Abstract
Follicle development is accompanied by proliferation of granulosa cells and increasing oocyte size. To obtain high-quality oocytes in vitro, it is important to understand the processes that occur in oocytes and granulosa cells during follicle development and the differences between in vivo and in vitro follicle development. In the present study, oocytes and granulosa cells were collected from early antral follicles (EAFs, 0.5–0.7 mm in diameter), small antral follicles (SAFs, 1–3 mm in diameter), large antral follicles (LAFs, 3–7 mm in diameter), and in vitro grown oocyte-and-granulosa cell complexes (OGCs), which were cultured for 14 days after collection from EAFs. Gene expression was analyzed comprehensively using the next-generation sequencing technology. We found top upstream regulators during the in vivo follicle development and compared them with those in in vitro developed OGCs. The comparison revealed that HIF1 is among the top regulators during both in vivo and in vitro development of OGCs. In addition, we found that HIF1-mediated upregulation of glycolysis in granulosa cells is important for the growth of OGCs, but the cellular metabolism differs between in vitro and in vivo grown OGCs. Furthermore, on the basis of comparison of upstream regulators between in vivo and in vitro development of OGCs, we believe that low expression levels of FLT1 (VEGFA receptor), SPP1, and PCSK6 can be considered causal factors of the suboptimal development under in vitro culture conditions.
Keywords: Early antral follicle, Granulosa cells, Oocytes
During development of an oocyte, its size rapidly increases from the early antral follicle (EAF) stage to the antral follicle (AF) stage; this process takes 2 weeks in pigs and cows. Follicle development is accompanied by a remarkable increase in the number of granulosa cells, such that EAFs contain only thousands of granulosa cells, whereas AFs contain at least one hundred thousand granulosa cells [1]. Granulosa cells support oocyte development in a paracrine and autocrine manner [2].
Because of structural limits, including the presence of theca layers, angiogenesis does not occur in the inner follicle milieu [3], and thus, the supply of energy and oxygen depends only on diffusion, which may make the inner follicular conditions hypoxic. The granulosa cell number is a key determinant of the follicular oxygen concentration [4], and in a mathematical model, the oxygen concentration in follicles was predicted to be low [5, 6]. In addition, the oxygen concentration in human follicular fluids was found to be 1.3–5.5% [7]. These results suggest that rapid proliferation of granulosa cells in follicles occurs during hypoxia.
Cellular proliferation requires energy, and the cellular energy depends on glycolysis for anaerobic production of ATP and pyruvate in the cytoplasm as well as on oxidative phosphorylation (OXPHOS) for aerobic production of ATP in mitochondria. Cells can vary the proportion of the methods of energy production such that some cancer cells switch the metabolic pathway to glycolysis due to a lack of angiogenesis [8]; this switch is a major factor that supports their proliferation [9]. In this context, however, it is unclear how the proliferation of granulosa cells during follicular development is supported.
Lately, immature follicles are considered useful genetic resources, and development of culture methods to induce immature oocytes to grow to the full size has been the goal of many researchers. Nonetheless, quality of oocytes grown in vitro and the efficiency of the culture system are still low. To shed light on the causal factors of the impaired oocyte development in vitro, there is a need for a comparison of gene expression patterns in granulosa cells and oocytes with the expression in their developing in vivo counterparts. In the present study, we showed the intrinsic changes in gene expression patterns of granulosa cells and oocytes during the development from EAFs to large AFs (LAFs) in swine. We also compared the gene expression patterns of oocytes grown in vitro and in the surrounding granulosa cells with such patterns in their counterparts that developed in vivo. The comparison revealed active glycolysis-mediated gene expression in granulosa cells during follicle development and identified the key factors that can improve development under in vitro culture conditions.
Materials and Methods
Reagents and media
All the reagents were purchased from Nacalai Tesque (Kyoto, Japan), unless otherwise stated. The medium used for the in vitro growth of oocyte-and-granulosa cell complexes (OGCs) was the α-minimum essential medium (αMEM; Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10 mM taurine, 1 μg/ml 17β-estradiol, 0.1 mAU/ml FSH (Kawasaki Mitaka, Tokyo, Japan), 2% polyvinylpyrrolidone-360K (Sigma-Aldrich), 2 mM hypoxanthine (Sigma-Aldrich), 1% insulin-transferrin-selenium (Gibco BRL, Grand Island, NY, USA), 3 mg/ml bovine serum albumin (Fraction V), and antibiotics (100 μg/ml streptomycin and 10 IU/ml penicillin).
Collection of ovaries
The porcine ovaries were collected from prepubertal gilts at a local abattoir and transported to the laboratory (at approximately 37ºC in phosphate-buffered saline [PBS] containing antibiotics) within 1 h.
Collection of OGCs from EAFs
The ovarian cortical tissues were excised from the ovarian surface, and OGCs were collected from EAFs (0.5–0.7 mm in diameter) under a dissection microscope. The OGCs were further cultured for 14 days or subjected to gene expression analysis. To prepare oocytes and granulosa cells for gene expression analysis, oocytes were removed from the OGCs with a narrow pulled pipet, and each oocyte and granulosa cell was subjected to massively parallel high-throughput RNA sequencing (RNA-Seq). A total of 200 oocytes and surrounding granulosa cells were prepared for the analysis.
Collection of cumulus cell-and-oocyte complexes (COCs) from small AFs (SAFs) and LAFs
Follicle contents were aspirated from SAFs (1–3 mm in diameter) and LAFs (3–5 mm in diameter) using a syringe connected to a 21-gauge needle. The same ovaries were used as those that we used for EAF collection. After aspiration, COCs were collected from the follicular contents, transferred to PBS containing 0.1% hyaluronidase, and vortexed for 5 min to obtain denuded oocytes and granulosa cells. A total of 200 oocytes and corresponding granulosa cells collected from each SAF and LAF were used for the gene expression analysis.
In vitro growth of the OGCs
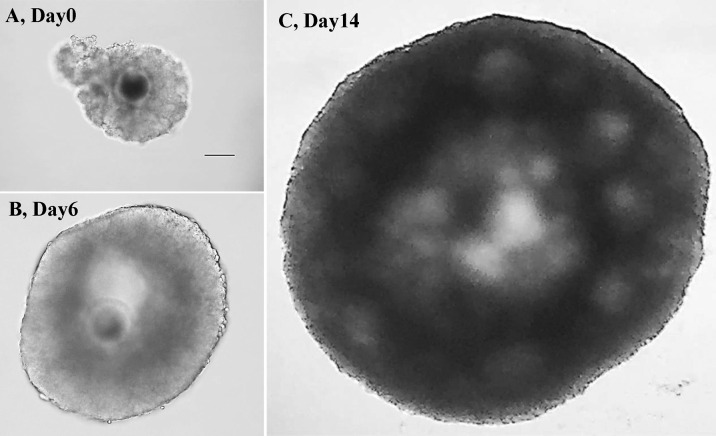
OGCs from EAFs (Fig. 1-A) were individually cultured in 200 µl of the culture medium in 96-well plates (Falcon 353072; BD Biosciences, Franklin Lakes, NJ, USA) for 14 days. A half of the medium was replaced with a fresh one at 4-day intervals. In vitro culture of OGCs was maintained at 38.5ºC in the atmosphere of 5% CO2 and 95% air. At the end of the culture period (14 days), OGCs with antrum cavities (Fig. 1-C) were further analyzed. The oocytes grown in vitro were removed from OGCs using narrow pulled pipettes, and the each oocyte and granulosa cell mass was subjected to the gene expression analysis. Two hundred oocytes and surrounding granulosa cells that developed in vitro were used for the gene expression analysis.
Fig. 1.
Representative pictures and images of oocyte-and-granulosa cell complexes (OGCs) cultured in vitro. OGCs were collected from early antral follicles (EAFs) (A) and cultured for 14 days. During the culture period, OGCs formed antrum like cavities and grew in size (panel B: Day 6 and panel C: Day 14). After 14 days, OGCs forming pseudo antrum cavities were selected (C). The scale bar is 100 µm.
Transcriptomic analysis
Total RNA was extracted using the RNAqueous kit (Life Technologies, Carlsbad, CA, USA) from oocytes or granulosa cells of EAFs, SAFs, LAFs, and in vitro grown OGCs (IVGs). A total of 20 gilts were used for collection of OGCs, and 200 oocytes were analyzed from each developmental stage. After an RNA quality check using a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), cDNA libraries were prepared using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). Quality and quantity of the libraries were examined using an Agilent 2100 Bioanalyzer and the KAPA Library Quantification Kit (KAPA Biosystems, Wilmington, MA, USA), respectively. From these libraries, clusters were generated on a cBot (Illumina), and two lanes for the eight groups were sequenced as 50-bp reads (single end) on a HiSeq 2500 (Illumina). Image analysis, base calling, and quality filtering were performed in the CASAVA software version 1.8.3 (Illumina) following the manufacturer’s instructions. The resulting sequence data were filtered to discard the adapter sequence, ambiguous nucleotides, and low-quality sequences. After that, the retained sequence data were aligned to the swine genome sequence (susScr3) to count the sequence reads. On the basis of the mapped sequence data, calculation of the expression value for each gene and statistical analysis of differentially expressed genes were performed. Filtering, mapping, and subsequent analysis were performed by means of the CLC genomics workbench (Qiagen, Redwood City, CA, USA). For prediction of upstream transcriptional regulators, genes that were significantly differentially expressed were analyzed using the Upstream Regulator function of Ingenuity Pathway Analysis (IPA, Qiagen). IPA determined how many known targets of each transcription regulator were present in the differentially-expressed-gene list, and calculated the overlapping P-value to measure the statistically significant overlap. Fisher’s exact test was used for the analysis enrichment of a gene set in the functional categories, and significance was generally assumed at P-values less than 0.01. IPA also compared the direction of the gene expression change to infer the activation status of the predicted transcriptional regulators. On the basis of the observed differential regulation of a gene in the differentially-expressed-gene set, the activation status of an upstream regulator was determined by the direction associated with the relation from the regulator to the gene. We then determined “Z-scores”, which determine whether an upstream transcription regulator has significantly more “activated” predictions than “inhibited” predictions (z > 0) or vice versa (z < 0). The data on the gene expression analysis were deposited in the DDBJ Sequence Read Archive (accession ID: DRA004449).
Quantitative reverse transcription real-time PCR
To validate the results of the next-generation sequencing (NGS) analysis, we conducted RT-PCR for FLT1, SPP1, HIF1, VEGF, and ACTN4 and compared relative expression levels among follicle developmental stages. On the basis of the NGS data, we found that the expression levels of almost all housekeeping genes (e.g., B-ACTIN, H2A family members, HPRT1, PGK1, GAPDH, and ATP5F1) changed significantly during follicular development. Thus, we selected GPX7 as the reference gene because of its high and constant expression level during follicular development from EAFs to LAFs. Fold changes between the different follicle stages were 1.001-fold for EAFs/SAFs (P = 0.88) and 0.845-fold for SAFs/LAFs (P = 0.35). Given that the expression level of GPX7 significantly differed between IVGs and SAFs and between LAFs and IVGs, and because we could not find other genes that showed high and constant expression levels at all stages (EAFs, SAFs, LAFs, and IVGs), we compared the relative expression levels of the genes in the EAFs, SAFs, and LAFs. Granulosa cells were collected from OGCs of 20 EAFs, SAFs, and LAFs as described above, and three samples were prepared from different ovary series. RNA was extracted from oocytes using an RNA isolation kit (RNAqueous-Micro, Ambion, Applied Biosystems, Austin, TX, USA). The extracted RNA was reverse-transcribed into cDNA using the Thermo script RT-PCR system (Invitrogen). The primer for the reverse transcription was the oligo(dT)20 included in the kit. Quantification of cDNA was then performed by real-time PCR using the Rotor-Gene 6500 system. The forward and reverse primers for FLT1, SPP1, HIF1, VEGF, ACTN4, and GPX7 were designed using DDBJ (http://arsa.ddbj.nig.ac.jp/top-j.html) and Primer3 (http://frodo.wi.mit.edu/primer3/). The primer sets were FLT1: 123-bp amplicon, CGTGGCTTCCAACAAAGTGG and GACAGCTTCAGGTCTTCCCC; SPP1: 138-bp amplicon, AGTCCAACGAAAGCCCTGAG and CGGAGTGATGAGACTCGTCG; HIF1: 144-bp amplicon, AGCCAGATGATCGTGCAACT and CCATTGATTGCCCCAGGAGT; VEGFA: 129-bp amplicon, TCGGAGCGGAGAAAGCATTT and CGGCTTGTCACATCTGCAAG; ACTN4: 132-bp amplicon, CGACCACTTGGCAGAGAAGT and TTGCGGATGAGGGCTTTGAT; and GPX7: 95-bp amplicon, GGGCGGAAAAGTCTAGCAGT and GGCTGGTGAATGACTGGTCA. Each PCR involved an initial denaturation step of 95°C for 1 min followed by 40 cycles of 98°C for 5 sec and 59°C for 11 sec. SYBR green fluorescence was measured at the end of each extension step (59°C). A melting curve was analyzed to check specificity of the PCR products, and agarose gel electrophoresis was carried out to verify the amplicon sizes. Relative gene expression levels of each gene of interest were calculated via normalization to the expression levels of the endogenous control: GPX7. The reactions were run in duplicate, and the experiments were repeated three times for each cellular series. For each PCR run, a standard curve was generated using serial 10-fold dilutions of the corresponding standard plasmid of a known concentration.
Statistical analysis
The ratios of the number of genes with significant up and downregulation associated with HIF1, glycolysis, and OXPHOS were compared using chi-squared tests. Comparison of the relative-gene-expression data obtained by RT-PCR between EAFs and SAFs and between SAFs and LAFs involved Student’s t-test. Differences with P-values less than 0.05 were considered statistically significant.
Results
Comparison of NGS data with those of real-time RT-PCR analysis
Relative expression levels of FLT1, SPP1, HIF1, VEGF, and ACTN4 as compared to the expression of GPX7 are shown in Table 1. We found that the expression patterns of the genes were almost identical between the RT-PCR data and NGS data.
Table 1. Comparison between NGS and RT-PCR.
| Feature ID | NGS | RT-PCR | ||
| Fold difference |
Fold difference |
|||
| SAFs/EAFs | LAFs/SAFs | SAFs/EAFs | LAFs/SAFs | |
| GPX7 | 1.00 | –1.11 | - | - |
| FLT1 | 6.45 * | –2.30 * | 6.27 * | –2.67 * |
| SPP1 | 7.66 * | –17.63 * | 21.85 * | –1.69 * |
| HIF1A | 1.54 * | 1.66 * | 1.68 * | 1.21 * |
| VEGFA | 7.03 * | 1.73 * | 3.38 * | 1.48 * |
| ACTN4 | –2.19 * | –1.15 * | –1.93 * | –1.42 * |
* Significant difference (P < 0.01). NGS; fold difference of RPKM value between SAFs and EAFs, and between LAFs and SAFs. RT-PCR; relative expression level to GPX7 was compared between SAFs and EAFs, and between LAFs and SAFs.
Top five upstream regulators in granulosa cells and oocytes during follicle development in vivo and in vitro
The gene expression of granulosa cells was compared between SAFs (in vivo grown OGCs from SAFs) and IVGs (in vitro grown OGCs) and between LAFs (in vivo grown OGCs from LAFs) and IVGs; 483 and 410 statistically significant upstream regulators were predicted using Ingenuity Upstream Regulator Analysis as implemented in IPA. The top five of these upstream regulators in granulosa cells are shown in Table 2. The comparison revealed that the common upstream regulators were TP53, HIF1A, and SP1. All of these factors are important for follicular development; thus, TP53, HIF1A, and SP1 are also predicted to be upstream regulators in both follicular developmental periods: from EAFs to SAFs and from SAFs to LAFs (Table 3). Table 4 shows the top five upstream regulators of in vitro and in vivo grown oocytes that were collected from SAFs and LAFs. HIF1A, SMAD4, and STAT3 are the top upstream regulators that are common for these datasets. These factors are also predicted to be key upstream regulators during the development from EAFs to SAFs, whereas only SMAD4 was predicted to be important during the development from SAFs to LAFs (Table 5).
Table 2. Top five upstream regulators in in vitro granulosa cells between SAFs and IVGs and between LAFs and IVGs.
| No. of best | SAFs - IVGs |
LAFs - IVGs |
||||||
| Upstream regulator |
Activation z-score |
P-value of overlap |
Predicted activation state |
Upstream regulator |
Activation z-score |
P-value of overlap |
Predicted activation state |
|
| 1 | SP1 | –1.56 | 3.26E-26 | SP1 | –0.47 | 9.92E-21 | ||
| 2 | TP53 | –4.02 | 4.80E-24 | Inhibited | TP53 | 0.18 | 1.39E-18 | |
| 3 | HIF1A | –1.24 | 2.76E-22 | CTNNNB1 | –1.68 | 5.08E-18 | ||
| 4 | SP3 | 0.46 | 7.96E-19 | HIF1A | –1.46 | 9.16E-14 | ||
| 5 | FOS | –0.01 | 1.40E-18 | AR | –0.85 | 2.01E-13 | ||
* The definitions of the terms (Activation z-score, P-value of overlap, and Predicted activation state) are described in the main text.
Table 3. Top five upstream regulators in granulosa cells during in vivo follicular development.
| No. of best | EAFs - SAFs |
SAFs - LAFs |
||||||
| Upstream regulator |
Activation z-score |
P-value of overlap |
Predicted activation state |
Upstream regulator |
Activation z-score |
P-value of overlap |
Predicted activation state |
|
| 1 | SP1 | –0.76 | 4.90E-22 | SP1 | –2.41 | 8.72E-22 | Inhibited | |
| 2 | TP53 | –2.94 | 2.49E-17 | Inhibited | CTNNB1 | –0.44 | 1.15E-19 | |
| 3 | HIF1A | 0.45 | 1.17E-14 | TP53 | –4.05 | 3.05E-19 | Inhibited | |
| 4 | NR3C1 | –1.48 | 2.83E-14 | SP3 | –0.27 | 2.54E-18 | ||
| 5 | STAT3 | 0.30 | 5.93E-14 | JUN | –1.64 | 4.03E-17 | ||
| others | HIF1A | –2.59 | 1.99E-09 | Inhibited | ||||
Table 4. Top five upstream regulators in oocytes between SAFs and IVGs and between LAFs and IVGs.
| No. of best | SAFs - IVGs |
LAFs - IVGs |
||||||
| Upstream regulator |
Activation z-score |
P-value of overlap |
Predicted activation state |
Upstream regulator |
Activation z-score |
P-value of overlap |
Predicted activation state |
|
| 1 | HIF1A | 1.02 | 2.58E-09 | HIF1A | 1.94 | 6.92E-08 | ||
| 2 | SMAD4 | 1.29 | 6.57E-08 | SMAD4 | 0.75 | 1.35E-06 | ||
| 3 | ELK4 | - | 2.10E-07 | TP53 | 3.59 | 2.02E-06 | activated | |
| 4 | STAT3 | 2.54 | 5.48E-07 | activated | STAT3 | 2.06 | 2.30E-06 | activated |
| 5 | VHL | –0.95 | 2.23E-06 | MAFG | –1.91 | 2.50E-06 | ||
Table 5. Upstream regulators including top five between EAFs and SAFs and between SAFs and LAFs.
| No. of best | EAFs - SAFs |
SAFs - LAFs |
||||||
| Upstream regulator |
Activation z-score |
P-value of overlap |
Predicted activation state |
Upstream regulator |
Activation z-score |
P-value of overlap |
Predicted activation state |
|
| 1 | TGFB1 | –1.36 | 3.01E-11 | Inhibited | ATF5 | –1.12 | 1.29E-05 | |
| 2 | TP53 | –3.12 | 8.38E-10 | Inhibited | WNT3A | –3.02 | 2.33E-05 | |
| 3 | ERBB2 | –4.84 | 2.29E-09 | Inhibited | 2-methoxyestradiol | - | 3.17E-05 | |
| 4 | SP1 | –1.61 | 5.52E-08 | Inhibited | retaspimycine | - | 7.59E-05 | |
| 5 | HIF1A | –1.19 | 6.07E-08 | Inhibited | leukotriene D4 | - | 1.18E-04 | |
| others | SMAD4 | –3.22 | 4.39E-06 | HIF1A | - | - | ||
| others | STAT3 | 1.36 | 1.36E-02 | Inhibited | SMAD4 | –1.80 | 3.29E-04 | |
| others | STAT3 | - | - | |||||
Expression of genes related to OXPHOS, glycolysis, and HIF1A
We found that HIF1 was commonly predicted as a top upstream factor in both granulosa cells and oocytes and as a key regulator of glycolysis and OXPHOS; therefore, we summarized the expression of genes related to glycolysis, OXPHOS, and HIF1 in Table 6. The genes were selected from the Kyoto Encyclopedia of Genes and Genomes pathway database of Sus scrofa (http://www.genome.jp/kegg/pathway.html) (Map No. 00010, 00190, and 04066, respectively). In addition, genes with certain expression levels (reads per kilobase of exon per million mapped sequence reads; RPKM > 1.0 during follicle development from EAFs to LAFs) were selected. The number of genes with significantly enhanced or reduced expression was counted for each period of follicle development (EAFs to SAFs, SAFs to LAFs, and SAFs to IVGs). In our preliminary experiments and previous reports involving the same culture methods [1, 10, 11], oocyte diameter (average 112 μm) and the ability to complete nuclear maturation (42%) were found to be similar to those in oocytes of SAFs. Accordingly, here, we compared gene expression between IVGs and SAFs.
Table 6. Rate of up- or down-regulated genes associated with glycolysis, OXPHOS, and HIF1 in granulosa cells of EAFs, SAFs, LAFs, and IVGs.
| Expression pattern | Follicle stages |
|||
| EAFs - SAFs | SAFs - LAFs | SAFs - IVGs | ||
| Glycolysis | Down | 5.6 (1/18) | 16.7 (3/18) | 11.1 (2/18) |
| Up | 83.3 (15/18) * | 72.2 (13/18) * | 83.3 (15/18) * | |
| OXPHOS | Down | 11.4 (8/70) | 31.4 (22/70) | 54.3 (38/70) |
| Up | 71.4 (50/70) * | 37.1 (26/70) | 24.3 (17/70) * | |
| HIF1 | Down | 34.1 (14/41) | 41.5 (17/41) | 26.8 (11/41) |
| Up | 53.7 (22/41) | 51.2 (21/41) | 53.7 (22/41) * | |
* Significant difference between number of up- and down-regulated genes. P < 0.05; (Chi-squared test).
The comparison of the number of differentially expressed genes in granulosa cells between EAFs and SAFs showed that 5.6% (1/18) of glycolysis-associated genes were significantly downregulated, whereas 83.3% (15/18) were upregulated, and the difference between the two values was significant (Chi-square test). This trend continued during the development from SAFs to LAFs (3/18 down versus 13/18 up) and was observed in the comparison between SAFs and IVGs (2/18 down versus 15/18 up). The expression level of HIF1A statistically significantly increased 1.5- and 1.7-fold respectively during both follicle development periods: from EAFs to SAFs and from SAFs to LAFs (Supplementary Table 1: online only). Certain HIF1-associated genes (34.1%, 14/41) were significantly downregulated, whereas 53.7% (22/41) were upregulated during the development from EAFs to SAFs; however, the difference was not statistically significant (P = 0.075, Table 6). In addition, expression of the genes associated with HIF1 was enhanced more strongly in IVGs than in SAFs (Table 6). The present analysis shows that 11.4% (8/70) of OXPHOS-associated genes were significantly downregulated, whereas 71.4% (50/70) were upregulated (Table 6). This trend diminished during the development from SAFs to LAFs (Table 6). In contrast to the in vivo development, expression levels of OXPHOS-associated genes were greater for IVGs than for SAFs.
In the case of oocytes, no obvious differences in expression of glycolysis-associated genes were observed between in vivo and in vitro grown oocytes (Table 7), whereas genes associated with HIF1 were significantly downregulated (EAFs–SAFs: down 64.7%, up 5.9%; SAFs–LAFs: down 38.2% and up 2.9%). In addition, the expression of the genes was low in IVGs as compared with oocytes in SAFs (down 32.4% and up 2.9%). Expression levels of OXPHOS-associated genes decreased during development from EAFs to SAFs (32/71 down, 18/71 up, P < 0.05) and increased during development from SAFs to LAFs (15/71 down, 27/71 up; Table 7).
Table 7. Rate of up- or down-regulated genes associated with glycolysis, OXPHOS, and HIF1 in oocytes of EAFs, SAFs, LAFs, and IVGs.
| Expression pattern | Follicle stages |
|||
| EAFs - SAFs | SAFs - LAFs | SAFs - IVGs | ||
| Glycolysis | Down | 46.7 (7/15) | 26.7 (4/15) | 20.0 (3/15) |
| Up | 26.7 (4/15) | 20.0 (3/15) | 33.3 (5/15) | |
| OXPHOS | Down | 45.1 (32/71) | 21.1 (15/71) | 21.1 (15/71) |
| Up | 25.4 (18/71) * | 38.0 (27/71) * | 33.8 (24/71) | |
| HIF1 | Down | 64.7 (22/34) | 38.2 (13/34) | 32.4 (11/34) |
| Up | 5.9 (2/34) * | 2.9 (1/34) * | 2.9 (1/34) * | |
* Significant difference between number of up- and down-regulated genes. P < 0.05; (Chi-squared test).
Comparison of upstream regulators in granulosa cells and oocytes between follicle development from EAFs to SAFs and from EAFs to IVGs
Table 8 shows the comparison of upstream regulators between in vivo and in vitro development of oocytes and granulosa cells of OGCs. According to comparison of granulosa cells between EAFs and SAFs (in vivo development), 16 regulators were significantly inhibited, nine of which were also identified as “inhibited” in the comparison between EAFs and IVGs (in vitro development). In addition, 12 regulators were predicted to be activated during in vivo development, and only three (ARNT, HDAC4, and TBX2) were identified as “activated” during in vitro development. In contrast, IRF5, IRF7, ETS2, and STAT2 were identified as “inhibited” between EAFs and IVGs (in vitro development, Table 8). To find out why for the factors were adversely affected in IVGs, expression of the genes associated with IRF5, IRF7, ETS2, and STAT2 (IRF5: CXCL10, IFIT1, IFIT2, IFIT3, IRF5, IRF7, ISG15, OAS1, OAS2, PMAIP1, and TNFSF10; IRF7: ATF4, CXCL10, DNAJA1, IFI35, IFIT1, IFIT2, IFIT3, IRF1, IRF7, IRF8, ISG15, MCL1, MX2, OAS1, OAS2, PARP14, PMAIP1, TNFSF10, and ZBP1; ETS2: CD34, CSF1R, FLT1, HPSE, MMP13, MMP3, MMP9, MSR1, MYC, PCSK6, PLAU, and PTHLH, SPP1; STAT2: BCL2, IFI35, IFIT1, IFIT2, IFIT3, IRF1, IRF5, IRF7, ISG15, OAS1, OAS2, and TNFSF10) was examined (Supplementary Table 1). We found that FLT1, SPP1, CD34, IFI35, OAS2, IFIT1, PCSK6, and ISG15 had a high expression level (RPKM > 1.0) at the EAF stage, which increased during development from EAFs to SAFs but significantly decreased during development from EAFs to IVGs.
Table 8. Comparison of up- and down-regulated upstream regulators between EAFs and SAFs and between EAFs and IVGs.
| Upstream regulator | EAFs - SAFs |
EAFs -IVGs |
||
| Predicted activation state | Activation z-score | Predicted activation state | Activation z-score | |
| IRF5 | Activated | 2.74 | Inhibited | –3.58 |
| SPI1 | Activated | 2.61 | 0.52 | |
| ARNT | Activated | 2.60 | Activated | 2.67 |
| HDAC4 | Activated | 2.41 | Activated | 2.62 |
| FHL2 | Activated | 2.40 | 0.75 | |
| TCF7L2 | Activated | 2.20 | 1.72 | |
| ETS2 | Activated | 2.16 | Inhibited | –2.36 |
| RUNX2 | Activated | 2.12 | 0.93 | |
| TBX2 | Activated | 2.11 | Activated | 2.50 |
| IRF7 | Activated | 2.06 | Inhibited | –5.14 |
| HDAC1 | Activated | 2.05 | 0.97 | |
| STAT2 | Activated | 2.04 | Inhibited | –2.43 |
| POU3F2 | Inhibited | –2.00 | - | |
| STAT4 | Inhibited | –2.07 | Inhibited | –2.78 |
| NEUROG1 | Inhibited | –2.13 | - | |
| BMI1 | Inhibited | –2.18 | –1.96 | |
| NEUROG3 | Inhibited | –2.18 | - | |
| NEUROD1 | Inhibited | –2.19 | - | |
| NUPR1 | Inhibited | –2.28 | Inhibited | –4.57 |
| ATF6 | Inhibited | –2.39 | –1.44 | |
| RXRB | Inhibited | –2.41 | - | |
| XBP1 | Inhibited | –2.49 | Inhibited | –2.13 |
| CREB1 | Inhibited | –2.56 | Inhibited | –3.19 |
| ATF4 | Inhibited | –2.64 | Inhibited | –2.79 |
| SMARCB1 | Inhibited | –2.69 | Inhibited | –4.53 |
| TP53 | Inhibited | –2.94 | Inhibited | –5.45 |
| NFE2L2 | Inhibited | –3.26 | Inhibited | –2.57 |
| CREM | Inhibited | –3.34 | Inhibited | –2.38 |
During follicular development from EAFs to SAFs, 54 upstream activators were detected in oocytes. Only two of them were found to be activated during the in vitro oocyte development, whereas none was identified as inhibited. In addition, 165 upstream suppressors were predicted during the in vivo oocyte development from EAFs to SAFs, 12 of which were identified as inhibited during in vitro OGC development, whereas none was identified as activated.
Discussion
The present analysis revealed several possible crucial regulators in granulosa cells and oocytes during in vivo follicle development and showed several casual factors of the poorer development of OGCs in vitro than in vivo.
The present comparison of gene expression patterns in granulosa cells and oocytes between in vivo and in vitro developing OGCs showed that TP53, SP1, and HIF1 and STAT3, SMAD4, and HIF1 are commonly predicted as top upstream regulators, respectively. The gene expression in bovine granulosa cells collected from three different phases of follicle development was examined previously, and TP53 and SP1 were shown to be the main upstream factors between the growth and plateau phases [12]. In the comparison of oocytes with expanded cumulus cells and those with compact cumulus cells, expression of STAT3 was detected in oocytes associated with high developmental competence [13]. Although it was reported that the deletion of SMAD4 using Gdf9-icre results in a slight but significant reduction in litter size in mice [14], the weaker contribution of this factor to bovine oocyte development was also reported [15]. Here, the expression level of SMAD4 was high during follicular development and gradually but significantly increased from EAFs to LAFs (RPKM values of EAFs, SAFs, and LAFs, are 6720, 6885, and 7473, respectively). The molecular significance of SMAD4 in follicle development still needs to be elucidated.
HIF1 is a basic helix-loop-helix transcription factor consisting of a heterodimeric complex of α (HIF1A) and β (ARNT) subunits. HIF1A is stable under hypoxic conditions but becomes unstable and is targeted by the ubiquitin-proteasome pathway in the presence of oxygen. HIF1 plays diverse roles in cellular adaptation for atmospheric conditions and changes the cellular metabolism mechanism from OXPHOS to glycolysis [9]. In addition, cellular adaptation to the culture milieu including atmospheric and energy substrates is a key factor for viability and development of cells. The present analysis showed that glycolysis-associated genes were upregulated in the granulosa cells during follicle development from EAFs to LAFs, suggesting that upregulation of glycolysis is a general trend for granulosa cells during development from EAFs to LAFs. Makanji et al. [16] reported that early-stage follicle development is supported by hypoxia-mediated glycolysis, and low-oxygen cultivation of follicles enhances the expression of genes associated with glycolytic enzymes (Pgk1, Hmox1, Hk2, Gpi, Pfkl, Pfkp, Aldoa, Gapdh, Pgam1, Eno1, Pkm, and Ldha). In the present comparison, of the 12 genes, we could not detect Aldoa, and the remaining 11 genes were significantly upregulated during development from EAFs to SAFs (Supplementary Table 1). In addition, HIF1 is known to activate the expression of GLUT 1 (SLC2A1) and GLUT3 (SLC2A3) in cancer cells [9]. Our results show that the expression level of SLC2A3 is very high in granulosa cells of SAFs (RPKM: 671.9), whereas expression levels of SLC2A1, GLUT 4_1 (SLC2A4_1), and GLUT 4_2 (SLC2A4_2) were low (RPKM: undetectable, 0.33, and 1.11, respectively; Supplementary Table 1). The expression levels significantly increased during development from EAFs to SAFs for GLUT 3, suggesting that high glucose uptake corresponds to high glycolytic activity. Hypoxia activates glycolysis [17], and the present gene analysis revealed that HIF1-associated genes tend to be upregulated during follicle development (P = 0.075). The granulosa cell number reportedly determines the concentration of oxygen in follicles [4]. Thus, we believe that during granulosa cell proliferation, dense follicular cells in SAFs may induce hypoxic conditions, which stabilize HIF1 expression and regulate downstream signaling, including that related to glycolysis. For example, VEGFA is a major downstream effector of HIF1A and participates in folliculogenesis [18, 19]. Our analysis showed that the expression levels of VEGFA increased 7.0- and 1.7-fold during the two follicle transition periods: from EAFs to SAFs and from SAFs to LAFs, respectively (Supplementary Table 1). Furthermore, Rico et al. reported that HIF1 activation is detectable for FSH-regulated VEGFA in mice [20]. Thus, FSH is also considered a causal factor of high glycolytic and hypoxic metabolism of granulosa cells. The present study showed that expression of genes associated with HIF1 was enhanced more strongly in IVGs than in SAFs (Table 6). Keeping in mind that there were no differences in the expression of HIF1-associated genes between SAFs and LAFs, we can hypothesize that granulosa cells cultured in vitro are more hypoxic than OGCs grown in vivo. Similarly, the expression of genes associated with glycolysis was significantly enhanced in IVGs as compared with SAFs (Table 6), and expression levels of VEGFA were significantly higher for IVGs than for SAFs (2.3-fold, P < 0.05). Therefore, enlargement of follicle size during angiogenesis in follicle development, well-orchestrated management of the granulosa cell number, oxygen provision, and the volume of the culture medium are crucial for OGC development if we consider the of granulosa cell proliferation.
The metabolic switch from OXPHOS to glycolysis is regulated by HIF1A [21], but the present analysis showed significant upregulation of OXPHOS-associated genes in granulosa cells during in vivo follicle development from EAFs to SAFs, and this trend diminished during the period SAFs–LAFs. The activation of both OXPHOS and glycolysis was unexpected, and the possible reason for this discrepancy is unclear. Nevertheless, the inner-follicle state of granulosa cells depends on the location [22], and some reports suggest that even cancer cells, which are believed to prefer glycolysis [23], show active oxidative phosphorylation [24]. In the present analysis, we could not determine the inner follicular location of OGCs. Thus, differential characteristics of granulosa cells are mixed, or active phosphorylation is also important for granulosa cells for production of ATP: these questions should be addressed in future studies. When the gene expression pattern of granulosa cells of IVGs was compared to that of SAFs, the genes associated with OXPHOS were found to significantly inactive (Table 6). Therefore, we believe that in vitro culture conditions lacking oxygen and energy substrate provision may hamper activation of OXPHOS.
In the case of oocytes, genes associated with HIF1 were significantly downregulated, and the expression of the genes was weak in IVGs in comparison with oocytes in SAFs (down 32.4% and up 2.9%). Therefore, inactivation of HIF1-associated genes seems to be a common feature of oocytes during follicle development. The activity of OXPHOS-related genes significantly decreased during follicle development from EAFs to SAFs but increased during development from SAFs to LAFs, pointing to changes in oocyte energy metabolism during follicle development.
Here, we compared upstream regulators between in vivo and in vitro development of oocytes and of granulosa cells of OGCs, and found that FLT1, SPP1, CD34, IFI35, OAS2IFIT1, PCSK6, and ISG15 have different expression patterns between the periods EAFs–SAFs and EAFs–IVGs. Thus, these factors can be considered key determinants of differences in the features of granulosa cells between in vivo and in vitro conditions. FLT1 (which belongs to the VEGF receptor family) reportedly improves follicle development [25, 26]. Given our finding that VEGFA expression increased during both periods (EAFs–SAFs and EAFs–IVGs: 7.0- and 2.3-fold, respectively), a decrease in the receptor level is a causal factor of impaired in vitro development of OGCs. Expression of SPP1 (osteopontin) increases during follicle development [27]. SPP1 increases interferon γ expression [28], and it is noteworthy that the above-mentioned genes that were adversely affected under in vitro conditions are interferon-associated genes. These genes were significantly downregulated during follicular development from EAFs to IVGs but upregulated in the period EAFs–SAFs. Lédée et al. [29] reported that interferon γ in follicular fluid is highly expressed for human embryos that cleaved early. These results as well as the previous report indicate that low expression levels of the genes related to interferons may be the major determinants of the poor in vitro development of OGCs. PCSK6, a member of the proprotein convertase family, is involved in inhibition of apoptosis of granulosa cells in preovulatory human follicles [30], and PCSK6-mutant mice show a loss of ovarian function [31]; these data are suggestive of low expression of the genes associated with impaired development of OGCs.
During follicular development from EAFs to SAFs, we identified a large number of upstream regulators in in vivo developing oocytes but observed a similar trend in a small fraction of the upstream factors during period EAFs–IVGs. The extent of differences between in vivo and in vitro conditions indicates that the conditions of oocytes grown in vitro differ strongly from those of in vivo grown oocytes.
In conclusion, follicle development from EAFs to AFs requires granulosa proliferation, which is likely supported by activation of the glycolysis pathway; cultivation of OGCs in vitro changes the pattern of gene expression (in comparison with those in in vivo developing OGCs).
Supplementary
Acknowledgments
This study was supported by the Promotion and Mutual Aid Corporation for Private Schools of Japan and the Ministry of Education, Culture, Sports, Science, and Technology (Grant-in-Aid for Scientific Research # S0801025) and by the Grant-in-Aid for Scientific Research C (KAKENHI, grant No. 25450400) from the Japan Society for the Promotion of Science.
References
- 1.Oi A, Tasaki H, Munakata Y, Shirasuna K, Kuwayama T, Iwata H. Effects of reaggregated granulosa cells and oocytes derived from early antral follicles on the properties of oocytes grown in vitro. J Reprod Dev 2015; 61: 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orisaka M, Tajima K, Tsang BK, Kotsuji F. Oocyte-granulosa-theca cell interactions during preantral follicular development. J Ovarian Res 2009; 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Wezel IL, Rodgers RJ. Morphological characterization of bovine primordial follicles and their environment in vivo. Biol Reprod 1996; 55: 1003–1011. [DOI] [PubMed] [Google Scholar]
- 4.Li D, Redding GP, Bronlund JE. Oxygen consumption by bovine granulosa cells with prediction of oxygen transport in preantral follicles. Reprod Fertil Dev 2013; 25: 1158–1164. [DOI] [PubMed] [Google Scholar]
- 5.Redding GP, Bronlund JE, Hart AL. Mathematical modelling of oxygen transport-limited follicle growth. Reproduction 2007; 133: 1095–1106. [DOI] [PubMed] [Google Scholar]
- 6.Redding GP, Bronlund JE, Hart AL. Theoretical investigation into the dissolved oxygen levels in follicular fluid of the developing human follicle using mathematical modelling. Reprod Fertil Dev 2008; 20: 408–417. [DOI] [PubMed] [Google Scholar]
- 7.Van Blerkom J, Antczak M, Schrader R. The developmental potential of the human oocyte is related to the dissolved oxygen content of follicular fluid: association with vascular endothelial growth factor levels and perifollicular blood flow characteristics. Hum Reprod 1997; 12: 1047–1055. [DOI] [PubMed] [Google Scholar]
- 8.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell 2008; 13: 472–482. [DOI] [PubMed] [Google Scholar]
- 9.Courtnay R, Ngo DC, Malik N, Ververis K, Tortorella SM, Karagiannis TC. Cancer metabolism and the Warburg effect: the role of HIF-1 and PI3K. Mol Biol Rep 2015; 42: 841–851. [DOI] [PubMed] [Google Scholar]
- 10.Tasaki H, Iwata H, Sato D, Monji Y, Kuwayama T. Estradiol has a major role in antrum formation of porcine preantral follicles cultured in vitro. Theriogenology 2013; 79: 809–814. [DOI] [PubMed] [Google Scholar]
- 11.Itami N, Shirasuna K, Kuwayama T, Iwata H. Resveratrol improves the quality of pig oocytes derived from early antral follicles through sirtuin 1 activation. Theriogenology 2015; 83: 1360–1367. [DOI] [PubMed] [Google Scholar]
- 12.Douville G, Sirard MA. Changes in granulosa cells gene expression associated with growth, plateau and atretic phases in medium bovine follicles. J Ovarian Res 2014; 7: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammadi-Sangcheshmeh A, Held E, Rings F, Ghanem N, Salilew-Wondim D, Tesfaye D, Sieme H, Schellander K, Hoelker M. Developmental competence of equine oocytes: impacts of zona pellucida birefringence and maternally derived transcript expression. Reprod Fertil Dev 2014; 26: 441–452. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Tripurani SK, James R, Pangas SA. Minimal fertility defects in mice deficient in oocyte-expressed Smad4. Biol Reprod 2012; 86: 1–6. [DOI] [PubMed] [Google Scholar]
- 15.Kaune H, Peyrache E, Williams SA. Oocyte-derived Smad4 is not required for development of the oocyte or the preimplantation embryo. Theriogenology 2015; 83: 897–903. [DOI] [PubMed] [Google Scholar]
- 16.Makanji Y, Tagler D, Pahnke J, Shea LD, Woodruff TK. Hypoxia-mediated carbohydrate metabolism and transport promote early-stage murine follicle growth and survival. Am J Physiol Endocrinol Metab 2014; 306: E893–E903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yee Koh M, Spivak-Kroizman TR, Powis G. HIF-1 regulation: not so easy come, easy go. Trends Biochem Sci 2008; 33: 526–534. [DOI] [PubMed] [Google Scholar]
- 18.Grasselli F, Basini G, Bussolati S, Tamanini C. Effects of VEGF and bFGF on proliferation and production of steroids and nitric oxide in porcine granulosa cells. Reprod Domest Anim 2002; 37: 362–368. [DOI] [PubMed] [Google Scholar]
- 19.Doyle LK, Walker CA, Donadeu FX. VEGF modulates the effects of gonadotropins in granulosa cells. Domest Anim Endocrinol 2010; 38: 127–137. [DOI] [PubMed] [Google Scholar]
- 20.Rico C, Dodelet-Devillers A, Paquet M, Tsoi M, Lapointe E, Carmeliet P, Boerboom D. HIF1 activity in granulosa cells is required for FSH-regulated Vegfa expression and follicle survival in mice. Biol Reprod 2014; 90: 135. [DOI] [PubMed] [Google Scholar]
- 21.Chen CT, Hsu SH, Wei YH. Mitochondrial bioenergetic function and metabolic plasticity in stem cell differentiation and cellular reprogramming. Biochim Biophys Acta 2012; 1820: 571–576. [DOI] [PubMed] [Google Scholar]
- 22.Russo V, Berardinelli P, Martelli A, Di Giacinto O, Nardinocchi D, Fantasia D, Barboni B. Expression of telomerase reverse transcriptase subunit (TERT) and telomere sizing in pig ovarian follicles. J Histochem Cytochem 2006; 54: 443–455. [DOI] [PubMed] [Google Scholar]
- 23.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov 2012; 2: 881–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez-Enríquez S, Hernández-Esquivel L, Marín-Hernández A, El Hafidi M, Gallardo-Pérez JC, Hernández-Reséndiz I, Rodríguez-Zavala JS, Pacheco-Velázquez SC, Moreno-Sánchez R. Mitochondrial free fatty acid β-oxidation supports oxidative phosphorylation and proliferation in cancer cells. Int J Biochem Cell Biol 2015; 65: 209–221. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu T, Jiang JY, Iijima K, Miyabayashi K, Ogawa Y, Sasada H, Sato E. Induction of follicular development by direct single injection of vascular endothelial growth factor gene fragments into the ovary of miniature gilts. Biol Reprod 2003; 69: 1388–1393. [DOI] [PubMed] [Google Scholar]
- 26.Celik-Ozenci C, Akkoyunlu G, Kayisli UA, Arici A, Demir R. Localization of vascular endothelial growth factor in the zona pellucida of developing ovarian follicles in the rat: a possible role in destiny of follicles. Histochem Cell Biol 2003; 120: 383–390. [DOI] [PubMed] [Google Scholar]
- 27.Skinner MK, Schmidt M, Savenkova MI, Sadler-Riggleman I, Nilsson EE. Regulation of granulosa and theca cell transcriptomes during ovarian antral follicle development. Mol Reprod Dev 2008; 75: 1457–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan M, Yuan X, Song LZ, Roberts L, Zarinkamar N, Seryshev A, Zhang Y, Hilsenbeck S, Chang SH, Dong C, Corry DB, Kheradmand F. Cigarette smoke induction of osteopontin (SPP1) mediates T(H)17 inflammation in human and experimental emphysema. Sci Transl Med 2012; 4: 117ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lédée N, Lombroso R, Lombardelli L, Selva J, Dubanchet S, Chaouat G, Frankenne F, Foidart JM, Maggi E, Romagnani S, Ville Y, Piccinni MP. Cytokines and chemokines in follicular fluids and potential of the corresponding embryo: the role of granulocyte colony-stimulating factor. Hum Reprod 2008; 23: 2001–2009. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Wang XH, Fan DX, Zhang Y, Li MQ, Wu HX, Jin LP. PCSK6 regulated by LH inhibits the apoptosis of human granulosa cells via activin A and TGFβ2. J Endocrinol 2014; 222: 151–160. [DOI] [PubMed] [Google Scholar]
- 31.Mujoomdar ML, Hogan LM, Parlow AF, Nachtigal MW. Pcsk6 mutant mice exhibit progressive loss of ovarian function, altered gene expression, and formation of ovarian pathology. Reproduction 2011; 141: 343–355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.