they have not yet been studied in detail.
In Drosophila we also observed defects in Dhc and Grk localization upon overexpression and mis-localization of eGFP-Bsg25D mRNA in developing oocytes, which in turn led to an altered distribution of eGFP-Bsg25D protein. Polarization of microtubules within the developing oocyte is critical for transport of mRNAs necessary for axis determination in the early embryo. Our data suggest that overexpression and/or mislocalization of Bsg25D during oogenesis may affect microtubule-dependent localization processes, such as grk (MacDougall et al., 2003), and Dhc localization (Li et al., 1994), within the oocyte. Consistent with this, ubiquitous expression of Bsg25D with actin or tubulin Gal4 drivers results in early pupal lethality, also demonstrating that overexpression of Bsg25D is deleterious (Zheng et al., 2016).
Analogous results have been obtained for Nin and Nlp in mammalian cells. For example, in mammalian cultured cells overexpression of Nlp recruits γ-tubulin and hGCP4, a component of the γ-tubulin ring complex (γ-TURC) to ectopic loci, resulting in off-site microtubule nucleation and spindle formation (Casenghi et al., 2003). Overexpression of Nin has also been reported to lead to mis-localization of γ-tubulin in cultured human cells (Stillwell et al., 2004). Nlp overexpression is also frequently associated with cancer, including head and neck squamous cell carcinomas and ovarian cancer (Qu et al., 2008; Yu et al., 2009). In one study Nlp was found to be overexpressed in 80% of human breast and lung carcinomas that were investigated, and its overexpression led to tumorigenesis in transgenic mice (Shao et al., 2010).
In conclusion, our study of the dynamics of Bsg25D in vitro and of the consequences of its mutation or ectopic expression in an intact metazoan establish Drosophila as a model system for studying Ninein family proteins. Further work in this system will help reveal potential mechanisms through which loss or gain of function of Nin and Nlp might result in human disease.
MATERIALS AND METHODS
Drosophila strains
Oregon-R was used as wild-type for all experiments. The deficiency spanning Bsg25D (Df(2L)Exel6011, BL#7497) was received from the Bloomington Drosophila Stock Center. Truncated and null alleles of Bsg25D were generated using the ends-out gene targeting method, using the pw25.5 vector (generously provided by Dr David R. Hipfner; Maggert et al., 2008). Primers used to produce Bsg25D N from D. melanogaster genomic DNA were: left arm Forward Not1-tkv-Bsg 5′-GCGGCCGCCATCGACGCGGTATCGATATTC-3′ and Acc651-tkv-Bsg 5′-GGTACCCTAACAGAGGAGAGCCCTCG-3′, and right arm Asc1-Bsg-Bsg 5′-GGCGCGCCCCACGGCAAGCAAAGCCAC-3′ and Reverse Asc1-Bsg-Bsg 5′-GGCGCGCCGCGATAGAAACGTGTTGTTGGG-3′. Primers for Bsg25D Null were: left arm Forward Acc651-Bsg-Bsg 5′-GGTACCGGTAGCCACCTAAGATCCATAC-3′ and Reverse Not1-Bsg-Bsg 5′-GCGGCCGCCAATCGGCTATCTCTCCCTC-3′, and right arm Forward Asc1-Bsg-Bub1 5′-GGCGCGCCGGTTACGGATAATGGAGGTATC-3′ and Reverse Asc1-Bsg-Bub1 5′-GGCGCGCCCTTGAGCGCCACTACATTGC-3′. UASp-eGFP-Bsg25D transgenic flies were generated as described in Iampietro, et al. (2014). Briefly, the Bsg25D _CR_3′UTR (Coding Region+3′ UTR), Bsg25D _CR (Coding region) and Bsg25D _3′UTR (3′ UTR) sequences were amplified by PCR using Drosophila Gene Collection bacterial cDNA clones (dBsg25D=LD21844) as template and primers listed below containing the restriction sites required for the insertion in the vector. PCR fragments were ligated into the pGem4-GFP vector to generate in-frame fusion cassettes with the GFP coding region in 5′ to Bsg25D sequences. The GFP-fusion cassettes were then sub-cloned into the pUASTp-attb plasmid (generously provided by Dr Howard Lipshitz, Molecular Genetics Department, University of Toronto, Ontario, Canada) by using the EcoRI restriction enzyme to obtain the transgenesis vectors. All vector sequences were confirmed by sequencing and injected into syncytial stage embryos of the attP-3B acceptor fly line (stock number BL24871) using a Leica DMIL microinjection microscope. Subsequent selection of transgenic progeny was performed as described previously (Bischof et al., 2007; Venken et al., 2006), using primers Bsg25D CR Fw- Kpn1 5′-ATTAGGTACCATGGAGGTATCCGCCGATCCG-3′ and Bsg25D CR Rv EcoR1 5′-ATTAGAATTCCTAAGGCATGCCAGGCAGTCC-3′, and Bsg25D 3′UTR Fw Kpn1: 5′-ATTAGGTACCTAGTTTGCCCCACCGGCAAAC-3′ and Bsg25D 3′UTR Rv EcoR1: 5′-ATTAGAATTCTCGAAAGTATTGATTTAAGCACTGA-3′.
Immunoblots
Ovaries for immunoblotting were dissected from 2-5-day-old females, lysed in ovary lysis buffer (1× PBS, 1× Halt proteinase inhibitor, 1% PMSF in water) and loaded on an 8-15% gradient gel. The primary antibodies for immunoblotting were: guinea pig anti-Bsg25D (Iampietro et al., 2014; 1:20,000), mouse anti-α-tubulin (Sigma #T6199, 1:10,000), mouse anti-Dynein heavy chain (Developmental Studies Hybridoma Bank #2C11-2, 1:1000), and mouse anti-GFP (Molecular Probes monoclonal antibody 3E6, 1:2500). Secondary antibodies used were donkey anti-guinea pig (Jackson Labs #706-035-148, 1:2500) and anti-mouse (GE Healthcare, #NA931, 1:5000 dilution).
Immunofluorescence and in situ hybridization
Embryos were collected as previously described (Lécuyer et al., 2008). Primary antibodies used for immunostaining were; guinea pig anti-Bsg25D (1:2000), mouse anti-Dynein heavy chain (1:500), rabbit anti-γ-Tubulin (Sigma #T0950, 1:50), rat anti-α-Tubulin (AdB Serotec #MCA78G, 1:50), and rabbit anti-Grk (Dehghani and Lasko; 2015; 1:500). For DNA staining DAPI (Invitrogen #D3571) was used at 10 μg/ml. Secondary antibodies were goat anti-guinea pig (Abcam Dylight 488 #ab96959, 1:500) goat anti-mouse, rabbit and rat (Thermo Fisher Scientific, A11030, A11010 and A-11081, 1:500). Images were collected on the Zeiss LSM510 confocal laser-scanning microscope at the CIAN, Department of Biology, McGill University.
In situ hybridization and co-staining was performed as previously described (Lécuyer et al., 2008; Iampietro et al., 2014). Bsg25D anti-sense RNA probe was synthesized from clone LD21844 of the Drosophila gene collection library and a full-length probe was used.
Embryos and ovaries for immunostaining or in situ were fixed with 4% paraformaldehyde, except for embryos stained for anti-γ-tubulin and anti-α-tubulin. For γ- and α-tubulin stainings embryos were dechorionated with bleach and shaken in heptane for 30 s, followed by shaking for 30 s in 50% heptane:50% methanol to crack the vitelline membranes. Finally, embryos were placed on a nutator for 1 h in methanol, then rehydrated for staining or kept at −20°C for future use.
Embryo quantification
For hatching counts virgin females were mated to young males on grape juice plates containing yeast. Total eggs from overnight collections were counted and then aged for 48 h at 25°C. After 48 h unhatched eggs were counted. For DNA damage counts embryos were collected for 2, 3 and 16 h, fixed and stained with DAPI (Lécuyer et al., 2008). Embryos were counted manually on a Leica DM6000B microscope under a 20× objective.
In vitro microtubule assays
Bovine tubulin was purified and tetramethylrhodamine labeled as previously described (Noujaim et al., 2014; Wieczorek et al., 2015). Paclitaxel stabilized microtubules used for microtubule pull-down assay were polymerized as described previously (Noujaim et al., 2014). TIRF imaging of microtubules was performed on a Zeiss Axiovert Z1 microscope chassis, using a 100×1.45 NA Plan-apochromat objective lens, and Zeiss TIRF III slider.
GST-tagged full length Dlc90f (39 kDa) and His-tagged N-terminal Bsg25D (43 kDa) protein was expressed and purified from bacteria. Both proteins were fluorescently labeled with Alexa Fluor 488 TFP ester (Thermo Fisher Scientific Cat: A37570). A size exclusion column (Amicon Ultra-0.5 30K, UFC503024) was used to concentrate dye-protein conjugates.
A microtubule pull-down assay modified from (Amrute-Nayak and Bullock, 2012; Lindesmith et al., 2001) was used to purify motor proteins from Drosophila embryo lysate. Drosophila embryos were collected for 4 h, lysed in BXB buffer (Amrute-Nayak and Bullock, 2012), and mixed with purified fluorescent protein along with an AMP analog, AMP-PNP. Isolation of microtubule-motor protein complexes was completed by centrifugation in a Beckman Airfuge at maximum speed, the pellet containing these complexes was washed with assay buffer and the supernatant with unbound protein was discarded. Assay buffer containing ATP resulted in a release of motor protein from microtubules. These isolated motor protein complexes were then mixed with anti-bleach buffer and imaged by TIRF microscopy (Amrute-Nayak and Bullock, 2012). Rat Kinesin-1 430-GFP and purified bovine brain tetramethylrhodamine-labeled α- and β-tubulin were prepared as previously described (Noujaim et al., 2014). MetaMorph was used to record live imaging and analysis. Velocities were calculated using MetaMorph and shown as mean±s.e.m.
Acknowledgements
We are grateful to Beili Hu for Drosophila embryo microinjections and to Hong Han for providing antibodies.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
M.K., J.B., and M.W. performed the experiments, M.K. and P.L. wrote the paper, G.B. and É.L. contributed to the analysis and interpretation of the data, and to the final draft of the paper.
Funding
This work was supported by the Canadian Institutes of Health Research (CIHR) [operating grant IOP-107945 to P.L., grants MOP-161111 and MOP-137096 to E.L, and grant MOP-137055 to G.B.]; Natural Sciences and Engineering Research Council of Canada (NSERC) [grant RGPIN-2014-03791 to G.B.]. J.B. and E.L. were supported by salary awards from the Fonds de Recherche du Québec - Santé, G.B. is a CIHR New Investigator, M.W. received an Alexander Graham Bell Graduate Scholarship from NSERC, and P.L. is a James McGill Professor.
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/doi/10.1242/bio.019638.supplemental
References
- Amrute-Nayak M. and Bullock S. L. (2012). Single-molecule assays reveal that RNA localization signals regulate Dynein-dynactin copy number on individual transcript cargoes. Nat. Cell Biol. 14, 416-423. 10.1038/ncb2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka-Taguchi M., Yamada M., Nakamura A., Hanyu K. and Kobayashi S. (1999). Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1, 431-437. 10.1038/15666 [DOI] [PubMed] [Google Scholar]
- Azimzadeh J. and Bornens M. (2007). Structure and duplication of the centrosome. J. Cell Sci. 120, 2139-2142. 10.1242/jcs.005231 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F. and Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312-3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bober M. B., Khan N., Kaplan J., Lewis K., Feinstein J. A., Scott C. I. and Steinberg G. K. (2010). Majewski osteodysplastic primordial Dwarfism type II (MOPD II): expanding the vascular phenotype. Am. J. Med. Genet. 152A, 960-965. 10.1002/ajmg.a.33252 [DOI] [PubMed] [Google Scholar]
- Boyer P. D., Mahoney P. A. and Lengye J. A. (1987). Molecular characterization of bsg25D: a blastoderm-specific locus of Drosophila melanogaster. Nucleic Acids Res. 15, 2309-2325. 10.1093/nar/15.5.2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. D. M. and Moore M. W. (2012). The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm. Genome 23, 632-640. 10.1007/s00335-012-9427-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceras L. and Nilson L. A. (2005). Production of gurken in the nurse cells is sufficient for axis determination in the Drosophila oocyte. Development 132, 2345-2353. 10.1242/dev.01820 [DOI] [PubMed] [Google Scholar]
- Callaini G. and Riparbelli M. G. (1990). Centriole and centrosome cycle in the early Drosophila embryo. J. Cell Sci. 97, 539-543. [DOI] [PubMed] [Google Scholar]
- Callaini G., Whitfield W. G. F. and Riparbelli M. G. (1997). Centriole and centrosome dynamics during the embryonic cell cycles that follow the formation of the cellular blastoderm in Drosophila. Exp. Cell Res. 234, 183-190. 10.1006/excr.1997.3618 [DOI] [PubMed] [Google Scholar]
- Casenghi M., Meraldi P., Weinhart U., Duncan P. I., Körner R. and Nigg E. A. (2003). Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell 5, 113-125. 10.1016/S1534-5807(03)00193-X [DOI] [PubMed] [Google Scholar]
- Casenghi M., Barr F. A. and Nigg E. A. (2005). Phosphorylation of Nlp by Plk1 negatively regulates its Dynein-dynactin-dependent targeting to the centrosome. J. Cell Sci. 118, 5101-5108. 10.1242/jcs.02622 [DOI] [PubMed] [Google Scholar]
- Chen C.-H., Howng S.-L., Cheng T.-S., Chou M.-H., Huang C.-Y. and Hong Y.-R. (2003). Molecular characterization of human ninein protein: two distinct subdomains required for centrosomal targeting and regulating signals in cell cycle. Biochem. Biophys. Res. Comm. 308, 975-983. 10.1016/S0006-291X(03)01510-9 [DOI] [PubMed] [Google Scholar]
- Dahlgaard K., Raposo A. A. S. F., Niccoli T. and St Johnston D. (2007). Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev. Cell 13, 539-553. 10.1016/j.devcel.2007.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber A., LaFranchi S. H., Maliga Z., Lui J. C., Moon J. E., McDeed C., Henke K., Zonana J., Kingman G. A., Pers T. H. et al. (2012). Novel Microcephalic Primordial Dwarfism Disorder associated with variants in the centrosomal protein Ninein. J. Clin. Endocr. Metab. 97, E2140-E2151. 10.1210/jc.2012-2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghani M. and Lasko P. (2015). In vivo mapping of functional regions of the DEAD-box helicase Vasa. Biol. Open 4, 450-457. 10.1242/bio.201410579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N., Sillibourne J. and Bornens M. (2005). Microtubule nucleation and anchoring at the centrosome are independent processes linked by Ninein function. J. Cell Sci. 118, 1565-1575. 10.1242/jcs.02302 [DOI] [PubMed] [Google Scholar]
- Dinkel H., Van Roey K., Michael S., Kumar M., Uyar B., Altenberg B., Milchevskaya V., Schneider M., Kühn H., Behrendt A. et al. (2016). ELM 2016–data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 44, D294-D300. 10.1093/nar/gkv1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V. E. and Alberts B. M. (1983). Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell. Sci. 61, 31-70. [DOI] [PubMed] [Google Scholar]
- Fu J. and Glover D. M. (2012). Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2, 120104 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell C., Bormuth V., Brouhard G. J., Cohen D. N., Diez S., Friel C. T., Helenius J., Nitzsche B., Petzold H., Ribbe J. et al. (2010). Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 95, 221-245. 10.1016/S0091-679X(10)95013-9 [DOI] [PubMed] [Google Scholar]
- Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E. et al. (2003). A protein interaction map of Drosophila melanogaster. Science 302, 1727-1736. 10.1126/science.1090289 [DOI] [PubMed] [Google Scholar]
- Gonzalez C., Tavosanis G. and Mollinari C. (1998). Centrosomes and microtubule organisation during Drosophila development. J. Cell Sci. 111, 2697-2706. [DOI] [PubMed] [Google Scholar]
- Hanyu-Nakamura K., Sonobe-Nojima H., Tanigawa A., Lasko P. and Nakamura A. (2008). Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature 451, 730-733. 10.1038/nature06498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y.-R., Chen C.-H., Chuo M.-H., Liou S.-Y. and Howng S.-L. (2000). Genomic organization and molecular characterization of the human Ninein gene. Biochem. Biophys. Res. Comm. 279, 989-995. 10.1006/bbrc.2000.4050 [DOI] [PubMed] [Google Scholar]
- Howard J., Hudspeth A. J. and Vale R. D. (1989). Movement of microtubules by single kinesin molecules. Nature 342, 154-158. 10.1038/342154a0 [DOI] [PubMed] [Google Scholar]
- Iampietro C., Bergalet J., Wang X., Cody N. A. L., Chin A., Lefebvre F. A., Douziech M., Krause H. M. and Lécuyer E. (2014). Developmentally regulated elimination of damaged nuclei involves a chk2-dependent mechanism of mRNA nuclear retention. Dev. Cell 29, 468-481. 10.1016/j.devcel.2014.03.025 [DOI] [PubMed] [Google Scholar]
- Jones J. and Macdonald P. M. (2015). Neurl4 contributes to germ cell formation and integrity in Drosophila. Biol. Open 4, 937-946. 10.1242/bio.012351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongens T. A., Hay B., Jan L. Y. and Jan Y. N. (1992). The germ cell-less gene product: a posteriorly localized component necessary for germ cell development in Drosophila. Cell 70, 569-584. 10.1016/0092-8674(92)90427-E [DOI] [PubMed] [Google Scholar]
- Kadyrova L. Y., Habara Y., Lee T. H. and Wharton R. P. (2007). Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134, 1519-1527. 10.1242/dev.002212 [DOI] [PubMed] [Google Scholar]
- Kimura M., Yoshioka T., Saio M., Banno Y., Nagaoka H. and Okano Y. (2013). Mitotic catastrophe and cell death induced by depletion of centrosomal proteins. Cell Death Dis. 4, e603 10.1038/cddis.2013.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani S., Tugendreich S., Fujii M., Jorgensen P.-M., Watanabe N., Hoog C., Hieter P. and Todokoro K. (1998). PKA and MPF-activated polo-like kinase regulate anaphase-promoting complex activity and mitosis progression. Mol. Cell 1, 371-380. 10.1016/S1097-2765(00)80037-4 [DOI] [PubMed] [Google Scholar]
- Lantz V., Chang J. S., Horabin J. I., Bopp D. and Schedl P. (1994). The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 8, 598-613. 10.1101/gad.8.5.598 [DOI] [PubMed] [Google Scholar]
- Lasko P. (2012). mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb. Perspect. Biol. 4, a012294 10.1101/cshperspect.a012294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S., Hasegan M., Gupta G. D. and Pelletier L. (2012). Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14, 1148-1158. 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- Letunic I., Copley R. R., Pils B., Pinkert S., Schultz J. and Bork P. (2006). SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34, D257-D260. 10.1093/nar/gkj079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T. R., Tomancak P. and Krause H. M. (2007). Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174-187. 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Lécuyer E., Parthasarathy N. and Krause H. M. (2008). Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol. Biol. 420, 289-302. 10.1007/978-1-59745-583-1_18 [DOI] [PubMed] [Google Scholar]
- Lerit D. A. and Gavis E. R. (2011). Transport of germ plasm on astral microtubules directs germ cell development in Drosophila. Curr. Biol. 21, 439-448. 10.1016/j.cub.2011.01.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., McGrail M., Serr M. and Hays T. S. (1994). Drosophila cytoplasmic dynein, a microtubule motor that is asymmetrically localized in the oocyte. J. Cell Biol. 126, 1475-1494. 10.1083/jcb.126.6.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-C., Cheng T.-S., Hsu C.-M., Wu C.-H., Chang L.-S., Shen Z.-S., Yeh H.-M., Chang L.-K., Howng S.-L. and Hong Y.-R. (2006). Characterization and functional aspects of human ninein isoforms that regulated by centrosomal targeting signals and evidence for docking sites to direct gamma-tubulin. Cell Cycle 5, 2517-2527. 10.4161/cc.5.21.3404 [DOI] [PubMed] [Google Scholar]
- Lindesmith L. C., Kumar J. and Sheetz M. P. (2001). Identification of kinesin-associated proteins. Methods Mol. Biol. 164, 205-212. 10.1385/1-59259-069-1:205 [DOI] [PubMed] [Google Scholar]
- MacDougall N., Clark A., MacDougall E. and Davis I. (2003). Drosophila gurken (TGFα) mRNA localizes as particles that move within the oocyte in two Dynein-dependent steps. Dev. Cell 4, 307-319. 10.1016/S1534-5807(03)00058-3 [DOI] [PubMed] [Google Scholar]
- Maggert K. A., Gong W. J. and Golic K. G. (2008). Methods for homologous recombination in Drosophila. Methods Mol. Biol. 420, 155-174. 10.1007/978-1-59745-583-1_9 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., Geer R. C., He J., Gwadz M., Hurwitz D. I. et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222-D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss D. K., Bellett G., Carter J. M., Liovic M., Keynton J., Prescott A. R., Lane E. B. and Mogensen M. M. (2007). Ninein is released from the centrosome and moves bi-directionally along microtubules. J. Cell Sci. 120, 3064-3074. 10.1242/jcs.010322 [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg F. S. and Schüpbach T. (1996). The Drosophila TGF-α-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech. Dev. 59, 105-113. 10.1016/0925-4773(96)00567-9 [DOI] [PubMed] [Google Scholar]
- Noujaim M., Bechstedt S., Wieczorek M. and Brouhard G. J. (2014). Microtubules accelerate the kinase activity of Aurora-B by a reduction in dimensionality. PLoS ONE 9, e86786 10.1371/journal.pone.0086786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I. M. and St Johnston D. (2002). Kinesin light chain-independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localization in the Drosophila oocyte. Development 129, 5473-5485. 10.1242/dev.00119 [DOI] [PubMed] [Google Scholar]
- Parton R. M., Hamilton R. S., Ball G., Yang L., Cullen C. F., Lu W., Ohkura H. and Davis I. (2011). A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J. Cell Biol. 194, 121-135. 10.1083/jcb.201103160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M. J., Deng W., Wang Z. and Lin H. (2001). The arrest gene is required for germline cyst formation during Drosophila oogenesis. Genesis 29, 196-209. 10.1002/gene.1024 [DOI] [PubMed] [Google Scholar]
- Paschal B. M., Shpetner H. S. and Vallee R. B. (1987). MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has Dynein-like properties. J. Cell Biol. 105, 1273-1282. 10.1083/jcb.105.3.1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu D., Qu H., Fu M., Zhao X., Liu R., Sui L. and Zhan Q. (2008). Increased expression of Nlp, a potential oncogene in ovarian cancer, and its implication in carcinogenesis. Gynecol. Oncol. 110, 230-236. 10.1016/j.ygyno.2008.04.015 [DOI] [PubMed] [Google Scholar]
- Read D., Levine M. and Manley J. L. (1992). Ectopic expression of the Drosophila tramtrack gene results in multiple embryonic defects, including repression of even-skipped and fushi tarazu. Mech. Dev. 38, 183-195. 10.1016/0925-4773(92)90052-L [DOI] [PubMed] [Google Scholar]
- Reck-Peterson S. L., Yildiz A., Carter A. P., Gennerich A., Zhang N. and Vale R. D. (2006). Single-molecule analysis of Dynein processivity and stepping behavior. Cell 126, 335-348. 10.1016/j.cell.2006.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roark M., Mahoney P. A., Graham M. L. and Lengyel J. A. (1985). Blastoderm-differential and blastoderm-specific genes of Drosophila melanogaster. Dev. Biol. 109, 476-488. 10.1016/0012-1606(85)90473-7 [DOI] [PubMed] [Google Scholar]
- Ross J. L., Wallace K., Shuman H., Goldman Y. E. and Holzbaur E. L. F. (2006). Processive bidirectional motion of Dynein-dynactin complexes in vitro. Nat. Cell Biol. 8, 562-570. 10.1038/ncb1421 [DOI] [PubMed] [Google Scholar]
- Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., Eaton M. L., Landolin J. M., Bristow C. A., Ma L., Lin M. F. et al. (2010). Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787-1797. 10.1126/science.1198374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Hayashi Y., Ninomiya Y., Shigenobu S., Arita K., Mukai M. and Kobayashi S. (2007). Maternal Nanos represses hid/skl-dependent apoptosis to maintain the germ line in Drosophila embryos. Proc. Natl. Acad. Sci. USA 104, 7455-7460. 10.1073/pnas.0610052104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S., Liu R., Wang Y., Song Y., Zuo L., Xue L., Lu N., Hou N., Wang M., Yang X. et al. (2010). Centrosomal Nlp is an oncogenic protein that is gene-amplified in human tumors and causes spontaneous tumorigenesis in transgenic mice. J. Clin. Invest. 120, 498-507. 10.1172/JCI39447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H., Sakayori N., Takayashi M. and Osumi N. (2013). Ninein is essential for the maintenance of the cortical progenitor character by anchoring the centrosome to microtubules. Biol. Open 2, 739-749. 10.1242/bio.20135231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C. J. A., de Castro E., Cerutti L., Cuche B. A., Hulo N., Bridge A., Bougueleret L. and Xenarios I. (2012). New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344-D347. 10.1093/nar/gks1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Benison G., Nyarko A., Hays T. S. and Barbar E. (2007). Potential role for phosphorylation in differential regulation of the assembly of Dynein light chains. J. Biol. Chem. 282, 17272-17279. 10.1074/jbc.M610445200 [DOI] [PubMed] [Google Scholar]
- Stillwell E. E., Zhou J. and Joshi H. C. (2004). Human Ninein is a centrosomal autoantigen recognized by CREST patient sera and plays a regulatory role in microtubule nucleation. Cell Cycle 3, 921-928. 10.4161/cc.3.7.947 [DOI] [PubMed] [Google Scholar]
- Tomancak P., Berman B. P., Beaton A., Weiszmann R., Kwan E., Hartenstein V., Celniker S. E. and Rubin G. M. (2007). Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8, R145 10.1186/gb-2007-8-7-r145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk E., Kersten F. F. J., Kartono A., Mans D. A., Brandwijk K., Letteboer S. J. F., Peters T. A., Märker T., Yan X., Cremers C. W. et al. (2009). Usher syndrome and Leber congenital amaurosis are molecularly linked via a novel isoform of the centrosomal ninein-like protein. Hum. Mol. Genet. 18, 51-64. 10.1093/hmg/ddn312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., He Y., Hoskins R. A. and Bellen H. J. (2006). P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747-1751. 10.1126/science.1134426 [DOI] [PubMed] [Google Scholar]
- Vorobjev I. A. and Nadezhdina E. S. (1987). The centrosome and its role in the organization of microtubules. Int. Rev. Cytol. 106, 227-293. 10.1016/S0074-7696(08)61714-3 [DOI] [PubMed] [Google Scholar]
- Wang C. and Lehmann R. (1991). Nanos is the localized posterior determinant in Drosophila. Cell 66, 637-647. 10.1016/0092-8674(91)90110-K [DOI] [PubMed] [Google Scholar]
- Wang Y. and Zhan Q. (2007). Cell cycle-dependent expression of centrosomal Ninein-like protein in human cells is regulated by the Anaphase-promoting Complex. J. Biol. Chem. 282, 17712-17719. 10.1074/jbc.M701350200 [DOI] [PubMed] [Google Scholar]
- Wasbrough E. R., Dorus S., Hester S., Howard-Murkin J., Lilley K., Wilkin E., Polpitiya A., Petritis K. and Karr T. L. (2010). The Drosophila melanogaster sperm proteome-II (DmSP-II). J. Proteomics 73, 2171-2185. 10.1016/j.jprot.2010.09.002 [DOI] [PubMed] [Google Scholar]
- Wieczorek M., Bechstedt S., Chaaban S. and Brouhard G. J. (2015). Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat. Cell. Biol. 17, 907-916. 10.1038/ncb3188 [DOI] [PubMed] [Google Scholar]
- Winslow G. M., Carroll S. B. and Scott M. P. (1988). Maternal-effect genes that alter the fate map of the Drosophila blastoderm embryo. Dev. Biol. 129, 72-83. 10.1016/0012-1606(88)90162-5 [DOI] [PubMed] [Google Scholar]
- Woodruff J. B., Wueseke O. and Hyman A. A. (2014). Pericentriolar material structure and dynamics. Philos. Trans. R. Soc. Lond B Biol. Sci. 369, 20130459 10.1098/rstb.2013.0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Song Y., Zhang Q. and Zhan Q. (2009). Ninein-like protein is overexpressed in head and neck squamous cell carcinoma and contributes to cancer growth and resistance to apoptosis. Oncol. Rep. 22, 789-798. 10.3892/or_00000501 [DOI] [PubMed] [Google Scholar]
- Zhai B., Villen J., Beausoleil S. A., Mintseris J. and Gygi S. P. (2008). Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 7, 1675-1682. 10.1021/pr700696a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Mennella V., Marks S., Wildonger J., Elnagdi E., Agard D. and Megraw T. L. (2016). The Seckel syndrome and centrosomal protein Ninein localizes asymmetrically to stem cell centrosomes but is not required for normal development, behavior, or DNA damage response in Drosophila. Mol. Biol. Cell 27, 1740-1752. 10.1091/mbc.E15-09-0655 [DOI] [PMC free article] [PubMed] [Google Scholar]