ABSTRACT
Hypoxia is a general event in solid tumor growth. Therefore, induced cellular responses by hypoxia are important for tumorigenesis and tumor growth. MicroRNAs (miRNAs) have recently emerged as important regulators of hypoxia induced cellular responses. Here we report that miR-147a is a novel and crucial hypoxia induced miRNA. HIF-1α up-regulates the expression of miR-147a, and miR-147a in turn stabilizes and accumulates HIF-1α protein via directly targeting HIF-3α, a dominant negative regulator of HIF-1α. Subsequent studies in xenograft mouse model reveal that miR-147a is capable of inhibiting tumor growth. Collectively, these data demonstrate a positive feedback loop between HIF-1α, miR-147a and HIF-3α, which provide a new insight into the mechanism of miR-147a induced cell proliferation arrest under hypoxia.
KEYWORDS: Cell proliferation, HIF-1α, HIF-3α, miR-147a
Abbreviations
- miRNA
microRNA
- CCK-8
cell counting kit-8
- HIF-1α
hypoxia-inducible factor 1 α subunit
- HIF-3α
hypoxia-inducible factor 1 α subunit
- NC
negative control
- MUT
mutated
- UTR
untranslated region
Introduction
MicroRNAs (miRNAs or miRs) are a kind of small non-coding RNAs with a length of about 20–24 nucleotides. They participate in a vast majority of physiological and pathological processes, including cell proliferation, differentiation, apoptosis, autophagy, angiogenesis, metabolism and cancer.1-7 Especially, miRNAs have recently emerged as important regulators of cellular responses under hypoxia. Hypoxia-induced miR-26 production influences myogenic differentiation of embryonic stem cells by targeting HDAC6.8 MiR-133a attenuates hypoxia-induced apoptosis via inhibition of TAGLN2 expression 9 while miR-195 and miR-26b enhance apoptosis by directly targeting HIF-1α and PTEN respectively.10,11 MiR-101a exerts anti-fibrotic effects via targeting TGFβRI.12 MiR-21, miR-322, miR-20s and miR-130/301 influence hypoxia-induced proliferation of pulmonary artery smooth muscle cells.13-16 We have earlier reported a group of miRNAs which respond to hypoxia induction.17 Among these, miR-210 and miR-155 are related to cell cycle, proliferation and autophagy.18,19
Hsa-miR-147a is related to hsa-miR-210 closely, which differs by one nucleotide in the seed region. MiR-147a was discovered in mouse spleen tissue 20 and subsequently in humans 21 by Thomas Tuschl. Like miR-210, miR-147a inhibits cell proliferation by downregulating cell cycle proteins, such as pRB, CycB, CycA, Cdk6.22 MiR-147a is upregulated in Squamous Cell Carcinoma of tongue, human gastric cancer, small cell lung cancer and hepatocellular carcinoma.23-26 Some reports demonstrate that miR-147a affects cell development, migration and invasion, but has no influence on apoptosis.5,22,27
Here we report that miR-147a is a novel hypoxia-inducible miRNA and participates in cell hypoxia response processing. MiR-147a is up-regulated by HIF-1α in Hela cells under hypoxia condition, and then miR-147a increases stabilization of HIF-1α protein via targeting HIF-3α, which is a dominant negative regulator of HIF-1α. Thus, miR-147a plays an important role in inhibiting cell proliferation, one of cellular responses under hypoxic stress. Moreover, overexpressing miR-147a significantly inhibits tumor growth in xenograft mouse tumor models. Collectively, these data demonstrate that the HIF-1α-miR-147a-HIF-3α axis serves as a new mechanism underlying the role of miR-147a in regulating cell proliferation under hypoxia.
Materials and methods
Cell culture
HeLa (human cervical cancer, ATCC, CCL-2) cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Gibco, 12800–017) supplemented with 10% fetal bovine serum (FBS, PAA, A15–101). Cells were grown under normoxic (21% O2) or hypoxic (1% O2) conditions at 37°C in a humidified 5% CO2 incubator.
Plasmid, miRNA and siRNA transfection
HeLa cells were transiently transfected with Lipofectamine 2000 (Invitrogen, 11668–019) according to the manufacturer's instructions. Plasmid pEGFP-N1-FIH1 (plasmid # 21403) and HA-HIFalpha-pcDNA3 (plasmid # 18949) were purchased from Addgene. A mock-vehicle pcDNA3 and a small RNA with a random sequence were used as negative controls. The plasmid was added to adherent cells in a 6-well plate at a final concentration of 1uM; siRNA or miRNA was suspended for transfection at a final concentration of 100 pmol. MiR-147a mimics, miR-147a inhibitors, negative control (NC) and siRNAs were synthesized and purchased from Shanghai GenePharma (Shanghai, China). The sequences of siRNAs used were:
Si HIF-3α#1: 5′-CCACCACGCCCGACAGUAA-3′,
5′-UUACUGUCGGGCGUGGUGG-3′
Si HIF-3α#2: 5′-GUGCUGGGAUUACUGGUAU-3′,
5′-AUACCAGUAAUCCCAGCAC-3′
Si HIF-1α: 5′-AACUAACUGGACACAGUGUGU-3′
5′-ACACACUGUGUCCAGUUAGUU3′
miRNA targets prediction
We used two conventional online programs to predict the potential targets and precise binding sites of miR-147a: Targetscan (www.targetscan.org) and Findtar (bio.sz.tsinghua.edu.cn).
RNA isolation, reverse transcription and quantitative real-time PCR
After transfection (48h), cells were washed in 1XPBS and RNA was isolated using RNAiso Plus (Takara, D9108B) according to the manufacturer′s instructions. Total mRNA was reverse transcribed using a reverse transcription kit (Takara, D6130) with random primers and quantitative real-time PCR of cDNA was performed using SYBR Green Real-time PCR Master Mix (Toyobo, QPK-201). qRT-PCRs of miRNAs were performed according to the manufacturer′s instructions using a TaqMan MicroRNA Reverse Transcription Kit (ABI.4366597) and Real-time PCR Master Mix (Toyobo, QPK-101). miRNA probes were purchased from ABI: hsa-miR-147a (ABI, 000469), RNU6 (ABI, 001093). mRNA and miRNA expression levels were normalized to GAPDH and RNU6 respectively.
Western blotting
HeLa cells were lysed with an ice-cold cell lysis buffer (50mM Tris-HCl pH8.0, 4M Urea and 1% Triton X-100), containing protease and phosphatase inhibitors (Roche, 04693132001). Whole cell lysates were equalized and separated by SDS-PAGE and then transferred onto nitrocellulose membrane. Membranes were blocked with 5% milk in Tris-buffered saline plus 0.02% Tween-20 (TBST). Primary antibodies used in this paper were: HIF-1α (BD 610958); HIF-3α (Novusbio 100–2529), ACTB (CST 3700). After incubation with horseradish peroxidase-coupled secondary anti-mouse (KPL 074–1806) or anti-rabbit antibodies (KPL 474–1506), the membranes were washed and protein bands were detected with ECL blotting detection reagents (KPL 547100).
Luciferase activity assay
HeLa cells were seeded in a 24-well plate. The plasmids pmirGLO-HIF-3α1–3′UTR–WT or pmirGLO-HIF-3α1–3′UTR–MUT were constructed by inserting the wild-type or mutated 3′UTR fragments of HIF-3α into pmirGLO Dual-Luciferase miRNA Target Expression Vectors (Promega, E1330). To detect the reciprocity of miR-147a and HIF-3α1 3′UTR, HeLa cell were co-transfected with WT or MUT UTR luciferase vectors and miRNAs using lipofectamine 2000 according to reference. About 36h later, the cells were collected, firefly and Renilla Luciferase activities were measured by Dual-Luciferase Reporter assay system (Promega, E1960), and normalized to the ratio of firefly and Renilla luciferases. Each experiment was repeated in triplicate.
Cell proliferation assay and colony formation
Cell proliferation was evaluated using cell counting kit-8 (CCK-8 Kit, Dojindo, CK04). HeLa cells were seeded in 96-well plates in triplicate with 2000 cells per well and transfected with miR-NC, miR-147a or siRNA and the number of viable cells was determined by measuring OD450 using microplate reader (Epoch, BioTek). For the colony formation assay, HeLa cells were plated on a 6-well plate and transfected with miR-NC, miR-147a or siRNA once in every 3 d after cell adhesion. After culturing for 10 days, cells were fixed with cold methanol and stained with 1% crystal violet for 30 min. The number of colonies was counted assuming >50 cells/colony.
Mice and tumor formation assays
Four week old male BALB/c nude mice were purchased from Guangdong Experimental Animal Center (Guangzhou, China) and kept in a pathogen-free environment for a week at Tsinghua University Shenzhen Graduate School. Approximately 5 × 105 well-conditioned HeLa cells were injected into dorsal flank of nude mice subcutaneously. Ten days after tumor implantation, mice were randomly divided into 3 groups for treatment by either miR-147a, miR-NC. Transferrin-polyethylenimine (TF-PEI) / miRNA-mimics complex were delivered through subcutaneous injections every alternate day. At the end of the experiment (after 5 injections), the mice were killed by breaking the neck, individual tumors isolated and the weight of each animal was recorded.
Statistical analysis
The bar graphs represent mean ± SD from at least 3 independent experiments. Student's t-test was applied for significant difference between 2 groups of data. The results were statistically significant if p-value was <0.05.
Results
Hypoxia induces upregulation of miR-147a
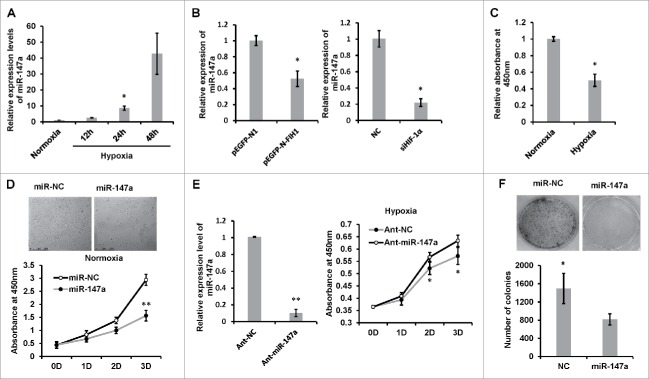
To determine the expression of miR-147a in hypoxia, we cultured HeLa cells at 1% O2, and subsequently collected the cells for qRT-PCR. The expression level of endogenous miR-147a increased nearly 10 folds after 24h in hypoxia (Fig. 1A). To investigate whether hypoxia-induced upregulation of miR-147a is due to HIF-1α, we transfected cells with pEGFP-N-FIH1, which can inhibit HIF-1α transcription activity.28 Overexpression of FIH1 dramatically reduced the expression of miR-147a. Knocking down endogenous HIF-1α showed the similar effect (Fig. 1B).
Figure 1.
Hypoxia-induced miR-147 inhibits cell proliferation and colony formation. (A) Hypoxia induces miR-147a expression. HeLa cells were exposed to 1% oxygen for 12, 24 and 48 h; real-time PCR analysis of miR-147a levels in HeLa cells was done and compared with normoxia (21% O2). U6 small nuclear RNA was used as an internal control. Data shown are mean ± SD of 3 independent experiments. *P < 0.05. (B) The expression of miR-147a is regulated by HIF-1α. HeLa cells were transfected with pEGFP-N-FHI plasmid or siHIF-1α for 48h in hypoxia (1% O2); miR-147a levels in HeLa cells were detected by Real-time PCR. pEGFP-N1 or NC were used as negative controls. Data shown are mean ± SD of 3 independent experiments. *P < 0.05. (C) Hypoxia inhibits cell proliferation. HeLa cells were cultivated in hypoxia (1% O2) or normoxia (21% O2) for 48h and viabilities were determined by CCK-8 assay. Data shown are mean ± SD of 3 independent experiments. *P < 0.05. (D) Overexpression of miR-147a inhibits cell proliferation. HeLa cells were transfected with miR-147a or miR-NC in normoxia (21% O2) for 1, 2 and 3 d and viabilities were determined by CCK-8 assay every day. Representative phase contrast images taken after 48h are shown. Data shown are mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.001. (E) Knockdown of endogenous miR-147a recovery from hypoxia-induced cell growth inhibition. HeLa cells were transfected with single strand ant-miRNA in hypoxia (1% O2); real-time PCR analysis shows that endogenous miR-147a expression was blocked; viabilities were determined by CCK-8 assay every day. Data shown are mean ± SD of 3 independent experiments. **P < 0.001. (F) Clonogenic assay of HeLa cells transfected with miR-NC or miR-147a. Cells were cultured for 10 d in normoxia and stained with Crystal Violet. The number of colonies were quantified and calculated as mean ± SD of 3 independent experiments (Student t test, *p < 0.05).
miR-147a inhibits cell proliferation and colony formation
Hypoxia appears to be strongly associated with tumor propagation and malignant progression, thus we were interested to know whether hypoxia-inducible miR-147a can regulate cell proliferation. Hypoxic stress caused cell growth retardation to nearly half of that observed in normoxia (Fig. 1C). Overexpression of exogenous miR-147a remarkably inhibited cell growth in normoxia (Fig. 1D). Conversely, inhibition of endogenous miR-147a accelerated cell proliferation in hypoxia (Fig. 1E). The effect of miR-147a on cell proliferation was also evaluated by colony formation assay. MiR-147a transfected cells formed significantly fewer colonies than the negative control (Fig. 1F).
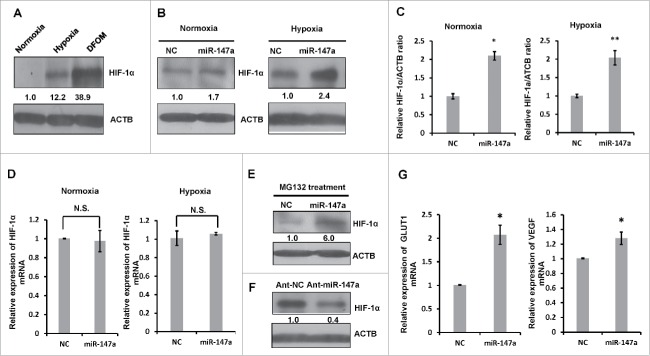
Overexpression of miR-147a leads to accumulation of HIF-1α
We were keen to understand whether miR-147a mediated cell proliferation inhibition is related to HIF-1α, which is directly involved in hypoxia-induced cell proliferation arrest.29 HIF-1α is a critical transcriptional regulator and specifically accumulates in low oxygen conditions (Fig. 2A). Overexpression of exogenous miR-147a significantly increased the protein level of HIF-1α (Fig. 2B, 2C), but not the mRNA levels of HIF-1α in both normoxia and hypoxia conditions (Fig. 2D). We also used MG132, a proteasome inhibitor to increase the stability of HIF-1α in normoxia. MiR-147a increased accumulation of HIF-1α even in the presence of MG132 (Fig. 2E). Conversely, blocking endogenous miR-147a decreased the protein expression of HIF-1α (Fig. 2F). Then, we examined the expression of VEGF and GLUT1, which are downstream targets of HIF-1α. MiR-147a increased the expression of VEGF and GLUT1, owing to the accumulation of HIF-1α (Fig. 2G).
Figure 2.
Overexpression of miR-147 leads to accumulation of HIF-1α. (A) Hypoxia induces HIF-1α expression and accumulation. HeLa cells were cultivated in hypoxia (1% O2) or treated with or without DFOM for 24 h in normoxia (21%O2); cells were harvested for western blot. Relative HIF-1α/ACTB ratios were determined by Image J densitometric analysis. (B) Overexpression of miR-147a leads to accumulation of HIF-1α protein. HeLa cells were transfected with NC, miR-147a in normoxia (21%O2) or hypoxia (1% O2). Western blotting was performed to analyze the status of HIF-1α and ACTB. (C) Relative HIF-1α / ACTB ratios from (B) were determined by Image J densitometric analysis. Data shown are mean ± SD of 3 independent experiments, *P < 0.05, **P < 0.01. (D) Overexpression of miR-147a doesn't effect HIF-1α transcription. HeLa cells were transfected with NC, miR-147a in normoxia (21%O2) or hypoxia (1% O2). Cells were harvested at 48h for qRT-PCR. N.S., not significant. (E) miR-147a enhanced HIF-1α accumulation of MG132 in 21% O2. HeLa cells were transfected with miR-147a or NC. After 36h of transfection, HeLa cells were treated with MG132 (10 μμ). The protein level of HIF-1α and ACTB were detected by protein gel blot. Relative HIF-1α/ACTB ratios were determined by Image J densitometric analysis. (F) Blockage of endogenous miR-147a leads to HIF-1α protein reduction. HeLa cells were transfected with Ant-NC or Ant-miR-147a in hypoxia (1% O2). Western blotting was performed to analyze the status of HIF-1α and ACTB. Relative HIF-1α/ACTB ratios were determined by Image J densitometric analysis. (G) Overexpression of miR-147a enhances the mRNA levels of VEGF and GLUT1 in 21% O2, which are regulated by HIF-1α.
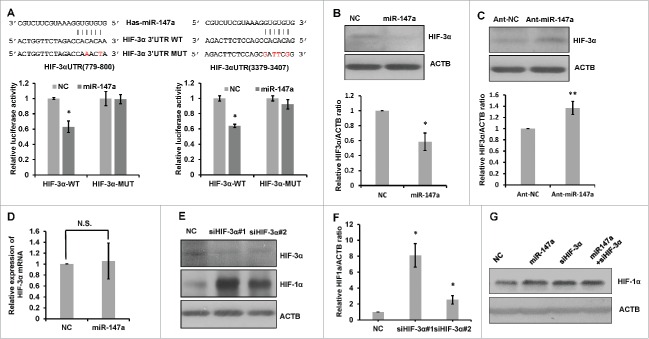
miR-147a upregulates HIF-1α via targeting HIF- 3α
We used bioinformatics approach 17 to identify the putative miR-147a targets. HIF-3α 3′ UTR contains 2 potential miR-147 binding sites (Fig. 3A). To determine if miR-147a regulates the expression of HIF-3α via binding to its 3′UTR, we performed luciferase activity assay. We generated luciferase reporters by inserting the 3′UTR fragments of HIF-3α with wild-type (WT) or mutated (MUT) miR-147a binding sites into a pmirGLO-luciferase vector. The constructs were then introduced into the cells for luciferase activity assay. As shown in Fig. 3A, miR-147a significantly suppressed luciferase activity when co-transfected with the reporter constructs with wild-type binding sites of miR-147a. By contrast, the reporter vectors carrying mutated binding sites of miR-147a rescued the repressive effects of miR-147a on luciferase activity. Overexpression of exogenous miR-147a significantly reduced HIF-3α protein expression, but had no effect on HIF-3α mRNA (Fig. 3B, 3D). Conversely, inhibition of endogenous miR-147a increased HIF-3α protein level (Fig. 3C). Similar to miR-147, HIF-3α siRNA also increased protein expression of HIF-1α (Fig. 3E, 3F). To determine whether the stabilization of HIF-1α by miR-147a is dependent on its regulation on HIF-3α, we firstly knockdown HIF-3α with HIF-3α siRNA, then, introduced miR-147a into the cells. Compared with HIF-3α knockdown cells, adding miR-147a cannot further increase the accumulation of HIF-1α (Fig. 3G). This result suggests that miR-147a stabilize HIF-1α via HIF-3α.
Figure 3.
miR-147a upregulates HIF-1α via targeting HIF- 3α. (A) miR-147a directly targets the 3′UTR of HIF-3α. Predicted binding sequences between miR-147a and seed matches in HIF-3α 3′UTR. Luciferase reporter assay of wild type (WT) or mutated (MUT) HIF-3α 3′UTR vector co-transfected with NC, miR-147a respectively in normoxia (21%O2). Data shown are mean ± SD of 3 independent experiments, *P < 0.05. (B) Western blot analysis of HIF-3α and ACTB proteins in HeLa cells transfected with NC or miR-147a in normoxia (21%O2). Relative HIF-3α/ACTB ratios were determined by Image J densitometric analysis. Data shown are mean ± SD of 3 independent experiments, *P < 0.05. (C) Western blot analysis of HIF-3α and ACTB proteins in HeLa cells transfected with ant-miR-147a or anti-NC in hypoxia (1% O2). Relative HIF-3α/ACTB ratios were determined by Image J densitometric analysis. Data shown are mean ± SD of 3 independent experiments, **P < 0.01. (D) Overexpression of miR-147a doesn't effect the mRNA level of HIF-3α. HeLa cells were transfected with NC, miR-147a. Cells were harvested at 48h in normoxia (21%O2) for qRT-PCR. N.S., not significant. (E) HIF-1α accumulates when HIF-3α expression is inhibited. HeLa cells transfected with NC or 2 designed HIF-3α siRNAs in hypoxia (1% O2). Western blotting was performed to analyze the protein levels of HIF-1α and ACTB. (F) Relative HIF-1α / ACTB ratios from (E) were determined by Image J densitometric analysis. Data shown are mean ± SD of 3 independent experiments, *P < 0.05, student 2-tailed t test. (G) miR-147a stabilized HIF-1α through HIF-3α. HeLa cells transfected with HIF-3α siRNA firstly and then introduced miR-147a into the cells in hypoxia (1% O2). Western blot analysis of HIF-1α and ACTB proteins.
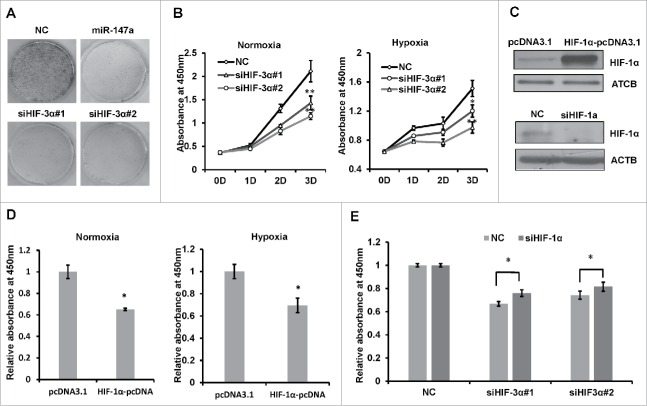
HIF- 3α regulates cell proliferation
To examine whether miR-147a regulate cell proliferation through targeting HIF-3α, we performed colony formation assay. Less colonies were observed in cells transfected with miR-147a or HIF-3α siRNAs (Fig. 4A). HIF-3α siRNAs also inhibited cell proliferation under both normoxia and hypoxia (Fig. 4B). Since siRNA for HIF-1α was reported to accelerate cellular proliferation,35 we wonder if exogenous HIF-1α increased cell proliferation. We firstly examined the availability of siHIF-1α and HIF-1α pcDNA3.1 (Fig. 4C). In agreement with these observations, exogenous HIF-1α increased cell proliferation, under normoxic and hypoxia conditions (Fig. 4D). We wondered if HIF-1α siRNA can rescue HIF-3α siRNA induced inhibition of cell proliferation. When siHIF-3α and siHIF-1α were co-transfected into HeLa cells, siHIF-3α-induced inhibition of cell proliferation was partially rescued (Fig 4E). These data suggested that HIF-3α may regulate cell proliferation via HIF-1α.
Figure 4.
HIF- 3α regulates cell proliferation. (A) Clonogenic assay of HeLa cells transfected with NC, miR-147a or siHIF-3α in normoxia. (B) Inhibition of HIF-3α expression inhibits HeLa cell proliferation. HeLa cells transfected with 2 designed HIF-3α siRNAs or NC for 1, 2 or 3 days; viabilities were determined with CCK-8 assay every day. Data shown are mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.001. (C) Transfection with HIF-1α siRNA reduced HIF-1α synthesis in 1% O2and transfection with HIF-1α plasmid increased HIF-1α synthesis in 21% O2 by western blot. (D) Overexpression HIF-1α inhibits cell proliferation in normoxia or hypoxia. HeLa cells were transfected with pcDNA or HIF-1α-pcDNA in 21% O2or 1%O2. Expression of HIF-1α was confirmed by protein gel blot and cell viabilities were determined by CCK-8 assay after 3 d. Data shown are mean ± SD of 3 independent experiments. *P < 0.05. (E) CCK-8 assay of HeLa cells co-transfected with siRNAs or NC; cell viability (OD490 nm absorbance) was examined at 72h after transfection in hypoxia. Data shown are mean ± SD of 3 independent experiments, *P < 0.05, student 2-tailed t test.
miR-147a suppresses tumor growth in nude mice
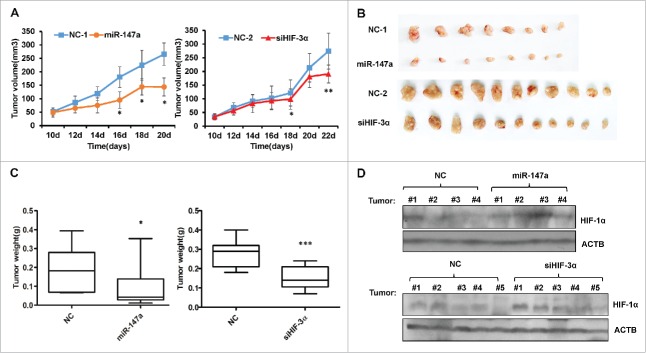
In order to determine the anticancer potential of miR-147a and siHIF-3α, we generated xenografts mouse model by subcutaneously injecting HeLa cells into nude mice. Tumor volumes grew sustainably and significantly in the NC group, whereas miR-147a and siHIF-3α groups showed slow tumor growth (Fig. 5A). At the end of the experiments, the nude mice were sacrificed and the tumors were separated to measure tumor mass. Compared with NC (negative control), miR-147a suppressed tumor weight to 43% (P < 0.005), siHIF-3α also moderately decreased tumor mass (Fig. 5B, 5C). Tumor samples with different treatments were collected for Western Blotting assay, the result demonstrated that both miR-147a and siHIF-3α increased protein expression of HIF-1α (Fig. 5D).
Figure 5.
miR-147a suppresses tumor growth in nude mice. (A) Tumor volumes were calculated by the length and width measurement using Vernier calipers every 2 d. The xenografts were established through subcutaneous injection of Hela cells in the flank of nude mice. When the tumors had reached an average volume of 40–50 mm3, tumor-bearing nude mice were treated with miR-147a, HIF-3α siRNAs and NC. Group miR-147a and NC-1 have 5 mice bearing 8 tumors respectively. siHIF-3α and NC-2 have 7 mice bearing 11 tumors respectively. *P < 0.05, **P < 0.001, student 2-tailed t test. (B) Images of tumors from (A). (C) The weight of tumors shown in (B). *P < 0.05, ***P < 0.0001, student 2-tailed t test. (D) Western blot analysis of HIF-1α in tumors treated with NC, miR-147a or siHIF-3α.
Discussion
In this study, we reported that hypoxia-induced miR-147a inhibits cancer cell proliferation, both in vitro and in vivo. It has been reported that miR-147a inhibits cell growth via downregulation of cell cycle proteins, including pRB, cyclin B, cyclin A, CDK2,CDK4 and CDK6.22,27 But in this study, we find that miR-147a induced cell growth arrest under hypoxia condition also related to HIF-1α accumulation. As a transcription factor, HIF-1α is involved in the regulation for cell growth, angiogenesis, metabolism, apoptosis.30 Furthermore, there has been reported that HIF-1α accumulation inhibits cell cycle progression and leads to arrest 29 through upregulating cell cycle inhibitors p21 and p27.31,32 Thus, our results suggest that miR-147a induced cell proliferation arrest could be due to the accumulation and upregulation of HIF-1α protein, which activates the relevant downstream pathway of HIF-1α.
The hypoxia-inducible factors (HIFs) family is an important group of regulators for the cellular oxygen tension response. There are 4 principal isoforms of HIF (HIF-1α, HIF-2α, HIF-3α and HIF-1β). HIF-1 is a heterodimer containing HIF-1α and HIF-1β, and its expression is regulated by cellular oxygen tension.33 HIF-3α is a third α-class hypoxia-induced factor isoform. There is cell-specific Characteristics for the expression of HIF-3α, only several cells such as CaKi-1 and HeLa expressing HIF-3α 34 HIF-3α is a negative regulator of hypoxia-inducible gene expression.35,36 HIF-3α has the capacity of competing with HIF-1α for binding to HIF-1β and causes the degradation of HIF-1α via the ubiquitin pathway. When HIF-3α is silenced, there are more HIF-1β molecules to bind with HIF-1α, thereby increasing stability of HIF- 1α.37 In this investigation, we discovered that miR-147a induced accumulation of HIF-1α via targeting HIF-3α. Through suppressing HIF-3α expression, miR-147a positively regulates hypoxia-inducible gene expression, including HIF-1α, and finally inhibits cell proliferation.
Our results indicate that the siHIF-3α treatment can increase the level of HIF-1α to a comparable extent as miR-147a treatment does (Fig. 3F). However, the inhibitory effect of siHIF-3α on tumor growth is not as robust as that caused by miR-147a (Fig. 5A). This is due to multi-targets, the prominent feature of miRNAs. In addition to HIF-3α, miR-147a also directly targets some cell cycle-related genes. Thus, the inhibitory effect of siHIF-3α on tumor growth is not as robust as that caused by miR-147a. To further confirm that the suppressive effect of HIF-3α on cell proliferation is related to HIF-1α accumulation, we completed a rescuing experiment via co-transfecting siHIF-3α and siHIF-1α into HeLa cells. The knockdown of HIF-1α can only partially rescue the suppressive effect of siHIF-3α on cell proliferation (Fig. 4E), suggesting there are other mechanisms/pathways that are involved in the siHIF-3α-induced suppression. It was reported that in addition to negatively regulating HIF-1α expression in protein level, HIF-3α also positively regulates expression of HIF-2α. HIF-2α can stimulate proliferation under hypoxic conditions via regulating expression of NANOG.38 NANOG is involved in cervical tumorigenesis.39 CDK6 and CDC25A are the downstream cell cycle effectors of NANOG during the transition from G1 to S.40 Therefore, the knockdown of HIF-1α only partially rescue the suppressive effect of siHIF-3α on cell proliferation.
Under hypoxia condition, HIF-1 binds to hypoxia response elements (HREs) in the promoter region of target genes and activates transcription. Although miR-147a is upregulated during hypoxic stress, we did not find any HRE upstream of miR-147a gene up to 10 kb. Thus, we hypothesized the expression of miR-147a was regulated by HIF-1α indirectly under hypoxic condition. Using ChIPBase (deepbase.sysu.edu.cn/chipbase/index.php), a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data, we found that HNF4A was a direct transcription factor of miR-147a from chip-seq data.41 Hypoxia induced the expression of HNF4A and HIF-1α, so HNF4A and HIF-1α were upregulated in hypoxia.42 HNF4A interacts with HIF-1α to exert transcription activity.43 Chip-seq data demonstrated that HNF4A was a direct transcription factor of miR-147a. Taken together, miR-147a is a hypoxia-induced miRNA and regulate by HIF-1α indirectly. Hypoxia induced HIF-1α increases the expression of miR-147a through HNF4A. MiR-147a promotes the stability of HIF-1α by targeting HIF-3α, thus forming a positive feedback loop. The accumulation of HIF-1α increases the expression of downstream gene, which contribute to cell proliferation arrest (Fig. 6). These discoveries provide a new insight into the mechanism of miR-147a induced cell proliferation arrest under hypoxia.
Figure 6.
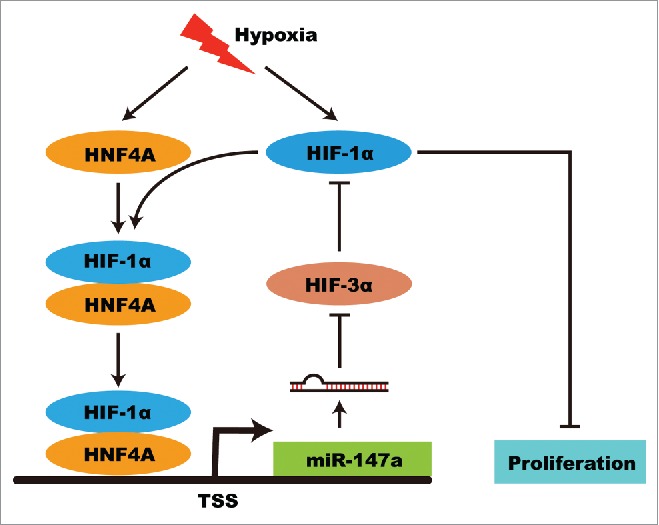
Model of hypoxia-induced miR-147a in regulating cell proliferation and its role in positive feedback. Hypoxia induced HIF-1α and HNF4A expression. HIF-1α increases the expression of miR-147a through interacting with HNF4A. MiR-147a promotes the stability of HIF-1α by targeting HIF-3α. The accumulation of HIF-1α increases the expression of downstream gene, which contribute to cell proliferation arrest.
According to our results and other peoples' reports, HIF-1α inhibits cell proliferation in vitro experiments, but promotes tumor growth and angiogenesis in vivo experiments.44 This is due to that HIF-1α can up-regulate the expression of p21, p27, p53, VEGF, and other gene.45 In vitro experiment, upregulated p21, p27 and p53 inhibit cell proliferation.46-48 In vivo experiment, p21, p27, and p53 may also be up-regulated. However, angiogenesis induced by VEGF and other genes play more important roles, and finally promote tumor growth and progression.49 HIF-1α promotes angiogenesis to improve the blood supply and changes the metabolic model of solid tumor for adapting hypoxia environment in vivo. Proliferation arrest may be necessary to survival under the initial stage of hypoxia condition before the cellular functional alteration derived by HIF-1α.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Natural Science Foundation of China (No.31371315,No.31301117) and International Cooperation Grant of Shenzhen (No. GJHZ20140416153718941).
References
- 1.Wu J, Lv Q, He J, Zhang H, Mei X, Cui K, Huang N, Xie W, Xu N, Zhang Y. MicroRNA-188 suppresses G1/S transition by targeting multiple cyclin/CDK complexes. Cell Commun Signal 2014; 12:66; PMID:25304455; http://dx.doi.org/ 10.1186/s12964-014-0066-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Y, Liu X, Wang Y. MicroRNA-378 regulates neural stem cell proliferation and differentiation in vitro by modulating Tailless expression. Biochem Biophys Res Commun October 2015; 466(2):214-220; doi:http://dx.doi.org/ 10.1016/j.bbrc.2015.09.011 [DOI] [PubMed] [Google Scholar]
- 3.Tu H, Wei G, Cai Q, Chen X, Sun Z, Cheng C, Zhang L, Feng Y, Zhou H, Zhou B, et al.. MicroRNA-212 inhibits hepatocellular carcinoma cell proliferation and induces apoptosis by targeting FOXA1. Onco Targets Ther 2015; 8:2227-35; PMID:26347321; http://dx.doi.org/ 10.2147/OTT.S87976 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Huang N, Wu J, Qiu W, Lyu Q, He J, Xie W, Xu N, Zhang Y. MiR-15a and miR-16 induce autophagy and enhance chemosensitivity of Camptothecin. Cancer Biol Ther 2015; 16(6):941-8; PMID:25945419; http://dx.doi.org/ 10.1080/15384047.2015.1040963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seok JK, Lee SH, Kim MJ, Lee YM. MicroRNA-382 induced by HIF-1alpha is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res 2014; 42(12):8062-72; PMID:24914051; http://dx.doi.org/ 10.1093/nar/gku515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deiuliis JA. MicroRNAs as regulators of metabolic disease: Pathophysiologic significance and emerging role as biomarkers and therapeutics. Int J Obes (Lond) 2015; 40(1):88-101; PMID:26311337; http://dx.doi.org/ 10.1038/ijo.2015.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamfjord J, Stangeland AM, Hughes T, Skrede ML, Tveit KM, Ikdahl T, Kure EH. Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS One 2012; 7(4):e34150; PMID:22529906; http://dx.doi.org/ 10.1371/journal.pone.0034150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SW, Yang J, Kim SY, Jeong HK, Lee J, Kim WJ, Lee EJ, Kim HS. MicroRNA-26a induced by hypoxia targets HDAC6 in myogenic differentiation of embryonic stem cells. Nucleic Acids Res 2015; 43(4):2057-73; PMID:25662604; http://dx.doi.org/ 10.1093/nar/gkv088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li AY, Yang Q, Yang K. miR-133a mediates the hypoxia-induced apoptosis by inhibiting TAGLN2 expression in cardiac myocytes. Mol Cell Biochem 2015; 400(1–2):173-81; PMID:25421410; http://dx.doi.org/ 10.1007/s11010-014-2209-x [DOI] [PubMed] [Google Scholar]
- 10.Bai R, Zhao AQ, Zhao ZQ, Liu WL, Jian DM. MicroRNA-195 induced apoptosis in hypoxic chondrocytes by targeting hypoxia-inducible factor 1 α. Eur Rev Med Pharmacol Sci 2015; 19(4):545-51; PMID:25753868; http://www.europeanreview.org/article/8545 [PubMed] [Google Scholar]
- 11.Wang X, Li C, Dai Q. Down-regulation of microRNA-26b rescued hypoxia-induced apoptosis in cultured neonatal rat cardiac myocytes by regulating PTEN. Int J Clin Exp Med 2015; 8(3):4073-9; PMID:26064312; http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4443146/ [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao X, Wang K, Liao Y, Zeng Q, Li Y, Hu F, Liu Y, Meng K, Qian C, Zhang Q, et al.. MicroRNA-101a inhibits cardiac fibrosis induced by hypoxia via targeting TGFbetaRI on cardiac fibroblasts. Cell Physiol Biochem 2015; 35(1):213-26; PMID:25591764; http://dx.doi.org/ 10.1159/000369689 [DOI] [PubMed] [Google Scholar]
- 13.Green DE, Murphy TC, Kang BY, Searles CD, Hart CM. PPAR gamma ligands attenuate hypoxia-induced proliferation in human pulmonary artery smooth muscle cells through modulation of MicroRNA-21. PLoS One 2015; 10(7):e0133391; PMID:26208095; http://dx.doi.org/ 10.1371/journal.pone.0133391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng Y, Liu H, Kang K, Wang Z, Hui G, Zhang X, Zhong J, Peng W, Ramchandran R, Raj JU, et al.. Hypoxia inducible factor-1 mediates expression of miR-322: potential role in proliferation and migration of pulmonary arterial smooth muscle cells. Sci Rep 2015; 5:12098; PMID:26166214; http://dx.doi.org/ 10.1038/srep12098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng Y, Pan Y, Liu H, Kang K, Wu Y, Hui G, Peng W, Ramchandran R, Raj JU, Gou D. MiR-20a regulates the PRKG1 gene by targeting its coding region in pulmonary arterial smooth muscle cells. FEBS Lett 2014; 588(24):4677-85; PMID:25447536; http://dx.doi.org/ 10.1016/j.febslet.2014.10.040 [DOI] [PubMed] [Google Scholar]
- 16.Bertero T, Lu Y, Annis S, Hale A, Bhat B, Saggar R, Saggar R, Wallace WD, Ross DJ, Vargas SO, et al.. Systems-level regulation of microRNA networks by miR-130/301 promotes pulmonary hypertension. J Clin Invest 2014; 124(8):3514-28; PMID:24960162; http://dx.doi.org/ 10.1172/JCI74773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, et al.. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 2006; 1:e116; PMID:17205120; http://dx.doi.org/ 10.1371/journal.pone.0000116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He J, Wu J, Xu N, Xie W, Li M, Li J, Jiang Y, Yang BB, Zhang Y. MiR-210 disturbs mitotic progression through regulating a group of mitosis-related genes. Nucleic Acids Res 2013; 41(1):498-508; PMID:23125370; http://dx.doi.org/ 10.1093/nar/gks995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan G, Xie W, Liu Z, Xu W, Lao Y, Huang N, Cui K, Liao M, He J, Jiang Y, et al.. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy 2014; 10(1):70-9; PMID:24262949; http://dx.doi.org/ 10.4161/auto.26534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol 2002; 12(9):735-9; PMID:12007417; http://dx.doi.org/ 10.1016/S0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- 21.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al.. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007; 129(7):1401-14; PMID:17604727; http://dx.doi.org/ 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertero T, Grosso S, Robbe-Sermesant K, Lebrigand K, Hénaoui IS, Puisségur MP, Fourre S, Zaragosi LE, Mazure NM, Ponzio G, et al.. “Seed-Milarity” confers to hsa-miR-210 and hsa-miR-147b similar functional activity. PLoS One 2012; 7(9):e44919; PMID:23028679; http://dx.doi.org/ 10.1371/journal.pone.0044919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res 2008; 14(9):2588-92; PMID:18451220; http://dx.doi.org/ 10.1158/1078-0432.CCR-07-0666 [DOI] [PubMed] [Google Scholar]
- 24.Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L, Zhang WG, Nan KJ, Song TS, Huang C. MicroRNA profiling of human gastric cancer. Mol Med Rep 2009; 2(6):963-70; PMID:21475928; http://dx.doi.org/ 10.3892/mmr_00000199 [DOI] [PubMed] [Google Scholar]
- 25.Yi Z, Fu Y, Ji R, Li R, Guan Z. Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PLoS One 2012; 7(8):e43184; PMID:22900099; http://dx.doi.org/ 10.1371/journal.pone.0043184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han ZB, Zhong L, Teng MJ, Fan JW, Tang HM, Wu JY, Chen HY, Wang ZW, Qiu GQ, Peng ZH. Identification of recurrence-related microRNAs in hepatocellular carcinoma following liver transplantation. Mol Oncol 2012; 6(4):445-57; PMID:22552153; http://dx.doi.org/ 10.1016/j.molonc.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlmann S, Mannsperger H, Zhang JD, Horvat EÁ, Schmidt C, Küblbeck M, Henjes F, Ward A, Tschulena U, Zweig K, et al.. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol Syst Biol 2012; 8:570; PMID:22333974; http://dx.doi.org/ 10.1038/msb.2011.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang BH, Zheng JZ, Leung SW, Roe R, Semenza GL. Transactivation and inhibitory domains of hypoxia-inducible factor 1alpha. Modulation of transcriptional activity by oxygen tension. J Biol Chem 1997; 272(31):19253-60; PMID:9235919; http://dx.doi.org/ 10.1074/jbc.272.31.19253 [DOI] [PubMed] [Google Scholar]
- 29.Hanze J, Eul BG, Savai R, Krick S, Goyal P, Grimminger F, Seeger W, Rose F. RNA interference for HIF-1alpha inhibits its downstream signalling and affects cellular proliferation. Biochem Biophys Res Commun 2003; 312(3):571-7; PMID:14680803; http://dx.doi.org/ 10.1016/j.bbrc.2003.10.153 [DOI] [PubMed] [Google Scholar]
- 30.Moniz S, Biddlestone J, Rocha S. Grow(2): the HIF system, energy homeostasis and the cell cycle. Histol Histopathol 2014; 29(5):589-600; PMID:24407868; http://dx.doi.org/ 10.14670/HH-29.10.589 [DOI] [PubMed] [Google Scholar]
- 31.Hackenbeck T, Knaup KX, Schietke R, Schödel J, Willam C, Wu X, Warnecke C, Eckardt KU, Wiesener MS. HIF-1 or HIF-2 induction is sufficient to achieve cell cycle arrest in NIH3T3 mouse fibroblasts independent from hypoxia. Cell Cycle 2009; 8(9):1386-95; PMID:19342889; http://dx.doi.org/ 10.4161/cc.8.9.8306 [DOI] [PubMed] [Google Scholar]
- 32.Goda N, Ryan HE, Khadivi B, McNulty W, Rickert RC, Johnson RS. Hypoxia-inducible factor 1alpha is essential for cell cycle arrest during hypoxia. Mol Cell Biol 2003; 23(1):359-69; PMID:12482987; http://dx.doi.org/ 10.1128/MCB.23.1.359-369.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor-1 is a basic-helix-loop-helix-pas heterodimer regulated by cellular O-2 tension. Proc Natl Acad Sci U S A 1995; 92(12):5510-14; PMID:7539918; http://dx.doi.org/ 10.1073/pnas.92.12.5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka T, Wiesener M, Bernhardt W, Eckardt KU, Warnecke C. The human HIF (hypoxia-inducible factor)-3 α gene is a HIF-1 target gene and may modulate hypoxic gene induction. Biochem J 2009; 424:143-51; PMID:19694616; http://dx.doi.org/ 10.1042/BJ20090120 [DOI] [PubMed] [Google Scholar]
- 35.Hara S, Hamada J, Kobayashi C, Kondo Y, Imura N. Expression and characterization of hypoxia-inducible factor (HIF)-3alpha in human kidney: suppression of HIF-mediated gene expression by HIF-3alpha. Biochem Biophys Res Commun 2001; 287(4):808-13; PMID:11573933; http://dx.doi.org/ 10.1006/bbrc.2001.5659 [DOI] [PubMed] [Google Scholar]
- 36.Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, Tanaka H, Cao Y, Berkenstam A, Poellinger L. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature 2001; 414(6863):550-4; PMID:11734856; http://dx.doi.org/ 10.1038/35107085 [DOI] [PubMed] [Google Scholar]
- 37.Heikkila M, Pasanen A, Kivirikko KI, Myllyharju J. Roles of the human hypoxia-inducible factor (HIF)-3 α variants in the hypoxia response. Cell Mol Life Sci 2011; 68(23):3885-901; PMID:21479871; http://dx.doi.org/ 10.1007/s00018-011-0679-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forristal CE, Wright KL, Hanley NA, Oreffo RO, Houghton FD. Hypoxia inducible factors regulate pluripotency and proliferation in human embryonic stem cells cultured at reduced oxygen tensions. Reproduction 2010; 139(1):85-97; PMID:19755485; http://dx.doi.org/ 10.1530/REP-09-0300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding Y, Yu AQ, Li CL, Fang J, Zeng Y, Li DS. TALEN-mediated Nanog disruption results in less invasiveness, more chemosensitivity and reversal of EMT in Hela cells. Oncotarget 2014; 5(18):8393-401; PMID:25245189; http://dx.doi.org/ 10.18632/oncotarget.2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Neganova I, Przyborski S, Yang C, Cooke M, Atkinson SP, Anyfantis G, Fenyk S, Keith WN, Hoare SF, et al.. A role for NANOG in G1 to S transition in human embryonic stem cells through direct binding of CDK6 and CDC25A. J Cell Biol 2009; 184(1):67-82; PMID:19139263; http://dx.doi.org/ 10.1083/jcb.200801009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang JH, Li JH, Jiang S, Zhou H, Qu LH. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res 2013; 41(Database issue):D177-87; PMID:23161675; http://dx.doi.org/ 10.1093/nar/gks1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamdan FH, Zihlif MA. Gene expression alterations in chronic hypoxic MCF7 breast cancer cell line. Genomics 2014; 104(6):477-81; PMID:25449175; http://dx.doi.org/ 10.1016/j.ygeno.2014.10.010 [DOI] [PubMed] [Google Scholar]
- 43.Tsuchiya T, Kominato Y, Ueda M. Human hypoxic signal transduction through a signature motif in hepatocyte nuclear factor 4. J Bio Chem 2002; 132(1):37-44; PMID:12097158; http://dx.doi.org/ 10.1093/oxfordjournals.jbchem.a003196 [DOI] [PubMed] [Google Scholar]
- 44.Maxwell PH, Dachs GU, Gleadle JM, Nicholls LG, Harris AL, Stratford IJ, Hankinson O, Pugh CW, Ratcliffe PJ. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc Natl Acad Sci U S A 1997; 94(15):8104-9; PMID:9223322; http://dx.doi.org/ 10.1073/pnas.94.15.8104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, et al.. Role of HIF-1[α] in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 1998; 394(6692):485-90; PMID:9697772; http://dx.doi.org/ 10.1038/28867 [DOI] [PubMed] [Google Scholar]
- 46.Gartel AL, Serfas MS, Tyner AL. p21-negative regulator of the cell cycle. Proc Soc Exp Biol Med 1996; 213(2):138-49; PMID:8931660; http://dx.doi.org/ 10.3181/00379727-213-44046 [DOI] [PubMed] [Google Scholar]
- 47.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 1994; 78(1):67-74; PMID:8033213; http://dx.doi.org/ 10.1016/0092-8674(94)90573-8 [DOI] [PubMed] [Google Scholar]
- 48.Ge Q, Wang C, Ruan Y, Chen Z, Liu J, Ye Z. Overexpression of p53 activated by small activating RNA suppresses the growth of human prostate cancer cells. Onco Targets Ther 2016; 9:231-41; PMID:26811691; http://dx.doi.org/ 10.2147/OTT.S96710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al.. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 1998; 394(6692):485-90; PMID:9697772; http://dx.doi.org/ 10.1038/28867 [DOI] [PubMed] [Google Scholar]