Abstract
Although c-kit+ cardiac progenitor cells (CPCs) are currently used in clinical trials there remain considerable gaps in our understanding of the molecular mechanisms underlying their proliferation and differentiation. G-protein coupled receptors (GPCRs) play an important role in regulating these processes in mammalian cell types thus we assessed GPCR mRNA expression in c-kit+ cells isolated from adult mouse hearts. Our data provide the first comprehensive overview of the distribution of this fundamental class of cardiac receptors in CPCs and reveals notable distinctions from that of adult cardiomyocytes. We focused on GPCRs that couple to RhoA activation in particular those for sphingosine-1 -phosphate (S1P). The S1P2 and S1P3 receptors are the most abundant S1P receptor subtypes in mouse and human CPCs while cardiomyocytes express predominantly S1P1 receptors. Treatment of CPCs with S1P, as with thrombin and serum, increased proliferation through a pathway requiring RhoA signaling, as evidenced by significant attenuation when Rho was inhibited by treatment with C3 toxin. Further analysis demonstrated that both S1P- and serum- induced proliferation are regulated through the S1P2 and S1P3 receptor subtypes which couple to Gα12/13 to elicit RhoA activation. The transcriptional co-activator MRTF-A was activated by S1P as assessed by its nuclear accumulation and induction of a RhoA/MRTF-A luciferase reporter. In addition S1P treatment increased expression of cardiac lineage markers Mef2C and GATA4 and the smooth muscle marker GATA6 through activation of MRTF-A. In conclusion, we delineate an S1P–regulated signaling pathway in CPCs that introduces the possibility of targeting S1P2/3 receptors, Gα12/13 or RhoA to influence the proliferation and commitment of c-kit+ CPCs and improve the response of the myocardium following injury.
Keywords: Sphingosine-1-phosphate, GPCR, cardiac progenitor cells, MRTF-A, RhoA
1. Introduction
The heart was traditionally thought to be a terminally differentiated post-mitotic organ incapable of cardiac myocyte regeneration. However the accumulation of experimental evidence, including the occurrence of natural cardiomyocyte apoptosis [1, 2], telomere shortening of cardiomyocytes [3, 4], and histological images of myocyte replication [5] changed the perception of the heart to a dynamic self-renewing organ that has the potential of undergoing cardiomyocyte turnover. Endogenous multipotent cardiac progenitor cells (CPCs), positive for the stem cell lineage marker c-kit and negative for hematopoietic markers, were discovered in the adult mammalian heart, including the mouse, rat and human [6–8]. In addition, these endogenous CPCs have been observed to respond to cardiac injury in vivo [8]. In mouse models, c-kit+ CPCs were shown to undergo population expansion along the border zones of the infarct region following myocardial infarction (MI) [8]. Although these endogenous CPCs exist and localize in areas of damaged myocardium, very little is known about the molecular mechanisms that regulate their biology, or why they fail to spontaneously regenerate cardiac tissue in the diseased heart.
Sphingosine-1-phosphate (S1P), a sphingolipid metabolite, is a bioactive signaling molecule that binds to specific G-protein coupled receptors (GPCRs) and has been implicated in regulating cellular processes such as maintenance of pluripotency, differentiation, proliferation, survival, and migration in a wide variety of cell types including stem cells [9–19]. The release of S1P from the mouse heart is increased following myocardial infarction and ischemia/reperfusion (I/R) [20, 21] serving to acutely protect the heart against ischemic injury [20, 22–26]. In addition S1P acts as a chemoattractant for hematopoietic stem cells following MI [27]. The effect of S1P on the endogenous c-kit+ CPCs has not, however, been investigated.
There are five high-affinity S1P receptor subtypes (S1PR1–5) that are differentially expressed in various cells and tissues. In the heart, only S1PR1, S1PR2, and S1PR3 are expressed at detectable levels, with S1PR1 being the most abundant in cardiomyocytes [28–30]. S1P receptors can couple to and signal through several heterotrimeric G-proteins (Gαi, Gαq/11, Gαs, and Gα12/13) but the S1PR1 subtype is limited in coupling only to Gαi [31]. The S1P2 and S1P3 receptors which, in contrast to S1PR1, couple to both the Gαq and Gα12/13 proteins are effective activators of the low molecular weight G-protein RhoA [32, 33]. We have attributed the cardioprotective effect of S1P in the mouse heart following I/R to activation of S1P2 and S1P3 receptors [22] and more recently to activation of RhoA [25]. RhoA activation promotes actin polymerization, a response originally associated with altered contractile function but subsequently shown to also result in the accumulation of myocardin-related transcription factor A (MRTF-A) in the nucleus [34–38]. MRTF-A is a transcriptional co-activator which binds to serum response factor (SRF) to induce gene transcription [39–42]. The activation of MRTF-A in mesenchymal stem cells (MSC) had been shown to play a role in MSC-mediated angiogenesis and induce MSC differentiation into several cell types, including endothelial and contractile smooth muscle cells [43–45]. Studies using MRTF-A knockdown implicate it in myotube formation and SRF-dependent activation of muscle genes in vitro [41] and overexpression of MRTF-A in bone marrow stem cells has been shown to be protective following myocardial infarction in rats (MI) [46].
In the present study we assess the possibility that the predominance of S1PR2/3 on CPCs might allow them to be targeted for expansion without affecting cardiomyocyte physiology through S1PR1 activation. In particular given the unique ability of S1PR2/3 receptor subtypes to couple to and signal trough RhoA, we investigated the involvement of this signalling pathway in responses of c-kit+ CPCs to S1P. We establish that S1P activates the Gα12/13 coupled S1P2 and S1P3 receptors to elicit RhoA activation, and that this pathway leads to increased proliferation and MRTF-A regulated cardiac gene transcription.
2. Materials and Methods
2.1 Animals
All procedures were performed in accordance with NIH Guide for the Care and use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee at the University of California San Diego. Male 8–12wk old FVB/N, C57BL/6J, S1PR2 and-S1 PR3- KO mice (kindly provided by Dr. Jerold Chun) were used for the isolation of c-kit+ cardiac progenitor cells.
2.2 Cardiac c-kit+ Cell Isolation
Adult c-kit+ cardiac progenitor cells (CPCs) cells were isolated from 8–12 week old FVB/N, C57BL/6J WT, S1PR2-and S1PR3- KO mice as previously described [7] with modification. Briefly, mice were euthanized with CO2 followed by cervical dislocation. The heart was cannulated on a Langendorff apparatus and perfused with warmed basic buffer followed by digestion with collagenase II solution. All buffers were made as described in Beltrami et al. The cell suspension was passed through a 30-micron filter (Miltenyi Biotech, Bergisch Gladbach, Germany) and incubated with CD117-conjugated Miltenyi magnetic microbeads to sort for c-kit+ cells. Mouse cells were grown in Growth Medium, which consisted of Dulbecco’s Modified Eagle’s Medium and Ham’s F12 (ratio 1:1), basic fibroblast growth factor (10 ng/ml), epidermal growth factor (20 ng/ml), leukemia inhibitory factor (10 ng/ml), embryonic stem cell fetal bovine serum (20%), and insulin-transferrin-selenite. Medium was changed 5–7 days later and the adherent cells were passaged using standard trypsinization protocols. For each isolation, the purity of the CPC population was assessed by FACs analysis at early passages. The typical % of c-kit+ cells was 60–80%, while endothelial (CD31) and hematopoietic (CD45 and CD34) markers were present on the surface of less than 5–10% of the cell population. After continued passaging, c-kit immunofluorescence was assessed; nearly 100% were c-kit+ cells by passage 15. Cells used for experiments were from passage 15 to 23 (stemness decreases beyond passage 23). In all experiments with agonists cells were cultured in serum-free medium for 24 hours before treatment. For longer term experiments with S1P and dexamethasone (Dex), CPCs were plated at a very low density (20,00 cells/ 10cm plate) and treated for only 3 days with S1P or Dex alone (no serum) to avoid confounding effects of cell contact resulting from increased proliferation. Human c-kit+ cardiac progenitor cells were isolated as previously described [47].
2.3 GPCR Expression Analysis
To define the complement of GPCRs in mouse c-kit+ CPCs RNA (1µg) was isolated with Trizol (Life Technologies, Carlsbad, CA) following the manufacturers protocol and cDNA was synthesized from RNA with random hexamer primers using Verso cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Cells were used at passage 15. Using a TaqMan GPCR mouse array (4378718) from Applied Biosystems (Life Technologies) the relative levels of GPCR transcripts present were determined and normalized to internal controls. To analyze S1PR mRNA expression in human and mouse c-kit+ CPCs and adult cardiomyocytes, cDNA was synthesized with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and qPCR was performed using the TaqMan Universal MasterMix II with UNG (Applied Biosystems) according to the manufacturer’s protocol. IDT TaqMan probes for human and mouse S1PR1, S1PR2, and S1PR3 were used.
2.4 Transfections in CPCs
CPCs were transfected with siRNA or cDNA using DharmaFECT-1 transfection reagent (Thermo Scientific) based on manufacturer’s instruction and as previously described [25]. siRNA or cDNA and DharmaFECT-1 (in a 1:3 ratio, respectively) were individually incubated in Opti-MEM media (GIBCO) at room temperature for 5 min, mixed and incubated at room temperature for 20 min. The siRNA or cDNA/DharmaFECT-1 mixtures (1 ml/dish) were added to cultures in Opti-MEM (GIBCO). After 4 hours the cells were washed and fresh Growth Media added overnight.
For gene knockdown, predesigned mouse On-Targetplus siRNA for S1P1, S1P2, S1P3 receptors, RhoA , Gα12, Gα13, Gαi , Gαq, and MRTF-A was purchased from Thermo Scientific. All siRNAs had a knockdown efficiency of ~80–100% when transfected 1 µg/well. In particular, the double knockdown of siGα12 together with siGα13 had an efficiency of ~80% for both G proteins, while the single knockdown of one had no effect on the other isoform.
For cardiac lineage gene expression analysis upon MRTF-A overexpression, CPCs were transfected with 500ng of a cDNA construct encoding MRTF-A, serum starved for 24 hours and harvested 48 hours later.
For luciferase assay, CPCs were plated on 12 well tissue culture plates (12,000 cells / well) in Growth Media. The following day, cells were transfected with an SRE.L reporter (500ng) as well as a Renilla plasmid (50ng) to normalize the luminescence signal.
After transfections, cells were starved for 24 hours and either proliferation, qPCR, or luciferase assay were performed as described in 2.5, 2.6 and 2.7 respectively.
2.5 Proliferation Assay
To measure cell growth, CPCs were subjected to a CyQUANT NF Cell Proliferation Assay (Invitrogen, C35011). CPCs were plated on a transparent 48 well plate (8,000 cells/well), cultured overnight with Growth Media and then maintained in serum free Media for 24 hours. CPCs isolated from FVB/N and used for most studies were stimulated with S1P (3µM), thrombin (0.5U/ml), lysophosphatidic acid (LPA, 10µM) or serum (20%) for 24 hours. CPCs from S1PR2 - and S1PR3- KOs in a C57BL/6J background were stimulated with serum (20%) for 24, 48 and 72 hours. For inhibition of Rho, cells were pretreated with C3 toxin (1µg/ml) for 6 hours prior to the addition of agonist. For knockdown of S1P receptors, cells were transfected as described in 2.4. Following stimulation, the plates were assayed according to the manufacturers’ protocol and the fluorescence intensity determined using a Tecan Spectrophotometer as previously described [48].
2.6 Cardiac and smooth muscle lineage marker expression
For gene expression analyses RNA was isolated as described above, cDNA synthesized with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems ABI) and real-time qPCR performed with TaqMan Universal Master Mix II, with UNG (Applied Biosystems ABI). To analyze gene expression in mouse c-kit+ CPCs treated with S1P (3µM) or Dex (10nM) IDT TaqMan probes for mouse Mef2C, GATA4, GATA6 and GAPDH were used. To analyze cardiac lineage gene expression upon MRTF-A activity inhibition, starved cells were treated with the pharmacological inhibitor CCG-293971[49], kindly provided by Dr. Neubig’s lab, for 1 hour before S1P treatment. For knockdown or overexpression of MRTF-A cells were transfected as described in 2.4.
2.7 Luciferase Assay
CPCs were transfected with SRE.L and Renilla as described in 2.4. Following 24 hours starvation, cells were treated with S1P (3µM) or serum (20%) for 6 hours. In the case of inhibition of Rho with C3-toxin (1µg/ml), cells were pretreated for 6 hours prior to agonist stimulation. For knockdown of G-proteins cells were transfected as described in 2.4.
Reporter activity was measured using the Promega Dual-Luciferase Reporter Assay based on manufacturer’s protocol.
2.8 Immunocytochemistry
CPCs were plated on 6-well tissue culture plates (10,000 cells / well) in Growth Media and serum starved the following day for 24 hours. Cells were treated with S1P (3µM) or serum (20%) in the presence or absence of C3-toxin (1µg/ml) for 30 minutes, fixed in 4% paraformaldehyde, permeabilized with 10% Triton X-100, and incubated over-night with MRTF-A antibody 1:200 (Santa Cruz Biotechnology, sc-32909). The following day, cells were washed and incubated with the secondary antibody, Alexa-fluor 488 dye (ThermoFisher Scientific) for 1 hour. After washing, the slides were mounted with Vectashield with DAPI (Vector Laboratories) and analyzed using a Leica SP5 confocal microscopy (40x oil immersion objective). Images were acquired at a focal distance of 0.49 µm and z-stacks were compressed to provide a composite image (ImageJ software analysis, plugins Bio-format). Images from independent experiments were analyzed and the percentage of cells with nuclear MRTF-A localization was calculated for each (10 fields / experiment).
2.9 Statistical Analyses
Researchers were blinded to the treatment group during analyses. Data are represented as mean ± SEM. Differences of p<0.05 were considered statistically significant and were assessed using unpaired Student’s t-test (for two groups), one-way (for multiple comparisons) or two-way (for multiple comparisons with more than one variable) ANOVA with post-hoc Tukey analysis using GraphPad Prism software (GraphPad, La Jolla, CA, USA).
3. Results
3.1 GPCR profiling in CPCs
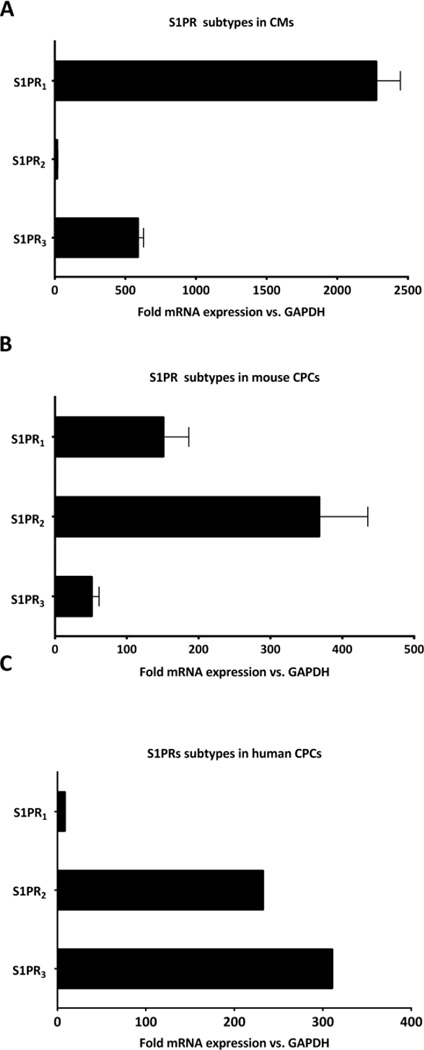
To determine the complement of GPCRs present on c-kit+ cardiac progenitor cells (CPCs) we analysed the levels of mRNA in undifferentiated mouse CPCs via a TaqMan GPCR array. Over 200 GPCRs were expressed at levels 2 times higher than GAPDH and those most commonly studied for their roles in cardiac function are shown in Fig. S.I.1A. Interestingly, the relative expression of receptor subtypes in CPCs was different from that of cardiomyocytes for several GPCRs. Of interest, the expression of sphingosine-1 phosphate (S1P) receptor isoforms in CPCs differed from that reported for cardiomyocytes [29, 30], as confirmed by qPCR analysis. Specifically whereas S1PR1 is the predominant receptor subtype in mouse cardiomyocytes (Fig. 1A), S1PR2 and S1PR3 are the more highly expressed subtypes in mouse and human CPCs (Fig. 1B and C). S1PR4 and S1PR5 were not detected in mouse CPCs. Since the CPCs we isolate and study are undifferentiated we also examined the expression of S1P receptor isoforms following treatment with dexamethasone, an intervention routinely used to induce differentiation and expression of cardiac lineage markers in CPCs [50]. Although S1PR1, S1PR2 and S1PR3 mRNA levels were increased, we did not observe a switch in the predominance of S1PR isoforms (Fig. S.I.1B) to resemble that of differentiated adult cardiomyocytes. This finding is consistent with other evidence that CPCs do not fully differentiate into functional cardiomyocytes in culture. We also noted that mRNAs for LPAR1 and LPAR2 subtypes, and for the thrombin receptor PAR1, all of which can activate RhoA [51–53], were highly expressed in mouse CPCs (Fig. S.I.1A).
Figure 1. Expression of S1 PR subtypes in adult cardiomyocytes, and in mouse and human cardiac progenitor cells.
S1P receptor mRNA levels were analyzed by qPCR and expressed as fold versus GAPDH. (A) Cardiomyocytes (CMs) were isolated from adult mouse heart as described [28] and cardiac progenitor cells (CPCs) (B) mouse and (C) human isolated as described in Methods. S1PR1 was the highest receptor in CMs while S1PR2 and S1PR3 were the highest in mouse and human CPCs respectively. n=4 in A, n = 5 in B, n=1 in C.
3.2 The GPCR agonist S1P induces CPC proliferation in a RhoA dependent manner
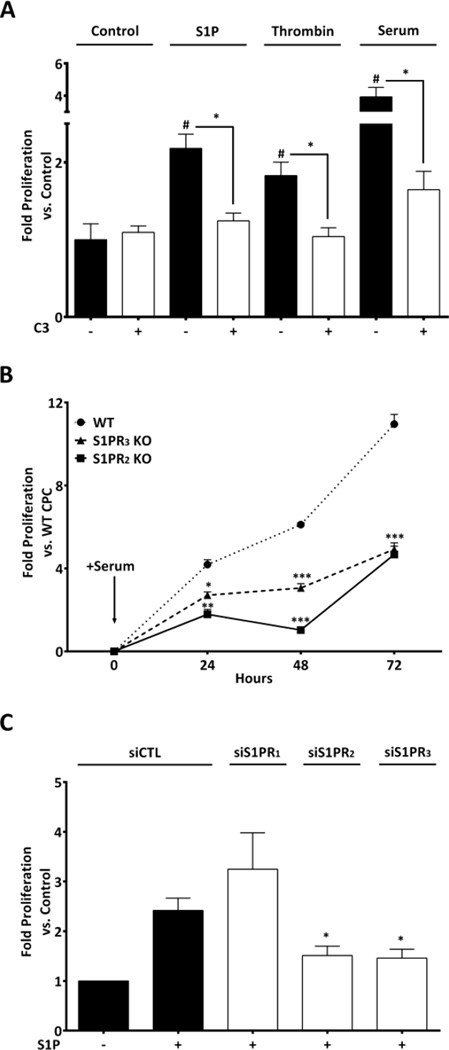
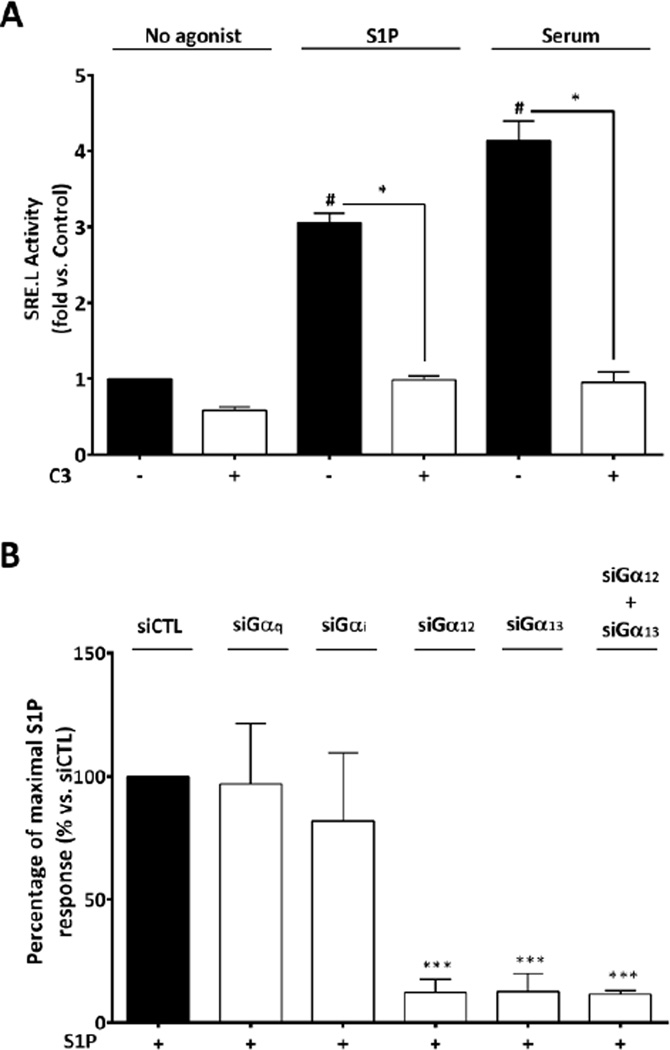
S1P, LPA, and thrombin have been shown to promote proliferation of various types of stem cells [11–19]. Thus, we asked if treatment with these agonists increased CPC proliferation and whether this occurred in a RhoA-dependent manner. CPCs cultured in serum-free medium and stimulated with S1P, thrombin (Fig. 2A) or LPA (Fig. S.I.2) for 24 hours showed a significant increase in proliferation (2.2, 1.8 and 3.2 fold respectively). As a control we utilized 20% serum, which increased proliferation 4 fold. In order to determine if the agonist-stimulated proliferation was RhoA-dependent, we pretreated CPCs with the C3 exoenzyme which ribosylates and functionally inactivates Rho. Thrombin and S1P- induced proliferation were significantly inhibited by C3 treatment and notably serum- induced proliferation was also fully inhibited (Fig. 2A). In contrast, the proliferative response to LPA was not blocked by C3 but was inhibited by treatment with pertussis toxin (Fig. S.I.2), indicating that it is mediated through Gαi signaling. Strikingly serum-induced proliferation was fully inhibited by C3 but not pertussis toxin (Fig.2A, Fig. S.I.2) suggesting that the S1P, thrombin or other ligands that activate RhoA are components of serum responsible for serum-induced proliferation.
Figure 2. S1P induced proliferation in CPCs is RhoA dependent and mediated through S1PR2 and S1PR3.
(A) Serum starved mouse CPCs were stimulated with S1P (3µM), thrombin (0.5U/ml) or 20% serum for 24 hours and proliferation was measured using the CyQUANT assay. Pre-treatment with C3 toxin (1µg/ml) blocked proliferation induced by S1P (3µM), thrombin and serum. #p≤0.05 vs. control, *p≤0.05 vs. treated by one-way ANOVA; n=15, 7 independent experiments (B) Serum starved CPCs isolated from WT, S1PR2 KO and S1PR3 KO mice stimulated with 20% serum. Proliferation was significantly lower in S1PR2 or S1PR3 KO CPCs than in WT. *p≤0.05, **p≤0.01, ***p≤0.005 vs. WT by two-ways ANOVA; n=6, 3 independent experiments (C) CPCs were transfected with control, S1P1, S1P2 or S1P3 receptor siRNA for 48 hours prior to addition of S1P (3µM) for 24 hours. Knockdown of S1P receptor mRNA assessed by qPCR was ~80–90% for all three receptor subtypes (data not shown). *p≤0.05 vs. S1P treated Control (siCTL) by one-way ANOVA; n=4, 2 independent experiments.
To further elucidate the role of S1P receptor signalling on CPC proliferation we isolated c-kit+ CPCs from S1PR2 and S1PR3 knockout (KO) mice (S1PR1 deletion is embryonic lethal) and treated them with 20% serum. There was a significantly lower rate of serum-induced proliferation in CPCs from either S1PR2 or S1PR3 KOs compared to those from wild-type (WT) mice (Fig.2B). In addition, we used siRNA-mediated knockdown of all three S1P receptor subtypes in wild type CPCs (knockdown ~80–90% for each) to examine the receptor subtypes through which S1P induces proliferation. S1P–induced proliferation was inhibited by knockdown of S1PR2 or S1PR3, while knockdown of S1PR1 did not affect S1P–induced proliferation (Fig. 2C). Taken together these results suggest that S1P, alone or as a component of serum, promotes CPC proliferation through activation of S1P receptor subtypes that couple to RhoA.
3.3 RhoA dependent activation of MRTF-A by S1P in CPCs
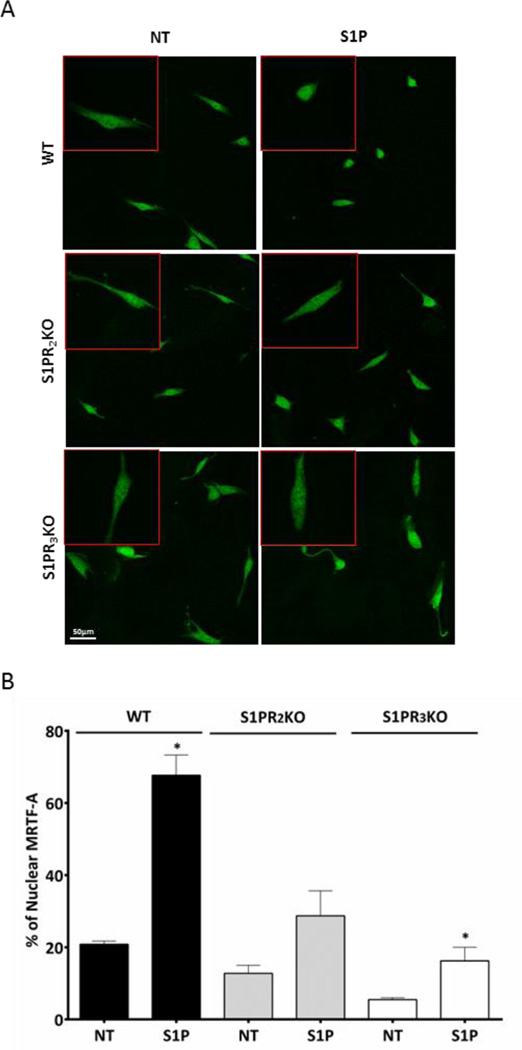
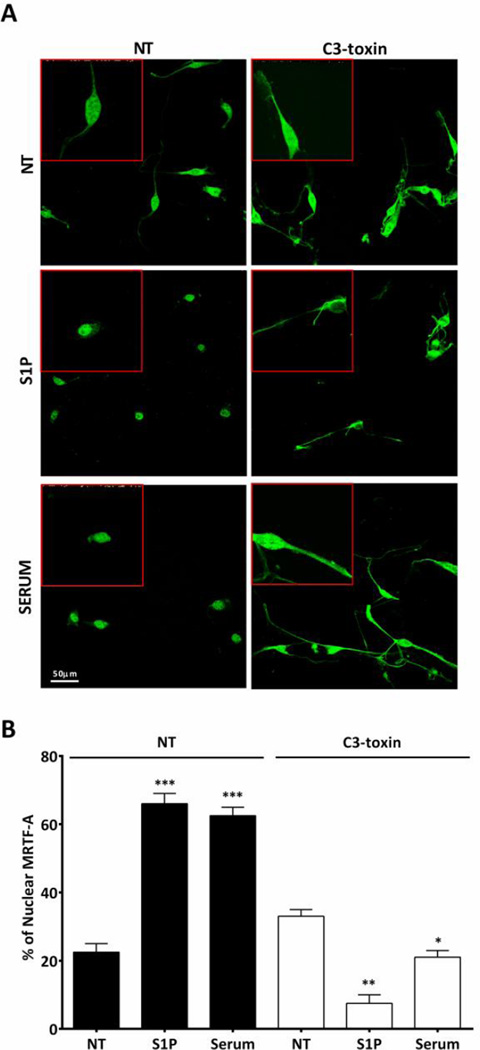
Activation of RhoA in mesenchymal stem cells regulates SRF dependent gene transcription through its co-transcriptional activator, MRTF-A [43]. MRTF-A is also involved in transcriptional activation downstream of RhoA in other cell types [39–41]. We asked whether MRTF-A was activated downstream of S1P–induced RhoA signaling in CPCs. MRTF-A activation was assessed using immunofluorescence to examine increases in its nuclear accumulation. The percent of WT CPCs with nuclear MRTF-A staining increased significantly with S1P or serum treatment (Fig. 3A, B and 4A, B). In CPCs from S1PR2 or S1PR3 KO mice MRTF-A nuclear accumulation was partially diminished in response to S1P (Fig. 3A and B). Furthermore inhibition of Rho by pre-treatment with C3 toxin attenuated the nuclear accumulation of MRTF-A elicited by S1P and notably also that induced by serum (Fig. 4A and B). These findings demonstrate that S1P activates MRTF-A in CPCs through S1PR2 and S1PR3 and in a Rho-dependent mechanism.
Figure 3. MRTF-A nuclear accumulation is diminished in S1PR KO CPCs.
Serum starved WT, S1PR2 and S1PR3 KO mouse CPCs were stimulated with S1P (3µM) for 30 minutes. MRTF-A was localized by immunofluorescence; z-stack confocal images were compressed for analysis. The percentage of cells with nuclear MRTF-A was quantified by counting 10 fields in 3 independent experiments (n=50 cells). (A) Representative immunofluorescence of MRTF-A localization Green- MRTF-A; (B) Quantification of MRTF-A nuclear accumulation. *p≤0.05 vs. non-treated (NT) by unpaired t-test.
Figure 4. S1P induces MRTF-A accumulation in the nucleus in a Rho dependent manner.
Serum starved mouse CPCs were stimulated with S1P (3µM) or serum (20%) with or without pre-treatment with C3 toxin (1ug/ml). MRTF-A was localized by immunofluorescence; z-stack confocal images were compressed for analysis. The percentage of cells with nuclear MRTF-A was quantified by counting 10 fields in 3 independent experiments (n=50 cells). (A) Representative immunofluorescence of MRTF-A localization Green- MRTF-A (B) Quantification of MRTF-A nuclear accumulation. *p≤0.05 **p≤0.01, and *** p≤0.005 vs. non-treated (NT) by unpaired t-test
3.4 S1P induces SRF-dependent transcriptional activity in CPCs
The accumulation of MRTF-A in the nucleus should lead to increased SRF-dependent transcriptional activity. To demonstrate that this occurs, and as further evidence of RhoA activation, we measured MRTF-A/SRF regulated activation of an SRE.L luciferase reporter gene in CPCs. As demonstrated in Fig. 5A, SRE.L luciferase activity was increased following 6 hours of S1P or serum stimulation and both responses were abolished by pre-treatment with C3 toxin or by siRNA mediated knockdown of RhoA (data not shown). We also tested G-protein involvement and observed that S1P-induced SRE.L activity was significantly diminished by knockdown of Gα12 and Gα13, but not of Gαq or Gαi (knockdown ~80–100% for all G proteins) (Fig. 5B). These findings demonstrate that S1P induces SRF-dependent transcriptional activation in CPCs through a RhoA/MRTF-A mediated signaling pathway and imply that Gα12 and Gα13 are both required.
Figure 5. S1P induces RhoA mediated gene expression through Gα12/13.
Mouse CPCs were transfected with an SRE.L luciferase reporter construct (0.5 µg) for 48 hrs (A) Cells were stimulated with S1P (µM) or 20% serum for 6 hours with or without pre-treatment with C3 toxin (1ug/ml). #p≤0.05 vs. non-treated (NT); *p≤0.05 vs. agonist alone by one-way ANOVA; n=12, 4 independent experiments (B) Cells were transfected with siControl or siRNA targeting various G protein α subunits. Knockdown of G protein mRNAs assessed by qPCR was ~80% for Gα12, ~100% for Gα13, ~90% for Gαq and Gαi . SRE.L luciferase activity was normalized to Renilla and expressed as percentage of maximal S1P induction with control siRNA. ***p≤0.005 vs. Max S1P response (100%) by unpaired t-test; n=4 independent experiments.
3.5 S1P upregulates cardiac lineage markers through a RhoA/MRTF-A dependent pathway
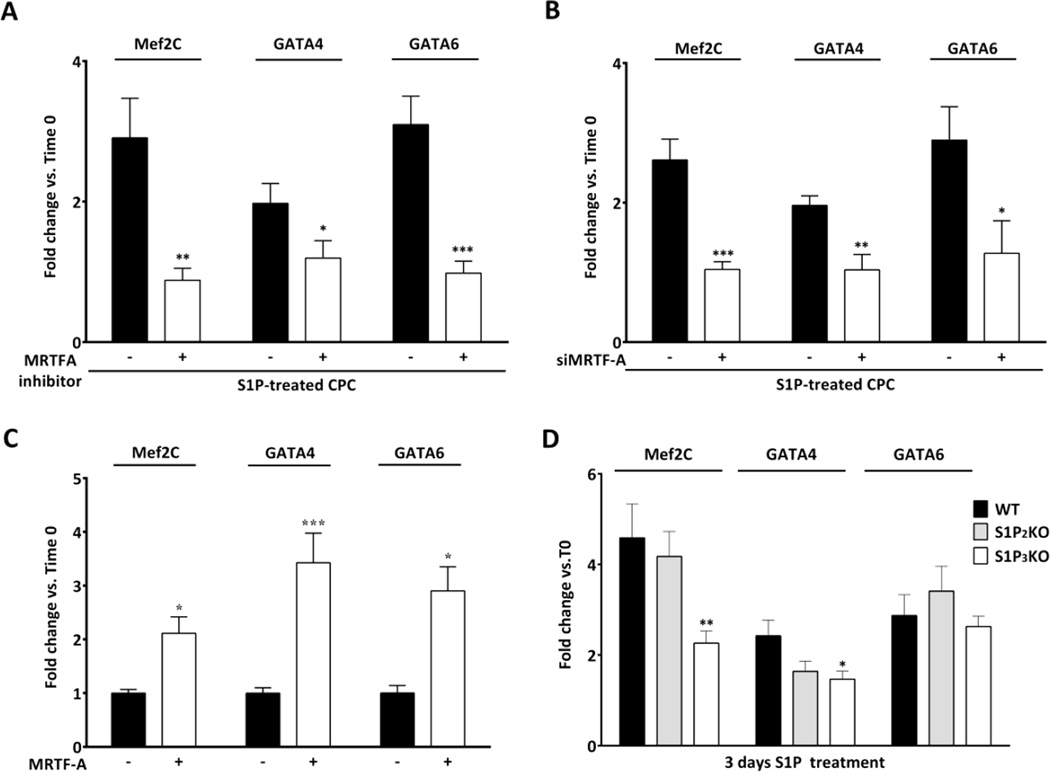
Understanding the signalling pathways that regulate expression of genes important for differentiation could enable use of the innate capacity of c-kit+ CPCs to repair the heart. Since it is known that SRF/MRTF-A plays a fundamental role in myogenesis [41, 42], we asked whether S1P affects expression of known cardiac or smooth muscle lineage marker genes. The extent of gene regulation by S1P was compared to that induced by dexamethasone. CPCs isolated from FVBN WT mice were treated with S1P or Dex in absence of serum and harvested after 3 days. The mRNA levels of two cardiomyocyte markers (GATA4 and Mef2C) and a smooth muscle cell marker (GATA6) were analysed by qPCR. As shown in Fig. S.I.3, treatment of CPCs with S1P increased mRNA for GATA4 (~2 fold), Mef2C and GATA6 (both ~3 fold). Cells cultured for 3 days under the same culture conditions but in the absence of S1P or Dex did not show significant increases in gene expression relative to time zero (data not shown). S1P induced increases in gene expression (Fig.6A and B) were nearly equivalent to those of Dex (S.I. Fig.3). Notably, induction of these genes by S1P was significantly attenuated by addition of a pharmacological inhibitor of MRTF-A activation, CCG-293971 [49] or by silencing MRTF-A through siRNA-mediated knockdown (~90% knockdown) (Fig. 6A and B). Furthermore we determined that overexpression of MRTF-A was sufficient to induce GATA4, Mef2C and GATA6 gene expression in CPCs (Fig. 6C). This transcriptional response to MRTF-A was not attributable to changes in cell density since neither MRTF-A overexpression nor its inhibition affected CPC proliferation (data not shown). To determine the contribution of S1PR2 or S1PR3 to S1P induced cardiac gene changes in CPCs, we analysed mRNA levels in CPCs isolated from C57Bl/6 WT, S1PR2 and S1PR3 KO mice treated with S1P. We confirmed that cardiac lineage marker genes were upregulated by S1P in the C57Bl/6 background as it was in FVBN. Genetic deletion of S1PR3 significantly reduced S1P induced Mef2C and GATA4, while S1PR2 deletion tended towards inhibition of GATA4 only (Fig. 6D). Taken together our data demonstrate that prolonged treatment of CPCs with S1P stimulates the expression of cardiac lineage markers through activation of an S1PR/RhoA/SRF/MRTF-A signaling pathway.
Figure 6. S1P induces cardiac lineage gene expression in a MRTF-A dependent manner.
Starved mouse CPCs were treated with S1P (3µM) for 3 days and Mef2C, GATA4 and GATA6 expression levels were measured by qPCR. Fold changes over control at time 0 are graphed. Pharmacological inhibition (10µM, CCG-293971) (A) or silencing (B) of MRTF-A attenuate the effect of S1P on cardiac gene expression. *p≤0.05, **p≤0.01, ***p≤0.005 vs. S1P treated by unpaired t-test; n=8, 3 independent experiments. (C) CPCs were transfected with a plasmid encoding MRTF-A (0.5 µg). Overexpression of MRTF-A for 48 hours induced cardiac gene expression in CPCs. n=12, 4 independent experiments. *p≤0.05 and ***p≤0.005 vs. control at time 0 by unpaired T-test. (D) qPCR analysis of gene expression in S1PR KO CPCs following 3 days of S1P stimulation *p≤0.05 **p≤0.01 vs. WT S1P treated by unpaired t-test; n = 9, 3 independent experiments
4. Discussion
During cardiac injury the heart attempts to maintain its ability to pump blood throughout the body. The persistence of the insult leads, however, to cardiac remodeling resulting from cardiomyocyte (CMs) apoptosis and fibroblast proliferation, and this ultimately leads to cardiac dysfunction and death. In contrast to skeletal muscle where stem cells, called satellite cells, reside in the tissue and are activated upon injury to regenerate muscle [54], the mammalian heart is incapable of spontaneous regeneration. On the other hand it has become evident in the last decade that adult c-kit+ cardiac progenitor cells (CPCs), are present in the heart [55] and that exogenous delivery of these cells has salutary effect on cardiac performance. In fact, CPCs are currently being used in clinical trials for the treatment of heart failure in the SCIPIO (Stem Cell Infusion in Patients with Ischemic cardiomyopathy) trial with striking results [56, 57] supporting the need to elucidate the signaling pathways influencing CPC function. Ideally the endogenous c-kit+ cells would be utilized to repair the heart, thus recent work has considered means of enhancing their innate ability to proliferate, migrate and differentiate both in vitro and in vivo [58–61]. In this context, our research has focused on identifying GPCR signaling pathways that regulate CPC function and which could be modulated or engaged in response to external stimuli or ligands.
G-protein coupled receptors (GPCRs) are one of the largest known classes of receptors and highly drugable targets. Delineating the complement of GPCRs on CPCs and determining how their agonists affect CPC function could provide insights into ways to alter their function. The GPCRs present on CPCs have not been previously screened, and we determined here that the mRNA for more than 200 GPCRs are expressed in mouse c-kit+ CPCs. Notably the relative expression levels of some physiologically important GPCR subtypes in the CPCs are distinct from those reported for CMs. For example, in contrast with the well-known higher abundance of the β1 versus the β2 adrenergic receptor (βARs) in CMs [62], β2AR mRNA was more abundant in CPCs. The functional significance of the βAR subtype differences in CPCs and adult CMs was evidenced by the demonstration that β2AR receptor stimulation plays a role in protecting CPCs which is lost following a shift to β 1AR during differentiation [63]. Similarly, the Angiotensin II (AT) receptor subtypes AT1 and AT2 show different relative expression levels with AT1 expressed at higher levels in adult cardiomyocytes [64], whereas AT2 are predominant on c-kit+ CPCs. Notably c-kit+ AT2 receptor containing cells in the rat heart have been shown to increase after acute ischemic injury and to contribute to cardioprotection [65]. The fact that βARs and ATRs are important drug targets in heart failure (HF) and that the mechanisms by which β-blockers exert beneficial effects in the clinical treatment of HF is not fully understood, suggests that effects of these drugs on endogenous CPCs merits consideration; the potentially maladaptive changes in βAR and ATR subtypes during differentiation towards CMs could be a factor limiting CPC-mediated cardiac healing.
Signaling through and shifts in S1P receptor expression could likewise affect the cardiac response to injury. Accordingly, our work focused on the sphingosine-1 phosphate (S1P) receptors and in particular the unique role played by S1PR2 and S1PR3 in CPCs. We and others have shown that S1P plays a protective role against damage caused by ischemia/reperfusion (I/R) in the heart [20, 22, 26, 66]. Ex vivo studies have shown that perfusing the mouse heart with S1P either prior to or post ischemia, reduces the infarct area when compared to untreated hearts [20, 25]. Our lab previously attributed the protective effects of S1P to S1PR2 and S1PR3 activation in studies comparing in vivo I/R injury in WT versus S1PR2 and S1PR3 knockout mice [22]. Mice lacking both S1PR2 and S1PR3 had an increase in infarct area relative to WT mice, implying that endogenous S1P released during MI serves a cardioprotective role through S1PR2 and S1PR3 signaling in the adult heart. This data is of particular interest considering that S1PR2 and S1PR3 are highly expressed in CPCs isolated from both mouse and human hearts and that these receptors mediate the effects of S1P and at least in part the effect of serum on CPC proliferation. Thus the predominance of S1P2 and S1P3 receptors may be important for enhancing CPC expansion. This has also been suggested to be the case in other stem cells, as in skeletal muscle, where S1P has been shown to be a key regulator in satellite cell biology, promoting cell proliferation and migration [67–69]. In particular S1PR2 has been shown to promote muscle regeneration [70] and together with S1PR3 has been implicated in the transmission of S1P–induced cell proliferation in satellite cells [67], in line with our findings in CPCs.
While increasing proliferation of CPCs is critical to their participation in cardiac repair, the ability of these cells to regulate genes necessary for commitment or for paracrine signaling could further enhance their utility. Thus we investigated the effects of S1P on gene expression and involvement of RhoA, which is known to activate several downstream transcriptional regulators including the transcriptional co-activator MRTF-A. MRTF-A, together with myocardin and MRTF-B, are members of a family of co-activators that regulates gene expression by their interaction with the transcription factor SRF [71]. Of note, while myocardin is constitutively localized in the nucleus, MRTF-A and -B associate with G-actin in the cytoplasm and translocate to the nucleus to activate SRF upon RhoA-dependent actin polymerization [71] . MRTF-A and B are thus responsible for transduction of the RhoA/actin signals to gene transcription. We demonstrate here that in S1P–treated CPCs, MRTF-A accumulates in the nucleus, and that this is dependent on both S1PR2 and S1PR3. This nuclear accumulation is blocked by C3-toxin, supporting its dependence on Rho signalling. This is concordant with our finding that the SRF-dependent transcriptional activity induced by S1P in CPCs is dependent on Gα12/13, which couples to RhoA activation, but not on either Gαi, or Gαq. Thus while LPA elicits CPC proliferation through Gαi, and it has been reported that Gαi mediates ERK activation in response to S1P in embryonic stem cells [10], the predominant S1P signalling pathway for both proliferation and gene expression of CPCs is through coupling of the more highly expressed S1P2 and S1P3 receptors to Gα12/13 and RhoA .
MRTF-A has been shown to regulate genes involved in muscle cell differentiation, cardiovascular development, and epithelial-mesenchymal transition [40, 43, 44, 72]. MRTF-A has also been shown to induce the expression of SRF-dependent genes involved in smooth muscle cell differentiation [72], and in undifferentiated embryonic stem cells overexpression of MRTF-A induces expression of genes encoding markers of SMC differentiation [73]. Similarly, a sphingosine metabolite, sphingosylphosphorylcholine (SPC), was shown to induce differentiation of mesenchymal stem cells into contractile smooth muscle cells through RhoA/Rho kinase-dependent nuclear translocation of MRTF-A [44]. In agreement with the role of Rho/MRTF-A in smooth muscle differentiation, we found that S1P was able to induce expression of GATA6 in CPCs; in addition we demonstrated effects on cardiac lineage markers Mef2C and GATA4. In this regard the response to S1P was similar to the well-described response of CPCs to dexamethasone. The transcriptional effect of S1P was dependent on the activation of MRTF-A, as evidenced by either MRTF-A gene silencing or pharmacological inhibition. Not only was MRTF-A necessary for S1P induced gene transcription but it was sufficient to induce cardiac lineage genes when transiently over-expressed in CPCs. The finding that genetic deletion of S1PR2 or S1PR3 only partially diminished S1P induced gene expression in CPCs is likely due to the redundant function of these receptors as mediators of RhoA activation [74], and is supported by the fact that, neither of the S1PR KOs completely abolished MRTF-A nuclear accumulation. The low level of MRTF-A that still accumulates in the nucleus when either receptor is deleted may be sufficient for the induction of GATA6 explaining why it is not reduced.
In summary, the present study demonstrates for the first time that S1P increases proliferation and gene expression of CPCs, and that this occurs by activating S1PR2/3 coupling to Gα12/13 and RhoA and subsequently to SRF/MRTF-A dependent transcriptional response. The finding that S1P regulates CPC gene transcription through an MRTF-A dependent mechanism may have implications not only for lineage commitment but also additionally for regulation of genes and gene products that provide a more permissive environment for CPCs or other cells in the heart. For example, we and others have reported that S1P–dependent RhoA and MRTF-A activation in cardiomyocytes lead to the expression and subsequent secretion of the matricellular protein CCN1, a protein that mediates angiogenesis and cell survival [75, 76]. In addition recent work showed a functional role for RhoA and MRTF-A activation in regulating integrin expression in mesenchymal stem cells [45]. Future studies will attempt to identify factors that are regulated through S1P /RhoA / MRTF-A signaling in CPCs and could influence not only their function and survival but also that of surrounding cells in the heart.
5. Conclusion
There is general agreement that the endogenous population of CPCs is, at the present, unable to repair the heart following injury. However, we have delineated a signaling cascade through which an endogenous GPCR agonist, S1P can enhance CPC proliferation and gene expression. A better understanding of the genes and processes regulated by S1P, a ligand generated by the heart following injury, should inform future approaches to enhance the therapeutic potential of CPCs.
Supplementary Material
Highlights.
S1P receptor subtypes expressed on c-kit+ CPCs differ from those on cardiomyocytes
S1P stimulates RhoA-mediated proliferation of CPCs through S1P2/3
S1P induces RhoA-mediated transcriptional activation through Gα12/13 and MRTF-A.
Acknowledgments
This work was supported by grants from the National Heart, Lung and Blood Institute Grants: HL085577 (J.H.B, N.H.P and M.A.S.), HL028143 (J.H.B.), and HL114949 (N.H.P)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferrari R, Ceconi C, Campo G, et al. Mechanisms of remodelling: a question of life (stem cell production) and death (myocyte apoptosis) Circ J. 2009;73:1973–1982. doi: 10.1253/circj.cj-09-0573. [DOI] [PubMed] [Google Scholar]
- 2.Piccoli MT, Gupta SK, Thum T. Noncoding RNAs as regulators of cardiomyocyte proliferation and death. J Mol Cell Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Fuster JJ, Andres V. Telomere biology and cardiovascular disease. Circ Res. 2006;99:1167–1180. doi: 10.1161/01.RES.0000251281.00845.18. [DOI] [PubMed] [Google Scholar]
- 4.Wong LS, van der Harst P, de Boer RA, et al. Aging, telomeres and heart failure. Heart Fail Rev. 2010;15:479–486. doi: 10.1007/s10741-010-9173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltrami AP, Urbanek K, Kajstura J, et al. Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med. 2001;344:1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- 6.Quaini F, Urbanek K, Beltrami AP, et al. Chimerism of the transplanted heart. N Engl J Med. 2002;346:5–15. doi: 10.1056/NEJMoa012081. [DOI] [PubMed] [Google Scholar]
- 7.Beltrami AP, Barlucchi L, Torella D, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 8.Fransioli J, Bailey B, Gude NA, et al. Evolution of the c-kit-positive cell response to pathological challenge in the myocardium. Stem Cells. 2008;26:1315–1324. doi: 10.1634/stemcells.2007-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyne S, Pyne NJ. Translational aspects of sphingosine 1-phosphate biology. Trends Mol Med. 2011;17:463–472. doi: 10.1016/j.molmed.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Rodgers A, Mormeneo D, Long JS, et al. Sphingosine 1-phosphate regulation of extracellular signal-regulated kinase-1/2 in embryonic stem cells. Stem Cells Dev. 2009;18:1319–1330. doi: 10.1089/scd.2009.0023. [DOI] [PubMed] [Google Scholar]
- 11.Harada J, Foley M, Moskowitz MA, et al. Sphingosine-1-phosphate induces proliferation and morphological changes of neural progenitor cells. J Neurochem. 2004;88:1026–1039. doi: 10.1046/j.1471-4159.2003.02219.x. [DOI] [PubMed] [Google Scholar]
- 12.Kang S, Han J, Song SY, et al. Lysophosphatidic acid increases the proliferation and migration of adiposederived stem cells via the generation of reactive oxygen species. Mol Med Rep. 2015 doi: 10.3892/mmr.2015.4023. [DOI] [PubMed] [Google Scholar]
- 13.Smadja DM, Cornet A, Emmerich J, et al. Endothelial progenitor cells: characterization, in vitro expansion, and prospects for autologous cell therapy. Cell Biol Toxicol. 2007;23:223–239. doi: 10.1007/s10565-007-0177-6. [DOI] [PubMed] [Google Scholar]
- 14.He X, H’Ng SC, Leong DT, et al. Sphingosine-1-phosphate mediates proliferation maintaining the multipotency of human adult bone marrow and adipose tissue-derived stem cells. J Mol Cell Biol. 2010;2:199–208. doi: 10.1093/jmcb/mjq011. [DOI] [PubMed] [Google Scholar]
- 15.Donati C, Cencetti F, Nincheri P, et al. Sphingosine 1-phosphate mediates proliferation and survival of mesoangioblasts. Stem Cells. 2007;25:1713–1719. doi: 10.1634/stemcells.2006-0725. [DOI] [PubMed] [Google Scholar]
- 16.Avery K, Avery S, Shepherd J, et al. Sphingosine-1-phosphate mediates transcriptional regulation of key targets associated with survival, proliferation, and pluripotency in human embryonic stem cells. Stem Cells Dev. 2008;17:1195–1205. doi: 10.1089/scd.2008.0063. [DOI] [PubMed] [Google Scholar]
- 17.Lu W, Xiu X, Zhao Y, et al. Improved Proliferation and Differentiation of Bone Marrow Mesenchymal Stem Cells Into Vascular Endothelial Cells With Sphingosine 1-Phosphate. Transplant Proc. 2015;47:2035–2040. doi: 10.1016/j.transproceed.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 18.Price MM, Kapitonov D, Allegood J, et al. Sphingosine-1-phosphate induces development of functionally mature chymase-expressing human mast cells from hematopoietic progenitors. FASEB J. 2009;23:3506–3515. doi: 10.1096/fj.08-128900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitson SM, Pebay A. Regulation of stem cell pluripotency and neural differentiation by lysophospholipids. Neurosignals. 2009;17:242–254. doi: 10.1159/000231891. [DOI] [PubMed] [Google Scholar]
- 20.Vessey DA, Li L, Honbo N, et al. Sphingosine 1-phosphate is an important endogenous cardioprotectant released by ischemic pre- and postconditioning. Am J Physiol Heart Circ Physiol. 2009;297:H1429–H1435. doi: 10.1152/ajpheart.00358.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sattler KJ, Elbasan S, Keul P, et al. Sphingosine 1-phosphate levels in plasma and HDL are altered in coronary artery disease. Basic Res Cardiol. 2010;105:821–832. doi: 10.1007/s00395-010-0112-5. [DOI] [PubMed] [Google Scholar]
- 22.Means CK, Xiao CY, Li Z, et al. Sphingosine 1-phosphate S1P2 and S1P3 receptor-mediated Akt activation protects against in vivo myocardial ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2007;292:H2944–H2951. doi: 10.1152/ajpheart.01331.2006. [DOI] [PubMed] [Google Scholar]
- 23.Theilmeier G, Schmidt C, Herrmann J, et al. High-density lipoproteins and their constituent, sphingosine-1-phosphate, directly protect the heart against ischemia/reperfusion injury in vivo via the S1P3 lysophospholipid receptor. Circulation. 2006;114:1403–1409. doi: 10.1161/CIRCULATIONAHA.105.607135. [DOI] [PubMed] [Google Scholar]
- 24.Jin ZQ, Karliner JS, Vessey DA. Ischaemic postconditioning protects isolated mouse hearts against ischaemia/reperfusion injury via sphingosine kinase isoform-1 activation. Cardiovasc Res. 2008;79:134–140. doi: 10.1093/cvr/cvn065. [DOI] [PubMed] [Google Scholar]
- 25.Xiang SY, Ouyang K, Yung BS, et al. PLCepsilon, PKD1, and SSH1L transduce RhoA signaling to protect mitochondria from oxidative stress in the heart. Sci Signal. 2013;6:ra108. doi: 10.1126/scisignal.2004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy S, Kane KA, Pyne NJ, et al. Targeting sphingosine-1-phosphate signalling for cardioprotection. Curr Opin Pharmacol. 2009;9:194–201. doi: 10.1016/j.coph.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Seitz G, Boehmler AM, Kanz L, et al. The role of sphingosine 1-phosphate receptors in the trafficking of hematopoietic progenitor cells. Ann N Y Acad Sci. 2005;1044:84–89. doi: 10.1196/annals.1349.011. [DOI] [PubMed] [Google Scholar]
- 28.Means CK, Miyamoto S, Chun J, et al. S1P1 receptor localization confers selectivity for Gi-mediated cAMP and contractile responses. J Biol Chem. 2008;283:11954–11963. doi: 10.1074/jbc.M707422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Means CK, Brown JH. Sphingosine-1-phosphate receptor signalling in the heart. Cardiovasc Res. 2009;82:193–200. doi: 10.1093/cvr/cvp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Honbo N, Goetzl EJ, et al. Signals from type 1 sphingosine 1-phosphate receptors enhance adult mouse cardiac myocyte survival during hypoxia. Am J Physiol Heart Circ Physiol. 2007;293:H3150–H3158. doi: 10.1152/ajpheart.00587.2006. [DOI] [PubMed] [Google Scholar]
- 31.Kihara Y, Maceyka M, Spiegel S, et al. Lysophospholipid receptor nomenclature review: IUPHAR Review 8. Br J Pharmacol. 2014;171:3575–3594. doi: 10.1111/bph.12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siehler S, Manning DR. Pathways of transduction engaged by sphingosine 1-phosphate through G protein-coupled receptors. Biochim Biophys Acta. 2002;1582:94–99. doi: 10.1016/s1388-1981(02)00142-7. [DOI] [PubMed] [Google Scholar]
- 33.Gohla A, Schultz G, Offermanns S. Role for G(12)/G(13) in agonist-induced vascular smooth muscle cell contraction. Circ Res. 2000;87:221–227. doi: 10.1161/01.res.87.3.221. [DOI] [PubMed] [Google Scholar]
- 34.Miralles F, Posern G, Zaromytidou AI, et al. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 35.Cen B, Selvaraj A, Prywes R. Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression. J Cell Biochem. 2004;93:74–82. doi: 10.1002/jcb.20199. [DOI] [PubMed] [Google Scholar]
- 36.Guettler S, Vartiainen MK, Miralles F, et al. RPEL motifs link the serum response factor cofactor MAL but not myocardin to Rho signaling via actin binding. Mol Cell Biol. 2008;28:732–742. doi: 10.1128/MCB.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuwahara K, Barrientos T, Pipes GC, et al. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol Cell Biol. 2005;25:3173–3181. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu OM, Brown JH. G Protein-Coupled Receptor and RhoA-Stimulated Transcriptional Responses: Links to Inflammation, Differentiation, and Cell Proliferation. Mol Pharmacol. 2015;88:171–180. doi: 10.1124/mol.115.097857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwasaki K, Hayashi K, Fujioka T, et al. Rho/Rho-associated kinase signal regulates myogenic differentiation via myocardin-related transcription factor-A/Smad-dependent transcription of the Id3 gene. J Biol Chem. 2008;283:21230–21241. doi: 10.1074/jbc.M710525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staus DP, Weise-Cross L, Mangum KD, et al. Nuclear RhoA signaling regulates MRTF-dependent SMC-specific transcription. Am J Physiol Heart Circ Physiol. 2014;307:H379–H390. doi: 10.1152/ajpheart.01002.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selvaraj A, Prywes R. Megakaryoblastic leukemia-1/2, a transcriptional co-activator of serum response factor, is required for skeletal myogenic differentiation. J Biol Chem. 2003;278:41977–41987. doi: 10.1074/jbc.M305679200. [DOI] [PubMed] [Google Scholar]
- 42.Lockman K, Hinson JS, Medlin MD, et al. Sphingosine 1-phosphate stimulates smooth muscle cell differentiation and proliferation by activating separate serum response factor co-factors. J Biol Chem. 2004;279:42422–42430. doi: 10.1074/jbc.M405432200. [DOI] [PubMed] [Google Scholar]
- 43.Wang N, Zhang R, Wang SJ, et al. Vascular endothelial growth factor stimulates endothelial differentiation from mesenchymal stem cells via Rho/myocardin-related transcription factor--a signaling pathway. Int J Biochem Cell Biol. 2013;45:1447–1456. doi: 10.1016/j.biocel.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 44.Jeon ES, Park WS, Lee MJ, et al. A Rho kinase/myocardin-related transcription factor-A-dependent mechanism underlies the sphingosylphosphorylcholine-induced differentiation of mesenchymal stem cells into contractile smooth muscle cells. Circ Res. 2008;103:635–642. doi: 10.1161/CIRCRESAHA.108.180885. [DOI] [PubMed] [Google Scholar]
- 45.Zhang R, Wang N, Zhang M, et al. Rho/MRTF-A-Induced Integrin Expression Regulates Angiogenesis in Differentiated Multipotent Mesenchymal Stem Cells. Stem Cells Int. 2015;2015:534758. doi: 10.1155/2015/534758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong Z, Hu JQ, Wu XD, et al. Myocardin-related transcription factor-A-overexpressing bone marrow stem cells protect cardiomyocytes and alleviate cardiac damage in a rat model of acute myocardial infarction. Int J Mol Med. 2015;36:753–759. doi: 10.3892/ijmm.2015.2261. [DOI] [PubMed] [Google Scholar]
- 47.Khan M, Mohsin S, Toko H, et al. Cardiac progenitor cells engineered with betaARKct have enhanced beta-adrenergic tolerance. Mol Ther. 2014;22:178–185. doi: 10.1038/mt.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cottage CT, Bailey B, Fischer KM, et al. Cardiac progenitor cell cycling stimulated by pim-1 kinase. Circ Res. 2010;106:891–901. doi: 10.1161/CIRCRESAHA.109.208629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson LA, Rodansky ES, Haak AJ, et al. Novel Rho/MRTF/SRF inhibitors block matrix-stiffness and TGF-beta-induced fibrogenesis in human colonic myofibroblasts. Inflamm Bowel Dis. 2014;20:154–165. doi: 10.1097/01.MIB.0000437615.98881.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fischer KM, Cottage CT, Konstandin MH, et al. Pim-1 kinase inhibits pathological injury by promoting cardioprotective signaling. J Mol Cell Cardiol. 2011;51:554–558. doi: 10.1016/j.yjmcc.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pulinilkunnil T, An D, Ghosh S, et al. Lysophosphatidic acid-mediated augmentation of cardiomyocyte lipoprotein lipase involves actin cytoskeleton reorganization. Am J Physiol Heart Circ Physiol. 2005;288:H2802–H2810. doi: 10.1152/ajpheart.01162.2004. [DOI] [PubMed] [Google Scholar]
- 52.Otani H, Yoshioka K, Nishikawa H, et al. Involvement of protein kinase C and RhoA in protease-activated receptor 1-mediated F-actin reorganization and cell growth in rat cardiomyocytes. J Pharmacol Sci. 2011;115:135–143. doi: 10.1254/jphs.10197FP. [DOI] [PubMed] [Google Scholar]
- 53.Soto AG, Smith TH, Chen B, et al. N-linked glycosylation of protease-activated receptor-1 at extracellular loop 2 regulates G-protein signaling bias. Proc Natl Acad Sci U S A. 2015;112:E3600–E3608. doi: 10.1073/pnas.1508838112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu X, Wang H, Hu P. Stem cell activation in skeletal muscle regeneration. Cell Mol Life Sci. 2015;72:1663–1677. doi: 10.1007/s00018-014-1819-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milasinovic D, Mohl W. Contemporary perspective on endogenous myocardial regeneration. World J Stem Cells. 2015;7:793–805. doi: 10.4252/wjsc.v7.i5.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chugh AR, Beache GM, Loughran JH, et al. Administration of cardiac stem cells in patients with ischemic cardiomyopathy: the SCIPIO trial: surgical aspects and interim analysis of myocardial function and viability by magnetic resonance. Circulation. 2012;126:S54–S64. doi: 10.1161/CIRCULATIONAHA.112.092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong KU, Bolli R. Cardiac stem cell therapy for cardiac repair. Curr Treat Options Cardiovasc Med. 2014;16:324. doi: 10.1007/s11936-014-0324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fabrizi C, Angelini F, Chimenti I, et al. Thrombin and thrombin-derived peptides promote proliferation of cardiac progenitor cells in the form of cardiospheres without affecting their differentiation potential. J Biol Regul Homeost Agents. 2011;25:S43–S51. [PubMed] [Google Scholar]
- 59.Konoplyannikov M, Haider KH, Lai VK, et al. Activation of diverse signaling pathways by ex-vivo delivery of multiple cytokines for myocardial repair. Stem Cells Dev. 2013;22:204–215. doi: 10.1089/scd.2011.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim HJ, Kim MH, Kim JT, et al. Intracellular transduction of TAT-Hsp27 fusion protein enhancing cell survival and regeneration capacity of cardiac stem cells in acute myocardial infarction. J Control Release. 2015;215:55–72. doi: 10.1016/j.jconrel.2015.07.026. [DOI] [PubMed] [Google Scholar]
- 61.Tufan H, Zhang XH, Haghshenas N, et al. Cardiac progenitor cells engineered with Pim-1 (CPCeP) develop cardiac phenotypic electrophysiological properties as they are co-cultured with neonatal myocytes. J Mol Cell Cardiol. 2012;53:695–706. doi: 10.1016/j.yjmcc.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steinberg SF. The molecular basis for distinct beta-adrenergic receptor subtype actions in cardiomyocytes. Circ Res. 1999;85:1101–1111. doi: 10.1161/01.res.85.11.1101. [DOI] [PubMed] [Google Scholar]
- 63.Khan M, Mohsin S, Avitabile D, et al. beta-Adrenergic regulation of cardiac progenitor cell death versus survival and proliferation. Circ Res. 2013;112:476–486. doi: 10.1161/CIRCRESAHA.112.280735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sadoshima J, Izumo S. Molecular characterization of angiotensin II--induced hypertrophy of cardiac myocytes, hyperplasia of cardiac fibroblasts Critical role of the AT1 receptor subtype. Circ Res. 1993;73:413–423. doi: 10.1161/01.res.73.3.413. [DOI] [PubMed] [Google Scholar]
- 65.Altarche-Xifro W, Curato C, Kaschina E, et al. Cardiac c-kit+AT2+ cell population is increased in response to ischemic injury and supports cardiomyocyte performance. Stem Cells. 2009;27:2488–2497. doi: 10.1002/stem.171. [DOI] [PubMed] [Google Scholar]
- 66.Hofmann U, Burkard N, Vogt C, et al. Protective effects of sphingosine-1-phosphate receptor agonist treatment after myocardial ischaemia-reperfusion. Cardiovasc Res. 2009;83:285–293. doi: 10.1093/cvr/cvp137. [DOI] [PubMed] [Google Scholar]
- 67.Calise S, Blescia S, Cencetti F, et al. Sphingosine 1-phosphate stimulates proliferation and migration of satellite cells: role of S1P receptors. Biochim Biophys Acta. 2012;1823:439–450. doi: 10.1016/j.bbamcr.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 68.Sassoli C, Frati A, Tani A, et al. Mesenchymal stromal cell secreted sphingosine 1-phosphate (S1P) exerts a stimulatory effect on skeletal myoblast proliferation. PLoS One. 2014;9:e108662. doi: 10.1371/journal.pone.0108662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fortier M, Figeac N, White RB, et al. Sphingosine-1-phosphate receptor 3 influences cell cycle progression in muscle satellite cells. Dev Biol. 2013;382:504–516. doi: 10.1016/j.ydbio.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Germinario E, Peron S, Toniolo L, et al. S1P2 receptor promotes mouse skeletal muscle regeneration. J Appl Physiol (1985) 2012;113:707–713. doi: 10.1152/japplphysiol.00300.2012. [DOI] [PubMed] [Google Scholar]
- 71.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 72.Parmacek MS. Myocardin-related transcription factors: critical coactivators regulating cardiovascular development and adaptation. Circ Res. 2007;100:633–644. doi: 10.1161/01.RES.0000259563.61091.e8. [DOI] [PubMed] [Google Scholar]
- 73.Du KL, Chen M, Li J, et al. Megakaryoblastic leukemia factor-1 transduces cytoskeletal signals and induces smooth muscle cell differentiation from undifferentiated embryonic stem cells. J Biol Chem. 2004;279:17578–17586. doi: 10.1074/jbc.M400961200. [DOI] [PubMed] [Google Scholar]
- 74.Mutoh T, Rivera R, Chun J. Insights into the pharmacological relevance of lysophospholipid receptors. Br J Pharmacol. 2012;165:829–844. doi: 10.1111/j.1476-5381.2011.01622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hanna M, Liu H, Amir J, et al. Mechanical regulation of the proangiogenic factor CCN1/CYR61 gene requires the combined activities of MRTF-A and CREB-binding protein histone acetyltransferase. J Biol Chem. 2009;284:23125–23136. doi: 10.1074/jbc.M109.019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao X, Ding EY, Yu OM, et al. Induction of the matricellular protein CCN1 through RhoA and MRTF-A contributes to ischemic cardioprotection. J Mol Cell Cardiol. 2014;75:152–161. doi: 10.1016/j.yjmcc.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.