Abstract
Muscle proteolysis in CKD is stimulated when the ubiquitin-proteasome system is activated. Serum glucocorticoid–regulated kinase 1 (SGK-1) is involved in skeletal muscle homeostasis, but the role of this protein in CKD–induced muscle wasting is unknown. We found that, compared with muscles from healthy controls, muscles from patients and mice with CKD express low levels of SGK-1. In mice, SGK-1-knockout (SGK-1-KO) induced muscle loss that correlated with increased expression of ubiquitin E3 ligases known to facilitate protein degradation by the ubiquitin-proteasome, and CKD substantially aggravated this response. SGK-1-KO also altered the phosphorylation levels of transcription factors FoxO3a and Smad2/3. In C2C12 muscle cells, expression of dominant negative FoxO3a or knockdown of Smad2/3 suppressed the upregulation of E3 ligases induced by loss of SGK-1. Additionally, SGK-1 overexpression increased the level of phosphorylated N-myc downstream–regulated gene 1 protein, which directly interacted with and suppressed the phosphorylation of Smad2/3. Overexpression of SGK-1 in wild-type mice with CKD had similar effects on the phosphorylation of FoxO3a and Smad2/3 and prevented CKD–induced muscle atrophy. Finally, mechanical stretch of C2C12 muscle cells or treadmill running of wild-type mice with CKD stimulated SGK-1 production, and treadmill running inhibited proteolysis in muscle. These protective responses were absent in SGK-1-KO mice. Thus, SGK-1 could be a mechanical sensor that mediates exercise-induced improvement in muscle wasting stimulated by CKD.
Keywords: chronic kidney disease, Cell Signaling, nutrition, SGK-1
CKD is frequently complicated by muscle atrophy because of stimulation of muscle protein degradation mediated by increased activity of both caspase-3 and the ubiquitin-proteasome system (UPS).1 In muscles of rodents with CKD, caspase-3 stimulates cleavage of the complex structure of muscle protein to provide substrates for degradation by the UPS; it also stimulates proteolytic activity of the 26S proteasome.2 The UPS can selectively degrade myofibrillar protein by stimulating the expression of ubiquitin E3 ligases, Atrogin1 and MuRF1. These ligases conjugate ubiquitin to proteins targeted for protein degradation, resulting in their degradation in the 26S proteasome.3 Conditions associated with CKD that initiate loss of muscle mass include metabolic acidosis, inflammation, excess angiotensin II, myostatin expression, and insulin resistance.1 A prominent mechanism causing muscle wasting in insulin-resistant conditions involves a reduction in the phosphorylation of insulin-receptor substrate 1. The degradation of insulin-receptor substrate 1 impairs activities of phosphatidylinositol 3-kinase (PI3K) and the Akt.4 The reduced phosphorylation of these signaling proteins results in decreased phosphorylation of forkhead transcription factors (FoxOs) with activation of caspase-3 and expression of the ubiquitin E3 ligases, Atrogin1 and MuRF1. The result is stimulation of muscle protein degradation.5–7 Akt also can block Smad2/3 activation, and this process is associated with blocking muscle wasting.8,9 Activation of the Smad2/3 pathway might serve as a stimulator of proteolysis by inducing Atrogin1 and MuRF1.
Serum glucocorticoid–regulated kinase 1 (SGK-1) is a downstream target of PI3K, and it has a 54% sequence identity with Akt.10,11 SGK-1 regulates certain physiologic functions and participates in the development of hypertrophy and hyperplasia in different cell types.12 For example, hibernating hamsters do not lose muscle mass as long as SGK-1 is expressed in muscle. SGK-1 also regulates skeletal muscle metabolism by inactivating FoxO3a while stimulating the mammalian target of rapamycin pathway. Although SGK-1 can influence skeletal muscle homeostasis,13 its role in CKD–induced muscle wasting is unknown.
In this study, we found that CKD decreases SGK-1 expression in muscle. Knockout (KO) of SGK-1 was found to significantly increase protein degradation via the UPS. Our goal is to identify the pathways causing these catabolic responses.
Results
CKD Inhibits Expression and Activation of SGK-1
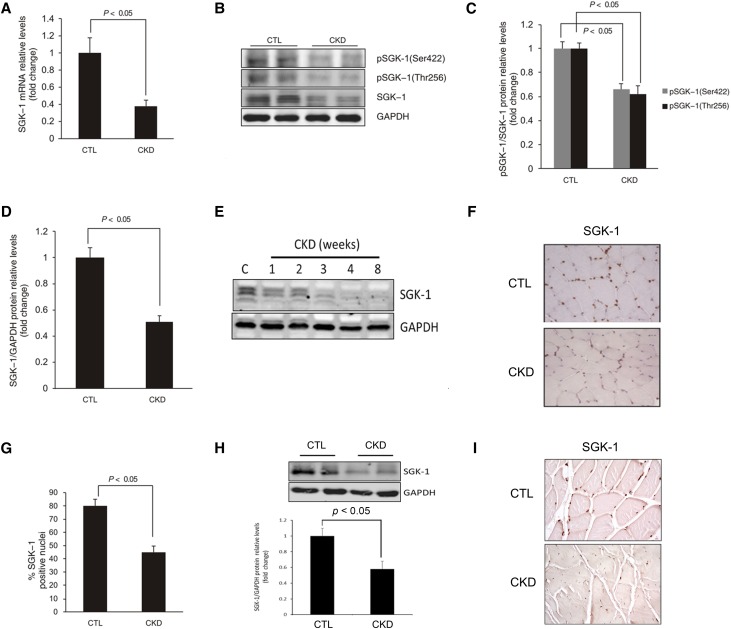
SGK-1 mRNA was decreased in muscle from CKD mice compared with results in control mice (Figure 1A). In addition, the levels of total SGK-1 and phosphorylated SGK-1 on Ser422 and Thr256 were significantly inhibited in muscle of CKD mice (Figure 1, B–D). The SGK-1 protein in muscles was decreased in a time-dependent manner by CKD, beginning as early as 2 weeks after creation of CKD (Figure 1E). Similar reductions in SGK-1 were found in the liver, lung, and heart but not found in the stomach (Supplemental Figure 1). Results of immunostaining of SGK-1 revealed it in the nuclei of myofibers, and CKD decreased the SGK-1 signal (Figure 1, F and G).
Figure 1.
CKD suppresses SGK-1 expression and activation. TA muscles of CKD and pair–fed control mice were analyzed by (A) RT-PCR and (B) Western blot. (C and D) The intensity analysis of SGK-1 protein and its phosphorylation are summarized. (E) TA muscle were collected from mice at different time points after the removal of the second kidney, and the expression of SGK-1 protein was determined. (F) Immunohistologic staining of SGK-1 in TA muscle (brown nuclei are SGK-1 positive). (G) The percentages of SGK-1–positive nuclei in TA muscles of control or CKD mice are shown. (H and I) SGK-1 expression in patients with CKD was detected by (H) Western blot and (I) immunostaining; the density analysis was performed and is shown in H, lower panel. For A, C, D, and G, five mice in each group were examined; for E, three mice in each group were examined (P<0.05). CTL, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
In muscle biopsies from patients with CKD, the expression of SGK-1 was decreased (Figure 1H); immunohistochemistry revealed that SGK-1 expression in muscles of these patients is decreased compared with results from healthy adults (Figure 1I). Because CKD causes inflammation and increases cytokine production,9,14 we examined how inflammation changes SGK-1 expression in C2C12 muscle cells. Both the expression and activation of SGK-1 were significantly decreased in cells that were treated with cytokines versus control cells (Supplemental Figure 2).
SGK-1-KO Stimulates CKD-Induced Atrogin1 and MuRF1, Promoting Muscle Wasting
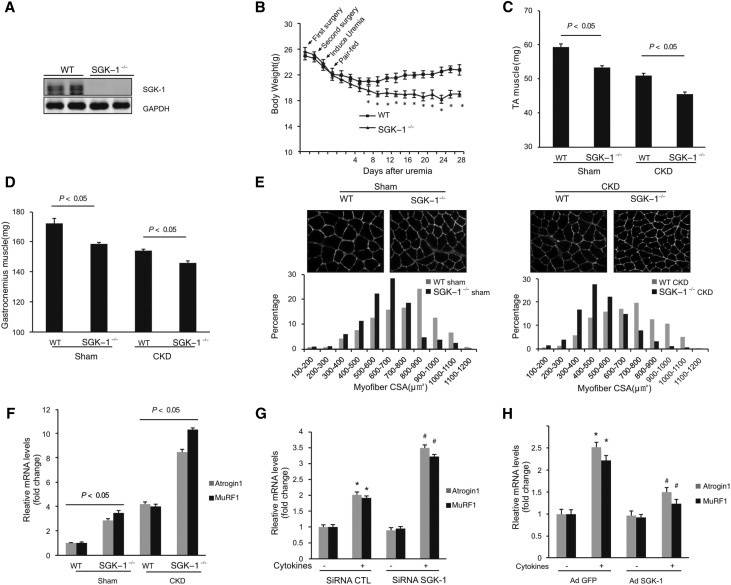
SGK-1-KO mice with CKD had a greater loss of body weight (Figure 2, A and B) and weights of tibialis anterior (TA) and gastrocnemius muscles versus responses in wild-type (WT) mice with CKD (Figure 2, C and D). Moreover, the distributions of the cross-sectional areas of myofibers of the TA muscle were left shifted and smaller in SGK-1-KO mice versus results in myofibers in muscles of WT mice (Figure 2E, left panel). This was also true in mice with CKD (Figure 2E, right panel). The losses of body and muscle mass were linked to muscle protein degradation in the UPS, because there was an increase in expression of the ubiquitin E3 ligases, Atrogin1 and MuRF1.15 SGK-1-KO also amplified the expression of these ubiquitin E3 ligases (Figure 2F).
Figure 2.
SGK-1-KO increases CKD–induced muscle wasting and activation of Atrogin1 and MuRF1. (A) Western blot analysis of SGK-1 expression in TA muscle in WT and SGK-1-KO mice (n=5). (B) Changes in body weight of WT and SGK-1-KO mice over 4 weeks after creation of CKD. *P<0.05 versus WT mice. (C and D) Weights of (C) TA and (D) gastrocnemius muscle from WT and SGK-1-KO mice after 4 weeks after creation of CKD (n=5). (E) Myofiber sizes (cross-sectional areas [CSAs]) in WT and SGK-1-KO mice, including sham and CKD mice, were studied. (E, upper panels) Cross-sections of TA muscles of WT and SGK-1-KO mice were immunostained with antilaminin. (E, lower panels) The distribution of myofiber sizes was calculated as the percentage of the number of myofibers in a designated area divided by the total number of myofibers in the area. (F) E3 ubiquitin ligases Atrogin1 and MuRF1 mRNAs in TA muscles were determined in WT and SGK-1-KO mice, including both control and CKD mice (n=5 pairs of mice). (G and H) C2C12 cells were (G) transfected with control scrambled or SGK-1 siRNA for 72 hours or (H) infected with adenovirus expressing SGK-1. Cells were treated with cytokine cocktail (IL-6 at 2 ng/ml, TNF-α at 2 ng/ml, and TGF-β1 at 2 ng/ml) for 24 hours. The expressions of Atrogin1 and MuRF1 were detected by RT-PCR. Representative data are shown from three repeated experiments. CTL, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; siRNA, short interfering RNA. *P<0.05 versus no treatment; #P<0.05 versus CTL siRNA or AdGFP (n=3).
Similar responses were obtained in cultured C2C12 myoblasts exposed to cytokines. Specifically, C2C12 myoblasts treated with cytokines had increased expressions of Atrogin1 and MuRF1. When SGK-1 was knocked down with SGK-1 siRNA, the expressions of Atrogin1 and MuRF1 increased even more. (Figure 2G). Overexpression of SGK-1 suppressed cytokine-induced expression of the two E3 ubiquitin ligases (Figure 2H).
SGK-1-KO Increases the Activation of FOXO3a and Smad2/3
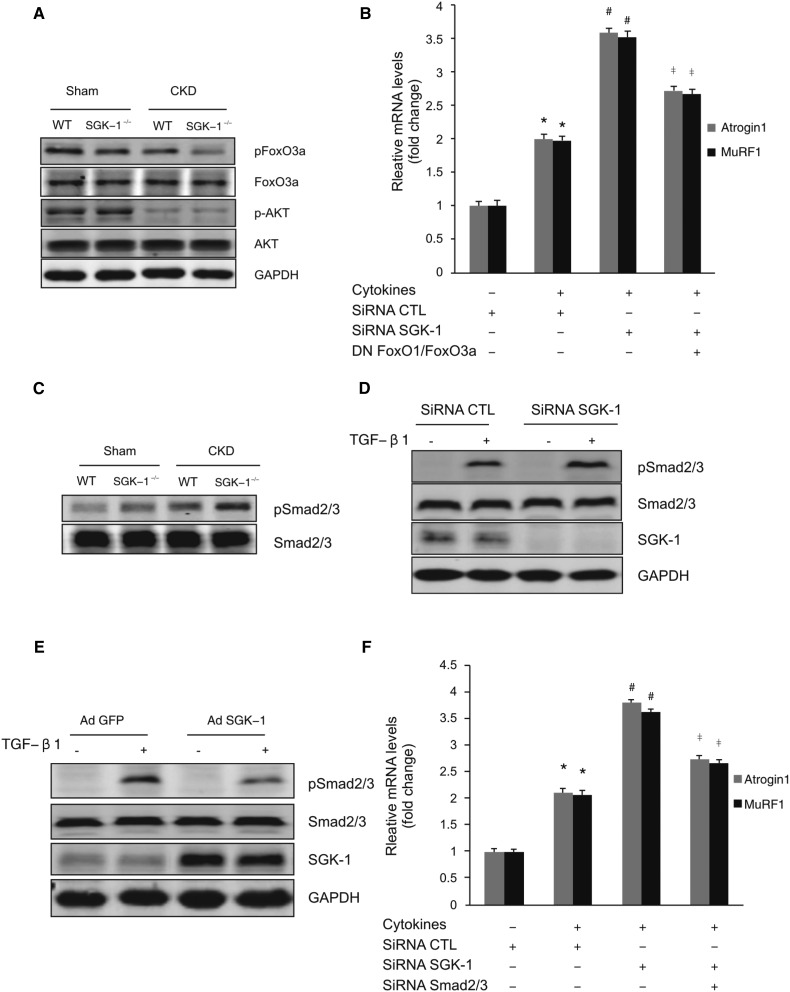
The FoxO3a transcription factor enhances Atrogin1 and MuRF1 expression in muscle.7 Although Akt can phosphorylate FoxO3a,16,17 the role of SGK-1 in CKD-induced dephosphorylation of FoxO3a is unknown. In muscles of SGK-1-KO mice with CKD, there is a reduction in phosphorylation of FoxO3a compared with results in WT mice. SGK-1-KO did not affect muscle levels of Akt and pAkt (Figure 3A). These results indicate that SGK-1 contributes to the phosphorylation of FoxO3a in muscle. Because Akt1 has been reported to regulate Foxo3a phosphorylation and muscle wasting, we compared the expressions of Atrogin1 and MuRF1 in C2C12 muscle cells that were treated by inhibitors of SGK-1 and/or Akt1. In both cases, the cytokine-induced expression of Atrogin1 and Murf1 in C2C12 cells was reduced when SGK-1 or Akt1 were suppressed. This response was enhanced by adding the two inhibitors together (Supplemental Figure 3A). Other than these results, we studied cells overexpressing SGK-1 or Akt1. Each kinase suppressed approximately 50% of the cytokine-induced expression of atrogin1/Murf1 as well as the phosphorylation of Foxo3a (Supplemental Figure 3, B and C). Our results indicate that both SGK-1 and AKT are involved in controlling FoxO3a phosphorylation and the expression of the ubiquitin E3 ligases.
Figure 3.
SGK-1 inhibits activation of Atrogin1 and MuRF1 via FoxO and Smad2/3 signaling pathways. (A) Representative Western blot indicates levels of signaling proteins in TA muscles of WT and SGK-1-KO mice after 4 weeks. (n=5 per group). (B) C2C12 cells were transfected with either an siRNA against SGK-1 or dominant negative FoxO1/FoxO3a plasmids; they then were treated with the cytokine cocktail. Atrogin1 and MuRF1 mRNAs were analyzed by RT-PCR. (C) Western blots analysis of Smad2/3 expression and phosphorylation in muscles of control and CKD in WT and SGK-1-KO mice (n=5). (D) C2C12 cells were transfected with control or SGK-1 siRNA, or (E) they were infected with control or AdSGK-1 adenovirus; the TGF-β1 addition (2 ng/ml at 30 minutes) induced pSmad2/3. (F) C2C12 cells were treated as indicated, and Atrogin1 and MuRF1 were determined by RT-PCR. CTL, control; DN, dominant negative; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; siRNA, short interfering RNA. *P<0.05 versus control; #P<0.05 versus control siRNA; ‡P<0.05 versus SGK-1 siRNA only (n=3 repeats).
To test whether FoxO1/3 is the sole mediator of the increased expression of Atrogin1 and MuRF1 that is induced by SGK-1-KO, we studied C2C12 cells expressing dominant negative FoxO1/FoxO3. The dominant negative proteins partial suppressed expressions of Atrogin1 and MuRF1 (48.6% and 46.8%, respectively) induced by SGK-1-KO (Figure 3B). Could another pathway add to SGK-1/Atrogin1/MuRF1 signaling? Because Smad regulates Atrogin1 and MuRF1 expression in muscle, we examined whether SGK-1 changes phosphorylation of Smad2/3. In muscles of sham and CKD mice, SGK-1-KO increased Smad2/3 phosphorylation without changes in total Smad2/3 levels (Figure 3C). In TGF-β1–treated C2C12 cells, SGK-1 knockdown promoted Smad2/3 phosphorylation (Figure 3D), whereas SGK-1 overexpression suppressed this response (Figure 3E). Moreover, we found that SGK-1 knockdown stimulated Atrogin1 and MuRF1 expression, and this response was reduced by 60.9% and 56.3%, respectively, when Smad2/3 was knocked down by siRNA (Figure 3F). Therefore, the inhibition of Atrogin1/MuRF1 expression by SGK-1 can be regulated through both the FoxO1/3 and Smad2/3 signaling pathways.
Phosphorylated NDRG1 Interacts with Smad2/3 to Regulate Its Phosphorylation
To determine if SGK-1 interacts with Smad2/3, we studied C2C12 cells and immunoprecipitated them with an SGK-1 antibody. There was no interaction between SGK-1 and Smad2/3. Because SGK-1 overexpression led to NDGR1 phosphorylation (Figure 4B) and because NDRG1 reportedly interferes with Smad activation,18–20 next, we examined whether there was an interaction of NDRG1 and Smad. In C2C12 cells infected with AdGFP or AdSGK-1 and then treated with TGF-β1, we found a direct interaction between NDRG1 and Smad2/3. Overexpression of SGK-1 promoted these interactions (Figure 4C). When NDRG1 was knocked down, the Smad2/3 phosphorylation was increased (Figure 4D). These results suggest that SGK-1 phosphorylates NDRG1 to enhance the interaction between NDRG1 and Smad2/3. This resulted in inhibition of Smad2/3 phosphorylation. When NDRG1 was knocked down, Smad2/3 phosphorylation increased in the presence of SGK-1 (Figure 4E). With the loss of NDRG1, the expression of Atrogin1 and MuRF1 increased (Figure 4F).
Figure 4.
NDRG1 mediates SGK-1 suppression of Smad2/3. (A) C2C12 cells were treated with TGF-β1, and interactions between SGK-1 and Smad2/3 were determined by immunoprecipitation with anti–SGK-1 antibodies. The precipitated proteins were detected by Western blot using indicated antibodies. (B) Phosphorylation of NDRG1 was determined in C2C12 cells infected with control vector or AdSGK-1. (C) The interactions between NDGR1 and Smad2/3 were determined by immunoprecipitation of C2C12 cells with anti-NDRG1 antibodies. C2C12 cells, infected with AdGFP or AdSGK-1, were treated with TGF-β1 for 1 hour, and the interaction of Smad2/3 with NDRG1 was detected. (D) NDRG1 knockdown increases TGF-β1–induced Smad2/3 phosphorylation. Cells were treated with NDRG1 siRNA for 72 hours and then exposed to TGF-β1 for 30 minutes. The phosphorylation of Smad2/3 was determined. (E and F) Knockdown of NDRG1 suppresses SGK-1–mediated inhibition of Smad2/3 phosphorylation and Atrogin1/MuRF1 expression. C2C12 cells were infected with control or NDRG1 siRNA in the presence or absence of AdSGK-1. TGF-β1–induced (E) Smad2/3 phosphorylation and (F) Atrogin1/MuRF1 expression were determined by Western blot and RT-PCR, respectively. Data represent three repeats of the experiments. AdGFP, GFP adenovirus; CTL, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IB, immunoblot; IP, immunoprecipitation; siRNA, short interfering RNA.
Overexpression of SGK-1 Reverses CKD–Induced Muscle Atrophy
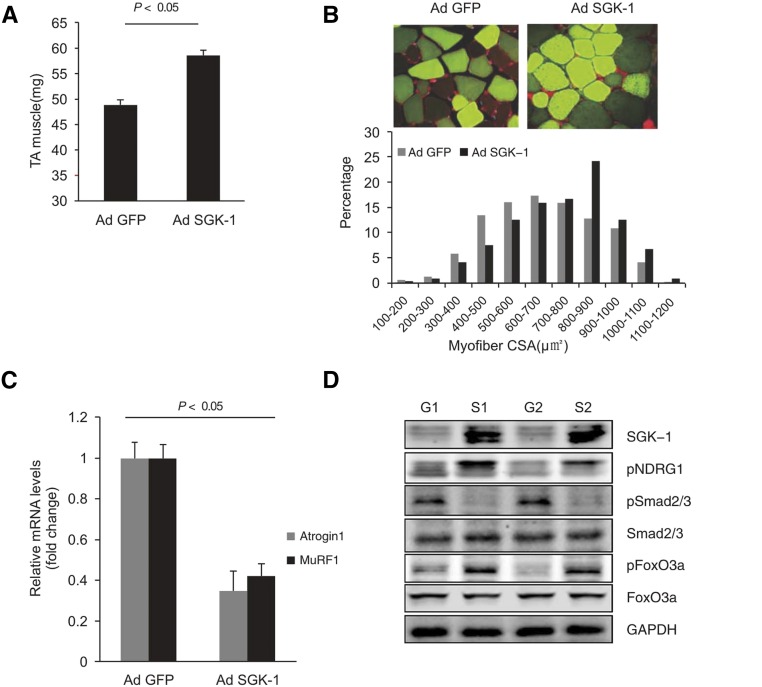
TA muscles were injected with control or SGK-1 adenoviruses. Overexpression of SGK-1 increased muscle weight (Figure 5A) and the size of myofibers in TA muscles. In addition, the distribution of the cross-sectional area was shifted to the right versus results in control mice (Figure 5B). Importantly, the expressions of Atrogin1 and MuRF1 were decreased in muscles overexpressing SGK-1 (Figure 5C). Overexpression of SGK-1 also increased phosphorylation of NDRG1 and FoxO3a but decreased Smad2/3 phosphorylation (Figure 5D). Thus, a higher SGK-1 level increases muscle size while reducing Atrogin1 and MuRF1.
Figure 5.
Overexpression of SGK-1 reverses CKD–induced muscle atrophy. (A) TA muscles were injected with AdGFP or AdSGK-1, and after 1 month, TA muscle weights were measured (n=10 pairs of mice; P<0.05 versus GFP mice). (B) Cross-sections of TA muscles were stained with GFP (green) and laminin (red). Myofiber areas were measured, and their distribution was calculated as in Figure 2. (C) The expressions of Atrogin1 and MuRF1 were determined by RT-PCR (n=5 pairs of mice; P<0.05 versus GFP mice). (D) Western blots was performed to examine the indicated molecules (n=5). AdGFP, GFP adenovirus; CSA, cross-sectional area; G1, sample 1 with adGFP; G2, sample 2 with AdGFP; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; S1, sample 1 with AdSGK-1; S2, sample 2 with AdSGK-1.
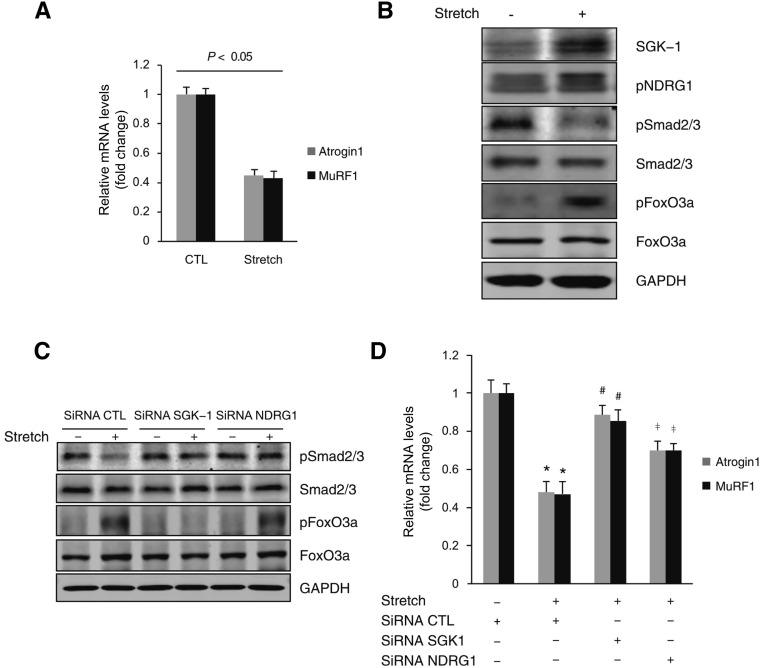
Mechanical Stretch through SGK-1 Decreases Atrogin1/MuRF1 Expression
SGK-1 is a sensor of mechanical stretch,21,22 and we found that mechanical stretch of C2C12 cells induced SGK-1 expression and inhibited Atrogin1/MuRF1 expression (Figure 6, A and B). In these cells, mechanical stretch also induced phosphorylation of NDRG1 and FoxO3a, but Smad2/3 phosphorylation was decreased (Figure 6B). The decrease in phosphorylation of Smad2/3 in cells being stretched was reversed by knockdown of SGK-1 or NDRG1. Stretch also induced FoxO3a phosphorylation, which was abolished by knockdown of SGK-1 but not by knockdown of NDRG1 (Figure 6C). Thus, phosphorylation of FoxO3a and Smad2/3 is differentially regulated through SGK-1. Ultimately, knockdown of SGK-1 blocks mechanical stretch–induced inhibition of Atrogin1/MuRF1 (Figure 6D).
Figure 6.
Mechanical stretch decreases Atrogin1/MuRF1 expression through SGK-1. (A and B) C2C12 cells were exposed to mechanical stretch (12% elongation for 24 hours), and mRNA levels of Atrogin1/MuRF1 and SGK-1 and its downstream targets were determined by real–time RT-PCR and Western blot, respectively (P<0.05 versus no stretch; n=3 repeats). (C) C2C12 cells were transfected with SGK-1 siRNA or NDRG1 siRNA for 72 hours and then subjected to mechanical stretch for 24 hours. The phosphorylation of Smad2/3 and FoxO3a was determined by Western blot. (D) After treatment as in C, Atrogin1/MuRF1 mRNA was determined by RT-PCR. CTL, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; siRNA, short interfering RNA. *P<0.05 versus no stretch; #P<0.05 versus control siRNA; ‡P<0.05 versus SGK-1 siRNA (n=3 repeats).
Exercise Raises SGK-1 to Ameliorate CKD–Induced Muscle Wasting
Mice with CKD were subjected to treadmill exercise for 2 weeks, resulting in a significant increase in weights of TA and gastrocnemius muscles versus results in nonexercised mice. These responses were attenuated in SGK-1-KO mice (Figure 7, A and B). Exercise also increased the sizes of myofibers in TA muscles of mice with CKD. These beneficial effects were blocked by SGK-1-KO (Figure 7C). In WT mice with CKD, exercise increased SGK-1 and regulated phosphorylation of Smad2/3 and FoxO3a. These responses were blocked in SGK-1-KO mice (Figure 7D). Moreover, exercise decreased Atrogin1/MuRF1 expression to preserve muscle mass (Figure 7E).
Figure 7.
SGK-1 mediates exercise–induced muscle hypertrophy. In WT or SGK-1-KO mice, treadmill-running exercise was performed for 2 weeks after CKD was created. (A and B) The (A) TA and (B) gastrocnemius muscles were collected and weighted (n=6 pairs of mice; P<0.05 versus no exercise mice). (C) Myofiber areas were measured, and their distribution was calculated. (D) Western blot analysis showed the exercise-induced changes in the indicated proteins from WT and SGK-1-KO mice with CKD. (E) Atrogin1 and MuRF1 were analyzed by RT-PCR in TA muscles (n=6). CSA, cross-sectional area; CTL, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NS, non-significant.
Discussion
In addition to the traditional IGF-1/PI3K signaling and its influence on muscle homeostasis,23,24 SGK-1 is a new candidate for the regulation of CKD–induced muscle atrophy. We have shown that CKD suppresses SGK-1, which leads to increased muscle proteolysis by the UPS (assessed by measuring Atrogin1 and MuRF1 expression in muscle). In cultured myoblasts, we uncovered that SGK-1 activates at least two pathways that regulate protein metabolism: (1) SGK-1 phosphorylates FoxO3a, and (2) SGK-1 and its substrate, NDRG1, suppress Smad2/3 phosphorylation in muscle cells. We examined if these pathways are present in mice and learned that SGK-1 could counteract the muscle wasting stimulated by CKD. The importance of SGK-1 was confirmed when we showed that overexpressing SGK-1 in muscles of mice actually blocks CKD–induced muscle wasting. The mechanism for this response is that SGK-1 phosphorylates FoxO3a, which suppresses Atrogin1, MuRF1, and muscle proteolysis in the UPS.
To determine if an increase in SGK-1 would suppress CKD–induced muscle wasting, we initially studied a cell model of exercise consisting of stretching C2C12 myoblasts. This increased SGK-1 expression and the phosphorylated FoxO3a, leading to suppression of Atrogin1 and MuRF1 expression. Next, we tested mice using an exercise program: treadmill running. Exercise increased the expression of SGK-1, resulting in decreased expression of Atrogin1 and MuRF1. Importantly, genetic deletion of SGK-1 abolished these positive responses to exercise. We conclude that raising SGK-1 could be a new target for the prevention of muscle wasting stimulated by CKD.
We found that SGK-1-KO mice had a decrease in the masses of TA and gastrocnemius muscles versus results from WT mice (Figure 2, B–D). In contrast, the body weights of SGK-1-KO and WT mice were not different in normal condition. We suggest that the absence of difference in body weight was because of a gain in the mass of another tissue (e.g., potentially adipose tissue). This mechanism will be the subject of future studies.
In starvation or catabolic conditions (e.g., inflammation and CKD), muscle wasting is a clinical problem that contributes to morbidity and mortality.13,25 A signal that initiates muscle wasting is impairment in insulin or IGF-1 signaling pathways that phosphorylate a sequence of proteins, including SGK-1 and Akt.26–28 SGK-1 and Akt are activated by PI3K and mTORC2 phosphorylation at two separate sites (for SGK-1, Ser422 and Thr256 and for Akt, Ser473 and Thr308).29 SGK-1 and Akt phosphorylate specific substrates (FoxO3a and p27kip1, respectively),30 although SGK-1 protects against immobilization–induced muscle wasting, which is not regulated by its sister kinase, Akt.31 In contrast, our results show that both SGK-1 and Akt regulate FoxO3a phosphorylation and the expression of Atrogin1/MurF1 in C2C12 cells.
The influence of SGK-1 on skeletal muscle homeostasis has been tested in hibernating animals.13 In these studies, neither skeletal muscle size nor morphology was altered in hibernating hamsters. The demonstration that muscle mass is maintained in hibernating animals (bears, hamsters, et al.) when SGK-1 is active provides insights into the mechanism that maintains a stable muscle mass in these animals. Notably, the degradation of muscle protein is blocked by SGK-1, thereby maintaining muscle mass. This also explains why the urea concentration is low in these animals, because the absence of proteolysis will not provide amino acids to be used for urea synthesis. These considerations emphasize how SGK-1 can influence interlocking metabolic responses.
SGK-1-KO activates at least two signaling pathways. First, SGK-1-KO does not phosphorylate FoxO3a, thereby promoting its entry into the nucleus, which stimulates the expression of Atrogin1 and MuRF1. This explains our finding that Atrogin1 expression is reduced when SGK-1 is overexpressed or if there is a dominant negative FoxO3a/FoxO1 construct. Second, SGK-1 can indirectly interfere with Smad2/3 signaling. We showed that phosphorylation of NDRG1 suppresses the expression of Atrogin1 and MuRF1 by directly interacting with Smad2/3. Interactions among SGK-1, NDRG1, and Smad3 lead to decreased Smad2/3 phosphorylation. Other reports are consistent with the finding that NDRG1 is phosphorylated by SGK-1.19,32 Others report that phosphorylation of NDRG1 inhibits Smad–induced epithelial-mesenchymal transition.20,32 Phosphorylation of NDRG1 also can inhibit NF-κB transcriptional activities and the expression of CXC chemokines.18 Thus, phosphorylated NDRG1 not only suppresses inflammation but also, prevents muscle atrophy.
In skeletal muscle cells, vascular smooth muscle cells, or kidney collecting duct cells, SGK-1 mediates responses to mechanical stretch.33–35 Exercise also increases the IGF-1 level in rats or mice with CKD as well as patients on hemodialysis.36–38 This is relevant, because our results show that SGK-1 expression rises in response to treadmill running. Clinical studies suggest that exercise can reduce CKD–stimulated muscle wasting in patients. In summary, our results indicate that exercise training could potentially activate SGK-1, which would protect against the muscle atrophy caused by CKD.
Concise Methods
Animals
All studies were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine and performed in accordance with National Institutes of Health guidelines. Mice were housed in a conventional animal facility with a 12-hour light/dark cycle. WT mice were from The Jackson Laboratory (Bar Harbor, ME). SGK-1-KO mice were used as described.21,22
Reagents
Penicillin, streptomycin, DMEM, and FBS were obtained from Life Technologies (Carlsbad, CA). The protein assay kit was from Bio-Rad (Hercules, CA). Antibodies against SGK-1 were purchased from Sigma-Aldrich (St. Louis, MO). Although antibodies against FoxO3a, pFoxO3a (Th32), Akt, pAkt (S473), Smad2/3, pSmad2/3, SGK, and pNDRG1 were purchased from Cell Signaling Technology (Danvers, MA), antibodies against pSGK-1 (Thr256) were purchased from Upstate Biotechnology (Billerica, MA). Antibodies against pSGK-1 (Ser422), NDRG1, laminin, GAPDH, the secondary antibodies, and IgG-conjugated agarose were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Recombinant IL-6 and TNF-α were obtained from Millipore Cooperation (Billerica, MA). Recombinant human TGF-β1 was purchased from eBioscience (San Diego, CA). siRNAs of SGK-1, Smad2/3, and control siRNA were purchased from GE Dharmacon (Lafayette, CO); siRNA NDRG1 was purchased from Santa Cruz Biotechnology. SGK-1 inhibitor was from Sigma-Aldrich, and Akt inhibitor X was purchased from EMD Millipore (Billerica, MA). Plasmids of dominant negative FoxO1/FoxO3 were purchased from Addgene (Cambridge, MA).39,40 Adenovirus expressing SGK-1 was constructed by cloning human SGK-1 cDNA into the pAdTrack-CMV plasmid using the AdEasy System (Clontech Laboratories, Mountain View, CA).21 This SGK-1 adenovirus was injected into mouse TA and gastrocnemius muscles to stimulate SGK-1 expression; adenovirus that expresses GFP vector was used as control.
CKD Model
CKD was induced by subtotal nephrectomy in anesthetized mice as described.8,41 After matching for body weights, subtotal nephrectomy was performed in anesthetized mice (ketamine at 125 mg/kg body wt and xylazine at 6.4 mg/kg body wt) in a two–step surgery method. The left kidney was decapsulated to avoid ureter and adrenal damage. Approximately three quarters of the left kidney were removed. During recovery, the mice were given two doses of buprenorphine (0.1–2.5 mg/kg body wt) after surgery and 12 hours later. The diet was 6% Protein Rodent Diet Chow (Harlan Teklad, Madison, WI) ad libitum to reduce mortality and limit hypertrophy. The right kidney was removed 1 week later, and paired CKD and sham–operated control mice were pair fed 40% protein chow. The BUN was measured by the Comparative Pathology Laboratory Center at Baylor College of Medicine, and serum creatinine was measured using the QuantiChrom Creatinine Assay Kit (BioAssay Systems, Hayward, CA). The BUN and serum creatinine were significantly increased in both WT and SGK-1-KO mice (data not shown). Human CKD samples were examined as described.9 During placement of a peritoneal dialysis catheter in patients with CKD, the rectus abdominis muscle was biopsied, frozen at −80°C, and stored until analyzed. Control muscle biopsies were obtained from healthy subjects during abdominal surgeries.
Exercise Training
Mice were exercised by running (Model 1055MSD Exer-6M Treadmill; Columbus Instruments, Columbus, OH). Before subtotal nephrectomy, mice ran at 15 m/min for 30 minutes daily. The exercise intensity was progressively increased to reach 30 m/min. After a 1-week recovery from the subtotal nephrectomy, the exercise was reinitiated, and mice were exercised by daily running at 30 m/min for 30 minutes for 2 weeks. Age– and weight–matched control and pair-fed mice were exercised at the same time rate and intensity.
Cell Culture
C2C12 cells (American Type Culture Collection, Manassas, VA) were cultured as described.42 The cells were treated with a cytokine mixture of IL-6 (2 ng/ml), TNF-α (2 ng/ml), and TGF-β1 (2 ng/ml).4
Mechanical Stretch
C2C12 cells were plated on silicone elastomer–bottomed plates and subjected to mechanical stretch as described.33 Briefly, at 90% confluence, cells were exposed to a cyclic mechanical stretch with a computer–controlled mechanical Strain Unit (Flexcell 4000) set at cyclic (60 cycles per minute) stretch. This yields a uniform deformation with 15% elongation of the elastomer-bottomed plates. Cell lysates were collected after 24 hours of stretch to be analyzed by Western blot and RT-PCR.
Immunohistochemical Staining
Frozen serial transverse cryosections (8 mm) from the midbelly of TA muscles from control and CKD mice were mounted on glass slides. The slides were blocked with 10% goat serum (Vector Laboratories, Burlingame, CA) for 30 minutes and then, incubated with primary antibodies (SGK-1 at 1:300 and laminin at 1:300). Sections were washed in 0.5% Tween 20 in PBS and incubated at room temperature with a biotinylated secondary antibody (Vector Laboratories). After washing in 0.5% Tween 20 in PBS, sections were incubated with an Elite ABC Reagent (Vector Laboratories) following the instructions as described in a peroxidase substrate kit (Vector Laboratories). Sections were counterstained by hematoxylin. For immunofluorescence staining of samples, fluorescent secondary antibodies were applied to sections, and DAPI was used as a counterstain. Pictures were recorded using a Nikon Eclipse 80i Fluorescence Microscope (Nikon, Tokyo, Japan).
Real–Time RT-PCR
Total RNA from TA muscles or cells was isolated using the RNeasy Kit (Qiagen, Valencia, CA). Real–time RT-PCR was performed using the Opticon Real–Time RT-PCR Machine (MJ Research, Waltham, MA). GAPDH was used as an internal standard. The primers were designed for mouse SGK-1 (forward 5′-TTCGTTAGCCTTTGGTGGAGTTGC-3′ and reverse 5′-AGCACCACGTTGGAAGGAAGAGAA-3′), mouse Atrogin1 (forward 5′-GAGGCAGATTCGCAAGCGTTTGAT-3′ and reverse 5′-TCCAGGAGAGAATGTGGCAGTGTT-3′), mouse MuRF1 (forward 5′-AGTGTCCATGTCTGGAGGTCGTTT-3′ and reverse 5′-ACTGGAGCACTCCTGCTTGTAGAT-3′), and GAPDH (forward 5′-AGTGGGAGTTGCTGTTGAAATC-3′ and reverse 5′-TGCTGAGTATGTCGTGGAGTCTA-3′).
Western Blot and Immunoprecipitation
TA or gastrocnemius muscles or cells extracts were prepared in RIPA buffer; protein concentrations in the extracts were assayed using a Bradford Protein Assay Kit (Bio-Rad). About 30 μg proteins were separated by SDS-PAGE, and after transferring to nitrocellulose membranes, immunoblots were probed separately with specific primary antibodies. Fluorescence–labeled secondary antibodies were used to detect protein using the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
Immunoprecipitation was performed by adding 4 μg anti–SGK-1 or anti-NDRG1 antibodies to C2C12 lysates. Phosphatase and protease inhibitors were added, and after shaking overnight at 4°C, 30 ml Protein A/G Plus Beads were added. The mixture was agitated in a rotational shaker for 2 hours at 4°C, and after centrifugation at 1000×g, the supernatant was discarded. The beads were washed three times with PBS, and Western blots were performed.
Statistical Analyses
All data are presented as means±SEMs. Results were analyzed using t test when results from two groups were compared or two-way ANOVA when data from three or more groups were studied; P<0.05 was considered statistically significant.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank Dr. William E. Mitch for insight discussion and sharing the human CKD samples and Dr. Liping Zhang for sharing the human CKD samples. We also thank Hunter Cheng for helpful discussion.
This work was supported by American Heart Association grants 115GRNT25700209 and R01-DK095867 (to J.C.), National Institutes of Health grants R37 and DK37175, and a grant from Dr. and Mrs. Harold Selzman.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Glucocorticoid-Regulated Kinase: Linking Azotemia and Muscle Wasting in CKD,” on pages 2545–2547.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2015080867/-/DCSupplemental.
References
- 1.Mitch WE, Goldberg AL: Mechanisms of muscle wasting. The role of the ubiquitin-proteasome pathway. N Engl J Med 335: 1897–1905, 1996 [DOI] [PubMed] [Google Scholar]
- 2.Du J, Wang X, Miereles C, Bailey JL, Debigare R, Zheng B, Price SR, Mitch WE: Activation of caspase-3 is an initial step triggering accelerated muscle proteolysis in catabolic conditions. J Clin Invest 113: 115–123, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang XH, Mitch WE: Mechanisms of muscle wasting in chronic kidney disease. Nat Rev Nephrol 10: 504–516, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas SS, Dong Y, Zhang L, Mitch WE: Signal regulatory protein-α interacts with the insulin receptor contributing to muscle wasting in chronic kidney disease. Kidney Int 84: 308–316, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL: Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117: 399–412, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han HQ, Zhou X, Mitch WE, Goldberg AL: Myostatin/activin pathway antagonism: Molecular basis and therapeutic potential. Int J Biochem Cell Biol 45: 2333–2347, 2013 [DOI] [PubMed] [Google Scholar]
- 7.Lee SW, Dai G, Hu Z, Wang X, Du J, Mitch WE: Regulation of muscle protein degradation: Coordinated control of apoptotic and ubiquitin-proteasome systems by phosphatidylinositol 3 kinase. J Am Soc Nephrol 215: 1537–1545, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Rajan V, Lin E, Hu Z, Han HQ, Zhou X, Song Y, Min H, Wang X, Du J, Mitch WE: Pharmacological inhibition of myostatin suppresses systemic inflammation and muscle atrophy in mice with chronic kidney disease. FASEB J 25: 1653–1663, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Pan J, Dong Y, Tweardy DJ, Dong Y, Garibotto G, Mitch WE: Stat3 activation links a C/EBPδ to myostatin pathway to stimulate loss of muscle mass. Cell Metab 18: 368–379, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García-Martínez JM, Alessi DR: mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem J 416: 375–385, 2008 [DOI] [PubMed] [Google Scholar]
- 11.Yan L, Mieulet V, Lamb RF: mTORC2 is the hydrophobic motif kinase for SGK1. Biochem J 416: e19–e21, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Lang F, Böhmer C, Palmada M, Seebohm G, Strutz-Seebohm N, Vallon V: (Patho)physiological significance of the serum- and glucocorticoid-inducible kinase isoforms. Physiol Rev 86: 1151–1178, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Andres-Mateos E, Brinkmeier H, Burks TN, Mejias R, Files DC, Steinberger M, Soleimani A, Marx R, Simmers JL, Lin B, Finanger Hedderick E, Marr TG, Lin BM, Hourdé C, Leinwand LA, Kuhl D, Föller M, Vogelsang S, Hernandez-Diaz I, Vaughan DK, Alvarez de la Rosa D, Lang F, Cohn RD: Activation of serum/glucocorticoid-induced kinase 1 (SGK1) is important to maintain skeletal muscle homeostasis and prevent atrophy. EMBO Mol Med 5: 80–91, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Wang XH, Wang H, Du J, Mitch WE: Satellite cell dysfunction and impaired IGF-1 signaling cause CKD-induced muscle atrophy. J Am Soc Nephrol 21: 419–427, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas SS, Mitch WE: Mechanisms stimulating muscle wasting in chronic kidney disease: The roles of the ubiquitin-proteasome system and myostatin. Clin Exp Nephrol 17: 174–182, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori S, Nada S, Kimura H, Tajima S, Takahashi Y, Kitamura A, Oneyama C, Okada M: The mTOR pathway controls cell proliferation by regulating the FoxO3a transcription factor via SGK1 kinase. PLoS One 9: e88891, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahin P, McCaig C, Jeevahan J, Murray JT, Hainsworth AH: The cell survival kinase SGK1 and its targets FOXO3a and NDRG1 in aged human brain. Neuropathol Appl Neurobiol 39: 623–633, 2013 [DOI] [PubMed] [Google Scholar]
- 18.Murakami Y, Hosoi F, Izumi H, Maruyama Y, Ureshino H, Watari K, Kohno K, Kuwano M, Ono M: Identification of sites subjected to serine/threonine phosphorylation by SGK1 affecting N-myc downstream-regulated gene 1 (NDRG1)/Cap43-dependent suppression of angiogenic CXC chemokine expression in human pancreatic cancer cells. Biochem Biophys Res Commun 396: 376–381, 2010 [DOI] [PubMed] [Google Scholar]
- 19.Murray JT, Campbell DG, Morrice N, Auld GC, Shpiro N, Marquez R, Peggie M, Bain J, Bloomberg GB, Grahammer F, Lang F, Wulff P, Kuhl D, Cohen P: Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem J 384: 477–488, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z, Zhang D, Yue F, Zheng M, Kovacevic Z, Richardson DR: The iron chelators Dp44mT and DFO inhibit TGF-β-induced epithelial-mesenchymal transition via up-regulation of N-Myc downstream-regulated gene 1 (NDRG1). J Biol Chem 287: 17016–17028, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng J, Truong LD, Wu X, Kuhl D, Lang F, Du J: Serum- and glucocorticoid-regulated kinase 1 is upregulated following unilateral ureteral obstruction causing epithelial-mesenchymal transition. Kidney Int 78: 668–678, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng J, Wang Y, Ma Y, Chan BT, Yang M, Liang A, Zhang L, Li H, Du J: The mechanical stress-activated serum-, glucocorticoid-regulated kinase 1 contributes to neointima formation in vein grafts. Circ Res 107: 1265–1274, 2010 [DOI] [PubMed] [Google Scholar]
- 23.Lecker SH, Goldberg AL, Mitch WE: Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol 17: 1807–1819, 2006 [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Hu Z, Hu J, Du J, Mitch WE: Insulin resistance accelerates muscle protein degradation: Activation of the ubiquitin-proteasome pathway by defects in muscle cell signaling. Endocrinology 147: 4160–4168, 2006 [DOI] [PubMed] [Google Scholar]
- 25.Silva KA, Dong J, Dong Y, Dong Y, Schor N, Tweardy DJ, Zhang L, Mitch WE: Inhibition of Stat3 activation suppresses caspase-3 and the ubiquitin-proteasome system, leading to preservation of muscle mass in cancer cachexia. J Biol Chem 290: 11177–11187, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glass DJ: Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat Cell Biol 5: 87–90, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Mitch WE, Hu Z, Lee SW, Du J: Strategies for suppressing muscle atrophy in chronic kidney disease: Mechanisms activating distinct proteolytic systems. J Ren Nutr 15: 23–27, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Miyazaki M, McCarthy JJ, Esser KA: Insulin like growth factor-1-induced phosphorylation and altered distribution of tuberous sclerosis complex (TSC)1/TSC2 in C2C12 myotubes. FEBS J 277: 2180–2191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearce LR, Komander D, Alessi DR: The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol 11: 9–22, 2010 [DOI] [PubMed] [Google Scholar]
- 30.Park J, Leong ML, Buse P, Maiyar AC, Firestone GL, Hemmings BA: Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J 18: 3024–3033, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacDonald EM, Andres-Mateos E, Mejias R, Simmers JL, Mi R, Park JS, Ying S, Hoke A, Lee SJ, Cohn RD: Denervation atrophy is independent from Akt and mTOR activation and is not rescued by myostatin inhibition. Dis Model Mech 7: 471–481, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun J, Zhang D, Bae DH, Sahni S, Jansson P, Zheng Y, Zhao Q, Yue F, Zheng M, Kovacevic Z, Richardson DR: Metastasis suppressor, NDRG1, mediates its activity through signaling pathways and molecular motors. Carcinogenesis 34: 1943–1954, 2013 [DOI] [PubMed] [Google Scholar]
- 33.Cheng J, Du J: Mechanical stretch simulates proliferation of venous smooth muscle cells through activation of the insulin-like growth factor-1 receptor. Arterioscler Thromb Vasc Biol 27: 1744–1751, 2007 [DOI] [PubMed] [Google Scholar]
- 34.Chen L, Wei TQ, Wang Y, Zhang J, Li H, Wang KJ: Simulated bladder pressure stimulates human bladder smooth muscle cell proliferation via the PI3K/SGK1 signaling pathway. J Urol 188: 661–667, 2012 [DOI] [PubMed] [Google Scholar]
- 35.Hills CE, Bland R, Squires PE: Functional expression of TRPV4 channels in human collecting duct cells: Implications for secondary hypertension in diabetic nephropathy. Exp Diabetes Res 2012: 936518, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun DF, Chen Y, Rabkin R: Work-induced changes in skeletal muscle IGF-1 and myostatin gene expression in uremia. Kidney Int 70: 453–459, 2006 [DOI] [PubMed] [Google Scholar]
- 37.Kopple JD, Wang H, Casaburi R, Fournier M, Lewis MI, Taylor W, Storer TW: Exercise in maintenance hemodialysis patients induces transcriptional changes in genes favoring anabolic muscle. J Am Soc Nephrol 18: 2975–2986, 2007 [DOI] [PubMed] [Google Scholar]
- 38.Workeneh BT, Mitch WE: Review of muscle wasting associated with chronic kidney disease. Am J Clin Nutr 91: 1128S–1132S, 2010 [DOI] [PubMed] [Google Scholar]
- 39.Nakae J, Barr V, Accili D: Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J 19: 989–996, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ Jr., DiStefano PS, Chiang LW, Greenberg ME: DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science 296: 530–534, 2002 [DOI] [PubMed] [Google Scholar]
- 41.Liang A, Wang Y, Han G, Truong L, Cheng J: Chronic kidney disease accelerates endothelial barrier dysfunction in a mouse model of an arteriovenous fistula. Am J Physiol Renal Physiol 304: F1413–F1420, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Z, Lee IH, Wang X, Sheng H, Zhang L, Du J, Mitch WE: PTEN expression contributes to the regulation of muscle protein degradation in diabetes. Diabetes 56: 2449–2456, 2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.