Abstract
Rare stochastic mutations may accumulate during dormancy of stem-like cells, but technical limitations in DNA sequencing have limited exploring this possibility. In this study, we employed a recently established deep sequencing method termed Duplex Sequencing to conduct a genome-wide analysis of mitochondrial (mt) DNA mutations in a human breast stem cell model that recapitulates the sequential stages of breast carcinogenesis. Using this method, we found significant differences in mtDNA amongst normal stem cells, immortal/preneoplastic cells, and tumorigenic cells. Putative cancer stem-like cell (CSC) populations and mtDNA copy numbers increased as normal stem cells become tumorigenic cells. Transformed cells exhibited lower rare mutation frequencies of whole mtDNA than did normal stem cells. The predicted mtDNA rare mutation pathogenicity was significantly lower in tumorigenic cells than normal stem cells. Major rare mutation types in normal stem cells are C>T/G>A and T>C/A>G transitions, while only C>T/G>A are major types in transformed cells. We detected a total of 1220 rare point mutations, 678 of which were unreported previously. With only one possible exception (m10342T>C), we did not find specific mutations characterizing mtDNA in human breast CSC; rather, the mitochondrial genome of CSC displayed a decrease in rare mutations overall. Based on our work, we suggest that this decrease (in particular T>C/A>G transitions), rather than the presence of specific mitochondrial mutations, may constitute an early biomarker for breast cancer detection. Our findings support the hypothesis that the mitochondrial genome is altered greatly as a result of the transformation of normal stem cells to CSC, and that mtDNA mutation signatures may aid in delineating normal stem cells from CSC.
Keywords: Mitochondrial DNA, rare mutation, cancer stem cells, Duplex Sequencing, next generation sequencing
Introduction
The mutation burden in the nuclear genome is increased during tumorigenesis (1, 2). In our study, we tested whether the nuclear mutation instability of human tumors (3) is recapitulated in the mutations of the mitochondrial genome. We have utilized a human breast stem cell model that mimics the sequential stages of breast carcinogenesis (Fig. 1A: normal, immortal/preneoplastic, and tumorigenic stages) (4–10).
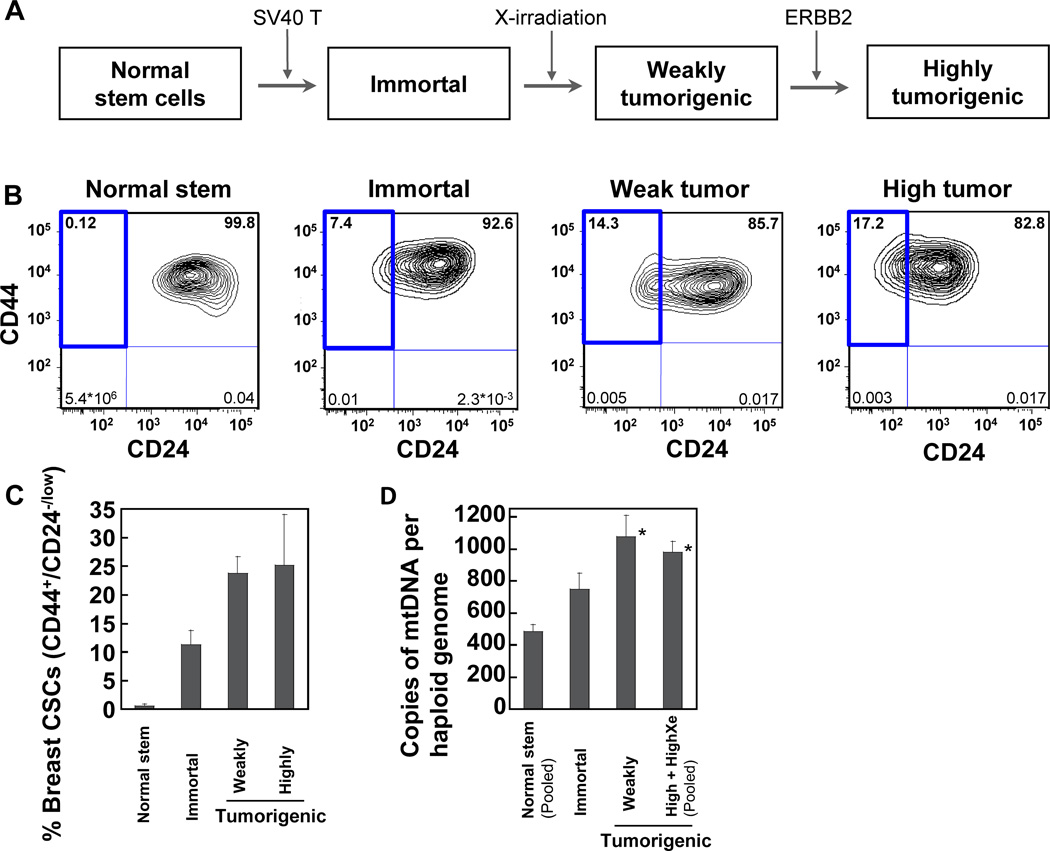
Figure 1.
Detection of the putative breast cancer stem cell (CSC) population and mtDNA copy numbers. (A) Derivation of transformed human breast epithelial cells (HBEC) from normal HBEC with stem cell characteristics. (B-C) The percentage of cells in each area (CD44+/CD24−/low, CD44+/CD24+, CD44−/CD24+, CD44−/CD24−) of four different sub-populations was determined using flow cytometry and the percentage of putative breast CSC population (CD44+/CD24−/low) was highlighted with a bold line in representative figures (B). (C) The average percentages of putative breast CSC populations (mean ± SEM) are quantified based on two to five independent cell culture experiments. (D) The numbers of mtDNA copies were quantified using QPCR. Data are from two (normal stem and highly tumorigenic xenograft cells) or three (immortal, weakly tumorigenic, and highly tumorigenic cells) independent culture experiments (mean ± SEM).
The abnormal mitochondrial function in cancer cells was first described as the Warburg effect: a phenomenon in which cancer cells forego oxidative phosphorylation even in normoxia and resort to glycolysis (11). Since then, conflicting results have been reported regarding the roles of somatic mtDNA mutations in cancer development and their relation to the Warburg effect (12–15).
Most studies involving DNA sequencing have investigated clonal mutations and have not explored sub-clonal and rare mutations. A major impediment for the detection of sub-clonal and rare mutations has been the lack of methods with sufficient accuracy to distinguish sub-clonal and rare mutations from artifacts. For example, conventional next generation sequencing (NGS) methods show high background error frequencies (10−2 to 10−3) (16, 17). We have established a new protocol referred to as Duplex Sequencing that is >10,000 fold more accurate (error rates 5×10−8 to 10−8) (16–22) than other currently available high-throughput sequencing methods. Unlike conventional sequencing technologies that sequence only a single strand of DNA, Duplex Sequencing sequences both strands of DNA and scores mutations only if they are present in both strands of the same DNA molecule as complementary substitutions. Here, by using Duplex Sequencing, we report the highly accurate mutation analysis for the entire mtDNA genome of a human breast carcinogenesis model derived from normal stem cells.
Materials and Methods
Development and culture of human breast epithelial cells (HBECs) and in vitro transformed HBEC
Breast tissues of healthy women at 21–29 years of age were obtained during reduction mammoplasty at Sparrow Hospital in Lansing, MI. Patients’ written consents were received and the use of HBEC was approved by the authors’ institutional review boards. The donors were not cancer patients and have never received chemotherapy or radiation therapy. The procedure for the development and culture of HBEC has been described (5, 9). Normal primary breast stem cells and transformed cells have been characterized using methods as described (4–10, 22–24). The cells were authenticated by short tandem repeat (STR) DNA profiling. Immortal, weakly tumorigenic, highly tumorigenic, and highly tumorigenic xenograft cells were derived sequentially from the same parental normal stem cells with treatments of SV40 large T-antigen, x-rays, and ERBB2 oncogene as described (4–6) (Fig. 1A). Highly tumorigenic cells were injected into nude mice and then the tumors formed in nude mice were collected and grown in culture to develop highly tumorigenic xenograft cells at Michigan State University. The cells used for experiments were cultured at the University of Washington for, on average, 23 days.
Flow cytometry for identification of breast cancer stem cell (CSC) population
Cells were cultured for two days after the cells were seeded. Then, the cells were collected and were incubated with antibodies labeled with fluorochromes: anti-CD24-PE and anti-CD44-APC (BD Biosciences, San Jose, CA). Cell sorting and immunofluorescence analysis were performed using BD FACS Aria or BD FACS Canto II (BD Immunocytometry Systems). After excluding non-viable cells by 7AAD viability dye and excluding debris and doublets using forward and side scatter functions of FACS instrument, the viable breast CSC population (CD44+/CD24−/low) (10, 25) was calculated using FlowJo version 9.5 program (Tree Star, Inc., Ashland, OR).
DNA extraction, mtDNA copy number, adapter synthesis, DNA library preparation, the sequencing data analysis, and pathogenicity of nonsynonymous mutations
DNA were extracted and mtDNA copy number was quantified as described (22). The synthesis of duplex adapters (18, 20), DNA library preparation (22), and Duplex Sequencing (DS) data processing (22) were carried out as described. Our DS software package can be downloaded from https://github.com/loeblab/Duplex-Sequencing. A script for amino acid changes (nonsynonymous and synonymous mutations) was described in Supplementary Methods. The GenBank (GB) frequency (%) of each identified mutation was calculated based on the previously reported mtDNA variant database (www.mitomap.org), which was derived from 29867 GenBank sequences with size greater than 15.4 kbp. MutPred web application tool (26) version 1.2 was used to predict pathogenicity for nonsynonymous mutations in the mitochondrial protein-coding genes (http://mutpred.mutdb.org) as described (22).
Statistical analysis
The mtDNA copy numbers were analyzed using one-way ANOVA. Differences in mutation frequencies and numbers of mutation contexts were analyzed by Chi-square test. Spearman correlation coefficients (rs) were applied to examine associations between mtDNA mutation signatures of tumorigenic cells and nuclear DNA mutation signatures of published tumor data. The g scores were compared using Kruskal-Wallis test. The size of mitochondrial protein coding genes and numbers of nonsynonymous mutations were analyzed by Pearson’s correlation coefficients. These statistical analyses were performed using Sigma Plot version 12.0 (Systat Software, San Jose, CA). Differences between the groups were considered significant when the p values were less than 0.05.
Results
Cells with breast cancer stem cell (CSC) features are increased during carcinogenesis
The normal human primary breast stem cells (referred to as ‘normal stem cells’ hereinafter) have been characterized by the ability to form ductal and terminal end bud-like structures on Matrigel, the ability to differentiate into basal and luminal epithelial cells, anchorage-independent growth, reduced expression of maspin, the expression of estrogen receptor-alpha, and the stem cell marker OCT4 (4–10, 23). Immortal (non-tumorigenic and preneoplastic), weakly tumorigenic, highly tumorigenic, and highly tumorigenic xenograft cells were derived sequentially from the same parental normal stem cells with oncogenic treatments (Fig. 1A) (4–6).
To study the association of CSCs with mtDNA mutations, we have identified and separated, by flow cytometry, pure populations of putative breast CSCs based on the breast CSC markers CD44+/CD24−/low (10, 25). Flow cytometry data from two to five independent cell culture experiments indicate that significantly higher percentages of breast CSC population are found among tumorigenic cells (average ~25%) than among normal stem cells and immortal cells (Fig. 1B, C). Putative CSC population is rarely present in normal stem cells (~0.1 to 0.5%).
Mitochondrial (mt) DNA copy numbers increase during breast carcinogenesis
The numbers of mtDNA genomes per cell were determined by quantifying the ratio of mtDNA to nuclear DNA using real-time quantitative PCR (QPCR). About 485 copies of mtDNA per haploid genome are found in normal stem cells. The mtDNA copy numbers per haploid genome of weakly tumorigenic, highly tumorigenic, and highly tumorigenic xenograft cells are about 2-fold significantly greater than those of normal stem cells (Fig. 1D).
Transformed cells show significantly lower frequencies of rare mutations and low-heteroplasmic mutations than do normal stem cells
The average number of nucleotides sequenced at each position of the entire mitochondria in all samples for single strand sequences (SS) was 97,915 X (Supplementary Table S1). Using the Duplex Sequencing method, we sequenced both strands of each DNA molecule individually. By doing so, we generated two single strand consensus sequence (SSCS) analyses and a duplex consensus sequence (DCS) analysis. The average depth of SSCS at each genome position in all samples was 9,069 X. The DCS was assembled by pairing complementary SSCS with each other and covered an average DCS depth of 1,509 X (Supplementary Table S1).
We determined rare variants as well as homoplasmic and heteroplasmic variants using Duplex Sequencing. Maternally inherited mitochondrial mutations arising during early embryonic development are more likely to be clonal (homoplasmic) (i.e., the same mutation existing at the same genome location in all or most mtDNA copies). Therefore, in this study, we focused on rare and low-heteroplasmic variants, as they most likely represent de novo somatic variants. In the current study, based on the mutation occurrence (clonality) at each genome position, we classified variants into those that are homoplasmic (95–100%), high-heteroplasmic (>20 to <95%), low-heteroplasmic (>0.5 to ≤ 20%), and rare (0.5% or less). Low-heteroplasmic variants and rare mutations, in particular, would not be accurately scored by conventional NGS, due to its high background error frequency (10−2 to 10−3) (16, 17). These variants, however, can be accurately detected using Duplex Sequencing.
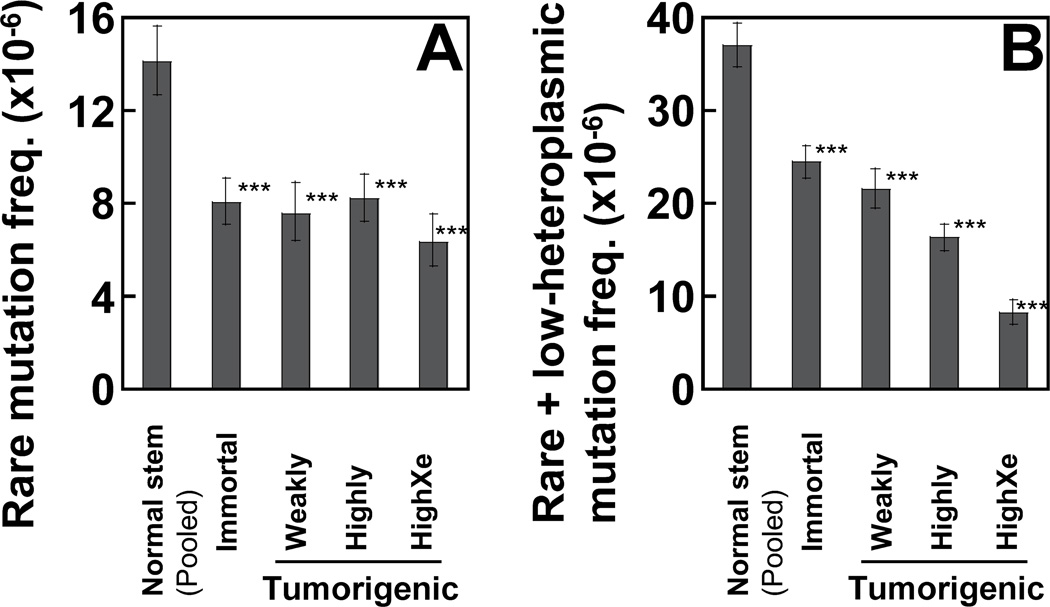
The overall frequencies of rare mutations are calculated as the total number of mutant nucleotides divided by the total number of sequenced DCS nucleotides. The frequencies of rare mutations detected by DCS analysis in all sets of transformed cells (average 7.5×10−6) are significantly lower than in those of normal stem cells (1.4×10−5) (Fig. 2A). The frequencies of combined rare and low-heteroplasmic mutations (Fig. 2B) are significantly reduced as tumorigenicity increases (Normal stem 3.7×10−5; Immortal 2.4×10−5; Tumorigenic (Pooled) 1.53×10−5).
Figure 2.
The frequency of rare and low-heteroplasmic point mutations of the whole mtDNA was determined using DCS analysis of Duplex Sequencing. The cutoffs of mutation clonality (%) used for rare and low-heteroplasmic mutations are: 0–0.5% and >0.5–20%, respectively. Data are from normal human breast stem cells (pooled from three women) and transformed cells (immortal, weakly tumorigenic, highly tumorigenic, and highly tumorigenic xenograft). (A, B) Error bars represent the Wilson Score 95% confidence intervals. Significant differences between normal stem cells and transformed cells are indicated (p values <0.05 (*), <5×10−4 (**), and <5×10−5 (***) by the Chi square test).
Independent experiments of Duplex Sequencing generate reproducible results
Duplex Sequencing has been used in recent publications from our laboratory (18–22). To further validate this methodology, we performed two independent experiments of Duplex Sequencing for each DNA sample of immortal cells and highly tumorigenic cells. The DNA library and duplex adapter synthesis were independently prepared and sequenced on different dates for the two independent sets of each sample (Supplementary Table S1, Supplementary Fig. S1). The Frequency (Supplementary Fig. S1A; Fig. 2B) and the proportion (%) of each mutation type (Supplementary Fig. S1B) of the combined rare and low-heteroplasmic mutations in these two samples obtained from independent experiments are similar to each other.
The transformation of normal stem cells to tumorigenic cells is accompanied by changes in mutation types
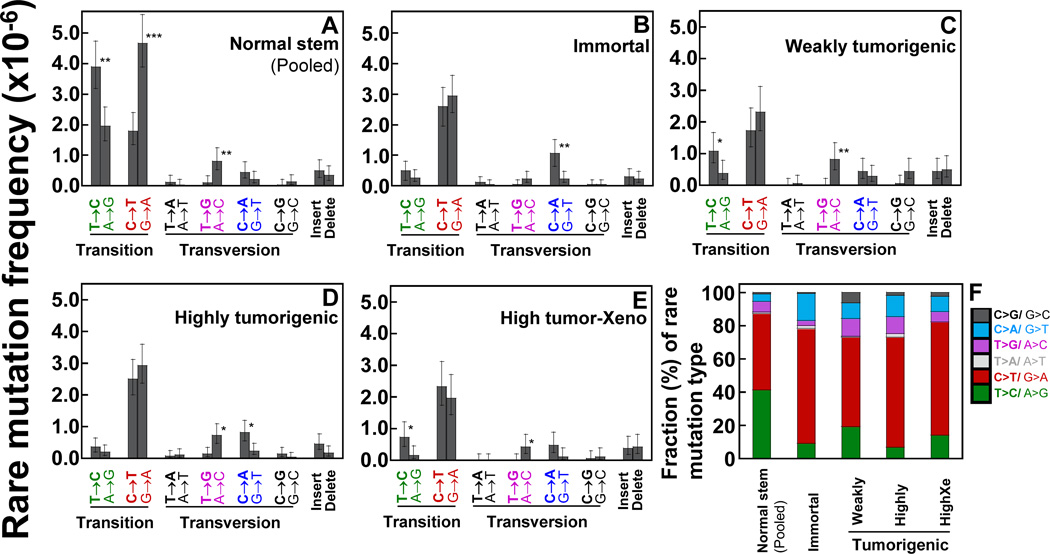
Frequencies and proportions (%) of each mutation type were analyzed. In normal stem cells, the C>T/G>A and T>C/A>G transitions are the prevalent types of rare mutations (Fig. 3A, F). However, only C>T/G>A are the major mutation types in transformed cells (Figs. 3B–F). Insertions and deletions were found infrequently. We have grouped 12 mutation types into six groups by pairing complementary sequences of point mutations. We present the data as a proportion (%) of the total number of mutations for each set of transformed cells and the pooled normal stem cells (Fig. 3F). The average fraction (%) of C>T/G>A is higher in transformed cells (63.3%) than in normal stem cells (46%), while the fraction of T>C/A>G is lower in transformed cells (11.8%) than in normal stem cells (38%). These differences indicate that the overall sequences of mutations in normal stem cell mitochondrial genome undergo profound changes during tumorigenesis.
Figure 3.
Types of rare point mutations and insertions and deletions in the whole mtDNA were determined using DCS analysis of Duplex Sequencing. Data are from normal human breast stem cells (A) and transformed cells [immortal (B), weakly tumorigenic (C), highly tumorigenic (D), and highly tumorigenic xenograft cells]. (A–E) Error bars represent the Wilson Score 95% confidence intervals. Significant differences between two groups are indicated (p values <0.05 (*), <5×10−4 (**), and <5×10−5 (***) by the Chi-square test). (F) Each mutation type is presented as a percentage (%) of overall rare mutations for each set of cells.
The strand-bias of rare mutations is lost during breast carcinogenesis
The two strands of mtDNA are designated as heavy (H) and light (L) strands (14). Our mtDNA Duplex Sequencing data are referenced to the revised Cambridge Reference Sequence (rCRS), which is designated as the L-strand. In the L-strand of normal stem cells, G>A transitions are significantly more prevalent than C>T; T>C transitions are more prevalent than A>G; and A>C transversions are more prevalent than T>G (Figs. 3A). This suggests a significant strand orientation bias in the normal stem cells. In contrast, this strand bias of higher prevalence of G>A on the L-strand is not found in any of the transformed cells (Figs. 3B–E). The strand bias of higher prevalence of T>C on the L-strand is not found in any of the transformed cells except in weakly tumorigenic cells (Figs. 3B–E).
Sequence context spectra of rare mutations change during breast carcinogenesis
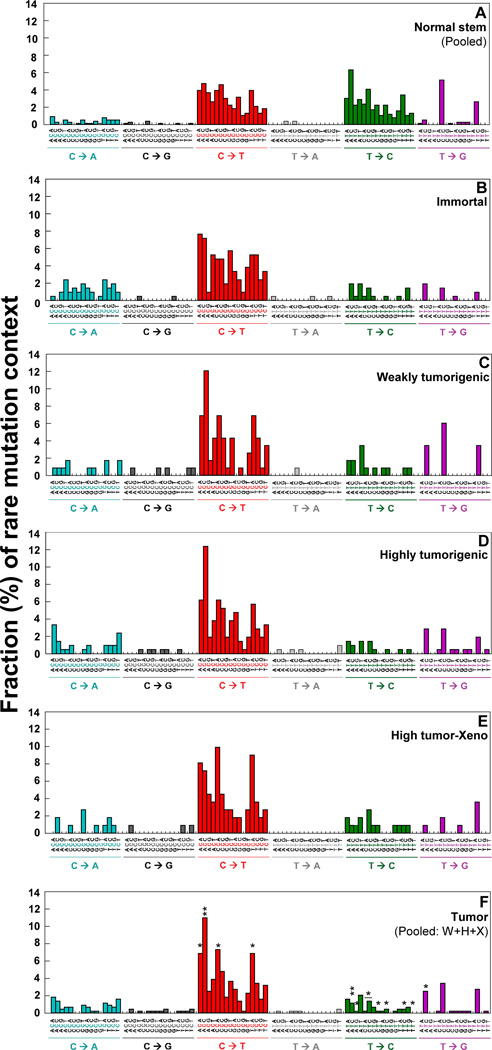
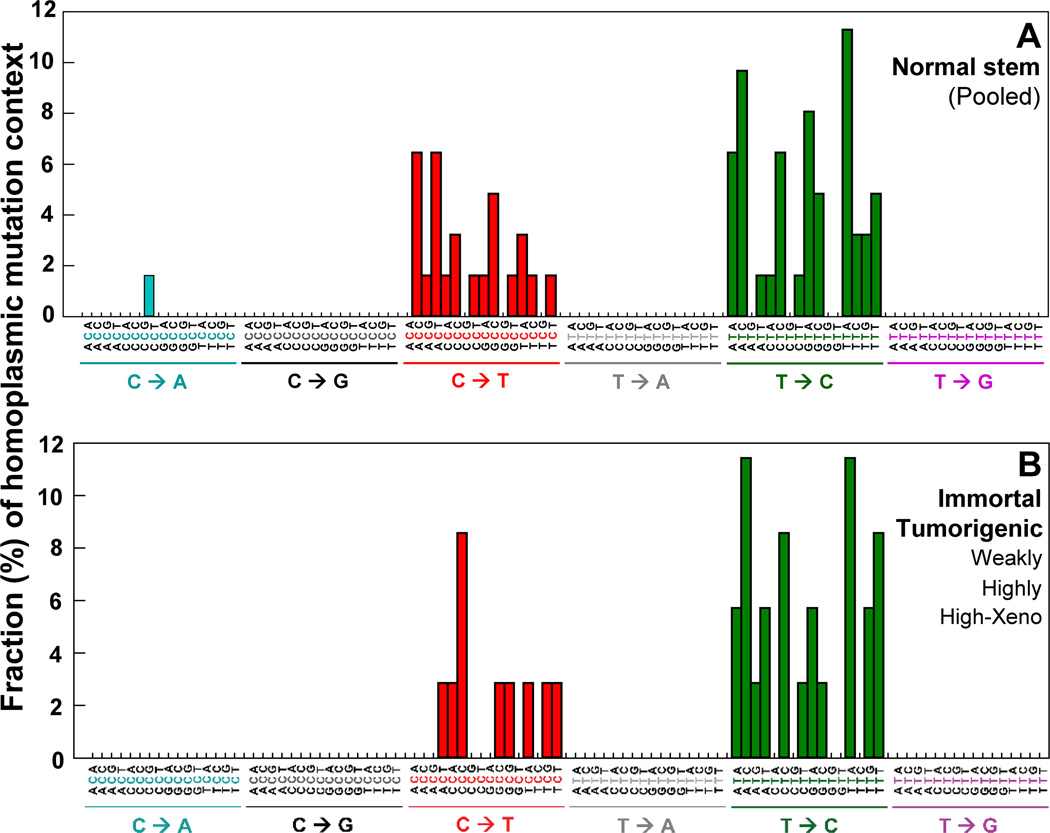
To investigate whether each type of rare point mutations occurs in a specific genome sequence context and to study how neighboring sequence context affects substitutions, we examined the bases immediately 5’ and 3’ to the mutated base (i.e. the mutation appears at the second position of each trinucleotide) on the 96 substitution classification (27). The identified substitution classifications (Fig. 4A–F) show the mutation context for every mutation from each cell type. The averages (%) of 16 possible mutation contexts for each of the six mutation types are presented (Supplementary Table S2A).
Figure 4.
DCS analysis of Duplex Sequencing was performed to determine the rare point mutation context spectrum in the whole mtDNA. (F) The significant differences in percentages of each mutation context between normal stem cells and tumorigenic cells (Pooled from weakly (W), highly (H), highly tumorigenic xenograft (X)) are indicated with * (p values <0.05 by the Chi-square test).
The rare mutation context signatures of normal stem cells (Fig. 4A) were compared with those of tumorigenic cells (pooled from weakly (W), highly (H), and highly tumorigenic xenograft (X) cells). We have identified 13 significantly different mutation contexts between normal stem cells and tumorigenic cells. The ACC for C>T transition and the ATC and TTC for T>C transition are the three most statistically significant contexts (Fig. 4F; Supplementary Table S2B). We have found 16 statistically different rare mutation contexts between normal stem cells and transformed cells (pooled from immortal, W, H, and X cells). The ATC, CTT, and TTC for T>C transitions and ACC for C>T transition are the four most statistically significant contexts (Supplementary Table S2C). These variations of rare mutation context spectra indicate distinctive frequencies and types of mutations that characterize normal stem and transformed cells and could aid in explaining their different phenotypes.
Mutation context signatures of mtDNA tumorigenic cells correlate with published nuclear DNA tumor mutation context signatures
To test whether rare mutation signatures of mtDNA share a common feature with those of nuclear (n) DNA, we compared the mtDNA mutation context signature of tumorigenic cells with published tumor data of nDNA mutation context signatures (27). Alexandrov et al (27) quantified percentages of trinucleotides (the bases immediately 5’ and 3’ to the mutated base) and identified 22 mutation signatures (ID #1A, #1B, and #2–21) that were overrepresented in nDNA of 7042 cancers of 30 different classes. Among the published 22 mutation signatures, signatures ID #1A, #1B, and #19 show the highest correlation (rs: ~0.6) with the mtDNA mutation signatures of breast tumorigenic cells (Supplementary Fig. S2). This modest correlation suggests that the high prevalence of C>T/G>A mutations is a common feature present in both mitochondrial and nuclear DNA in human cancers.
The signatures of ID #1A (12% of cancers) and #1B (61% of cancers) were found in the majority of human cancers. The signature #1B, in particular, was found in breast tumors (Supplementary Fig. S2). Both ID #1A and #1B signatures showed strong correlations between age of diagnosis and the number of mutations (27). Our results suggest that breast tumorigenic cells’ mitochondrial mutation context signature can reflect, at least in part, age-associated nDNA mutation context signatures that are present in the majority of cancers.
Duplex Sequencing identifies known and novel mitochondrial mutations
Using Duplex Sequencing, we identified a total of 1220 rare variants (841 nonsynonymous and 269 synonymous mutations) found in normal stem cells or transformed cells. We examined whether the variants were previously reported based on the mtDNA variant database (www.mitomap.org). Of the 1220 rare variants we detected, 678 are new variants (579 nonsynonymous and 79 synonymous mutations) that have not been reported by other studies. In contrast, all 71 homoplasmic variants found in our study have already been reported previously (Supplementary Table S3).
We compared all the positions of rare variants to identify common and unique variants between normal stem cells and transformed cells (Supplementary Table S4). Seven variants were found in all three sets of normal stem cells developed from three women (Supplementary Table S4B). Of these, three variants (m.13062A>G, m.13095T>C, m.13105A>G) were found in all sets of normal stem cells and transformed cells. We identified two rare mutations (m.8567T>C resulting two missense mutations S68P and I14T) that are found only in all sets of tumorigenic cells (weakly, highly, highly tumorigenic xenograft), but neither in normal stem cells nor in immortal cells (Supplementary Table S4G). There are 144 rare variants found only in immortal cells, but neither in normal stem cells nor in tumorigenic cells and 74 of these are new variants (Supplementary Table S4H).
We identified several specific nonsynonymous mutations that are clonally expanded or negatively selected during tumorigenesis (Supplementary Table S5). The m.10342T>C mutation is present in neither normal stem cells nor in immortal cells, but is found in tumorigenic cells, with its prevalence increasing with tumorigenicity. The mutation clonality percentages of the m.10342T>C mutation in weakly tumorigenic, highly tumorigenic, and highly tumorigenic xenograft cells are 12%, 30%, and 21%, respectively (Supplementary Table S5A). This finding suggests that the m.10342T>C mutation is clonally expanded during tumorigenesis and is selected toward tumorigenesis. Two other nonsynonymous mutations (m.6007T>C and m.15900T>C) are frequently mutated (~20%) in immortal cells, but are rarely or not mutated at all in tumorigenic cells (Supplementary Table S5B). This implies that both mutations are positively selected during immortalization and negatively selected during tumorigenesis. The m.10342T>C and m.6007T>C mutations are new mutations that have not been reported.
Nonsynonymous mutations occur by chance and the occurrence of nonsynonymous mutations correlates with the size of mitochondrial protein coding genes
The percentages of nonsynonymous rare mutations within the mutated codons are 76% in pooled normal stem cells and 72% in pooled transformed cells (Supplementary Table S6). The prevalence of nonsynonymous rare mutations within the mutated mitochondrial codons in the cells is close to the expected value of 75.7%, which is the prevalence of nonsynonymous mutations that would occur by chance (19, 28). This correspondence suggests that these rare variants are not subject to strong purifying selections and may in fact represent the accumulation of truly random mutations. Consistent with this correspondence, the size of each protein-coding gene positively correlates with the prevalence of nonsynonymous mutations in the protein-coding region (Supplementary Table S7).
Tumorigenic cells have significantly lower predicted pathogenicity scores than those of normal stem cells
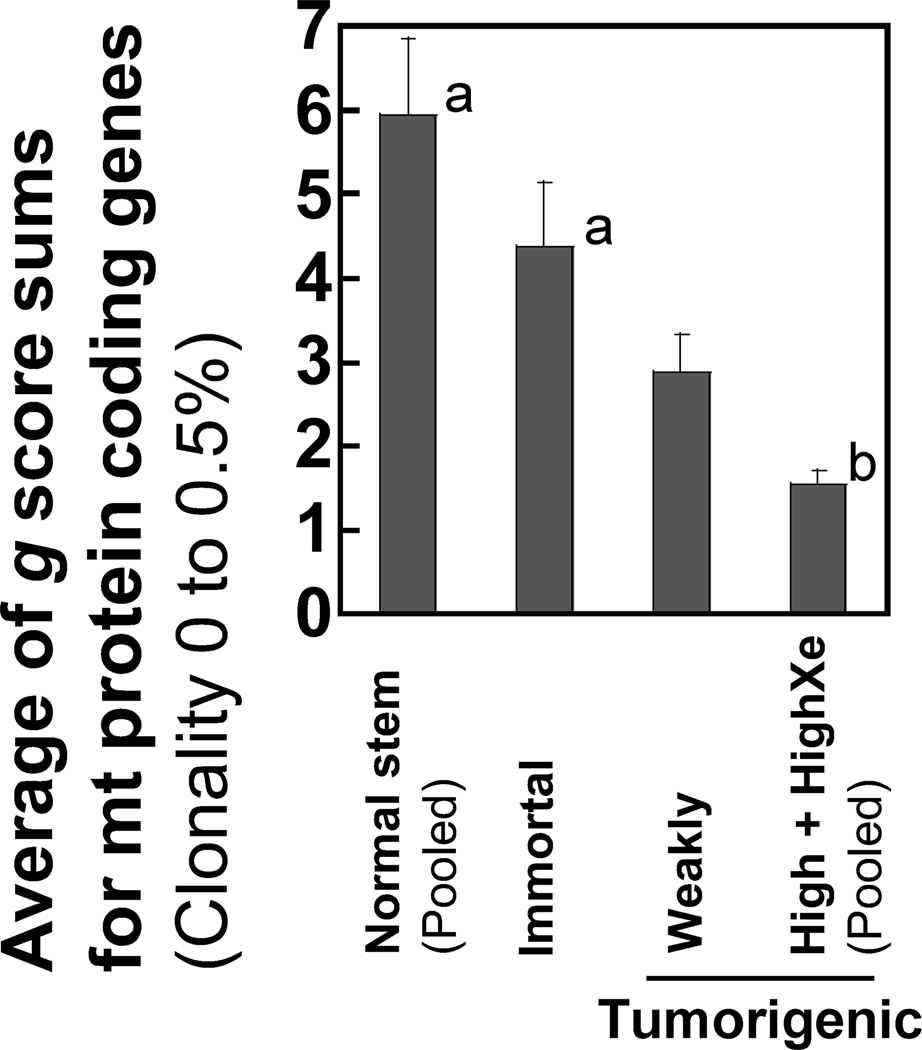
We examined whether the nonsynonymous rare mutations are likely to alter protein function and increase the predicted pathogenicity using the MutPred program (26). The g scores generated by MutPred analysis indicate deleterious substitutions with higher numerical values. The sum of g scores for each mitochondrial protein-coding gene in normal stem cells, immortal cells, and tumorigenic cells is presented (Supplementary Fig. S3). The average of the g score sums for all mitochondrial protein coding genes of highly tumorigenic cells is significantly lower (p<0.05) than those of either normal stem cells or immortal cells (Fig. 5), which indicates that nonsynonymous mutations in tumorigenic cells are of lower pathogenicity than those in normal stem cells or immortal cells.
Figure 5.
Predicted pathogenicity scores of nonsynonymous rare mutations of the mtDNA protein-coding regions. The average of the g score sums for mitochondrial protein coding genes is shown for normal stem, immortal, weakly tumorigenic, and the pooled highly tumorigenic cells (highly tumorigenic and highly tumorigenic xenograft).
Discussion
We report the first comprehensive study of mutations throughout the entire mitochondrial genome in normal human breast stem cells and compare them to mutations acquired during the transformation of normal stem cells to tumorigenic cells. Our data indicate that putative breast CSC populations are significantly increased as cells progressively acquire tumorigenic phenotypes (Figs. 1B, 1C). We demonstrate that the mtDNA copy number, relative to nuclear DNA, also increases significantly as normal stem cells are transformed to immortal cells and to tumorigenic cells (Fig. 1D). In previous studies by others, the levels of mtDNA copy numbers or mtDNA content in tumor cells/tissues compared to normal cells/tissues varied; the levels increased (29, 30), decreased (31, 32), or did not change in detectable amounts (13). In a prospective cohort study, a higher mtDNA copy number was associated with increased breast cancer risk (30). A progressive increase in mtDNA copy number with breast carcinogenesis suggests the changes in mtDNA replication or mtDNA turnover during transformation (33).
We focused our analysis on rare mutations of mtDNA because they are presumably acquired de novo somatic mutations during tumorigenesis. In contrast, homoplasmic mutations are more likely to be maternally inherited mtDNA mutations and unlikely to serve as markers for sequential stages of tumorigenesis. This is supported by our findings that all sets of transformed cells (immortal, weakly tumorigenic, highly tumorigenic, and highly tumorigenic xenograft) have the identical sequence context spectra of homoplasmic mutations of the whole mtDNA (Fig. 6B). These identical homoplasmic mutation contexts confirm that the transformed cells were derived from the same woman and indicate that no changes were introduced into the preexisting homoplasmic mutations during tumorigenesis.
Figure 6.
DCS analysis of Duplex Sequencing was performed to determine homoplasmic mutation context spectra in the whole mtDNA. The clonality cutoff used for homoplasmic mutations is 95–100%. Data are from normal human breast stem cells (A) and transformed cells (B) [immortal, weakly tumorigenic, highly tumorigenic, and highly tumorigenic xenograft]. All sets of transformed cells have identical sequence context spectra of homoplasmic mutations.
Our results indicate that rare point mutations occur stochastically (Supplementary Tables S6, S7). The lower accumulation of rare mutations in breast transformed cells than in normal stem cells contributes to lower predicted pathogenicity scores of nonsynonymous mutations. These decreases of both mtDNA mutations and predicted pathogenicity during breast carcinogenesis may reflect a regulatory response that more efficiently maintains the integrity of the mitochondria. Such response could remove mitochondria containing damaged DNA by mitophagy or employ negative selection to protect mitochondrial genomes of transformed cells from deleterious mutations.
One possible mechanism that could cause the lower mutation frequency in breast transformed cells (Fig. 2B) than in normal stem cells is the change in levels of glycolysis and oxidative phosphorylation. Ericson et al (13) reported that lower rare mutation frequencies of mtDNA at TaqI sites in human colon tumors than in normal colon tissues were associated with elevated glycolysis and reduced oxidative phosphorylation. This finding by Ericson et al (13) supports the Warburg effect in tumor cells, which suggests that increased glycolysis and reduced oxidative phosphorylation, decrease the likelihood of oxidative damage and therefore could lead to lower mutation frequencies in tumor cells (13, 34).
The frequencies and percentages of mtDNA mutation types in normal stem cells differ from those in transformed cells. In normal stem cells, the C>T/G>A and T>C/A>G transitions are the most frequent types of rare mutations (Fig. 3A). In contrast, the predominant substitutions in transformed cells are C>T/G>A. It was reported that transitions are more common than transversions in the mtDNA of humans (19, 35) and in 70 species of animals (36). Misincorporation by DNA polymerase γ and the deamination of cytosine to form uracil cause C>T/G>A transitions and have been demonstrated to be the main drivers of mtDNA mutation (37, 38). T>C/A>G mutations are probably due to the deamination of adenine to inosine or T-dGTP mispairing: a frequent base misinsertion made by DNA polymerase γ (39–41). Taken together, our results suggest that replication errors by DNA polymerase γ and base hydrolysis account for the majority of rare mutations in mtDNA of human breast epithelial cells.
The higher prevalence of G>A than C>T on the L-strand observed in rare mutations of normal stem cells indicates a strand bias (Fig. 3A). However, this strand bias of rare mutations reflected by asymmetric distributions is lost in transformed cells (Figs. 3B–E). Strand biases can occur during mtDNA asymmetric replication (42). The strand bias of mtDNA has been demonstrated in human population studies (36, 43) and experimental studies of human brain (19), putamen (44), tumors (32, 45), and fruit fly Drosophila melanogaster (46). The strand bias found in the whole mtDNA of human brain was detected using Duplex Sequencing (19). This disappearance of strand bias within rare mutations suggests extensive changes in the mtDNA or a prevalence of stochastic mutations due to DNA damage in cells during tumorigenesis.
The 8-oxo-2’-deoxyguanosine (8-oxo-dG) is known to be one of the predominant products of DNA oxidation (47, 48). However, in the human breast epithelial cell model we examined in the current study, G>T(/C>A) transversion produced by 8-oxo-dG (49) was not the major contributor of mtDNA mutations. Our finding reinforces a recent NGS tumor study which found that the G>T(/C>A) transversion constituted only 4% of mtDNA mutations identified from diverse tumors (45). A similar finding was reported in the human brain tissue Duplex Sequencing study (19). The low frequency of G>T(/C>A) mutations in the human whole mtDNA of breast cells (current study), tumors (45), and brain tissues (19) may imply three possible scenarios: 8-oxo-dG is hardly produced in these cells or tissues, mtDNA 8-oxo-dG lesions are rapidly repaired, or the damaged mtDNA is immediately degraded through mitophagy.
The nuclear DNA of human breast tumors have been sequenced extensively using NGS (50 cancergenome.nih.gov). It has been estimated that most tumors contain thousands of nucleotide changes in their nuclear genome, many of which are in oncogenes or tumor suppressor genes. These specific mutations of nuclear DNA have served as biomarkers for detection of breast cancer (50, cancergenome.nih.gov). In contrast, our results indicate that, in the mitochondrial genome, with the possible exception of the m10342T>C substitution, the presence of predominant mutation contexts, rather than specific substitutions, may prove to be a biomarker for breast CSCs. Features of mtDNA, including the relatively high mutation rate, the high copy number per cell, and maternal inheritance, render mitochondrial mutations as attractive candidates for tumor detection (14). These features have made studying mtDNA somatic mutations in body fluids useful for detection of pathological processes and forensic applications. Duplex Sequencing has enabled us to detect many rare point mutations that are not detectable by NGS. Our Duplex Sequencing data suggest that the overall decrease in rare mutations of mtDNA, in particular T>C/A>G transitions, might be germane for the detection of early breast malignancies.
Supplementary Material
Acknowledgments
We thank Prince J. Kim and Michael B. Lee for assistance in flow cytometry data analysis, Dr. James E. Trosko for obtaining HBEC, Brendan F. Kohrn for bioinformatics consultation, Dr. Rosa-Ana Risques for the mtDNA library capture set, Drs. Scott Kennedy, Marc J. Prindle, Michael W. Schmitt, and Edward J. Fox for comments on DS, Drs. Tom Walsh and Ming K. Lee for sequencing-technical assistance, and Drs. Raymond J. Monnat, George M. Martin, and Jesse J. Salk for critically reading this manuscript.
Grant Support
The research was supported by grants from the National Institutes of Health P01 AG001751 and R-33 CA 181771 (to LAL), National Institute of Environmental Health Sciences sponsored UW for Ecogenetics and Environmental Health and ITHS Grant #: NIH/NIEHS P30ES007033 (to EHA).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Conflicts of Interest: The authors declare no potential conflicts of interest.
Author’s Contributions
Conception and design: E.H. Ahn, L.A. Loeb
Development of methodology: E.H. Ahn, L.A. Loeb
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): E.H. Ahn, C.C. Chang, L.A. Loeb
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): E.H. Ahn, S.H. Lee, J.Y. Kim
Writing, review, and/or revision of the manuscript: E.H. Ahn, L.A. Loeb, J.Y. Kim, S.H. Lee, C.C. Chang
Administrative, technical, or material support (i.e. reporting or organizing data, constructing databases): E.H. Ahn, L.A. Loeb
Study supervision: E.H. Ahn, L.A. Loeb
References
- 1.Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61(8):3230–3239. [PubMed] [Google Scholar]
- 2.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer. 2011;11(6):450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Kang KS, Sun W, Nomata K, Morita I, Cruz A, Liu CJ, et al. Involvement of tyrosine phosphorylation of p185 (c-erbB2/neu) in tumorigenicity induced by x-rays and the neu oncogene in human breast ephithelial cells. Molecular Carcinogenesis. 1998;21(4):225–233. [PubMed] [Google Scholar]
- 5.Kao CY, Nomata K, Oakley CS, Welsch CW, Chang CC. Two types of normal human breast epithelial cells derived from reduction mammoplasty: phenotypic characterization and responses to SV40 transfection. Carcinogenesis. 1995;16(3):531–538. doi: 10.1093/carcin/16.3.531. [DOI] [PubMed] [Google Scholar]
- 6.Chang CC, Sun W, Cruz A, Saitoh M, Tai MH, Trosko JE. A human breast epithelial cell type with stem cell characteristics as target cells for carcinogenesis. Radiation Res. 2001;155(1 Pt 2):201–207. doi: 10.1667/0033-7587(2001)155[0201:ahbect]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 7.Park JS, Noh DY, Kim SH, Kim SH, Kong G, Chang CC, et al. Gene expression analysis in SV40-immortalized human breast luminal epithelial cells with stem cell characteristics using a cDNA microarray. Int J Oncology. 2004;24(6):1545–1558. [PubMed] [Google Scholar]
- 8.Tai MH, Chang CC, Kiupel M, Webster JD, Olson LK, Trosko JE. Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis. 2005;26(2):495–502. doi: 10.1093/carcin/bgh321. [DOI] [PubMed] [Google Scholar]
- 9.Ahn EH, Chang CC, Schroeder JJ. Evaluation of sphinganine and sphingosine as breast cancer chemotherapeutic and chemopreventive agents. Exp Biol Med. 2006;231(10):1664–1672. doi: 10.1177/153537020623101012. [DOI] [PubMed] [Google Scholar]
- 10.Wang KH, Kao AP, Chang CC, Lee JN, Hou MF, Long CY, et al. Increasing CD44+/CD24(−) tumor stem cells, and upregulation of COX-2 and HDAC6, as major functions of HER2 in breast tumorigenesis. Mol Cancer. 2010;9:288. doi: 10.1186/1476-4598-9-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 12.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;234(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ericson NG, Kulawiec M, Vermulst M, Sheahan K, O’Sullivan J, Salk JJ, et al. Decreased mitochondrial DNA mutagenesis in human colorectal cancer. PloS Genet. 2012;8(6):e1002689. doi: 10.1371/journal.pgen.1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol. 2013;5(11):a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yadav N, Chandra D. Mitochondrial DNA mutations and breast tumorigenesis. Biochim Biophys Acta. 2013;1836(2):336–344. doi: 10.1016/j.bbcan.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lou DI, Hussmannb JA, McBeea RM, Acevedoc A, Andinoc R, Press WH, et al. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc Natl Acad Sci USA. 2013;110(49):19872–19877. doi: 10.1073/pnas.1319590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox EJ, Reid-Bayliss KS, Emond MJ, Loeb LA. Accuracy of Next Generation Sequencing Platforms. Next Generat Sequenc & Applic. 2014;1(1):106. doi: 10.4172/jngsa.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci USA. 2012;109(36):14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PloS Genet. 2013;9(9):e1003794. doi: 10.1371/journal.pgen.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, et al. Detecting ultra-low frequency mutations by duplex sequencing. Nature Protocols. 2014;9(11):2586–2606. doi: 10.1038/nprot.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt MW, Fox EJ, Prindle MJ, Reid-Bayliss KS, True LD, Radich JP, et al. Sequencing small genomic targets with high efficiency and extreme accuracy. Nat Methods. 2015;12(5):423–425. doi: 10.1038/nmeth.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn EH, Hirohata K, Kohrn BF, Fox EJ, Chang CC, Loeb LA. Detection of ultra-rare mitochondrial mutations in breast stem cells by Duplex Sequencing. PloS One. 2015;10(8):e0136216. doi: 10.1371/journal.pone.0136216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang KS, Morita I, Cruz A, Jeon YJ, Trosko JE, Chang CC. Expression of estrogen receptors in a normal human breast epithelial cell type with luminal and stem cell characteristics and its neoplastically transformed cell lines. Carcinogenesis. 1997;18(2):251–257. doi: 10.1093/carcin/18.2.251. [DOI] [PubMed] [Google Scholar]
- 24.Ahn EH, Chang CC, Talmage DA. Loss of anti-proliferative effect of all-trans retinoic acid in advanced stage of breast carcinogenesis. Anticancer Res. 2009;29(8):2899–2904. [PubMed] [Google Scholar]
- 25.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25(21):2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenman C, Wooster R, Futreal PA, Stratton MR, Easton DF. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics. 2006;173(4):2187–2198. doi: 10.1534/genetics.105.044677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thyagarajan B, Wang R, Nelson H, Barcelo H, Koh W-P, Yuan J-M. Mitochondrial DNA copy number is associated with breast cancer risk. PloS one. 2013;8(6):e65968. doi: 10.1371/journal.pone.0065968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemnrau A, Brook MN, Fletcher O, Coulson P, Jones M, Tomczyk K, et al. Mitochondrial DNA copy number in peripheral blood cells and risk of developing breast cancer. Cancer Res. 2015;75(14):2844–2850. doi: 10.1158/0008-5472.CAN-14-1692. [DOI] [PubMed] [Google Scholar]
- 31.Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, et al. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life. 2007;59(7):450–457. doi: 10.1080/15216540701509955. [DOI] [PubMed] [Google Scholar]
- 32.McMahon S, LaFramboise T. Mutational patterns in the breast cancer mitochondrial genome, with clinical correlates. Carcinogenesis. 2014;35(5):1046–1054. doi: 10.1093/carcin/bgu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clay Montier LL, Deng JJ, Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genomics. 2009;36(3):125–131. doi: 10.1016/S1673-8527(08)60099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JW, Dang CV. Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66(18):8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 36.Belle EM, Piganeau G, Gardner M, Eyre-Walker A. An investigation of the variation in the transition bias among various animal mitochondrial DNA. Gene. 2005;355:58–66. doi: 10.1016/j.gene.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 37.Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287(5782):560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- 38.Spelbrink JN, Toivonen JM, Hakkaart GA, Kurkela JM, Cooper HM, Lehtinen SK, et al. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J Biol Chem. 2000;275(32):24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- 39.Nordmann PL, Makris JC, Reznikoff WS. Inosine induced mutations. Mol Gen Genet. 1988;214(1):62–67. doi: 10.1007/BF00340180. [DOI] [PubMed] [Google Scholar]
- 40.Song S, Pursell ZF, Copeland WC, Longley MJ, Kunkel TA, Mathews CK. DNA precursor asymmetries in mammalian tissue mitochondria and possible contribution to mutagenesis through reduced replication fidelity. Proc Nat Acad Sci USA. 2005;102(14):4990–4995. doi: 10.1073/pnas.0500253102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The fidelity of human DNA polymerase γ with and without exonucleolytic proofreading and the p55 accessory subunit. J Biol Chem. 2001;276(42):38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- 42.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28(4):693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 43.Reyes A, Gissi C, Pesole G, Saccone C. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol. 1998;15(8):957–966. doi: 10.1093/oxfordjournals.molbev.a026011. [DOI] [PubMed] [Google Scholar]
- 44.Williams SL, Mash DC, Zuchner S, Moraes CT. Somatic mtDNA mutation spectra in the aging human putamen. PloS Genet. 2013;9(12):e:1003990. doi: 10.1371/journal.pgen.1003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ju YS, Alexandrov LB, Gerstung M, Martincorena I, Nik-Zainal S, Ramakrishna M, et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife. 2014 doi: 10.7554/eLife.02935. 10.7554/eLife.029353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Itsara LS, Kennedy SR, Fox EJ, Yu S, Hewitt JJ, Sanchez-Contreras M, et al. Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PloS Genet. 2014;10(2):e1003974. doi: 10.1371/journal.pgen.1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FaSEB J. 2003;17(10):1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 48.Nishimura S. 8-Hydroxyguanine: From its discovery in 1983 to the present status. Proc Jpn Acad Ser B Phys Biol Sci. 2006;82(4):127–141. doi: 10.2183/pjab.82.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G>T and A>C substitutions. J Biol Chem. 1992;267(1):166–172. [PubMed] [Google Scholar]
- 50.Shih J, Bashir B, Gustafson KS, Andrake M, Dunbrack RL, Goldstein LJ, et al. Cancer Signature Investigation: ERBB2 (HER2)-Activating Mutation and Amplification-Positive Breast Carcinoma Mimicking Lung Primary. J Natl Compr Canc Netw. 2015;13(8):947–952. doi: 10.6004/jnccn.2015.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.