Abstract
HLA-B*27 is strongly associated with an inflammatory autoimmune disorder, the Ankylosing Spondylitis (AS) and plays a protective role in viral infections. The two aspects might be linked. In this work, we compared in B*2705/B*07 positive patients with AS, the CD8+ T cell responses to two immunodominant EBV-derived epitopes restricted for either the HLA-B*27 (pEBNA3C) or the HLA-B*07 (pEBNA3A). We have unexpectedly found that the HLA-B*07-restricted EBNA3A peptide is presented by both the B*0702 and the B*2705 but not by the non AS-associated B*2709, that differs from the AS-associated B*2705 for a single amino acid in the peptide-binding groove (His116Asp). We then analyzed 38 B*2705-positive/B*07-negative (31 AS-patients and 7 healthy donors) and 8 B*2709-positive/B*07-negative subjects. EBNA3A-specific CD8+ T lymphocytes were present in 55.3% of the HLA-B*2705 but in none of the B*2709 donors (p = 0.0049). TCR β-chain analysis identified common TCRBV and TCRBJ gene segments and shared CDR3β sequences in pEBNA3A-responsive CTLs of B*2705 carriers, suggesting the existence of a shared TCR repertoire for recognition of the uncanonical B*2705/pEBNA3A complex. These data highlight the plasticity of the AS-associated HLA-B*2705, which presents peptides with suboptimal binding motifs, possibly contributing both to its enhanced capacity to protect against pathogens and to predispose to autoimmunity.
INTRODUCTION
In the context of the adaptive immunity, HLA class I-restricted CD8+ cytotoxic T lymphocytes (CTL) are pivotal in controlling viral infections. They express αβ T-cell receptors (TCRs) recognizing short viral antigens that are presented on the infected cells in association with HLA encoded class I heavy chain and β2-microglobulin.
A major role of HLA-B alleles in mediating the most effective antiviral immunity has been reported (1,2). However, not all HLA-B alleles are equally protective and some, such as the HLA-B*27 and -B*57, rank at the top given their superior capacity of viral control (3–5).
Our understanding of their association with a slow progression to AIDS (4,5) and with a high rate of spontaneous clearance of hepatitis C virus is still incomplete (6,7). So far, either virological or immunological mechanisms have been postulated. Viral escape from HLA-B*27-restricted CTL responses directed against HIV- and HCV-immunodominant epitopes is unlike since it requires several and compensatory mutations to overcome structural and functional constraints, with high costs for the viral fitness (7–9). On the other hand, a number of immunological factors shape the virus-specific, HLA-B*27-restricted CTL responses including polyfunctionality, superior functional avidity, selection of specific TCR clonotypes associated with a higher cross-reactivity, lower sensitivity to regulatory T cell-mediated suppression as well as a rapid and efficient processing of the proper epitopes (2,6,10–13). Another aspect to consider is the level of promiscuity within the same or different supertypes (14). Few studies have extensively and systematically approached this topic (15,16). In addition, discordant results make difficult to determine whether epitope promiscuity by two or more HLA-B class I molecules is frequent (95% in ref 15) or relatively uncommon (3% in ref 16). HLA-B*27 and -B*57 have been correlated to a more “private” peptide repertoire (82% and 56%, respectively) in comparison to other HLA class I alleles (14) suggesting that a low degree of promiscuity has protective effects. Overall, these data indicate that HLA-B*27 and -B*57 alleles rarely share their own peptidome with other HLA molecules. Whether these alleles bind and present immunodominant peptides restricted by other HLA-class I molecules has never been convincingly explored.
Noteworthy, both HLA-B*27 and -B*57 family of alleles are associated with the susceptibility to autoimmune diseases, namely Ankylosing Spondylitis (AS) and Psoriasis, respectively (17–20). Therefore, it is conceivable that a common ground underlies the superior viral immune defense on the one hand and the propensity to autoimmunity on the other. The antigen presentation performed by such HLA-B supertypes could be a key event (7,21). A way to validate this hypothesis could be a functional comparison between very close pair of alleles, like as the B*2705 and the B*2709, which have a single amino acid substitution (Asp116His) (22) but are differently associated with AS (19,23–25).
Here, we show that the B*2705, but not the non-AS-associated HLA-B*2709, presents an EBV-derived immunodominant epitope (pEBNA3A; RPPIFIRRL) restricted by the HLA-B*0702 (26,27) and therefore devoid of the optimal B*27 binding motif, evoking a vigorous CD8+ T cell response in B*2705/B*07-double positive as well as in B*2705-single positive carriers, either AS-patients or healthy donors (HD). The occurrence of preferential TCRBV and TCRBJ gene segments and the sharing of similar CDR3β regions by the B*2705-restricted pEBNA3A-responsive CTLs support the involvement of the TCR in such recognition.
MATERIALS AND METHODS
Study Subjects
Four HLA-B*07/B*2705-double positive AS-patients, thirty-eight HLA-B*2705-positive subjects (31 AS-patients and seven HD) and eight HLA-B*2709-positive controls were enrolled in this study. Diagnosis of AS has been made according to modified New York criteria. The HLA-B*27 subtype was determined by serological analysis using ME1 mAb (specificity: HLA-B*27; -B*07; -B*42; -B*67; -B*73 and -B*w22) and genomic analysis. The positivity to HLA-B*07 was checked by BB7.1 (specificity: HLA-B*07). The study received the approval by the Ethics Committee of the University of Cagliari (365/09/CE). All subjects provided written informed consent prior to enrolment.
Synthetic Peptides and Chimeric Protein
pEBNA3C (RRIYDLIEL; aa 258-266) and pEBNA3A (RPPIFIRRL; aa 379–387) which are immunodominant EBV- epitopes restricted by HLA-B*27 (28) and by HLA-B*07 (26,27) respectively, have been used in this study. For the binding assay, a self-peptide restricted by HLA-B*27 (TIS, RRLPIFSRL) (29) and an immunodominant CMV-epitope HLA-A*02-restricted (pp65, NLVPMVATV) were included. A modified version of pEBNA3A (APPIFIRRL) has been also analyzed. Peptides (purity > 95%) were purchased by Aurogene. The chimeric protein containing EBNA3A epitope was generated using pTAT-HA expression vector according to the procedure already described (30). To generate the EBNA3A epitope, the following primers have been used:
forward: 5′-caag tactcgagaactgagaa gcaggccaccaatattcataaggaggctgaga-3′;
reverse: 5′-ccgaggaggccctctgttgattggtgt tccctccacttctcagcctccttatgaatat-3′.
Following DNA sequencing of the recombinant construct, the production, purification and detection of the chimeric protein have been done as previously described (31).
Cell Lines
TAP-defective CEM 174.T2 cells (ATCC number: CRL-1992™) (32) and HMy2.C1R cells (ATCC number: CRL-1993™) (33), stably expressing B*2705, B*2709 (T2 and C1R cells) or B*0702, A*0201 (C1R cells) were used. C1R.B*0702 transfectants were a gift from Dr. JA LÓpez De Castro.
Autologous B lymphoblastoid cell lines (B-LCLs) were generated by in vitro B cell immortalization with the standard type I EBV isolate B95.8 (22).
Cells were cultured in heat-inactivated 10% fetal bovine serum (FBS; Sigma Aldrich)/RPMI 1640 medium (Euroclone Spa) supplemented with 2 mmol/L L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin.
C1R and T2 stable transfectants were maintained in medium added with G418 (800 μg/mL) or hygromycin B (200 μg/mL), respectively.
PBMC Stimulation
PBMC (4 × 106) separated from whole blood by density gradient separation (Cedarlane Laboratories Ltd.) were incubated with the indicated peptides (20 μmol/L) and cultured for 12 d at 1.5 × 106 cells/mL in 5% FBS/RPMI complete medium, and 10 U/mL of human recombinant interleukin 2 (IL2) (Roche Applied Science). On d 3 and 9, fresh medium containing 20 U/mL of IL2 was replenished. After 12 d of antigen pulsing, PBMC were used for pentamer staining and, on d 14, used for intracellular IFNγ staining.
Pentamer Staining
B*0702/pEBNA3A and B*2705/pEBNA3C MHC class I pentamers phycoerythrin-conjugated were purchased by Proimmune. Twelve days after peptide stimulation, 0.5 × 106 PBMCs were treated with pentamers for 10’ at RT. Cells were then labeled with anti-CD8-FITC (mouse anti-human CD8-FITC-conjugated, BioLegend) for 20’ on ice. The samples were acquired by a FACSCalibur flow cytometer (Becton Dickinson) and analyzed by FlowJo software (Tree Star Inc.).
Intracellular IFNγ Staining
Briefly, C1R transfectants were incubated ON with pEBNA3A or pEBNA3C (30 μmol/L) or with medium alone and then plated with antigen-stimulated PBMC at 5:1 PBMC/APC ratio. After 2 h, the cells were treated with brefeldin A (10 μg/mL) at 37°C for 16h. Cells were stained with the anti-CD8-FITC mAb, 20’ on ice, fixed with 4% paraformaldehyde 20’ on ice, permeabilized with 1% BSA/0.1% saponin in PBS 1 × for 5′ at RT and then resuspended in 2% paraformaldehyde/1% BSA. Finally, cells were stained by an anti-IFNγ-APC mAb (BioLegend) 20’ RT and analyzed by FlowJo software.
CTL Generation
PBMC were stimulated with pEBNA3A or pEBNA3C (30 μmol/L) in 5% FBS/RPMI 1640. After 24 h, PBMC were washed and 10 U/mL of IL2 were added to the culture medium. On d 10, PBMC were depleted of the CD4+ T cell fraction by Dynabeads M-450 CD4+ (Dynal ASA) and the isolated CD8+ T cells were seeded at 1 × 105 cells/well in 96-well plates in the presence of antigen-pulsed (overnight at 30 μmol/L), γ-irradiated (200 Gy) autologous B-LCLs (CD8+ T cells/APC ratio 2:1) and 20 U/mL of IL2. After 3 rounds of stimulation, cell antigen specificity was assayed in a standard 51Cr-release assay as previously described (34).
TCR Gene Usage Analysis
Total RNA was isolated from CD8+ T cells using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Purified RNA was reverse-transcribed using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). cDNA was used to amplify the TCRBV family chains with 24 TRBV specific primers as already described (35). Specific products were purified from gel (QIAquick, Qiagen spa) and underwent direct sequencing. The IMGT nomenclature has been adopted for TCRBV (36).
T2.B*27 Stabilization Assay
ME1 mAb and an IgG1 isotype control antibody were employed to assess the capability of the peptide under study to stabilize the cell surface B*2705 or B*2709 molecules on T2 transfectants (32) according to the procedure previously described (34).
Statistics
Data obtained by IFNγ-production assays from antigen-stimulated PBMCs from B*2705-positive versus B*2709-positive carriers were compared by Fisher’s two-tailed exact test (Figure 3C) and Mann–Whitney U-test (Supplementary Figure S3). A p value < 0.05 was considered statistically significant.
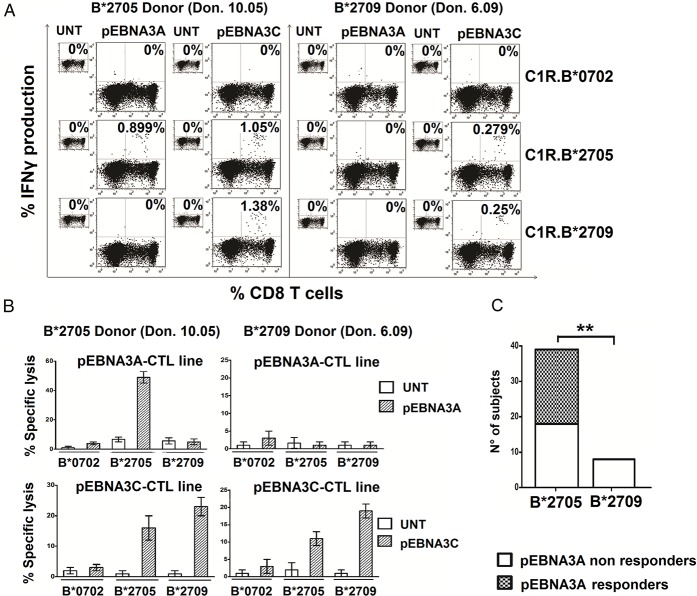
Figure 3.
B*2705 but not B*2709-positive carriers react toward the HLA-B*0702-restricted EBNA3A epitope. (A) Detection of intracellular IFNγ production by PBMCs from a B*2705 positive/B*07 negative subject (Don 10.05) (left) or a B*2709 positive/B*07 negative subject (Don 6.09) (right) activated with pEBNA3A or pEBNA3C peptide for 14 d and then re-stimulated by the indicated, untreated or peptide-pulsed C1R transfectants. Numbers represent the percentage of IFNγ-producing CD8+ T cells. (B) Cytotoxic activity by representative pEBNA3A- (top) and pEBNA3C- (bottom) stimulated CTL lines from the same individuals as in (A) toward C1R transfectants used as target cells. E/T ratio was 20:1. Bars represent the mean percentage of specific lysis ± SEM of three separate experiments. (C) CD8+ T cell responses to EBNA3A detected by IFNγ production analyzed in 38 B*2705 carriers (31 patients with AS and 7 healthy controls) and in 8 B*2709 healthy donors. Twenty-one out of 38 B*2705 and no subject out of 8 B*2709 individuals show reactivity against EBNA3A antigen (p value = 0.0049 calculated by Fisher’s two-tailed exact test).
All supplementary materials are available online at www.molmed.org.
RESULTS
Atypical Antigen Presentation by HLA-B*2705 Molecules
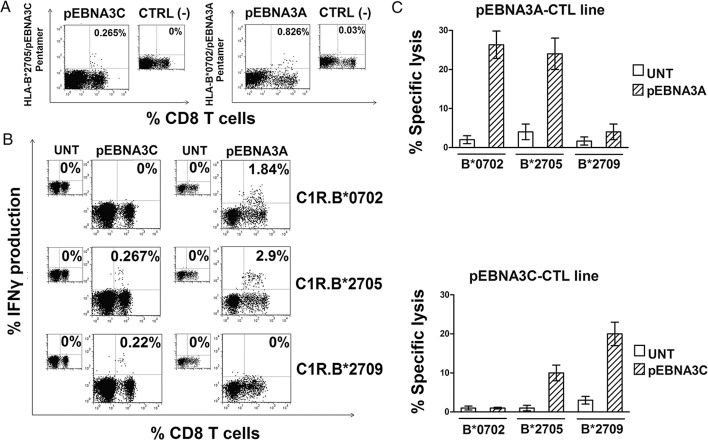
Our initial aim was to compare the immunological properties of HLA-B*2705-restricted versus HLA-B*0702-restricted CTLs. These two HLA class I alleles share 94% homology; nonetheless, the B*2705 is strongly AS-associated while the B*0702 plays a protective role (37). To overcome the inter-individual variability, we selected, a B*2705/B*0702-double positive patient with AS (Don 1.05.07). Freshly isolated PBMC were incubated with pEBNA3C or pEBNA3A, that are well known EBV-derived immunodominant epitopes restricted by HLA-B*2705 (28) or HLA-B*0702 (26,27), respectively. CD8+ T cells specific for both epitopes were present, as assessed using B*2705/pEBNA3C and B*0702/pEBNA3A pentamers (0.265% for pEBNA3C- and 0.826% for pEBNA3A-reactive cells) (Figure 1A).
Figure 1.
B*2705 can promiscuously present the B*0702-restricted EBNA3A epitope to CD8+ T cells from a B*2705/B*0702 positive AS patient (Don 1.05.07). (A) Flow cytometry plots for HLA-B*2705/pEBNA3C (left panel) and HLA-B*0702/pEBNA3A (right panel) pentamer staining within CD8+ T cells of PBMCs cultured for 12 d with pEBNA3C and pEBNA3A synthetic peptides (20 μmol/L), respectively. Numbers indicate the percentage of pentamer-positive CD8+ T cells. Smaller panels show flow cytometry profiles of pEBNA3C- (left) or pEBNA3A-stimulated (right) PBMCs for staining with an irrelevant pentamer. (B) Detection of intracellular IFNγ production by PBMCs stimulated with pEBNA3C or pEBNA3A for 14 d and then re-stimulated by the indicated, untreated or peptide-pulsed C1R transfectants (30 μmol/L). Numbers represent the percentage of IFNγ-producing CD8+ T cells. (C) Assessment of cytotoxicity by representative pEBNA3A- (top) and pEBNA3C- (bottom) stimulated CTL lines against HLA-B*0702, -B*2705, -B*2709 C1R transfectants used as target cells. E/T ratio was 20:1. Bars represent the mean percentage of specific lysis ± SEM of three separate experiments.
After 14 d, antigen-stimulated cells were re-exposed to the epitopes by co-culture with the peptide-pulsed C1R.B*0702, C1R.B*2705 or C1R.B*2709 transfectants and IFNγ-production was measured in the CD8+ T cell fraction. Figure 1B shows that pEBNA3C-exposed PBMCs were activated, as expected, only by pEBNA3C-prepulsed C1R.B*2705 and C1R.B*2709 cells, whereas no IFNγ-producing cells were detected when pEBNA3C-loaded C1R.B*0702 were used as control. In contrast, pEBNA3A-stimulated PBMCs produced IFNγ either when pEBNA3A was presented by either C1R.B*0702 cells (1.84% of IFNγ-producing CD8+ T cells) or C1R.B*2705 transfectants (2.9% of IFNγ-producing CD8+ T cells) but not when presented by C1R.B*2709 transfectants (Figure 1B). To further explore this unexpected finding, the cytotoxic activity of pEBNA3A-stimulated cells against C1R transfectants was verified. Cytotoxicity occurred upon presentation of pEBNA3A by HLA-B*0702 and B*2705 but not by B*2709 (Figure 1C). In parallel, we tested pEBNA3C-specific CTL lines which, as expected, lysed both the C1R.B*2705 and C1R.B*2709 transfectants but not the HLA-B*0702 (Figure 1C). The pEBNA3A- and pEBNA3C-responsive CTL lines, although oligoclonal, did not display reciprocal cross-reactivity (data not shown). To exclude that the atypical presentation of B*07-restricted epitopes by HLA-B*27 molecules was an occasional event, three more B*2705/B*07-double positive individuals were analyzed. Data from two other AS-patients, Don 2.05.07 and Don 3.05.07 (Figure 2 and Supplementary Figure S1, respectively) were consistent with the previous results. The fourth patient (Don 4.05.07) did not respond to pEBNA3A antigen either in B*07 or in B*27 context of presentation (data not shown). The results indicate that the promiscuous presentation of B*07 epitope is a common feature, occurring in 3/4 of the AS-patients analyzed. Moreover, it is selective for the AS-associated B*2705 molecule, since it is not shared by the closely related, non-disease associated B*2709.
Figure 2.
Uncanonical pEBNA3A presentation by HLA-B*2705 in a second B*2705/B*07- positive AS patient. (A) Measurement of IFNγ secretion in PBMCs from Don 2.05.07 stimulated with pEBNA3A for 14 d and then re-stimulated by untreated or pEBNA3A-pulsed C1R transfectants. Numbers represent the percentage of IFNγ-producing CD8+ T cells. (B) Schematic representation of the pEBNA3A-chimeric protein showing the location at the N-terminus of 6-His tag, the TAT transduction domain and the hemoagglutinin (HA) tag followed by the nucleoprotein of influenza A virus protein in which the sequence of EBNA3A epitope has been inserted. (C) 51Cr-release assay in which cytolytic effector functions of a representative pEBNA3A-stimulated CTL line was tested against C1R transfected cells, pre-pulsed with pEBNA3A provided as synthetic peptide (30 μmol/L) or transduced by the chimeric protein carrying EBNA3A epitope (1 μmol/L). E/T ratio was 20:1. Bars represent the mean percentage of specific lysis ± SEM of three separate experiments.
We have previously characterized an alternative antigen presentation pathway exploited by HLA-B*27, based on the use of intracellularly targeted chimeric proteins carrying viral epitopes (30,31). We asked whether pEBNA3A could be presented by HLA-B*27 through this endogenous pathway. Accordingly, C1R.B transfectants were transduced by the chimeric protein harbouring the pEBNA3A sequence (Figure 2B) and used as targets of pEBNA3A-stimulated CTLs in cytotoxicity assays. Again, the pEBNA3A-chimeric protein made both C1R.B*0702 and C1R.B*2705 transfectants susceptible to specific lysis whereas neither C1R.B*2709 or C1R.A*0201 could present the epitope (Figure 2C).
A possible explanation for the discordant behavior of B*2705 and B*2709 could reside in a different binding affinity. However, available MHC binding prediction tools (http://www.cbs.dtu.dk/services/Net MHCcons/) failed to identify any binding for pEBNA3A to either B*27 allele. Stabilization assays on T2.B*2705 or T2.B*2709 transfectants (32) incubated with increasing amount of pEBNA3A, or TIS as positive reference (29), confirmed no detectable binding of pEBNA3A to B*27 alleles (Supplementary Figure S2).
B*2705- But Not B*2709-Positive Carriers Elicit pEBNA3A-Driven CD8+ T Cell Reactivity
The results obtained in three out of four B*07/B*2705-double positive individuals clearly indicated a promiscuous presentation of pEBNA3A. We therefore asked whether this epitope could elicit a specific T cell response in B*2705-positive/B*07-negative individuals. PBMC from 38 HLA-B*2705-positive donors (31 patients with AS and 7 HD) were propagated in the presence of pEBNA3A. Following re-stimulation by pEBNA3A-pulsed C1R transfectants, antigen-specific IFNγ-production was measured. The results from one representative B*2705-positive individual (Don 10.05) revealed the presence of IFNγ-producing CD8+ T cells (0.899%) when the peptide was presented by B*2705 but no by B*2709 or B*0702 molecules (Figure 3A). A parallel stimulation with pEBNA3C yielded an expansion of IFNγ-producing CD8+ T cells in the B*27 context of presentation (1.05% by B*2705 and 1.38% by B*2709 allele). The representative B*2709 individual (Don 6.09) did not respond to pEBNA3A while displaying reactivity toward pEBNA3C presented by either B*2705 or B*2709 (Figure 3A).
Cytotoxicity assays performed with pEBNA3A- and pEBNA3C-specific CTLs mirrored the results obtained by intracellular IFNγ staining (Figure 3B). Twenty-one B*2705 carriers (18 AS-patients and 3 HD) out of 38 were found pEBNA3A-responsive (55.3%) exclusively in the B*2705 context. On the contrary, none of the eight B*2709 individuals analyzed showed pEBNA3A-reactivity making significant the difference between B*2705- and B*2709-positive EBNA3A responders (p = 0.0049) (Figure 3C). Functional data obtained by intracellular IFNγ staining after pEBNA3A stimulation in all B*2705 and B*2709 carriers analyzed are shown in supplementary figure S3. Thus, pEBNA3A could be efficiently presented by B*2705 molecules and the repertoire of B*2705-positive individuals, independently of the disease, comprises CD8+ T cell precursors triggered by the non-canonical pEBNA3A/B*2705 complex. The same does not occur in the B*2709 context of presentation.
To evaluate the functional avidity of pEBNA3A-specific CD8+ T cells of B*2705-positive/B*07-negative subjects, PBMC were expanded in presence of pEBNA3A or pEBNA3C, and analyzed in a dose-response intracellular IFNγ staining test upon re-stimulation with C1R.B*2705 transfectants. Figure 4A shows representative data obtained from an AS-patient (Don 15.05): at the higher peptide concentrations (3 and 30 μmol/L), the percentage of IFNγ-secreting CD8+ T cells is greater for pEBNA3A than for pEBNA3C (5.74% versus 3.42% and 3.29% versus 2.7%, respectively). This trend reversed at the lowest peptide concentration (0.3 μmol/L) (0.37% versus 2.1%) but yet, the detection of some pEBNA3A-responsive CD8+ T cells suggested that these virus-specific CTLs have a comparable functional avidity. These data have been further validated by cytotoxicity assays performed on these same cells following CD8+ T cell purification. Once again, at higher antigen concentrations (range 0.3–30 μmol/L), specific lysis was greater for pEBNA3A- than pEBNA3C-stimulated CTLs (Figure 4B) while the opposite occurred at the lowest concentration (0.03 μmol/L).
Figure 4.
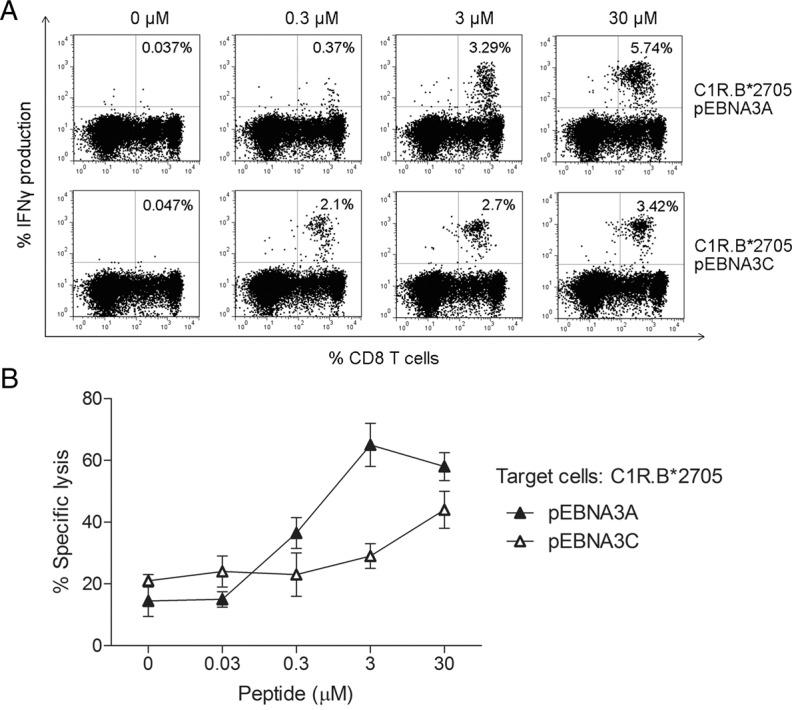
Functional avidity compared between pEBNA3A- and pEBNA3C-specific CD8+ T cells from B*2705 donors. (A) Flow cytometry plots reporting IFNγ production by PBMCs from a representative B*2705 AS patient (Don 15.05) expanded in presence of pEBNA3A or pEBNA3C (14 d) and exposed to C1R.B*2705 transfectants pulsed with increasing concentrations of each peptide before intracellular staining. Numbers indicate the percentage of IFNγ-secreting CD8+ T cells. (B) Assessment of the cytolytic activity of representative pEBNA3A- and pEBNA3C-stimulated CTL lines from the same donor as in (A). C1R.B*2705 transfectants pre-pulsed with serial amounts of the indicated peptide were used as targets. E/T ratio was 20:1. Bars represent the mean percentage of specific lysis ± SEM of three separate experiments.
To understand whether the N-terminal Arg in pEBNA3A acts as principal anchor fitting into the B-pocket, we tested a modified version of the peptide with pAla1 instead of pArg1. Functional data by EBNA3A-stimulated PBMC from a B*2705-positive patient with AS (Don 31.05) displayed substantially no production of IFNγ upon re-activation by C1R.B*2705 pulsed by the modified peptide pAla1-EBNA3A at increasing peptide concentrations (Figure S4). This result was replicated by PBMCs from other two B*2705-positive patients with AS (data not shown).
Identification of Shared TCR β-Chain Motifs Implicated In B*2705/pEBNA3A And B*0702/pEBNA3A Complex Recognition
We then verified whether common TCR β-chain motifs were used to recognize pEBNA3A either in association with B*2705 or B*07. The analysis was performed on repeatedly antigen-stimulated CD8+ T cell lines (four to five rounds of stimulation) that, although oligoclonal, were highly enriched for pEBNA3A-specific T cells. The assessment was conducted on four B*2705 carriers, two B*2705/B*07 subjects and three B*07-positive donors which were strongly pEBNA3A-reactive. For each subject, only the most abundantly expressed TCRBV chains underwent direct sequencing and a further selection was made to retrieve only unambiguous signals. Table 1 reports the most relevant sequences. T cells from two B*2705 AS-patients (Don 10.05 and Don 1.05) shared an almost identical TCR β-chain (TRBV24-1TRBJ2-1) with a single difference from Ser(S) to Gln(Q) in the CDR3 region (DLGS/QGVG). A similar TCR β-chain sequence was also expanded in T cells of a third B*2705 AS-patient (Don 17.05) showing an homologous CDR3 motif (DMGQN), a same BJ2-1 segment but BV4-1 instead of the BV24-1. Noteworthy, the BV4-1 segment was used by the public TCR β-chain (TRBV4-1TRBJ2-7) found in CTLs of a B*2705/B*07 AS-patient (Don 1.05.07) as well as in a B*07-positive HD (Don 2.07) and thus, presumably, selected as interacting with the canonical B*07/pEBNA3A complex. Consistent with these findings, we retrieved TCR β-chains similar to the previous one, bearing the same BV4-1 and CDR3 motifs, from two other B*07-positive HD (Don 3.07 and Don 4.07). It is noteworthy that almost all reported sequences have a negatively charged amino acid (D or E) in the CDR3 region.
Table 1.
TCR β-chain sequences of pEBNA3A-responsive CTL lines.
| Donor | TRBV | CDR3 | TRBJ |
|---|---|---|---|
| Pt B*27 (Don 10.05) | 24-1 | CATSDLGSGVGNEQF | 2-1 |
| Pt B*27 (Don 1.05) | 24-1 | CATSDLGQGVGNEQF | 2-1 |
| Pt B*27 (Don 17.05) | 4-1 | CASSQDMGQNNEQF | 2-1 |
| Pt B*27/B*07 (Don 1.05.07) | 4-1 | CASSQETGSYEQY | 2-7 |
| HD B*07 (Don 2.07) | 4-1 | CASSQETGSYEQY | 2-7 |
| HD B*07 (Don 3.07) | 4-1 | CASSQEGGTATYEQY | 2-7 |
| HD B*07 (Don 4.07) | 4-1 | CASSQETGIHTQY | 2-3 |
| Pt B*27 (Don 10.05) | 4-1 | CASSQDGAGGSTDTQY | 2-3 |
| Pt B*27 (Don 15.05) | 18 | CASSEGQVPSKEQF | 2-7 |
| Pt B*27 (Don 15.05) | 2 | CASSEDGENTDTQY | 2-3 |
| Pt B*27 (Don 10.05) | 6-1 | CASSEGAGGSYNEQF | 2-1 |
| Pt B*27/B*07 (Don 2.05.07) | 3-1 | CASSLGTDTQY | 2-3 |
Pt, patient with AS; HD, healthy donor.
The IMGT nomenclature has been used (36).
DISCUSSION
This study addresses the question whether the immunological mechanisms whereby HLA-B*27 is protective against viruses could overlap with those that make it strongly associated with autoimmunity. In spite of overwhelming data, the pathogenic role/s of HLA-B*27 in AS and related Spondyloarthritis remains uncertain (19).
Our findings show that an EBV-derived epitope immunodominant in the HLA-B*07 context, although lacking of an optimal B*27 binding motif, is presented by the HLA-B*2705 but not by the non-AS- associated B*2709 allele (17–19,23,24). Such atypical presentation by B*2705 evokes T cell-specific responses comparable in magnitude to those elicited by pEBNA3A presentation on B*07. It is therefore conceivable that during an EBV infection, HLA-B*07 and HLA-B*2705 could actively cooperate or even compete for this same epitope. Noteworthy, it has been recently shown that HLA-B*07 plays a protective role in AS (37). Our data allow to speculate that, in an HLA-B*2705 context, the co-presence of HLA-B*07 could play a favorable role subtracting “pathogenic” epitopes to HLA-B*27 (18,25). Conversely, it is questionable whether this epitope sharing is beneficial or detrimental to the antiviral immunity. This aspect would deserve further investigation in view of a vaccine design (38).
A further question is whether distinct CTL populations do exist, each recognizing pEBNA3A in a specific HLA context. Alternatively cross-reactive antigen-specific T cells could mediate the response. In this respect, in a B*07/B*2705-positive AS-patient (Don 2.05.07), we monitored, along the time, the cytotoxicity of pEBNA3A-stimulated CTLs that lost the capacity to kill pEBNA3A-pulsed C1R.B*0702 but not C1R.B*2705 targets (data not shown). This suggests the existence of CTLs with distinct specificities and a higher capacity of B*2705-restricted, pEBNA3A-responsive CTLs to survive. Presumably, a lower functional avidity of these CTLs decreases their susceptibility to exhaustion and activation-induced cell death (2,39).
Noteworthy, pEBNA3A-driven CD8+ T cell reactivity has been found in 55.3% B*2705-positive subjects analyzed and none of the B*2709 donors consistently with a sizeable event independent from the HLA-B*07.
Therefore, while in B*07/B*2705-positive individuals, the effect of the promiscuous presentation is unclear, in B*2705-positive/B*07-negative subjects the existence of long-lived pEBNA3A-responsive B*2705-restricted T cells would confer a net advantage on viral control. A further immunological basis for the enhanced protection of B*27 could be its engagement in a number of productive antiviral responses far greater than expected and characterized by a wide spectrum of functional avidities. Intriguingly, these data are consistent with a case of allele-dependent epitope promiscuity in which being a risk factor to autoimmunity parallels with an enhanced anti-viral immune response (7,17–19,21).
Several studies have showed that epitope binding affinity is not an absolute predictive factor for the strength and frequency of T cell responses (3,39). Paradoxically, predictive algorithms and stabilization experiments failed to show any pEBNA3A binding to B*2705 or B*2709 molecules. However, computational data indicated a higher number of hydrogen bonds lost by pEBNA3A/B*2709 compared with pEBNA3A/B*2705 complex during a 20 ns molecular dynamics simulation, hinting for a more loosely embedding of the peptide on the B*2709. Moreover, the pEBNA3A/B*2709 complex had a higher fluctuation in the α2-helix (residues 170–189) suggesting a higher entropic cost for TCR to interact with such complex (Prof. Rainer Böckmann, personal communication). Overall, although pEBNA3A is apparently a poor B*27 binder, nevertheless B*2705 but not B*2709 should be capable to form a complex stable enough to induce a CTL response. This is supported by the absence of pEBNA3A-specific CTLs in B*2709-positive individuals, by the lack of allo- and cross-recognition to pEBNA3A-B*2709 complexes in either B*07/B*2705- or B*2705-positive donors.
An obvious question is how pEBNA3A can fit into the B*2705 groove although devoid of pArg2, the B*27 dominant anchor residue (40,41). The presence of a pArg1 might allow the fitting into the B-pocket as strongly suggested by the abrogation of cell activation when the N-terminal Arg was replaced by an Ala. Accordingly, a comparable configuration was previously described by X-ray crystallography of an octameric peptide accommodating itself into the HLA-A2 groove leaving the A-pocket empty and prone to stabilization by water molecules (42). Very recently, the characterization of Behçet’s disease-associated B*5101 peptidome revealed that major peptide motifs are pPro2 or pAla2 (43). Interestingly, beside the canonical nonamers and, more rarely, decamers with pPro2 or pAla2, the peptide elution analysis showed the presence of N-terminal truncated versions with pPro1 or pAla1. For these peptides, it is conceivable a non-canonical binding mode with the P1 residue occupying the B-pocket. pEBNA3A has a C-terminal Leu often found, as PΩ anchor, in the B*2705 peptidome but even more frequently in that of B*2709 (40,41). However, the presence in pEBNA3A of two Arg preceding pLeu9 could favor the peptide stabilization in the F-pocket of B*2705 by direct or indirect interactions with Asp116. This would be not allowed for B*2709 possessing His116.
EBNA3A epitope has a proline at P2 (pPro2). Noteworthy, the endoplasmic reticulum aminopeptidase 1 and 2, ERAP1 and ERAP2, which trim N-terminally extended peptides into the ER, display a poor ability to cleave X-Pro bonds thus creating a considerable pool of peptides with pPro2 (44). Many HLA-B/C alleles bind pPro2 peptides, generated only into the ER by ERAP1 and ERAP2 processing since TAP molecules barely translocate them (44,45). Hence, the available amount of peptides such as pEBNA3A is heavily influenced by ERAP1 and ERAP2 enzymatic activity. ERAP1 is the second stronger gene associated with AS-susceptibility and the risk allelic variants are enzymatically more active than the protective ones (46–48). It must be considered that the association of ERAP1 with AS has been found only in B*27-positive patients, consistent with disease models in which an aberrant ERAP1 trimming and/or HLA-B*27 presentation to CD8+ T cells are involved in HLA-B*27-associated disease pathogenesis (46,48).
On CD8+ T cells, HLA-B*27, as classical trimeric complex, could be engaged by TCR as well as by innate immune receptors, such as KIR or LILR, but in the latter case it is unclear whether specific peptides are required (18). However, in our system, the TCR involvement is strongly supported by the shared TRBV24-1TRBJ2-1 usage and by an almost identical CDR3β expressed by pEBNA3A-responsive CD8+ T cells from B*2705 AS-patients. Furthermore, other TCRs from B*2705-restricted pEBNA3A-responsive CTLs are characterized by the TRBV4-1 segment usage in common with CTLs recognizing pEBNA3A in the B*07 context of presentation. It should be noticed that the majority of TCR β chains selected bona fide for recognition of pEBNA3A either in complex with B*07 or with B*2705, display an acidic amino acid (Asp or Glu) in the CDR3β region. Hence, it is likely that one of the two Arg, in P7 or in P8, depending on the positioning of pArg1 whether in A or B pocket, could be fully accessible to TCR, acting as docking site that selects CDR3β sequences having negative charge. Conversely, the N-terminus of peptide mainly contacted by the TCR α-chain could not exert the same imprinting effect and, consequently, from the TCR α-chain repertoire of pEBNA3A-responsive CTLs, no shared motifs emerged (data not shown).
CONCLUSION
Taken together, these data disclose the capability by B*2705 to behave as restriction element for a peptide discharged as binder by all algorithms but abundantly produced when ERAP1 is enzymatically fully active. This strong flexibility of B*2705 groove anticipated by biophysical methods and computational analysis (21,49,50) and here reinforced by T-cell functional data must be regarded as a key feature for its enhanced ability to mediate viral protection and for association with autoimmunity.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Ceschina Foundation to R Sorrentino and by Sapienza through Progetti di Ateneo to MT Fiorillo and R Sorrentino. We would like to thank Silvana Caristi and Federica Lucantoni for excellent technical assistance and all subjects participating in this study. The PhD program in Pasteurian Science, at Sapienza University, supporting Valentina Tedeschi is also acknowledged. We thank Dr. JA Lopez de Castro, Centro de Biologia Molecular Severo Ochoa, Universidad Autonoma, Madrid, Spain, for providing C1R.B*0702 transfectants, Prof. A Manfredi for critically reviewing the manuscript and Prof. R Böckmann for kindly sharing unpublished data.
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
Cite this article as: Tedeschi V, et al. (2016) The ankylosing spondylitis-associated HLA-B*2705 presents a B*0702-restricted EBV epitope and sustains the clonal amplification of cytotoxic T cells in patients. Mol. Med. 22:215–23.
REFERENCES
- Kiepiela P, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–75. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- Harari A, et al. Skewed association of polyfunctional antigen-specific CD8 T cell populations with HLA-B genotype; Proc Natl Acad Sci U S A; 2007. pp. 16233–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihl F, et al. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J Immunol. 2006;176:4094–101. doi: 10.4049/jimmunol.176.7.4094. [DOI] [PubMed] [Google Scholar]
- Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–51. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- Pereyra F, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–57. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J, et al. Rapid antigen processing and presentation of a protective and immunodominant HLA-B*27-restricted hepatitis C virus-specific CD8+ T-cell epitope. PLoS Pathog. 2012;8:e1003042. doi: 10.1371/journal.ppat.1003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann-Haefelin C. HLA-B27-mediated protection in HIV and hepatitis C virus infection and pathogenesis in spondyloarthritis: two sides of the same coin? Curr Opin Rheumatol. 2013;25:426–33. doi: 10.1097/BOR.0b013e328362018f. [DOI] [PubMed] [Google Scholar]
- McMichael AJ. Triple bypass: complicated paths to HIV escape. J Exp Med. 2007;204:2785–88. doi: 10.1084/jem.20072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidewind A, et al. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J Virol. 2008;82:5594–605. doi: 10.1128/JVI.02356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida JR, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–85. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger CT, et al. High-functional-avidity cytotoxic T lymphocyte responses to HLA- B-restricted Gag-derived epitopes associated with relative HIV control. J Virol. 2011;85:9334–45. doi: 10.1128/JVI.00460-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, et al. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol. 2012;13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S, et al. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat Med. 2011;17:989–95. doi: 10.1038/nm.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X, Hoof I, Costa AI, van Baarle D, Kesmir C. HLA class I allele promiscuity revisited. Immunogenetics. 2011;63:691–701. doi: 10.1007/s00251-011-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm N, et al. Extensive HLA class I allele promiscuity among viral CTL epitopes. Eur J Immunol. 2007;37:2419–33. doi: 10.1002/eji.200737365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen N, et al. Essential differences in ligand presentation and T cell epitope recognition among HLA molecules of the HLA-B44 supertype. Eur J Immunol. 2008;38:2993–3003. doi: 10.1002/eji.200838632. [DOI] [PubMed] [Google Scholar]
- Mathieu A, et al. The interplay between the geographic distribution of HLA-B27 alleles and their role in infectious and autoimmune diseases: a unifying hypothesis. Autoimmun Rev. 2009;8:420–25. doi: 10.1016/j.autrev.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Bowness P. HLA-B27. Annu Rev Immunol. 2015;33:29–48. doi: 10.1146/annurev-immunol-032414-112110. [DOI] [PubMed] [Google Scholar]
- Brown MA, Kenna T, Wordsworth BP. Genetics of ankylosing spondylitis—insights into pathogenesis. Nat Rev Rheumatol. 2015;12:81–91. doi: 10.1038/nrrheum.2015.133. [DOI] [PubMed] [Google Scholar]
- Feng BJ, et al. Multiple Loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genet. 2009;5:e1000606. doi: 10.1371/journal.pgen.1000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino R, Böckmann RA, Fiorillo MT. HLA-B27 and antigen presentation: at the crossroads between immune defense and autoimmunity. Mol Immunol. 2014;57:22–7. doi: 10.1016/j.molimm.2013.06.017. [DOI] [PubMed] [Google Scholar]
- Del Porto P, et al. Identification of a novel HLA-B27 subtype by restriction analysis of a cytotoxic gamma delta T cell clone. J Immunol. 1994;153:3093–100. [PubMed] [Google Scholar]
- D’Amato M, et al. Frequency of the new HLA-B*2709 allele in ankylosing spondylitis patients and healthy individuals. Dis Markers. 1995;12:215–17. doi: 10.1155/1994/394509. [DOI] [PubMed] [Google Scholar]
- D’Amato M, et al. Relevance of residue 116 of HLA-B27 in determining susceptibility to ankylosing spondylitis. Eur J Immunol. 1995;25:3199–201. doi: 10.1002/eji.1830251133. [DOI] [PubMed] [Google Scholar]
- Fiorillo MT, Sorrentino R. T-cell responses against viral and self-epitopes and HLA-B27 subtypes differentially associated with ankylosing spondylitis. Adv Exp Med Biol. 2009;649:255–62. doi: 10.1007/978-1-4419-0298-6_19. [DOI] [PubMed] [Google Scholar]
- Hill A, et al. Characterization of two Epstein–Barr virus epitopes restricted by HLA-B7. Eur J Immunol. 1995;25:18–24. doi: 10.1002/eji.1830250105. [DOI] [PubMed] [Google Scholar]
- Höllsberg P, Hansen HJ, Haahr S. Altered CD8+ T cell responses to selected Epstein-Barr virus immunodominant epitopes in patients with multiple sclerosis. Clin Exp Immunol. 2003;132:137–43. doi: 10.1046/j.1365-2249.2003.02114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JM, Murray RJ, Thomas WA, Kurilla MG, Rickinson AB. Different HLA-B27 subtypes present the same immunodominant Epstein-Barr virus peptide. J Exp Med. 1993;178:879–87. doi: 10.1084/jem.178.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurzia E, et al. Interaction pattern of Arg 62 in the A-pocket of differentially disease-associated HLA-B27 subtypes suggests distinct TCR binding mode. PLoS One. 2012;7:e32865. doi: 10.1371/journal.pone.0032865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettosini F, et al. The C terminus of the nucleoprotein of influenza A virus delivers antigens transduced by Tat to the trans-Golgi network and promotes an efficient presentation through HLA class I. J Virol. 2005;79:15537–46. doi: 10.1128/JVI.79.24.15537-15546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnacca A, et al. Characterization of a proteasome and TAP-independent presentation of intracellular epitopes by HLA-B27 molecules. J Biol Chem. 2012;287:30358–67. doi: 10.1074/jbc.M112.384339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorillo MT, et al. Susceptibility to ankylosing spondylitis correlates with the C-terminal residue of peptides presented by various HLA-B27 subtypes. Eur J Immunol. 1997;27:368–73. doi: 10.1002/eji.1830270205. [DOI] [PubMed] [Google Scholar]
- Zemmour J, Little AM, Schendel DJ, Parham P. The HLA-A, B “negative” mutant cell line C1R expresses a novel HLA-B35 allele, which also has a point mutation in the translation initiation codon. J Immunol. 1992;148:1941–48. [PubMed] [Google Scholar]
- Beltrami A, et al. Citrullination-dependent differential presentation of a self-peptide by HLA-B27 subtypes. J Biol Chem. 2008;283:27189–99. doi: 10.1074/jbc.M802818200. [DOI] [PubMed] [Google Scholar]
- Ruckert C, et al. Conformational dimorphism of self-peptides and molecular mimicry in a disease-associated HLA-B27 subtype. J Biol Chem. 2006;281:2306–16. doi: 10.1074/jbc.M508528200. [DOI] [PubMed] [Google Scholar]
- Giudicelli V, et al. IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 2006;34:D781–84. doi: 10.1093/nar/gkj088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes A, et al. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat Commun. 2015;6:7146. doi: 10.1038/ncomms8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akram A, Inman RD. Co-expression of HLA-B7 and HLA-B27 alleles is associated with B7-restricted immunodominant responses following influenza infection. Eur J Immunol. 2013;43:3254–67. doi: 10.1002/eji.201343597. [DOI] [PubMed] [Google Scholar]
- Dalla Santa S, et al. Functional avidity-driven activation-induced cell death shapes CTL immunodominance. J Immunol. 2014;193:4704–11. doi: 10.4049/jimmunol.1303203. [DOI] [PubMed] [Google Scholar]
- Lopez de Castro JA, et al. HLA-B27: a registry of constitutive peptide ligands. Tissue Antigens. 2004;63:424–45. doi: 10.1111/j.0001-2815.2004.00220.x. [DOI] [PubMed] [Google Scholar]
- Schittenhelm RB, Sian TC, Wilmann PG, Dudek NL, Purcell AW. Revisiting the arthritogenic peptide theory: quantitative not qualitative changes in the peptide repertoire of HLA-B27 allotypes. Arthritis Rheum. 2015;67:702–13. doi: 10.1002/art.38963. [DOI] [PubMed] [Google Scholar]
- Khan AR, Baker BM, Ghosh P, Biddison WE, Wiley DC. The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J Immunol. 2000;164:6398–405. doi: 10.4049/jimmunol.164.12.6398. [DOI] [PubMed] [Google Scholar]
- Guasp P, et al. The peptidome of the Behçet’s disease-associated HLA-B*51:01 includes two sub-peptidomes differentially shaped by ERAP1. Arthritis Rheum. 2015;68:505–15. doi: 10.1002/art.39430. [DOI] [PubMed] [Google Scholar]
- Fruci D, Romania P, D’Alicandro V, Locatelli F. Endoplasmic reticulum aminopeptidase 1 function and its pathogenic role in regulating innate and adaptive immunity in cancer and major histocompatibility complex class I-associated autoimmune diseases. Tissue Antigens. 2014;84:177–86. doi: 10.1111/tan.12410. [DOI] [PubMed] [Google Scholar]
- Reeves E, Elliott T, James E, Edwards CJ. ERAP1 in the pathogenesis of ankylosing spondylitis. Immunol Res. 2014;60:257–69. doi: 10.1007/s12026-014-8576-2. [DOI] [PubMed] [Google Scholar]
- Evans DM, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–67. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TM, Colbert RA. Endoplasmic reticulum aminopeptidase 1 and rheumatic disease: functional variation. Curr Opin Rheumatol. 2015;27:357–63. doi: 10.1097/BOR.0000000000000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonagle D, Aydin SZ, Gül A, Mahr A, Direskeneli H. ‘MHC-I-opathy’-unified concept for spondyloarthritis and Behçet disease. Nat Rev Rheumatol. 2015;11:731–40. doi: 10.1038/nrrheum.2015.147. [DOI] [PubMed] [Google Scholar]
- Fabian H, et al. HLA-B27 heavy chains distinguished by a micropolymorphism exhibit differential flexibility. Arthritis Rheum. 2010;62:978–87. doi: 10.1002/art.27316. [DOI] [PubMed] [Google Scholar]
- Narzi D, et al. Dynamical characterization of two differentially disease associated MHC class I proteins in complex with viral and self-peptides. J Mol Biol. 2012;415:429–42. doi: 10.1016/j.jmb.2011.11.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.