ABSTRACT
The NFAT family of transcription factors has been primarily related to T cell development, activation, and differentiation. Further studies have shown that these ubiquitous proteins are observed in many cell types inside and outside the immune system, and are involved in several biological processes, including tumor growth, angiogenesis, and invasiveness. However, the specific role of the NFAT1 family member in naive B cell proliferation remains elusive. Here, we demonstrate that NFAT1 transcription factor controls Cyclin E expression, cell proliferation, and tumor growth in vivo. Specifically, we show that inducible expression of NFAT1 inhibits cell cycle progression, reduces colony formation, and controls tumor growth in nude mice. We also demonstrate that NFAT1-deficient naive B lymphocytes show a hyperproliferative phenotype and high levels of Cyclin E1 and E2 upon BCR stimulation when compared to wild-type B lymphocytes. NFAT1 transcription factor directly regulates Cyclin E expression in B cells, inhibiting the G1/S cell cycle phase transition. Bioinformatics analysis indicates that low levels of NFAT1 correlate with high expression of Cyclin E1 in different human cancers, including Diffuse Large B-cell Lymphomas (DLBCL). Together, our results demonstrate a repressor role for NFAT1 in cell cycle progression and Cyclin E expression in B lymphocytes, and suggest a potential function for NFAT1 protein in B cell malignancies.
KEYWORDS: B lymphocytes, cell cycle, cyclin E, NFAT, Tumor
Introduction
The NFAT family of transcription factors encodes 4 different proteins (NFAT1-4) that are activated through the Calcium/Calcineurin signaling pathway.1,2 In resting cells, NFAT proteins are located in the cytoplasm in a hyperphosphorylated, inactive form. Calcium influx induced by different stimuli activates the serine/threonine phosphatase Calcineurin, which is able to dephosphorylate NFAT proteins, causing their nuclear translocation and DNA binding activity.3,4 Besides the canonical cytokines and cell surface markers regulated by NFAT transcription factors, several reports have shown that NFAT proteins control the activity of promoter and enhancer regions of many other inducible genes, such as FasL, Cox-2, and Vegf.5-7 NFAT transcription factors have also been shown to regulate the expression of cell cycle-related genes, such as Cdk-4, Cyclin A2, C-Myc and p15INK4b,8-11 indicating a central role for this family of transcription factors in cellular proliferation and tumorigenesis. Indeed, different NFAT family members have been related to tumor-associated processes, such as growth factor independency, apoptosis, angiogenesis, and invasiveness.12,13
Tumor development is driven by a succession of genetic changes that occur through aberrant cell division cycles, gradually converting normal cells into cancer cells.14 Cell cycle progression is precisely controlled by the activation of specific cyclin-dependent kinases (CDKs) and their partner cyclins.15-16 Cyclin E normally accumulates at the G1/S phase transition, where it promotes S phase entry and progression by binding to and activating CDK2. In normal cells, Cyclin E/CDK2 activity is associated with DNA replication-related functions.15-17 Ectopic expression of Cyclin E accelerates G1 progression, while inhibition of Cyclin E/CDK2 activity prevents S phase entry, indicating a positive role for this complex in G1/S transition.18-19 Overexpression of Cyclin E interferes with pre-replication complex assembly, causing replication stress and genomic instability.20-23 In malignant cells, deregulation of Cyclin E is associated with tumor development, aggressiveness, and poor outcome in different human cancers.24-28 Transgenic mice models expressing high levels of Cyclin E in T lymphocytes are predisposed to lymphoid hyperplasia and malignant transformation, indicating a causative role for Cyclin E oncoprotein in lymphomas.29,30 Finally, Cyclin E overexpression has been correlated to poor prognosis in several human lymphomas,25 as well as advanced stage and relapse in human B-cell leukemia patients.31
The NFAT family of transcription factors has been primarily described in T cells and further observed in many other cell types of the immune system. It has been shown that several NFAT family members, such as NFAT1 and NFAT2, are expressed in B lymphocytes and are required for B cell development and function upon activation.32-34 Together with NFAT4, NFAT1 has been described to be involved in the expansion of mature follicular B cells,35 and in B cell tolerance.36 However, the role of NFAT1 transcription factor in cell cycle control of B lymphocytes remains to be elucidated. Here, we show that NFAT1 protein controls cell proliferation and Cyclin E expression in B lymphocytes. We demonstrate that NFAT1-deficient naive B lymphocytes show increased cell cycle progression and high levels of Cyclin E1 and Cyclin E2 upon stimulation when compared to control cells. We show that NFAT1 transcription factor directly binds the human Cyclin E1 and E2 promoters, and NFAT1 activation represses Cyclin E levels and the G1/S phase transition in B cells. Finally, we observe that low levels of NFAT1 correlate with high expression of Cyclin E1 in different human cancers, including Diffuse Large B-cell Lymphomas (DLBCL). Taken together, our results indicate a repressor role for NFAT1 in cell cycle progression and Cyclin E expression in B lymphocytes, and suggest a protective role for NFAT1 protein in B cell lymphoma.
Results
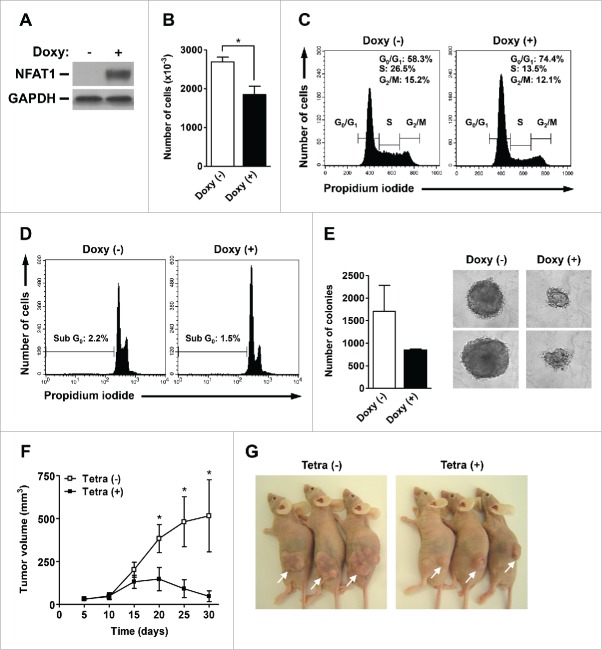
To address the role of NFAT1 transcription factor in cell cycle progression and tumorigenesis, we generated an NFAT1-inducible system in Chinese Hamster Ovary (CHO) cells stably-transfected with human NFAT1 cDNA. In this system, the NFAT1 protein is conditionally produced in the presence of tetracycline/doxycycline, which is able to inhibit the tetracycline repressor.9 As observed, CHO cells do not show detectable levels of NFAT1 protein, which is only induced upon doxycycline treatment (Fig. 1A). NFAT1 induction was associated with decreased number of cells and accumulation in G0/G1 cell cycle phases after doxycycline treatment (Fig. 1B and 1C, respectively). Accordingly, NFAT1 induction was followed by decreased number of cells in S and G2/M phases (Fig. 1C). Of note, doxycycline administration and NFAT1 expression did not lead to increased cell death as assessed by propidium iodide staining of Sub-G0 cells (Fig. 1D). These results indicate that NFAT1 transcription factor controls cell cycle progression in CHO cells, and suggest a repressor role for NFAT1 during G1/S phase transition.
Figure 1.
NFAT1 induces cell cycle arrest and tumor growth reduction in vivo. (A-E) CHO-NFAT1 cells were cultivated either in the absence or presence of doxycycline (1 μg/mL) for the indicated time points. (A) Total protein lysates were obtained after 48 hours of cell cultivation. NFAT1 and GAPDH protein levels were detected by protein gel blot. (B) CHO-NFAT1 cells were cultivated for 72 hours, harvested and counted using a Neubauer chamber. (C and D) CHO-NFAT1 cells were cultivated for 72 hours, labeled with propidium iodide, and DNA content was analyzed by flow cytometry for cell cycle phases (C) or sub-G0 profile (D). (E) CHO-NFAT1 cells were cultivated in 0.3% agarose-containing medium for 20 days, and colony unit formation was analyzed by phase-contrast microscopy. (F and G) CHO-NFAT1 cells (5 × 106 cells) were inoculated subcutaneously in BALB/c nude/nude mice previously treated or not with tetracycline (2 mg/mL) for 3 d. Tetracycline was administered or not during the entire experiment as indicated. Tumor dimensions were measured every 5 d for 30 d, and tumor volume was calculated as described. All results are representative of at least 2 independent experiments. Results are expressed as mean and error bars represent SD (B and E) or SEM (F). Asterisks indicate significance levels compared to controls (p < 0.05).
We next addressed the role of NFAT1 transcription factor in colony formation in vitro and tumor growth in vivo. In a semi-solid colony forming unit assay, NFAT1 induction caused a reduction in colony number and size when compared to CHO cells not treated with doxycycline (Fig. 1E). As observed before, the reduction in colony number and size upon doxycycline treatment was not followed by increased cell death (data not shown). We next investigated the role of NFAT1 transcription factor in tumor growth and progression in vivo. Immunodeficient nude mice were inoculated subcutaneously with CHO cells and treated or not with tetracycline for 30 d to induce NFAT1 expression (Fig. 1F and 1G). While non-treated mice showed an exponential increase in tumor volumes starting at day 10, NFAT1 induction by tetracycline administration was able to inhibit tumor growth in vivo as early as 15 d after cell inoculation (Fig. 1F). Furthermore, tetracycline-treated mice showed a 5-fold reduction in tumor volumes when compared to non-treated animals at 30 d after inoculation (Fig. 1F and 1G). Taken together, these results indicate a suppressor role for NFAT1 during cell cycle progression and tumor development in vivo.
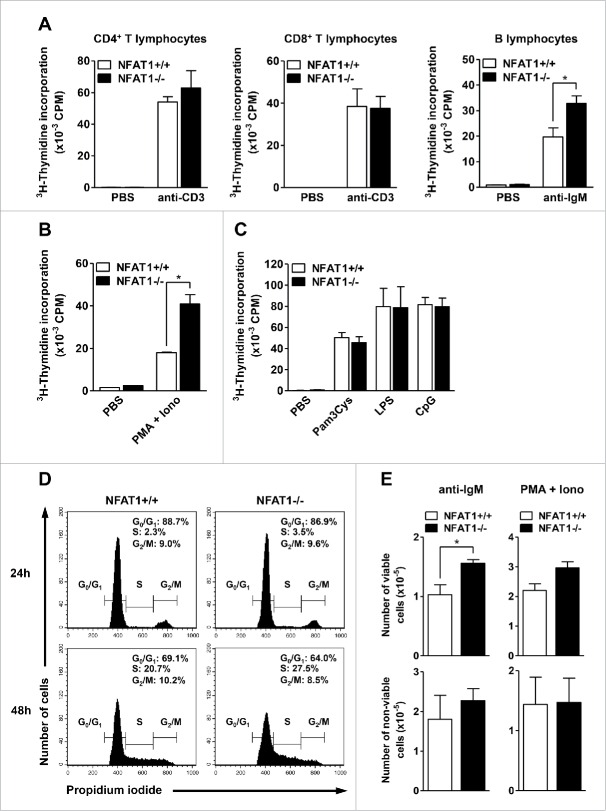
NFAT proteins have been related to cellular proliferation in different cell types and tissues. However, the specific role of the major NFAT family member, NFAT1, in naive lymphocyte proliferation remains to be elucidated. Therefore, we analyzed the contribution of NFAT1 transcription factor to lymphocyte proliferation in different cellular compartments. Absence of NFAT1 did not impair the proliferative responses of primary CD4+ or CD8+ T lymphocytes upon anti-CD3 stimulation as measured by 3H-thymidine incorporation, which indicates active DNA replication (Fig. 2A, left and middle panels). However, NFAT1 deficiency induced B cell hyperproliferation upon polyclonal stimulation with anti-IgM (Fig. 2A, right panel) or PMA plus Ionomycin (Fig. 2B) when compared to wild-type B cells. Interestingly, Toll-Like Receptor (TLR) stimulation did not induce B lymphocyte hyperproliferation in NFAT1-deficient cells (Fig. 2C). In fact, activation of NFAT proteins has been associated with TLR pathway only in macrophages so far.37 Cell cycle analyses indicated an increased number of NFAT1-deficient B lymphocytes in S phase when compared to wild-type cells upon PMA/Ionomycin stimulation (Fig. 2D). Consistently, stimulation of NFAT1-deficient B cells with polyclonal activator led to increased number of viable cells in culture, without affecting the number of non-viable cells when compared to controls (Fig. 2E). Furthermore, an increase in cell death rate was observed in NFAT1-deficient B lymphocytes upon PMA/Ionomycin stimulation as assessed by propidium iodide staining of Sub-G0 cells (data not shown), ruling out the possibility that impairment in cell death contributes to the B cell hyperproliferative phenotype observed in NFAT1-deficient lymphocytes. Together, these results indicate that NFAT1 transcription factor regulates primary B lymphocyte proliferation, suggesting a repressor role for NFAT1 in the G1/S cell cycle phase transition.
Figure 2.
NFAT1 controls cell proliferation in primary B lymphocytes. (A) CD4+, CD8+ T lymphocytes, and B lymphocytes were purified from naive NFAT1+/+ and NFAT1−/− mice by negative selection with magnetic beads. Cells were left unstimulated (PBS) or stimulated in vitro with plate-bound anti-CD3 (1 μg/mL) or anti-IgM (5 μg/mL) for 36 hours. (B and C) Purified B lymphocytes from naive NFAT1+/+ and NFAT1−/− mice were left unstimulated (PBS) or stimulated in vitro with PMA (10 nM) and Ionomycin (1 μM) (B) or Pam3Cys (0.1 μg/mL), LPS (1 μg/mL), and CpG (1 μg/mL) (C) for 36 hours. (A-C) After stimulation, cells were pulsed with 3H-thymidine (5 μCi/mL) for 8 hours. Cells were then harvested and 3H-thymidine incorporation was analyzed by β-spectrometer. CPM refers to counts per minute. (D and E) Purified B lymphocytes from naive NFAT1+/+ and NFAT1−/− mice were stimulated with PMA (10 nM) and Ionomycin (1 μM) or anti-IgM (5 μg/mL) as indicated. (D) Cells were stimulated with PMA/Ionomycin for the indicated time points, labeled with propidium iodide, and DNA content was analyzed by flow cytometry for cell cycle phases. (E) Cells were stimulated with anti-IgM (left panels) or PMA/Ionomycin (right panels) for 24 hours, and the numbers of viable (upper panels) and non-viable (lower panels) cells were assessed with Trypan Blue using a Neubauer chamber. All results were obtained from a pool of 3 mice, and are representative of at least 3 independent experiments. Results are expressed as mean and error bars represent SEM. Asterisks indicate significance levels compared to controls (p < 0.05).
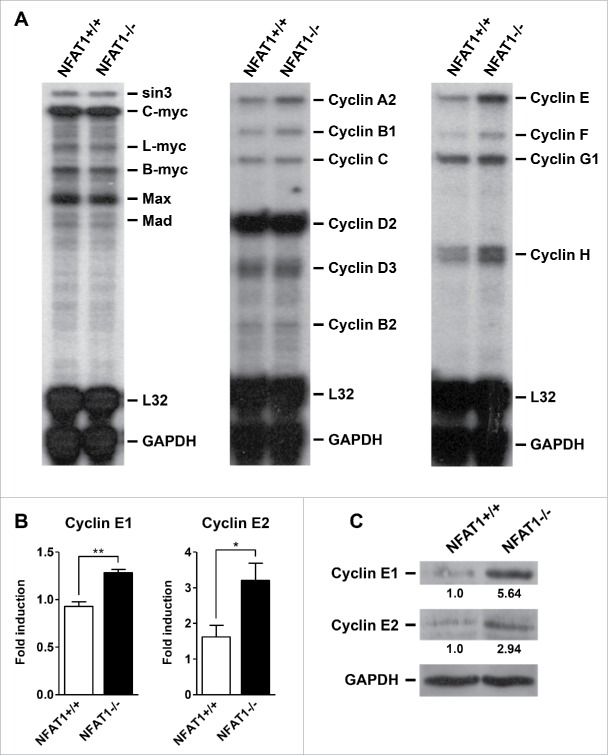
Deregulation of many cell cycle proteins has been associated with hyperproliferation in B cell malignancies. Among them, the proto-oncogene c-Myc is frequently upregulated in Burkitt's lymphomas and other B cell neoplasias. Therefore, we decided to investigate the expression levels of Myc family members in B lymphocytes from NFAT1-deficient mice by RNase Protection Assay (RPA). Interestingly, c-Myc expression was not altered in NFAT1-deficient B cells when compared to wild-type cells upon PMA/Ionomycin stimulation (Fig. 3A, left panel). In contrast, expression of Cyclins A2, B1, E, F, and H were upregulated at different levels in NFAT1-deficient B lymphocytes when compared to controls (Fig. 3A, middle and right panels). Of note, Cyclin E mRNA levels were pronouncedly higher in B cells that lack NFAT1.
Figure 3.
NFAT1 inhibits Cyclin E1 and E2 expression in primary B lymphocytes. (A-C) B lymphocytes were purified from naive NFAT1+/+ and NFAT1−/− mice and stimulated with PMA (10 nM) and Ionomycin (1 μM) for 24 hours (A and B) or 48 hours (C). (A) Total RNA was extracted and the expression of Myc family members (left panel) and Cyclin family members (middle and right panels) were analyzed by RNase Protection Assay. Transcript levels were analyzed by autoradiography, and RNA loading was estimated by measuring the intensities of 2 housekeeping genes (L32 and GAPDH). (B) Total RNA was extracted and Cyclin E1 and Cyclin E2 expression levels were analyzed by real-time PCR using TaqMan Gene Expression assays. HPRT expression was used for normalization. (C) Total protein lysates were obtained and Cyclin E1, Cyclin E2, and GAPDH protein levels were detected by western blot. Cyclin E protein levels were normalized to GAPDH and are indicated below blots. All results were obtained from a pool of 3 mice, and are representative of at least 2 independent experiments. Results are expressed as mean and error bars represent SEM. Asterisks indicate significance levels compared to controls (* p < 0.05; ** p < 0.005).
In mammals, Cyclin E is represented by 2 functionally redundant family members, named Cyclin E1 and E2 (CCNE1 and CCNE2, respectively), which are involved in cell cycle regulation, specifically in the G1/S phase transition. We then decided to further investigate the levels of Cyclin E1 and Cyclin E2 in NFAT1-deficient B lymphocytes. Both CCNE1 and CCNE2 mRNA and protein levels were increased in NFAT1-deficient B cells when compared to wild-type cells after PMA/Ionomycin stimulation (Fig. 3B and 3C, respectively). In fact, we have previously observed an overexpression of Cyclin E in total lymph nodes from NFAT1-deficient mice after ovalbumin challenge in vitro when compared to wild-type mice.38 However, Cyclin E overexpression was not detected in primary CD4+ or CD8+ T lymphocytes from NFAT1-deficient mice upon anti-CD3 stimulation in vitro when compared to wild-type mice (data not shown). These results suggest that increased proliferation and Cyclin E overexpression in the B cell compartment contribute to the lymphocyte hyperproliferation phenotype observed in NFAT1-deficient mice.39,40
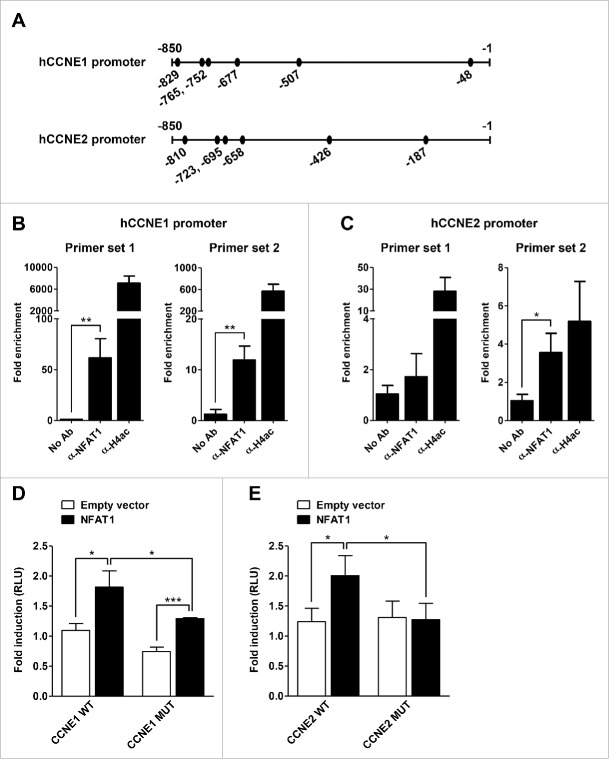
We next addressed whether NFAT1 was able to bind the human Cyclin E1 and E2 promoters in B cells. Bioinformatics analysis of the proximal promoter regions indicated 6 putative NFAT-binding sites at each human promoter (Fig. 4A). Chromatin immunoprecipitation (ChIP) assays showed that NFAT1 binds both promoters in Raji B lymphocytes stimulated with PMA/Ionomycin (Fig. 4B and 4C). NFAT1 binding on the human Cyclin E1 promoter occurred at a distal cluster of 4 binding sites located at positions −829, −765, −752, and −677 bp (Fig. 4A and 4B, hCCNE1 promoter - Primer sets 1 and 2; see Materials and Methods). We also detected NFAT1 binding on a distal cluster of NFAT-binding sites at the human Cyclin E2 promoter, located at positions −695 and −658 bp (Fig. 4A and 4C, hCCNE2 promoter - Primer set 2; see Materials and Methods). Due to the close vicinity on both promoters, we could not precisely map whether NFAT1 binds all or a subset of these sites. Unfortunately, we were not able to address the 2 most proximal NFAT-binding sites at both Cyclin E1 and Cyclin E2 promoters (sites −507, −48 bp and −426, −187 bp, respectively; Fig. 4A). These sites are located on high GC-rich promoter regions, which are impeditive to productive PCR-based amplifications. As a positive control, we were able to detect Histone H4 acetylation on both Cyclin E1 and E2 promoters, indicating a chromatin state that is accessible to the recruitment of transcription factors and gene regulation (Fig. 4B and 4C).
Figure 4.
NFAT1 binds and regulates human Cyclin E promoters. (A) Schematic representation of human Cyclin E1 and Cyclin E promoters (hCCNE1 and hCCNE2 promoters, respectively). Black circles represent putative binding sites for NFAT transcription factor. (B and C) Raji cells were stimulated with PMA (10 nM) and Ionomycin (1.0 μM) for 20 minutes. Chromatin was sonicated and immunoprecipitated with a set of NFAT1-specific antibodies (α-NFAT1), anti-acetyl-histone H4 antibody (α-H4ac) as a positive control, or no antibody as a negative control (No Ab). hCCNE1 (B) and hCCNE2 (C) promoters were analyzed by real-time PCR with specific primer sets as described in Materials and Methods. ChIP signals were normalized to the no-antibody control signal (No Ab) and were expressed as fold increase relative to the background (Fold enrichment). (D and E) Jurkat cells were co-transfected with the following plasmids: pcDNA5 (Empty vector) or pcDNA5-NFAT1 (NFAT1), and CCNE1 (D) or CCNE2 (E) reporter plasmids containing either the wild-type (WT) or NFAT-mutant (MUT) proximal promoter regions. After 24 hours, cells were stimulated with PMA (10 nM) and Ionomycin (1 μM) for 16 hours and luciferase activity was measured as described. RLU, Relative Light Units. All results are representative of at least 3 independent experiments. Results are expressed as mean and error bars represent SD. Asterisks indicate significance levels compared to controls (* p < 0.05; ** p < 0.005; *** p < 0.0005).
To determine whether NFAT1 transcription factor directly regulates Cyclin 1 and E2 expression, we evaluated human Cyclin E promoter activity using Luciferase reporter assays. Wild-type and NFAT-mutant CCNE1 and CCNE2 proximal promoter regions were co-transfected with an Empty or NFAT1-encoding vector into Jurkat cells stimulated with PMA/Ionomycin. Overexpression of NFAT1 upregulated the activity of both wild-type Cyclin E1 and Cyclin E2 promoters (Fig. 4D and 4E, respectively). Mutation of all 6 putative NFAT-binding sites on either promoter inhibited responsiveness to NFAT1 overexpression (Fig. 4D and 4E, black bars). CCNE1-mutant promoter, which lacks the 6 NFAT-binding sites predicted by our bioinformatics analysis, is still responsive to NFAT1 overexpression (Fig. 4D, right bars), suggesting the presence of other putative NFAT-binding sites on the human CCNE1 proximal promoter region. Our data however showed that NFAT1 overexpression does not inhibit the activity of human CCNE1 and CCNE2 proximal promoters using Luciferase transactivation assays in Jurkat cells as initially anticipated. It is widely known that NFAT transcription factors interact with different transcriptional partners (activators or repressors) to promote gene expression under different cellular contexts.2,13 Therefore, it will be important to dissect the impact of individual NFAT-binding sites on these promoters as well as the co-factors that associate with NFAT1 to regulate Cyclin E expression during B lymphocyte development and function. Together, our data indicate that activation of NFAT1 transcription factor directly regulates the expression of Cyclin E1 and E2 in lymphocytes.
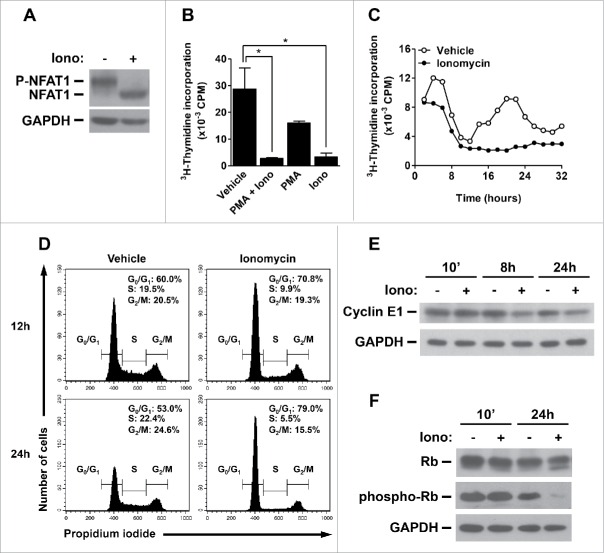
To further address the role of NFAT1 activation in the B cell compartment and its involvement in B cell malignancy, we took advantage of the B lymphocyte Burkitt's lymphoma Raji cell line. Raji cells show normal levels of NFAT1 transcription factor, which is dephosphorylated by Ionomycin-induced Calcium influx (Fig. 5A). Interestingly, Ionomycin treatment alone was sufficient to interfere with Raji cell proliferation as measured by 3H-thymidine incorporation (Fig. 5B). 3H-thymidine pulse-chase experiments further indicated that Ionomycin completely blocks S phase entry as observed after 12–18 hours of treatment (Fig. 5C). Cell cycle analysis showed that Raji cells arrest in G1 phase and do not progress through S phase after 12–24 hours of Ionomycin treatment (Fig. 5D). Importantly, Ionomycin-treated cells are completely viable as assessed by propidium iodide staining of Sub-G0 cells (data not shown). Furthermore, Ionomycin treatment induced Cyclin E1 downregulation in Raji cells, consistent with a G1-phase arrest in the cell cycle (Fig. 5E). To address the role of Cyclin E downregulation in cell cycle progression of B cells, we analyzed the status of Rb phosphorylation in Ionomycin-treated Raji cells. It is noteworthy that G1/S transition is characterized by phosphorylation of Rb protein on specific residues, which are modified by several Cyclin/CDK complexes, including Cyclin E/CDK2.41,42 In fact, Ionomycin treatment, which reduced the levels of Cyclin E1, decreased Rb phosphorylation in Raji cells after 24 hours (Fig. 5F). Consistently, Rb phosphorylation on Thr821 and Thr826, which have been demonstrated to be phosphorylated by Cyclin E/CDK2,41,42 was drastically impaired upon Ionomycin treatment (Fig. 5F). Taken together, our results suggest a repressor role for NFAT1 transcription factor in Cyclin E expression and cell cycle progression of B lymphocytes, indicating a cell type-specific proliferative capacity for NFAT1 in different lymphocyte compartments.
Figure 5.
NFAT1 induces cell cycle arrest and inhibits Cyclin E expression in B cell lymphoma. (A) Total protein lysates were obtained from Raji cells stimulated or not with Ionomycin (5 μM) in the presence of 2 mM of CaCl2 for 10 minutes. NFAT1 and GAPDH protein levels were detected by protein gel blot. (B) Raji cells were stimulated or not with PMA (10 nM) and/or Ionomycin (5 μM) for 24 hours. (C) Raji cells were stimulated or not with Ionomycin (5 μM) for 32 hours and analyzed every 2 hours. (B and C) After stimulation, cells were pulsed with 3H-thymidine (5 μCi/mL) for 2 hours. Cells were then harvested and 3H-thymidine incorporation was analyzed by β-spectrometer. CPM refers to counts per minute. (D) Raji cells were stimulated or not with Ionomycin (5 μM) for the indicated time points. Cells were then labeled with propidium iodide, and DNA content was analyzed by flow cytometry for cell cycle phases. (E and F) Total protein lysates were obtained from Raji cells stimulated or not with Ionomycin (5 μM) for the indicated time points. Cyclin E1, Rb, phospho-Rb (Thr821 and Thr826), and GAPDH protein levels were detected by western blot. All results are representative of at least 3 independent experiments. Results are expressed as mean and error bars represent SD. Asterisks indicate significance levels compared to controls (p < 0.05).
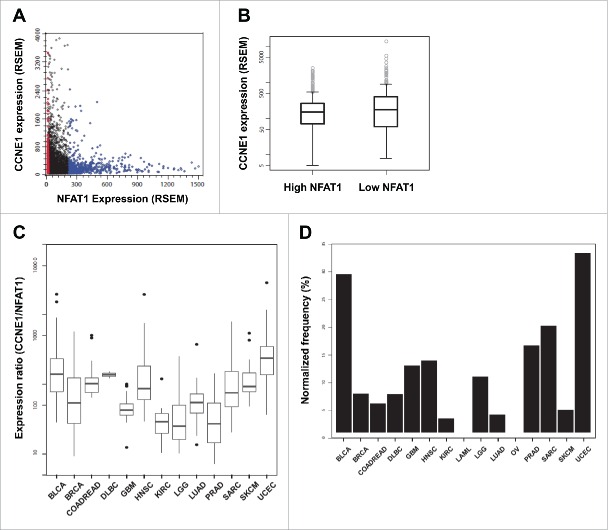
We next evaluated the relevance of our in vitro findings to human cancers. Expression data of 5,643 patients from 15 different tumor types were interrogated for mRNA expression levels of NFAT1 and Cyclin E1 (NFATC2 and CCNE1 genes, respectively) in The Cancer Genome Atlas (TCGA) database (Fig. 6A). According to NFAT1 expression, 2 groups of samples were created, each one represented by 1/10 of the entries with the lowest and highest levels of NFAT1 expression (“Low NFAT1” and “High NFAT1,” respectively; Fig. 6A). Analysis of Cyclin E1 expression levels between these 2 groups showed a slight but significant negative association with NFAT1 expression, suggesting a mutually exclusive expression pattern between these 2 genes (p = 0.01219, Student's t-test; Fig. 6B). A similar negative association pattern was observed between NFAT1 and Cyclin E2 expression levels, however it did not reach statistical significance (data not shown). We next evaluated the Cyclin E1/NFAT1 expression ratio among all tumor types to identify human cancers with pronounced differences between NFAT1 and Cyclin E1 expression. Interestingly, the higher CCNE1/NFAT1 expression ratios were observed in Uterine Corpus Endometrial Carcinoma (UCEC) and Diffuse Large B-Cell Lymphoma (DLBC), as well as Head and Neck Squamous Cell Carcinoma (HNSC), Bladder Urothelial Carcinoma (BLCA), and Sarcoma (SARC) (Fig. 6C). Samples in the “Low NFAT1” category are differentially represented in all analyzed tumors. UCEC and BLCA are the tumor types with the higher fraction of their samples classified as “Low NFAT1” (Fig. 6D). Taken together, our results indicate a repressor function for NFAT1 in cell cycle progression and Cyclin E expression in B lymphocytes, suggesting a potential role for NFAT1 in B cell lymphomas.
Figure 6.
NFAT1 shows a mutually exclusive expression pattern with Cyclin E1 in different human cancers. (A) Correlation between the expression (RSEM) of NFAT1 and CCNE1 for 5,643 samples from 15 different tumor types. Samples were stratified according to NFAT1 expression and classified as “Low NFAT1” (red dots corresponding to 1/10 of all samples with the lowest expression) and “High NFAT1” (blue dots corresponding to 1/10 of all samples with the highest expression). (B) Boxplot showing the distribution of CCNE1 expression for the “Low NFAT1” and “High NFAT1” groups. “Low NFAT1” group shows a median expression 1.47 higher than the “High NFAT1” group (t = 2.5106, df = 1103.8, and p = 0.01219). (C) Boxplot of the expression ratio of CCNE1 to NFAT1 for the samples in the “Low NFAT1” group with respect to the specified tumor types. (D) Normalized frequency (%) of samples from the “Low NFAT1” group per tumor type. The normalized frequency was calculated by dividing the number of “Low NFAT1” samples in a given tumor by the total number of samples from the same tumor.
Discussion
In B lymphocytes, NFAT activation occurs mostly through BCR or CD40 stimulation, which causes activation of the Calcium/Calcineurin signaling pathway, leading to NFAT dephosphorylation and nuclear translocation.43 In this work, we demonstrate that BCR-induced NFAT1 activation controls B cell proliferation through inhibition of the G1/S cell cycle phase transition and repression of Cyclin E expression. Our results show that NFAT1 deficiency causes hyperproliferation of primary B cells, inducing a higher frequency of cells in S phase (Fig. 2). Importantly, this phenotype was restricted to the B cell compartment and was not observed in CD4+ or CD8+ T lymphocytes. Accordingly, NFAT1 activation blocked S phase entry of the B lymphocyte Raji cell line, controlling cell proliferation and arresting cells in G1 (Fig. 5). In fact, NFAT1 has been indicated as a cell cycle repressor in different experimental models. Deficiency of NFAT1 caused lymphocyte hyperproliferation and increased cell cycle rates both in vitro and in vivo, leading to splenomegaly and follicular B cell expansion.38-40 However, most of these studies evaluated the responses from total lymph nodes or splenocytes to polyclonal stimuli and not specifically the stimulation of the B-cell compartment. Studies with purified B cells from NFAT1-deficient mice showed increased B-cell responsiveness and autoantibody production in the MD4/soluble hen egg lysozyme (sHEL) transgenic model, indicating that NFAT1 may play a negative role in B cell activation.36 Furthermore, NFAT1/NFAT4-double deficient B lymphocytes showed a hyperresponsiveness to BCR stimulation and increased numbers of mature follicular B cells.35 In that work, however, the authors have suggested that those effects were dependent on cell-extrinsic factors, such as the increased levels of Th2-type cytokines and the subsequent increase in IgG1 and IgE serum levels.35 Along the same lines, NFAT1 activation or overexpression has been demonstrated to induce cell cycle arrest or cell death in T lymphocytes, Burkitt's lymphoma, and NIH 3T3 fibroblasts.44-46 However, the molecular mechanisms by which NFAT1 induces cell cycle arrest in these cell types remain unknown. Together, our results support the notion that NFAT1 transcription factor works as a cell cycle repressor during the G1/S phase transition in B lymphocytes, and may contribute to controlling the tumorigenesis process.
NFAT proteins regulate the expression of many cell cycle-related genes, such as CDK4, CCNA2, C-Myc, and p15INK4b in different models.8-11 It has been shown that NFAT1 directly binds specific sites at the CDK4 and Cyclin A2 promoters and negatively regulates the expression of both genes, suggesting an inhibitory role for NFAT1 in cell cycle progression.8,9 However, the role of NFAT1 protein in cell cycle regulation of B lymphocytes has not been fully addressed. Here, we show that NFAT1 deficiency causes Cyclin E1 and E2 overexpression at the mRNA level in B cells, which is followed by increased protein levels (Fig. 3). Furthermore, NFAT1 activation downregulates Cyclin E1 levels in the Raji B cells, consistent with a G1-phase arrest (Fig. 5). In fact, human Cyclin E1 and E2 promoters showed each 6 putative NFAT-binding sites at the proximal promoter region, several of which are conserved in mice and rats (data not shown). NFAT1 transcription factor was able to bind both Cyclin E1 and E2 promoters upon PMA/Ionomycin stimulation in Raji cells (Fig. 4), supporting a repressive role for NFAT1 in Cyclin E/CDK2 activity and G1/S transition in B lymphocytes (Fig. 5).
Besides the upregulation of Cyclin E in NFAT1-deficient B lymphocytes, we have also observed that lack of NFAT1 caused an overexpression of other cyclin genes, notably Cyclin A2 and Cyclin H (Fig. 3A). In a previous work, we have demonstrated that NFAT1 binds the mouse Cyclin A2 promoter and represses Cyclin A2 expression levels in CD4+ T lymphocytes,9 another well-established activator of CDK2 function. NFAT proteins have not been related to Cyclin H regulation so far. Together with CDK7 and MAT1, Cyclin H forms a trimeric CDK-activating kinase (CAK) complex that directly phosphorylates and activates several CDKs, including CDK2.47,48 Therefore, it is possible that NFAT1 transcription factor controls CDK2-associated kinase activity through repression of Cyclin E, Cyclin A, and Cyclin H gene expression, while NFAT1-deficient B lymphocytes trigger a positive feedback loop for CDK2 activity that might contribute to B-cell hyperproliferation and malignancy. Interestingly, copy number gains at the 12q12–14 region, which contains the CDK2 gene, have been widely associated with human Diffuse Large B-Cell Lymphoma (DLBCL).49-51
Deregulation of NFAT signaling pathway has been associated with different human malignancies, including lymphomas and leukemias.52-54 Sustained activation of Calcineurin has been observed in both human B and T cell lymphomas, and showed intrinsic effects in the proliferation and survival of these cells, highlighting the importance of NFAT transcription factors in these malignancies.53 Specifically, it has been shown that NFAT2 preferentially localizes in the nucleus of Diffuse Large B-Cell Lymphomas (DLBCL) and Burkitt's lymphoma, potentially indicating NFAT2 activation in these malignancies.52 Also, it has been shown that constitutive activation of NFAT2 induces the expression of B-Lymphocyte Stimulator (BLyS, also known as BAFF), CD40L (also known as CD154), and C-Myc in aggressive B-cell non-Hodgkin lymphoma (NHL-B) cells, which contribute to cell growth and survival of B cell lymphomas.55-57
Even though there is substantial data regarding the oncogenic role of NFAT2 in B cell malignancies, the function of NFAT1 in normal and neoplastic B cells has not been extensively investigated. NFAT1-deficient mice older than 16 months spontaneously develop B cell neoplasms with features of anaplastic and/or plasmablastic plasmacytomas.58 However, the molecular mechanisms for preferential formation of B cell malignancies in age-associated NFAT1-deficient mice were not elucidated. Regarding the role of NFAT1 in other systems, it has been shown that lack of NFAT1 induces neoplastic transformation of cartilage cells and increased susceptibility to chemically-induced sarcoma formation in mouse models.46,59 Also, constitutively active NFAT1 (CA-NFAT1) was able to antagonize H-RasV12 oncogene-induced transformation of NIH 3T3 fibroblasts.46 Collectively, these data suggest a tumor suppressor role for NFAT1 in different cell types. On the other hand, oncogenic properties have also been associated to NFAT1. It has been shown that integrin-signaling pathway activates NFAT1, as well as NFAT5, promoting cell migration and invasion of human breast and colon carcinoma cell lines.60 More recently, in a model of intraepithelial pancreatic cancer, NFAT1 has been involved in the silencing of the p15INK4b tumor suppressor locus through sequential recruitment of the histone methyltransferase Suv39H1 and heterochromatin protein HP1γ, promoting cell cycle progression.11 It can be concluded from these data that NFAT1 transcription factor may present both oncogenic and tumor suppressor activities, depending on the cell type and cellular context considered. It will be important to determine the role of different NFAT family members, as well as their transcriptional partners (activators or repressors), in different cellular contexts, such as normal and neoplastic B lymphocytes.
In conclusion, we propose that NFAT1 is a repressor of cell cycle progression and Cyclin E expression in B cells. Indeed, we have observed a correlation between low levels of NFAT1 and high levels of Cyclin E1 in DLBCL, among other human tumors (Fig. 6). These results suggest that the loss of NFAT1 may contribute to B cell lymphoma development, and reestablishment of NFAT1 function may be an important therapeutic approach in B cell malignancies.
Materials and methods
Animals, cell cultures, and reagents
Control wild-type (NFAT1+/+), NFAT1-deficient (NFAT1−/−), and BALB/c nude/nude 8- to 12-week old mice were used in all experiments. Animals were bred and maintained in the Brazilian National Cancer Institute (INCA). All animal experiments were performed in accordance with the Brazilian Government´s ethical and animal experiment regulations. The experiments were approved and conducted according to the animal welfare guidelines of the Ethics Committee of Animal Experimentation from INCA. Primary lymphocytes were harvested in DMEM supplemented with 10% fetal calf serum (FCS), L-glutamine, streptomycin-penicillin, essential and nonessential amino acids, sodium pyruvate, vitamins, hepes, and 2-mercaptoethanol (all from Invitrogen). CHO cells (kindly provided by Dr. M. Bellio, UFRJ, Brazil) and Raji cells (ATCC) were harvested in RPMI supplemented with 10% fetal calf serum (FCS), L-glutamine, and streptomycin-penicillin. All cells were cultivated in 5% CO2 at 37°C. Primary lymphocytes (B, CD4+ and CD8+ T cells) were obtained from peripheral lymph nodes of naive animals (inguinal, brachial, axillary, and superficial cervical lymph nodes). Purified single-cell suspensions were isolated by negative selection with magnetic beads (Micro Beads, MACS technology), according to manufacturer's instructions (Miltenyi Biotec). Streptavidin magnetic beads were conjugated to specific-biotinylated antibodies (anti-CD4, anti-CD8, anti-B220/CD45R) to sort out undesired cell populations. Cell populations were analyzed on a FACSCalibur flow cytometer (BD Biosciences), and isolated to > 90% purity. The CHO-NFAT1 cell line was constructed by using the Flp-In T-Rex System (Invitrogen) as previously described.9 The anti-CD3 antibody (2C11) was purified from hybridoma supernatant by chromatography over protein-G (Amersham Biosciences) and the anti-IgM antibody was obtained from Jackson ImmunoResearch (Cat# 115–006-075). The TLR ligands Pam3Cys, LPS, and CpG were kindly provided by Drs. M. Bozza and A. Nobrega (UFRJ, Brazil). PMA, Ionomycin, and the antibiotics doxycycline and tetracycline were all obtained from Calbiochem.
Proliferation assays
For 3H-thymidine incorporation assays, primary lymphocytes (2 × 105 cells/well), CHO cells (5 × 103 cells/well), and Raji cells (3 × 104 cells/well) were left unstimulated (PBS or Vehicle) or stimulated for different periods of time in 96-well plates as indicated: plate-bound anti-CD3 antibody (1 μg/mL), anti-IgM antibody (5 μg/mL), PMA (10 nM), Ionomycin (1–5 μM), Pam3Cys (0.1 μg/mL), LPS (1 μg/mL) or CpG (1 μg/mL). Cells were pulsed with 3H-thymidine (5 μCi/mL) for 2 or 8 hours as indicated. At the indicated time points, cells were harvested and 3H-thymidine incorporation was analyzed by liquid scintillation in a LS 6000LL β-spectrometer (Beckman Instruments). For cell cycle phase analysis, primary B cells (1 × 106 cells), CHO cells (1 × 105 cells), and Raji cells (1 × 106 cells) were stimulated as indicated and collected at different time points. Cells were labeled in buffer containing 0.075 mM propidium iodide, 0.1% NP-40, and 700 U/L RNase for 15 minutes at 4°C and DNA content was analyzed on a FACSCalibur flow cytometer with CellQuest software (BD Biosciences). For cell count analysis, primary B cells (5 × 105 cells) were stimulated with anti-IgM (5 μg/mL) or PMA (10 nM) and Ionomycin (1 μM) for 24 hours. Viable and non-viable cells were scored with Trypan Blue solution using a Neubauer chamber. For colony unit forming assays, plates were coated with 0.6% agarose-supplemented RPMI. CHO cells (8 × 102 cells/well) were cultivated in 0.3% agarose-supplemented RPMI. On top of these 2 layers, fresh RPMI medium containing or not doxycycline (1 μg/mL) was added every 3 d for 20 d. Colony units were analyzed with a AxioVert S100 phase-contrast microscope (Carl Zeiss).
RNase protection assay (RPA)
For RNase Protection Assays, naive B lymphocytes (2 × 106 cells) from NFAT1+/+ and NFAT1−/− mice were stimulated with PMA (10 nM) and Ionomycin (1 μM) for 24 hours. Total RNA was extracted with TRIzol Reagent according to manufacturer's instructions (Invitrogen). The mRNA levels of c-MYC and the different Cyclins were analyzed with the mMYC, mCYC-1, and mCYC-2 multi-probe template sets by RPA kit according to manufacturer's instructions (RiboQuant, BD PharMingen). Transcript levels were analyzed by autoradiography, and RNA loading was estimated by measuring the intensities of 2 housekeeping genes (L32 and GAPDH).
Real-time PCR
For CCNE1 and CCNE2 relative expression assays, naive B lymphocytes (2 × 106 cells) from NFAT1+/+ and NFAT1−/− mice were stimulated with PMA (10 nM) and Ionomycin (1 μM) for 24 hours. Gene expression was assessed by total RNA extraction using RNeasy Mini Kit (Qiagen), followed by cDNA synthesis via RT-PCR using random primers and the SuperScript II Reverse Transcriptase (Thermo Fisher Scientific). Relative quantification of CCNE1 and CCNE2 transcripts was performed by real-time PCR using a 7500 Real Time PCR System (Applied Biosystems) and the following TaqMan Gene Expression Assay kits: CCNE1, Mm00432367_m1; CCNE2, Mm00438077_m1; and HPRT, Mm00446968_m1 (Thermo Fisher Scientific). All procedures were performed according to manufacturer's instructions. Cyclin E expression levels were normalized to HPRT and are relative to the experiment with the lowest absolute Ct value obtained for NFAT1+/+ B lymphocytes.
Western blot
To detect the presence of Cyclin E1, Cyclin E2 or NFAT1 proteins, naive B lymphocytes (2 × 106 cells) from NFAT1+/+ and NFAT1−/− mice were stimulated with PMA (10 nM) and Ionomycin (1 μM) for 48 hours. CHO-NFAT1 cells (4 × 105 cells) were maintained or not in the presence of doxycycline (1 μg/mL) for 48 hours. Raji cells (2 × 106 cells) were stimulated or not with Ionomycin (5 μM) for the indicated time points. For detection of dephosphorylated NFAT1, Raji cells were stimulated in the presence of 2 mM CaCl2. Total protein lysates were obtained as previously described.4 Briefly, cells were resuspended in lysis buffer (40 mM Tris pH 7.5, 60 mM Sodium Pyrophosphate, 10 mM EDTA, 5% SDS), followed by incubation at 100°C for 15 minutes. Total cell lysates were quantified using DC Protein Assay (Bio-Rad), resolved in SDS-PAGE, and separated proteins were transferred to Amersham Protran Premium 0.45 μm Nitrocellulose membrane (GE Healthcare). Antibodies were used as follows: Cyclin E1, polyclonal antibody M-20 (sc-481); Cyclin E2, polyclonal antibody N-20 (sc-9566); GAPDH, monoclonal antibody 6C5 (sc-32233) all from Santa Cruz; NFAT1, polyclonal antibody anti-67.1 or anti-T2B1 (Dr. Anjana Rao, La Jolla Institute for Allergy and Immunology); Rb, polyclonal antibody 851;61 and phospho-Rb Thr821/826, polyclonal antibody (sc-16669) from Santa Cruz. Immunodetection was performed with ECL western blotting detection kit (Amersham Biosciences). GAPDH protein levels were used to normalize protein loading on gel. Quantification of proteins was performed using LabWorks software (UVP). Cyclin E protein levels were normalized to GAPDH and are relative to the Cyclin E values obtained from NFAT1+/+ B lymphocytes.
CCNE promoter analysis
Genomic sequences from Homo sapiens CCNE1 and CCNE2 promoters were obtained from the National Center for Biotechnology Information (NCBI, Assembly GRCh38, Annotation Release ID: 107) on January 21, 2016. Accession numbers were as follows: NC_000019.10 (CCNE1) and NC_000008.11 (CCNE2). Putative NFAT binding sites on the proximal 850 bp from both promoters were analyzed using TRANSFAC Professional and Match 1.0 programs (BIOBASE Biological Databases).
Chromatin immunoprecipitation (ChIP) assay
ChIP experiments were performed as previously described before.9 Briefly, Raji cells (20 × 106 cells) were stimulated with PMA (10 nM) and Ionomycin (1.0 μM) for 20 minutes at 37°C. Then, cells were fixed directly with Formaldehyde solution for 20 minutes on ice, neutralized with Glycine for 5 minutes on ice, and lyzed with Lysis Buffer for 10 minutes on ice as previously described.9 Chromatin was sonicated 40 times of 30 seconds each, with 30-second intervals in between pulses to generate 200–500 bp DNA fragments (Bioruptor 300, Diagenode). Immunoprecipitation was performed overnight at 4°C with a set of NFAT1-specific antibodies (anti-67.1 and anti-T2B1), anti-acetyl-histone H4 antibody (Cat # 06–866, Upstate Biotechnology) or no antibody as a negative control. Immunocomplexes were captured with Protein A Sepharose CL-4B for 3 hours at 4°C (GE Healthcare). Formaldehyde crosslinks were reversed and DNA was purified by QIAquick Gel Extraction Kit according to manufacturer's instructions (Qiagen). Real-time PCR reactions were done with 1 μL of the immunoprecipitated DNA using SYBR Green PCR Master Mix (Applied Biosystems). PCR conditions were as follows: 10 min at 95°C, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C, completed by 1 min at 95°C. Melting curve was performed from 50°C to 90°C, followed by 1 min at 60°C (7500 Real Time PCR System, Applied Biosystems). Primers were used as follows: hCCNE1 Primer set 1: 5′ GCC TCA ACT GGA AGG CTT TA 3′ and 5′ GTG CAT TGG GTC GTT CT TC 3′ (167-bp product; NFAT sites −829, −765, and −752); hCCNE1 Primer set 2: 5′ GCA TGG AAG GAA CTC ACA GA 3′ and 5′ GCC TAG AAC CAA GGC TTC A 3′(140-bp product; NFAT sites −752 and −677); hCCNE2 Primer set 1: 5′ GTG GAG AAG AGA GGA AGC AA 3′ and 5′ AGG TAT GTA TCA ACC CTG CTT T 3′ (184-bp product; NFAT sites −810, −723, and −695); hCCNE2 Primer set 2: 5′ CGG AAA GCA GGG TTG ATA CA 3′ and 5′ GGA GTG GCT GCA GAA TGT AA 3′(174-bp product; NFAT sites −695 and −658) (IDT). ChIP signals were normalized to the no-antibody control signal and were expressed as fold increase of PCR triplicates relative to the background (Fold enrichment).
Plasmid construction
pcDNA5/FRT/TO (pcDNA5-Empty vector) and pcDNA5-NFAT1 expression plasmids were previously described.9 Human CCNE1 (−1 to −862) and CCNE2 (−1 to −838) proximal promoter regions (wild-type and NFAT-mutant constructs) were synthesized by GenScript, and later cloned into the pGL4.10 reporter plasmid (Promega) between XhoI and BglII restriction sites. In the CCNE1 and CCNE2 NFAT-mutant promoter constructs, all 6 putative NFAT binding sites identified on each promoter were substituted with 5′ ACTCT 3′ sequence.
Luciferase reporter assay
Jurkat cells (3 × 106 cells) were electroporated in a 0.2 cm GenePulser Cuvette (Bio-Rad Laboratories) using program X-005 of Amaxa Biosystems Nucleofactor II (Lonza). Cells were co-transfected with 3 different plasmids resuspended in 100 μl of buffer 3P,62 as follows: pcDNA5-Empty vector or pcDNA5-NFAT1 (6 μg); pGL4.10-CCNE1 or pGL4.10-CCNE2 reporter plasmids containing either the wild-type or NFAT-mutant promoter constructs (1 μg); and pRL-TK Renilla expression plasmid (0.1 μg) for normalization (Promega). After 24 hours, cells were stimulated for additional 16 hours with PMA (10 nM) and Ionomycin (1 μM). Then, luciferase activity was assessed with Dual-Glo Luciferase Assay System according to manufacturer's instructions (Promega) and measured using a Veritas Microplate Luminometer (Turner Biosystems). Firefly luciferase activities were normalized to Renilla luciferase activities and are expressed as relative light units (RLU) to the lowest value obtained from Jurkat cells co-transfected with pcDNA5-Empty vector and pGL4.10-CCNE wild-type promoter.
In vivo model of tumorigenesis
CHO-NFAT1 cells (5 × 106 cells) were resuspended in PBS and inoculated subcutaneously in BALB/c nude/nude mice, previously treated or not for 3 d with tetracycline (2 mg/mL) in the water. After cell inoculation, tetracycline continued to be administered or not during the entire experiment. Tumor dimensions were measured every 5 d for 30 d using calipers. Tumor volume (mm3) was calculated using the following formula: 0.52 × (L × W × H), where L is tumor length, W is weight, and H is height. Mice were euthanized and photographs were taken at 30 d after cell injection.
Bioinformatics analysis
Gene expression data derived from The Cancer Genome Atlas (TCGA) were downloaded on May 2015 from the cBioPortal for Cancer Genomics63 for the following tumors: BLCA, BRCA, COADREAD, DLBC, GBM, HNSC, KIRC, LAML, LGG, LUAD, OV, PRAD, SARC, SKCM, and UCEC. Tumor samples were selected based on the expression of CCNE1 and NFATC2 (NFAT1), which should show a RSEM expression higher than zero and different from “Not Available” (NA). As a result, 5,643 samples were selected, and their respective CCNE1 and NFAT1 expression data were loaded and plotted using in-house R scripts based on default and ggplot2 plotting libraries.64,65 Statistical t-test analysis between the distribution of CCNE1 expression of the “Low NFAT1” and “High NFAT1” groups was performed in R using a Welch Two Sample t-test.66
Statistical analysis
Statistical analyses between NFAT1+/+ and NFAT1−/− mice, and between control and treated groups, were determined using unpaired Student´s t test for single comparison. p < 0.05 was considered to be statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Dr. A. Nobrega, M. Bellio, M. Bozza, A. Bonomo, and H. Borges for kindly providing reagents, Dr. A. Rao for kindly providing NFAT1 mice and reagents, and members of our laboratory for helpful discussions.
Funding
This work was supported by grants from CNPq, (307296/2011-3 and 476314/2012-7), FAPERJ (112.056/2012, 102.308/2013, 110.794/2013 and 101.147/2013) and INCT-Câncer (573806/2008-0 and 170.026/2008) to J.P.B.V. and from CAPES (23038.004629/2014-19) to S.J.S. N.C. was supported by a CAPES fellowship, C.S. and J.E.K. were supported by CNPq fellowships, and D. C. D. and D.V.F. were supported by Brazilian Ministry of Health fellowships.
References
- [1].Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol 1997; 15:707-47; PMID:9143705; http://dx.doi.org/ 10.1146/annurev.immunol.15.1.707 [DOI] [PubMed] [Google Scholar]
- [2].Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol 2005; 5:472-84; PMID:15928679; http://dx.doi.org/ 10.1038/nri1632 [DOI] [PubMed] [Google Scholar]
- [3].Shaw KT, Ho AM, Raghavan A, Kim J, Jain J, Park J, Sharma S, Rao A, Hogan PG. Immunosuppressive drugs prevent a rapid dephosphorylation of transcription factor NFAT1 in stimulated immune cells. Proc Natl Acad Sci USA 1995; 92:11205-09; PMID:7479966; http://dx.doi.org/ 10.1073/pnas.92.24.11205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Loh C, Shaw KT, Carew J, Viola JP, Luo C, Perrino BA, Rao A. Calcineurin binds the transcription factor NFAT1 and reversibly regulates its activity. J Biol Chem 1996; 271:10884-91; PMID:8631904; http://dx.doi.org/ 10.1074/jbc.271.47.30233 [DOI] [PubMed] [Google Scholar]
- [5].Rengarajan J, Mittelstadt PR, Mages HW, Gerth AJ, Kroczek RA, Ashwell JD, Glimcher LH. Sequential involvement of NFAT and Egr transcription factors in FasL regulation. Immunity 2000; 12:293-300; PMID:10755616; http://dx.doi.org/ 10.1016/S1074-7613(00)80182-X [DOI] [PubMed] [Google Scholar]
- [6].Iniguez MA, Martinez-Martinez S, Punzon C, Redondo JM, Fresno M. An essential role of the nuclear factor of activated T cells in the regulation of the expression of the cyclooxygenase-2 gene in human T lymphocytes. J Biol Chem 2000; 275:23627-35; PMID:10816557; http://dx.doi.org/ 10.1074/jbc.M001381200 [DOI] [PubMed] [Google Scholar]
- [7].Chang C, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, Stankunas K, Graef IA, Crabtree GR. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell 2004; 118:649-63; PMID:15339668; http://dx.doi.org/ 10.1016/j.cell.2004.08.010 [DOI] [PubMed] [Google Scholar]
- [8].Baksh S, Widlund HR, Frazer-Abel AA, Du J, Fosmire S, Fisher DE, DeCaprio JA, Modiano JF, Burakoff SJ. NFATc2-mediated repression of cyclin-dependent kinase 4 expression. Mol Cell 2002; 10:1071-81; PMID:12453415; http://dx.doi.org/ 10.1016/S1097-2765(02)00701-3 [DOI] [PubMed] [Google Scholar]
- [9].Carvalho LD, Teixeira LK, Carrossini N, Caldeira AT, Ansel KM, Rao A, Viola JP. The NFAT1 transcription factor is a repressor of cyclin A2 gene expression. Cell Cycle 2007; 6:1789-95; PMID:17637565; http://dx.doi.org/ 10.4161/cc.6.14.4473 [DOI] [PubMed] [Google Scholar]
- [10].Buchholz M, Schatz A, Wagner M, Michl P, Linhart T, Adler G, Gress TM, Ellenrieder V. Overexpression of c-myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway. EMBO J 2006; 25:3714-24; PMID:16874304; http://dx.doi.org/ 10.1038/sj.emboj.7601246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Baumgart S, Glesel E, Singh G, Chen NM, Reutlinger K, Zhang J, Billadeau DD, Fernandez-Zapico ME, Gress TM, Singh SK, et al.. Restricted heterochromatin formation links NFATc2 repressor activity with growth promotion in pancreatic cancer. Gastroenterology 2012; 142:388-98; PMID:22079596; http://dx.doi.org/ 10.1053/j.gastro.2011.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Viola JP, Carvalho LD, Fonseca BP, Teixeira LK. NFAT transcription factors: from cell cycle to tumor development. Braz J Med Biol Res 2005; 38:335-44; PMID:15761612; http://dx.doi.org/ 10.1590/S0100-879X2005000300003 [DOI] [PubMed] [Google Scholar]
- [13].Mognol GP, Carneiro FR, Robbs BK, Faget DV, Viola JP. Cell cycle and apoptosis regulation by NFAT transcription factors: new roles for an old player. Cell Death Dis 2016; 7:e2199; PMID:27100893; http://dx.doi.org/ 10.1038/cddis.2016.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100:57-70; PMID:10647931; http://dx.doi.org/ 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- [15].Ekholm SV, Reed SI. Regulation of G1 cyclin-dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol 2000; 12:676-84; PMID:11063931; http://dx.doi.org/ 10.1016/S0955-0674(00)00151-4 [DOI] [PubMed] [Google Scholar]
- [16].Ciemerych MA, Sicinski P. Cell cycle in mouse development. Oncogene 2005; 24:2877-98; PMID:15838522; http://dx.doi.org/ 10.1038/sj.onc.1208608 [DOI] [PubMed] [Google Scholar]
- [17].Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene 2005; 24:2776-86; PMID:15838514; http://dx.doi.org/ 10.1038/sj.onc.1208613 [DOI] [PubMed] [Google Scholar]
- [18].Ohtsubo M, Roberts JM. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science 1993; 259:1908-12; PMID:8384376; http://dx.doi.org/ 10.1126/science.8384376 [DOI] [PubMed] [Google Scholar]
- [19].Resnitzky D, Gossen M, Bujard H, Reed SI. Acceleration of G1/S phase transition by expression of cyclins D1 and E with and inducible system. Mol Cell Biol 1994; 14:1669-79; PMID:8114703; http://dx.doi.org/ 10.1128/MCB.14.3.1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature 1999; 401:297-300; PMID:10499591; http://dx.doi.org/ 10.1038/45836 [DOI] [PubMed] [Google Scholar]
- [21].Hubalek MM, Widschwendter A, Erdel M, Gschwendter A, Fiegl HM, Muller HM, Goebel G, Mueller-Holzner E, Marth C, Spruck CH, et al.. Cyclin E dysregulation and chromosomal instability in endometrial cancer. Oncogene 2004; 23:4187-92; PMID:15048079; http://dx.doi.org/ 10.1038/sj.onc.1207560 [DOI] [PubMed] [Google Scholar]
- [22].Loeb KR, Kostner H, Firpo E, Norwood TD, Tsuchiya K, Clurman BE, Roberts JM. A mouse model for cyclin E-dependent genetic instability and tumorigenesis. Cancer Cell 2005; 8:35-47; PMID:16023597; http://dx.doi.org/ 10.1016/j.ccr.2005.06.010 [DOI] [PubMed] [Google Scholar]
- [23].Teixeira LK, Wang X, Li Y, Ekholm-Reed S, Wu X, Wang P, Reed SI. Cyclin E deregulation promotes loss of specific genomic regions. Curr Biol 2015; 25:1327-33; PMID:25959964; http://dx.doi.org/ 10.1016/j.cub.2015.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Porter PL, Malone KE, Heagerty PJ, Alexander GM, Gatti LA, Firpo EJ, Daling JR, Roberts JM. Expression of cell-cycle regulators p27Kip1 and cyclin E, alone and in combination, correlate with survival in young breast cancer patients. Nat Med 1997; 3:222-25; PMID:9018243; http://dx.doi.org/ 10.1038/nm0297-222 [DOI] [PubMed] [Google Scholar]
- [25].Erlanson M, Portin C, Linderholm B, Lindh J, Ross G, Landberg G. Expression of cyclin E and the cyclin-dependent kinase inhibitor p27 in malignant lymphomas - prognostic implications. Blood 1998; 92:770-77; PMID:9680343 [PubMed] [Google Scholar]
- [26].Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 2001; 413:316-22; PMID:11565034; http://dx.doi.org/ 10.1038/35095076 [DOI] [PubMed] [Google Scholar]
- [27].Keyomarsi K, Tucker SL, Buchholz TA, Callister M, Ding Y, Hortobagyi GN, Bedrosian I, Knickerbocker C, Toyofuku W, Lowe M, et al.. Cyclin E and survival in patients with breast cancer. N Engl J Med 2002; 347:1566-75; PMID:12432043; http://dx.doi.org/ 10.1056/NEJMoa021153 [DOI] [PubMed] [Google Scholar]
- [28].Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Inactivation of hCDC4 can cause chromosomal instability. Nature 2004; 428:77-81; PMID:14999283; http://dx.doi.org/ 10.1038/nature02313 [DOI] [PubMed] [Google Scholar]
- [29].Karsunky H, Geisen C, Schmidt T, Haas K, Zevnik B, Gau E, Moroy T. Oncogenic potential of cyclin E in T-cell lymphomagenesis in transgenic mice: evidence for cooperation between cyclin E and Ras but not Myc. Oncogene 1999; 18:7816-24; PMID:10618723; http://dx.doi.org/ 10.1038/sj.onc.1203205 [DOI] [PubMed] [Google Scholar]
- [30].Geisen C, Karsunky H, Yucel R, Moroy T. Loss of p27Kip1 cooperates with cyclin E in T-cell lymphomagenesis. Oncogene 2003; 22:1724-29; PMID:12642875; http://dx.doi.org/ 10.1038/sj.onc.1206340 [DOI] [PubMed] [Google Scholar]
- [31].Scuderi R, Palucka KA, Pokrovskaja K, Bjorkholm M, Wiman KG, Pisa P. Cyclin E overexpression in relapsed adult acute lymphoblastic leukemias of B-cell lineage. Blood 1996; 87:3360-67; PMID:8605353 [PubMed] [Google Scholar]
- [32].Ranger AM, Hodge MR, Gravallese EM, Oukka M, Davidson L, Alt FW, de la Brousse FC, Hoey T, Grusby M, Glimcher LH. Delayed lymphoid repopulation with defects in IL-4-driven responses produced by inactivation of NF-ATc. Immunity 1998; 8:125-34; PMID:9462518; http://dx.doi.org/ 10.1016/S1074-7613(00)80465-3 [DOI] [PubMed] [Google Scholar]
- [33].Peng SL, Gerth AJ, Ranger AM, Glimcher LH. NFATc1 and NFATc2 together control both T and B Cell activation and differentiation. Immunity 2001; 14:13-20; PMID:11163226; http://dx.doi.org/ 10.1016/S1074-7613(01)00085-1 [DOI] [PubMed] [Google Scholar]
- [34].Berland R, Wortis HH. Normal B-1a cell development requires B cell-intrinsic NFATc1 activity. Proc Natl Acad Sci USA 2003; 11:13459-64; http://dx.doi.org/ 10.1073/pnas.2233620100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Samanta DN, Palmetshofer A, Marinkovic D, Wirth T, Serfling E, Nitschke L. B cell hyperresponsiveness and expansion of mature follicular B cells but not of marginal zone B cells in NFATc2/c3 double-deficient mice. J Immunol 2005; 174:4797-802; PMID:15814705; http://dx.doi.org/ 10.4049/jimmunol.174.8.4797 [DOI] [PubMed] [Google Scholar]
- [36].Barrington RA, Borde M, Rao A, Carroll MC. Involvement of NFAT1 in B cell self-tolerance. J Immunol 2006; 177:1510-15; PMID:16849457; http://dx.doi.org/ 10.4049/jimmunol.177.3.1510 [DOI] [PubMed] [Google Scholar]
- [37].Herbst S, Shah A, Mazon Moya M, Marzola V, Jensen B, Reed A, Birrell MA, Saijo S, Mostowy S, Shaunak S, et al.. Phagocytosis-dependent activation of a TLR9-BTK-calcineurin-NFAT pathway co-ordinates innate immunity to Aspergillus fumigatus. EMBO Mol Med 2015; 7:240-58; PMID:25637383; http://dx.doi.org/ 10.15252/emmm.201404556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Caetano MS, Vieira-de-Abreu A, Teixeira LK, Werneck MB, Barcinski MA, Viola JP. NFATC2 transcription factor regulates cell cycle progression during lymphocyte activation: evidence of its involvement in the control of cyclin gene expression. FASEB J 2002; 16:1940-42; PMID:12368232 [DOI] [PubMed] [Google Scholar]
- [39].Xanthoudakis S, Viola JP, Shaw KT, Luo C, Wallace JD, Bozza PT, Curran T, Rao A. An enhanced immune response in mice lacking the transcription factor NFAT1. Science 1998; 272:892-95; http://dx.doi.org/ 10.1126/science.272.5263.892 [DOI] [PubMed] [Google Scholar]
- [40].Hodge MR, Ranger AM, de la Brousse FC, Hoey T, Grusby MJ, Glimcher LH. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity 1996; 4:397-405; PMID:8612134; http://dx.doi.org/ 10.1016/S1074-7613(00)80253-8 [DOI] [PubMed] [Google Scholar]
- [41].Zarkowska T, Mittnacht S. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J Biol Chem 1997; 272:12738-46; PMID:9139732; http://dx.doi.org/ 10.1074/jbc.272.19.12738 [DOI] [PubMed] [Google Scholar]
- [42].Connell-Crowley L, Harper JW, Goodrich DW. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol Biol Cell 1997; 8:287-301; PMID:9190208; http://dx.doi.org/ 10.1091/mbc.8.2.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Engelke M, Engels N, Dittmann K, Stork B, Wienands J. Ca2+ signaling in antigen receptor-activated B lymphocytes. Immunol Rev 2007; 218:235-46; PMID:17624956; http://dx.doi.org/ 10.1111/j.1600-065X.2007.00539.x [DOI] [PubMed] [Google Scholar]
- [44].Chuvpilo S, Jankevics E, Tyrsin D, Akimzhanov A, Moroz D, Jha MK, Schulze-Luehrmann J, Santner-Nanan B, Feoktistova E, Konig T, et al.. Autoregulation of NFATc1/A expression facilitates effector T cells to escape from rapid apoptosis. Immunity 2002; 16:881-95; PMID:12121669; http://dx.doi.org/ 10.1016/S1074-7613(02)00329-1 [DOI] [PubMed] [Google Scholar]
- [45].Kondo E, Harashima A, Takabatake T, Takahashi H, Matsuo Y, Yoshino T, Orita K, Akagi T. NF-ATc2 induces apoptosis in Burkitt's lymphoma cells through signaling via the B cell antigen receptor. Eur J Immunol 2003; 33:1-11; PMID:12594826; http://dx.doi.org/ 10.1002/immu.200390000 [DOI] [PubMed] [Google Scholar]
- [46].Robbs BK, Cruz AL, Werneck MB, Mognol GP, Viola JP. Dual roles for NFAT transcription factor genes as oncogenes and tumor suppressors. Mol Cell Biol 2008; 28:7168-81; PMID:18809576; http://dx.doi.org/ 10.1128/MCB.00256-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fisher RP, Morgan DO. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell 1994; 78:713-24; PMID:8069918; http://dx.doi.org/ 10.1016/0092-8674(94)90535-5 [DOI] [PubMed] [Google Scholar]
- [48].Makela TP, Tassan JP, Nigg EA, Frutiger S, Hughes GJ, Weinberg RA. A cyclin associated with the CDK-activating kinase MO15. Nature 1994; 371:254-57; PMID:8078587; http://dx.doi.org/ 10.1038/371254a0 [DOI] [PubMed] [Google Scholar]
- [49].Hough RE, Goepel JR, Alcock HE, Hancock BW, Lorigan PC, Hammond DW. Copy number gain at 12q12-14 may be important in the transformation from follicular lymphoma to diffuse large B cell lymphoma. Br J Cancer 2001; 84:499-503; PMID:11207045; http://dx.doi.org/ 10.1054/bjoc.2000.1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bea S, Zettl A, Wright G, Salaverria I, Jehn P, Moreno V, Burek C, Ott G, Puig X, Yang L, et al.. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood 2005; 106:3183-90; PMID:16046532; http://dx.doi.org/ 10.1182/blood-2005-04-1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Al-Assar O, Rees-Unwin KS, Menasce LP, Hough RE, Goepel JR, Hammond DW, Hancock BW. Transformed diffuse large B-cell lymphomas with gains of the discontinuous 12q12-14 Alicon display concurrent deregulation of CDK2, CDK4 and GADD153 genes. Br J Haematol 2006; 133:612-21; PMID:16704435; http://dx.doi.org/ 10.1111/j.1365-2141.2006.06093.x [DOI] [PubMed] [Google Scholar]
- [52].Marafiot T, Pozzobon M, Hansmann ML, Ventura R, Pileri SA, Roberton H, Gesk S, Gaulard P, Barth TF, Du MQ, et al.. The NFATc1 transcription factor is widely expressed in white cells and translocates from the cytoplasm to the nucleus in a subset of human lymphomas. Br J Haematol 2005; 128:333-42; PMID:15667535; http://dx.doi.org/ 10.1111/j.1365-2141.2004.05313.x [DOI] [PubMed] [Google Scholar]
- [53].Medyouf H, Alcalde H, Berthier C, Guillemin MC, dos Santos NR, Janin A, Decaudin D, de Thé H, Ghysdael J. Targeting calcineurin activation as a therapeutic strategy for T-cell acute lymphoblastic leukemia. Nat Med 2007; 13:736-41; PMID:17515895; http://dx.doi.org/ 10.1038/nm1588 [DOI] [PubMed] [Google Scholar]
- [54].Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, et al.. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA 2008; 105:13520-25; PMID:18765795; http://dx.doi.org/ 10.1073/pnas.0804295105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pham LV, Tamayo AT, Yoshimura LC, Lin-Lee YC, Ford RJ. Constitutive NF-kB and NFAT activation in aggressive B-cell lymphomas synergistically activates the CD154 gene and maintains lymphoma cell survival. Blood 2005; 106:3940-47; PMID:16099873; http://dx.doi.org/ 10.1182/blood-2005-03-1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Fu L, Lin-Lee YC, Pham LV, Tamayo A, Yoshimura L, Ford RJ. Constitutive NF-kB and NFAT activation leads to stimulation of the BLyS survival pathway in aggressive B-cell lymphomas. Blood 2006; 107:4540-48; PMID:16497967; http://dx.doi.org/ 10.1182/blood-2005-10-4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Pham LV, Tamayo AT, Li C, Bueso-Ramos C, Ford RJ. An epigenetic chromatin remodeling role for NFATc1 in transcriptional regulation of growth and survival genes in diffuse large B-cell lymphomas. Blood 2010; 116:3899-906; PMID:20664054; http://dx.doi.org/ 10.1182/blood-2009-12-257378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].May SL, Zhou Q, Lewellen M, Carter CM, Coffey D, Highfill SL, Bucher CM, Matise I, Morse HC 3rd, O'Sullivan MG, et al.. Nfatc2 and Tob1 have non-overlapping function in T cell negative regulation and tumorigenesis. PLoS One 2014; 9:e100629; PMID:24945807; http://dx.doi.org/ 10.1371/journal.pone.0100629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ranger AM, Gerstenfeld LC, Wang J, Kon T, Bae H, Gravallese EM, Glimcher MJ, Glimcher LH. The nuclear factor of activated T cells (NFAT) transcription factor NFATp (NFATc2) is a repressor of chondrogenesis. J Exp Med 2000; 191:9-22; PMID:10620601; http://dx.doi.org/ 10.1084/jem.191.1.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jauliac S, López-Rodriguez C, Shaw LM, Brown LF, Rao A, Toker A. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol 2002; 4:540-44; PMID:12080349; http://dx.doi.org/ 10.1038/ncb816 [DOI] [PubMed] [Google Scholar]
- [61].Welch PJ, Wang JY. A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-Abl tyrosine kinase in the cell cycle. Cell 1993; 75:779-90; PMID:8242749; http://dx.doi.org/ 10.1016/0092-8674(93)90497-E [DOI] [PubMed] [Google Scholar]
- [62].Chicaybam L, Sodre AL, Curzio BA, Bonamino MH. An efficient low cost method for gene transfer to T lymphocytes. PLoS One 2013; 8:e60298; PMID:23555950; http://dx.doi.org/ 10.1371/journal.pone.0060298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al.. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2:401-04; PMID:22588877; http://dx.doi.org/ 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].R Development Core Team R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna; 2008. Available from: http://www.r-project.org [Google Scholar]
- [65].Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2009. [Google Scholar]
- [66].Welch BL. The generalisation of student's problems when several different population variances are involved. Biometrika 1947; 34:28-35; PMID:20287819 [DOI] [PubMed] [Google Scholar]