TO THE EDITOR:
Approximately, 46 cases of constitutional interstitial 5q deletion have been reported to date [Baekvad-Hansen et al., 2006; Malan et al., 2006; Tzschach et al., 2006; Cardoso et al., 2009]. The majority of these cases were characterized by standard karyotyping methods, hence, delineation of the precise cytogenetic defects in these patients is important to understand their clinical phenotype, implicate novel genes that underlie the pathogenesis, and differentiate disease-causing genomic imbalances from benign copy number variations in the human genome. In recent years, novel molecular cytogenetic methods such as fluorescent in situ hybridization (FISH) and array comparative genomic hybridization (aCGH) have been employed to accurately characterize chromosome abnormalities that cause human disease phenotype [Higgins et al., 2008].
We report on the clinical and molecular cytogenetic characterization of a 7.4Mb genomic deletion, 5q14.3-q21.1, in a patient with intellectual disability, attention deficit hyperactivity disorder, iris coloboma, hearing loss, dental anomaly, and dysmorphic facial features.
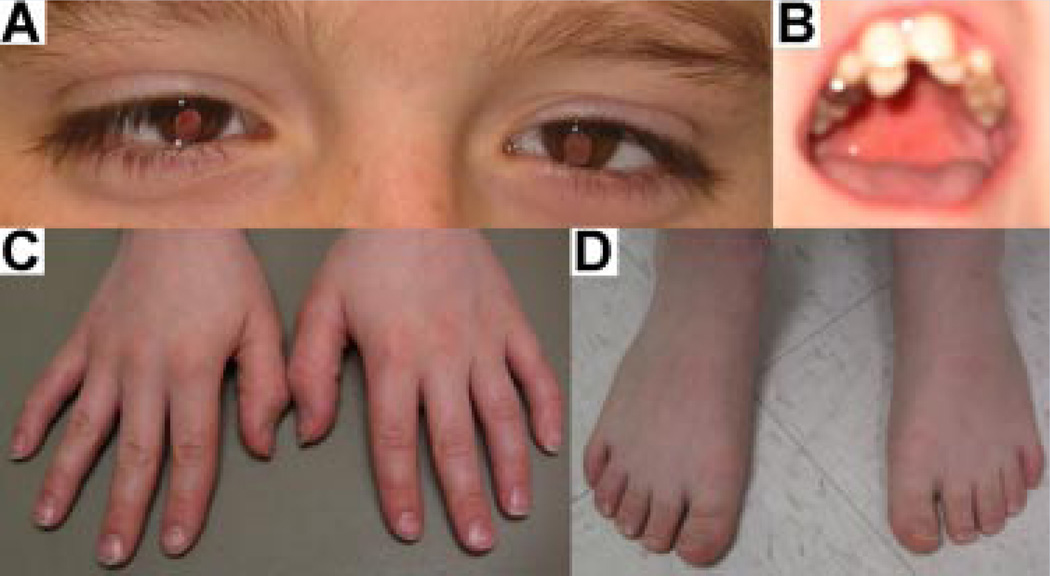
This 11-year-old boy was the only child from healthy parents with a negative family history for congenital malformations. The pregnancy was reported uneventful and consanguinity was denied. He presented with moderate intellectual disability, severe attention deficit, and hyperactivity disorder, as well as aggressive and stereotyped behaviors. He had a short stature (height <3rd centile) and a high frequency hearing loss, down-slanting palpebral fissures, bilateral iris coloboma with small optic nerves, cup-shaped ears, misplacement of frontal/lateral incisors, brachydactyly of hands, bilateral clinodactyly of the fifth fingers, and small feet (Fig. 1).
FIG. 1.
Multiple congenital anomalies in the patient. Panel A: Bilateral iris coloboma and down-slanting palpebral fissures; Panel B: Misalignment of frontal/lateral incisors; Panel C: Brachydactyly of hands and clinodactyly of fifth fingers; Panel D: Brachydactyly of feet, bilaterally. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
A brain MRI study showed mild delay in myelination but no structural anomaly. A head CT scan excluded defective formation of the ossicles in the middle ear. Anechocardiogram revealed a normal cardiac structure and function. An abdominal ultrasound did not detect structural anomalies including liver and kidneys. Metabolic workups including plasma amino acids, urine organic acids, and lactate were negative.
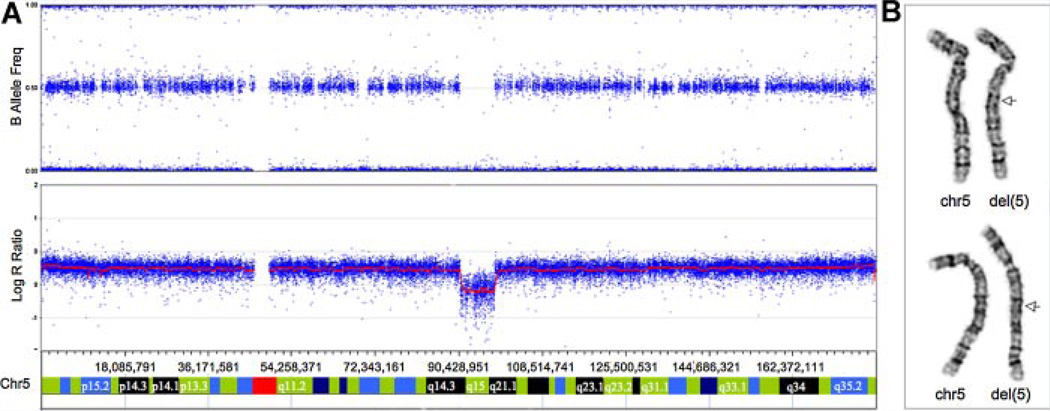
High-resolution karyotyping using PHA-stimulated peripheral lymphocytes was conducted following standard protocols at the Cytogenetics Laboratory of the Kennedy Krieger Institute (Baltimore, MD). For SNP analysis, we utilized the Illumina 610,000 array platform (Illumina, San Diego, CA). Samples were processed following the Illumina Infinium HD Assay Super manufacturer’s protocol. Beadchips were imaged on Illumina BeadArray reader. Allelic composition and signal intensity were analyzed with the KaryoStudio 1.0.3 Build 36.1 (Illumina). The SNP array identified a 7.4Mb deletion (90,787,099–98,232,469 bp) (Fig. 2A) which was confirmed by a high resolution karyotype (Fig. 2B), and FISH analysis (data not shown) in this patient. Parental samples were unavailable for these studies.
FIG. 2.
Panel A: full view of chromosome 5 analyzed by Illumina 610,000 SNP array showing signal intensity values (Log R ratio) and frequency of B allele; the red line represents the smoothed intensity data; the blue data points are the values of each individual SNP for the B allele frequency. Note absence of AB allele frequency calls in deleted region and decrease in intensity data. Panel B–G-banded chromosomes 5; note the 5q deletion indicated by an arrow. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Of the ~46 cases of constitutional interstitial 5q deletion reported in the literature, ~20 were found to have regions of the deletions that partially overlap with 5q14.3-q21. The majority of these cases were characterized by standard karyotyping methods and the sizes of individual deletionswere very variable [Malan et al., 2006]. The clinical features of individuals with these deletions included global developmental delay, congenital malformations, and nonspecific facial dysmorphisms [Cardoso et al., 2009].
Several individuals with overlapping 5q14.3-q21 deletions have been characterized using molecular cytogenetic techniques including FISH and array CGH [Malan et al., 2006]. Cardoso and colleagues reported the clinical and brain imaging features of three unrelated patients with de novo deletion of 5q14.3-15 as defined by microarray-based CGH [Cardoso et al., 2009]. These patients presented with epilepsy, mental retardation, and bilateral periventricular heterotopias (PH) in the walls of the temporal horns of lateral ventricles. One of these patients had an iris coloboma on the left. None of these patients was noted to have hearing impairment or dental anomaly. This study suggested the existence of a PH critical region of ~5.8 Mb as defined proximally by D5S1943 and distally by D5S2611 (88,833,718–94,826,030 bp) on chromosome 5q. Fourteen annotated genes were found within this region based on Human Genome Database (NCBI, Build 36.1). Semi-quantitative RT-PCR revealed that 13 of these genes are expressed in specialized brain regions during fetal and/or adult stages [Cardoso et al., 2009].
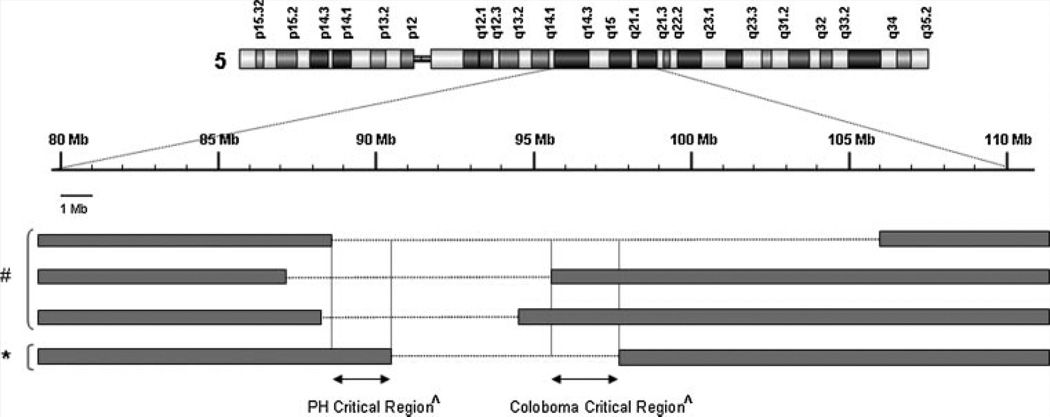
The size of the chromosomal deletion in our patient is 7.4 Mb (90,787,099–98,232,469 bp), which includes 27 annotated RefSeq genes (http://genome.ucsc.edu/). None of these genes is known to be a cause for dental anomaly, or iris coloboma. Since our patient did not have PH, we compared his deletion with those reported by Cardoso et al., which helped to narrow the putative PH critical region from 5.8 to 1.84 Mb (88,945,075–90,787,099 bp) including seven RefSeq annotated genes (http://genome.ucsc.edu/) (Fig. 3). Additionally, this patient and one of the three reported by Cardoso et al. were found to have iris coloboma. The estimated size of the chromosomal deletion shared by these two patients is 2.6 Mb (95,538,699–98,232,465 bp) involving nine RefSeq annotated genes (http://genome.ucsc.edu/), suggesting a putative genomic region for iris coloboma (Fig. 3). It is conceivable that additional genetic and environmental factors may play a role in the clinical phenotype of our patient.
FIG. 3.
Schematic diagram of the locations of the 5q14.3-q21.1 deletion and the putative PH and coloboma critical regions. The genomic location of the 5q14.3-q21.1 deletion (*) is compared to that of three deletions (#) reported by Cardoso et al. [2009]. The putative PH and coloboma critical regions (ffi) are indicated by arrows and vertical lines.
The association of iris coloboma, sensorineural hearing loss, and dental anomaly has been reported in a British family with otodental syndrome which is characterized by high frequency sensorineural hearing loss and a striking dental phenotype known as globodontia [Bloch-Zupan and Goodman, 2006]. The responsible loci for otodental syndrome has been mapped to 11q13 and further characterization of this region by Southern blot and quantitative expression analyses implicated a potential role for FGF3 and Fas-associated death domain (FADD) in otodental and iris coloboma phenotypes, respectively, in this syndrome [Gregory-Evans et al., 2007]. Our patient presented with iris coloboma and sensorineural hearing loss but the dental anomaly was not the classical presentation described in otodental syndrome. Because there is only one family described with otodental syndrome it is not clear whether our patient represents a variable expression of that syndrome or different condition. Either way the description of our case supports genetic heterogeneity for iris coloboma, dental anomaly, and sensorineural hearing loss in humans and suggests a smaller PH critical region and a new iris coloboma critical region.
ACKNOWLEDGMENTS
The authors wish to acknowledge the valuable contribution of the staff of the Cytogenetics Laboratory of the Kennedy Krieger Institute, Janet Biscoe, Barbara Jackson, Susan Morsey, Elizabeth Wohler and Linjie Wu. We also thank the patient and his family for their cooperation.
REFERENCES
- Baekvad-Hansen M, Tümer Z, Delicado A, Erdogan F, Tommerup N, Larsen LA. Delineation of a 2.2 Mb microdeletion at 5q35 associated with microcephaly and congenital heart disease. Am J Med Genet Part A. 2006;140A:427–433. doi: 10.1002/ajmg.a.31087. [DOI] [PubMed] [Google Scholar]
- Bloch-Zupan A, Goodman JR. Otodental syndrome. Orphanet J Rare Dis. 2006;1:5. doi: 10.1186/1750-1172-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso C, Boys A, Parrini E, Mignon-Ravix C, McMahon JM, Khantane S, Bertini E, Pallesi E, Missirian C, Zuffardi O, Novara F, Villard L, Giglio S, Chabrol B, Slater HR, Moncla A, Scheffer IE, Guerrini R. Periventricular heterotopia, mental retardation, and epilepsy associated with 5q14.3-q15 deletion. Neurology. 2009;72:784–792. doi: 10.1212/01.wnl.0000336339.08878.2d. [DOI] [PubMed] [Google Scholar]
- Gregory-Evans CY, Moosajee M, Hodges MD, Mackay DS, Game L, Vargesson N, Bloch-Zupan A, Rüschendorf F, Santos-Pinto L, Wackens G, Gregory-Evans K. SNP genome scanning localizes oto-dental syndrome to chromosome 11q13 and microdeletions at this locus implicate FGF3 in dental and inner-ear disease and FADD in ocular coloboma. Hum Mol Genet. 2007;16:2482–2493. doi: 10.1093/hmg/ddm204. [DOI] [PubMed] [Google Scholar]
- Higgins AW, Alkuraya FS, Bosco AF, Brown KK, Bruns GA, Donovan DJ, Eisenman R, Fan Y, Farra CG, Ferguson HL, Gusella JF, Harris DJ, Herrick SR, Kelly C, Kim HG, Kishikawa S, Korf BR, Kulkarni S, Lally E, Leach NT, Lemyre E, Lewis J, Ligon AH, Lu W, Maas RL, MacDonald ME, Moore SD, Peters RE, Quade BJ, Quintero-Rivera F, Saadi I, Shen Y, Shendure J, Williamson RE, Morton CC. Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet. 2008;82:712–722. doi: 10.1016/j.ajhg.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malan V, Martinovic J, Sanlaville D, Caillat S, Waill MC, Ganne ML, Tantau J, Attie-Bitach T, Vekemans M, Morichon-Delvallez N. Molecular characterisation of a prenatally diagnosed 5q15q21.3 deletion and review of the literature. Prenat Diagn. 2006;26:231–238. doi: 10.1002/pd.1386. [DOI] [PubMed] [Google Scholar]
- Tzschach A, Krause-Plonka I, Menzel C, Kalscheuer V, Toennies H, Scherthan H, Knoblauch A, Radke M, Ropers HH, Hoeltzenbein M. Molecular cytogenetic analysis of a de novo interstitial deletion of 5q23.3q31.2 and its phenotypic consequences. Am J Med Genet Part A. 2006;140A:496–502. doi: 10.1002/ajmg.a.31105. [DOI] [PubMed] [Google Scholar]