Abstract
Kinases are therapeutically actionable targets. Kinase inhibitors targeting vascular endothelial growth factor receptors (VEGFR) and mammalian target of rapamycin (mTOR) improve outcomes in metastatic clear cell renal cell carcinoma (ccRCC), but are not curative. Metastatic tumor tissue has not been comprehensively studied for kinase gene expression. Paired intra-patient kinase gene expression analysis in primary tumor (T), matched normal kidney (N) and metastatic tumor tissue (M) may assist in identifying drivers of metastasis and prioritizing therapeutic targets. We compared the expression of 519 kinase genes using NanoString in T, N and M in 35 patients to discover genes over-expressed in M compared to T and N tissue. RNA-seq data derived from ccRCC tumors in The Cancer Genome Atlas (TCGA) were used to demonstrate differential expression of genes in primary tumor tissue from patients that had metastasis at baseline (n = 79) compared to those that did not develop metastasis for at least 2 years (n = 187). Functional analysis was conducted to identify key signaling pathways by using Ingenuity Pathway Analysis. Of 10 kinase genes overexpressed in metastases compared to primary tumor in the discovery cohort, 9 genes were also differentially expressed in TCGA primary tumors with metastasis at baseline compared to primary tumors without metastasis for at least 2 years: EPHB2, AURKA, GSG2, IKBKE, MELK, CSK, CHEK2, CDC7 and MAP3K8; p<0.001). The top pathways overexpressed in M tissue were pyridoxal 5'-phosphate salvage, salvage pathways of pyrimidine ribonucleotides, NF-kB signaling, NGF signaling and cell cycle control of chromosomal replication. The 9 kinase genes validated to be over-expressed in metastatic ccRCC may represent currently unrecognized but potentially actionable therapeutic targets that warrant functional validation.
Background
Clear cell renal cell carcinoma (ccRCC) accounts for approximately 70–80% of kidney cancers [1]. At the time of diagnosis, 30% of RCC patients have metastatic lesions, and 30–50% will develop metastases during follow up after definitive surgery for localized disease. Metastatic ccRCC is generally incurable and long-term survival is observed in less than 10% of patients. While high-dose interleukin (IL)-2 remains an option due to durable complete remissions in ~7% of patients, toxicities preclude its administration in the vast majority of patients [2, 3]. Protein kinases are central-signaling molecules that affect proliferation, differentiation, motility, cell death and survival [4]. Several kinase inhibitors, including vascular endothelial growth factor receptor (VEGF) receptor tyrosine kinase inhibitors (TKIs), sunitinib, sorafenib, axitinib, pazopanib and cabozantinib, and mammalian target of rapamycin (mTOR) inhibitors, temsirolimus and everolimus, improve outcomes in metastatic RCC [5–11]. More recently, nivolumab, a programmed death (PD)-1 inhibitor immunotherapeutic, cabozantinib, a multitargeted kinase inhibitor, and the combination of lenvatinib plus everolimus, improved outcomes in patients progressing after treatment with VEGF inhibitors [11–13]. However, these agents are not curative, and intrinsic or acquired resistance result in a median overall survival of 2 to 3 years. Hence, a better understanding of tumor biology and new drug targets are critical to further advance ccRCC treatments.
Molecular analyses of tumor tissue have been used to address these needs. Comprehensive molecular profiling of many cancers, including ccRCC, has employed primary tumor tissue, as typified by The Cancer Genome Atlas (TCGA) Project and other smaller studies [14–16]. The tumor suppressor genes, SETD2, BAP1, and PBRM1 have been shown to be altered in addition to the VHL gene, although their therapeutic significance is unknown [17]. Although the PI3K pathway was recurrently mutated, the utility of inhibitors of this pathway heretofore has been modest [18, 19]. Few genomics studies of small numbers of patients have analyzed exome sequencing of metastatic ccRCC tumor tissue, but these have not focused on actionable molecules or studied gene expression [20, 21]. Kinase targets are particularly relevant because their inhibitors are both already approved for use in multiple malignancies and can be readily designed, facilitating therapeutic targeting[22]. In this study, we examined differential expression of kinase genes in matched and paired primary tumor tissue (T), adjacent normal kidney (N) and metastatic tumor tissue (M) from 35 ccRCC patients. For this work, we took advantage of the low quantity of nucleic acid material required by the NanoString nCounter platform to quantify gene expression from widely available formalin-fixed paraffin embedded (FFPE) tissue [23–27]. Furthermore, we explored evidence for the importance of these genes in the independent dataset from TCGA [14].
Materials and Methods
Patient and tumor selection
Treatment-naïve patients who underwent nephrectomy (total or partial) at UAB for ccRCC between years 2000 to 2013 were identified for the discovery dataset. Medical records were abstracted and clinical data including age at diagnosis, stage, tumor grade and metastatic tissue site were recorded. De-identified patients with available FFPE tissue from the metastatic tumor (M), primary tumor (T) and adjacent normal (N) kidney tissue were selected. Patients with bone as the metastatic tumor tissue site were excluded as demineralization during processing affects measurement of gene expression. The study was approved by the UAB Institutional Review Board (IRB).
Tumor dissection and RNA extraction
The tissue underwent central pathology review by a urologic pathologist. Tumor with predominantly clear cell component was demarcated for histologic macrodissection performed on 10 μm sections. RNA was isolated from dissected primary and metastatic tumor and normal kidney tissue using RNeasy FFPE kit (Qiagen, Valencia, CA). A tissue surface area of approximately 100 mm2 is adequate to harvest the necessary amount of RNA (~100 ng). RNA integrity was assessed via the 260/280 ratio using nanodrop.
NanoString platform for gene expression assay
RNA is input directly into the nCounterTM platform (NanoString Technologies, Seattle, WA) for the hybridization reaction containing color-coded molecular barcodes representing the gene 519 kinase genes and 8 internal reference genes (S1 Table) [23, 28]. A codeset specific to a 100-base region of the target mRNA was custom designed by NanoString Technologies using a 3’ biotinylated capture probe and a 5’ reporter probe tagged with a specific fluorescent barcode, creating two sequence-specific probes for each target transcript. Probes were hybridized to 100ng of total RNA for 19 hours at 65°C and then applied to the nCounterTM preparation station for automated removal of excess probe by immobilization of probe-transcript complexes on a streptavidin-coated cartridge.
Data processing
Data were collected using the nCounterTM Digital Analyzer by counting the individual barcodes.
Each codeset included probes for the 519 kinase genes, spiked-in external RNA consortium positive and negative controls, and 8 reference housekeeping genes. Background hybridization was determined using spiked-in negative controls. All signals below mean background plus 2 standard deviations were considered to be below the limits of detection, and set to mean background. A normalization factor was calculated from the spiked-in exogenous positive controls in each sample and applied to the raw counts from the nCounterTM output data.
Statistical and pathway analysis
The mean signals from N, T and M tissue were used to calculate change in gene expression, and a p-value by t-test <0.05 was considered significant. Change in mean intensity between tumor and metastatic site was used to derive top 10 kinases overexpressed in metastases compared to primary tumors. Functional analysis was conducted to identify key signaling pathways by using Ingenuity Pathway Analysis. The genes that had statistically significant (p<0.05) overexpression in the metastatic site vs primary tumor site were used for pathways analysis. Genes that were also significantly overexpressed in T vs. N tissue were eliminated in order to isolate kinases over-expressed in metastases only.
Heatmap generation
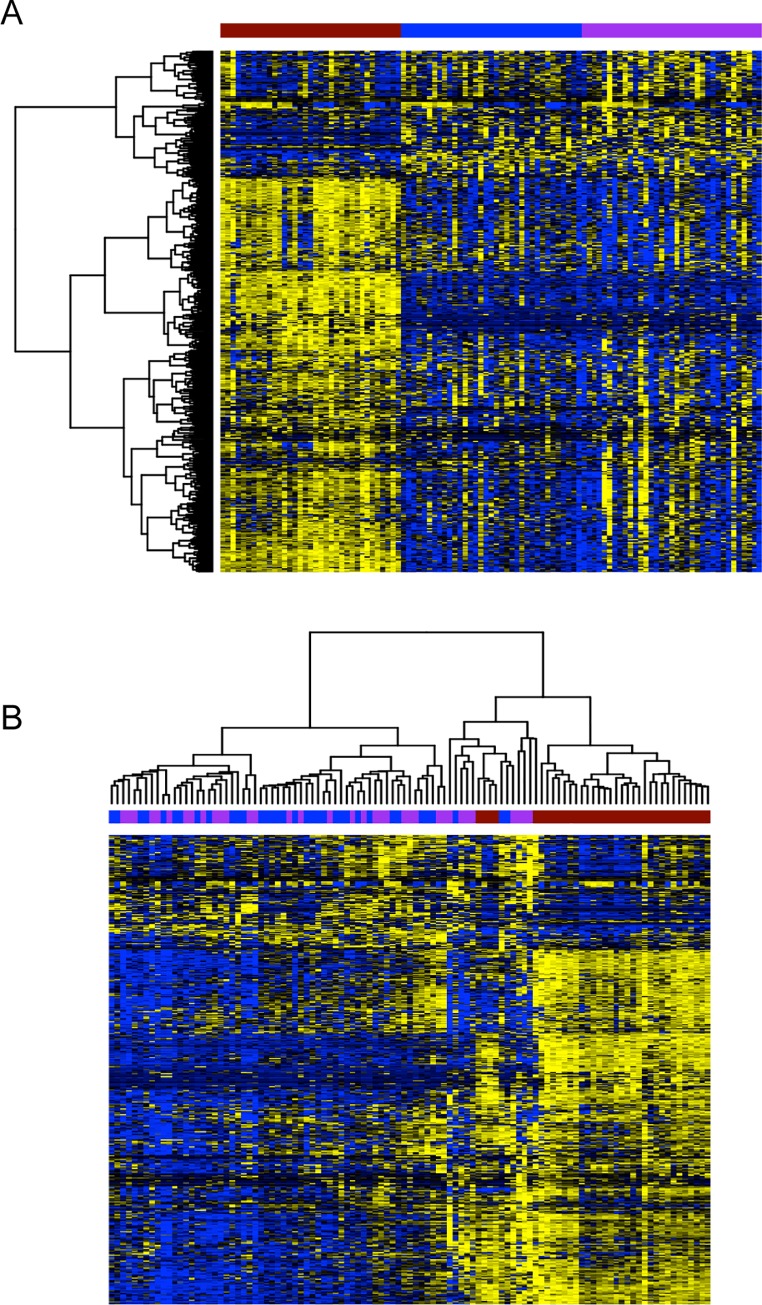
Fig 1 was generated in R (version 3.2.1) with RStudio (version 0.99.467, http://www.rstudio.com/). All genes with a normalized standard deviation less than 2 were removed and data was scaled using the scale() function before plotting. Heatmaps were generated with the heatmap.2 function with method = “ward.D2” from the gplots (version 2.17.0) package.
Fig 1.
Clustering of kinase genes showing A) normal tissue on the left (N, red), tumor in the middle (T, blue) and metastatic tumor tissue on the right (M, purple), and B) hierarchical clustering of all normal (N, red), tumor (T, blue), and metastatic (M, purple) tissues. Figure legend: X-axis represents the tissue sample, Y-axis represents the kinases. Yellow: up regulated kinases, Blue: down regulated kinases.
External validation in the TCGA dataset
For external validation, we downloaded the TCGA RNA-seq (UNC IlluminaHiSeq_RNASeqV2) dataset Level 3 results for all ccRCC patients on April 30, 2015 from http://cancergenome.nih.gov/. Primary tumors were defined as non-metastatic (M0) or metastatic (M1) based on stage at the time of entry to TCGA. Patients who had metastasis at baseline were compared with those that did not develop metastasis for at least 2 years for our primary external validation analysis. Secondarily, we also performed the following 2 comparisons: 1) those who developed metastasis vs. those who did not develop metastasis for at least 2 years, and 2) those who had no metastasis at baseline vs. those that did.
Analysis was conducted in R (version 3.2.1) with RStudio (version 0.99.467, http://www.rstudio.com/). Gene expression changes were examined by the R package DESeq2 (version 1.6.3), a method for differential analysis of sequence read count data [29]. All analysis used default settings, but employed likelihood ratio test (LRT) hypothesis testing, and removed non-convergent transcripts from subsequent analysis. False discovery rates (FDR) were estimated using the Benjamini-Hochberg (BH) algorithm[30].
Results
Patient population
For our discovery study, we identified 35 patients with adequate metastatic ccRCC tumor tissue (M), primary tumor (T) and adjacent normal (N) kidney. Adequate tissue was defined as a minimum of 100 mm2 of tumor in sections of 10 μM thickness. Clinical characteristics, including age at diagnosis, patient sex, patient race, nephrectomy type, clinical stage at diagnosis, Fuhrman grade, histology, and metastatic organ sites for this cohort are detailed in Table 1. The most common metastatic tumor sites were lungs and soft tissue/nodal followed by other visceral sites.
Table 1. Patient and tumor characteristics.
| Characteristic | No. of patients (%) |
|---|---|
| • Age at diagnosis, yearsMedianRange | • 5636–77 |
| Sex | • 30 (86%)5 (14%) |
| MaleFemale | |
| • RaceCaucasianAfrican AmericanHispanicOther | • 28 (80%)1 (2.8%)1 (2.8%)5 (14.2%) |
| • Type of nephrectomyComplete Partial | • 31 (88.5%)4 (11.4%) |
| • Clinical stage at diagnosis1234 | • 3 (8.6%)6 (17%)11 (31.4%)15 (42.8%) |
| • Fuhrman grade1234 | • 1 (2.8%)14 (40%)15 (42.8%)5 (14.3%) |
| • HistologyClear cell onlyClear cell with rhabdoidClear cell with sarcomatoidClear cell with papillaryClear cell with granular | • 29 (82.8%)1 (2.8%)3 (8.6%)1 (2.8%)1 (2.8%) |
| • Metastatic organ sites for studyLungs Soft tissue or lymph nodeAdrenal glandBrainPancreasLiver | • 13 (37.1%)10 (28.6%)5 (14.2%)4 (11.4%)2 (5.6%)1 (2.8%) |
Kinase gene expression in metastatic tumor and pathway analysis
Unsupervised and supervised clustering of the 519 kinase genes (S2 Table) showed increased gene expression of several kinase genes in M tumors compared to T tumors and N adjacent renal tissue (Fig 1A and 1B). The top 10 kinase genes over-expressed in M compared to T and N tissue (Table 2) were EPHB2, AURKA, GUCY2C, GSG2, IKBKE, MELK, CSK, CHEK2, CDC7 and MAP3K8 (p<0.05). A total of 33 genes exhibited >1.7 fold increased expression in M vs. T or N in at least a third of the 35 (n≥12) patients (Table 3). Using Ingenuity Pathway Analysis, the top pathways identified based on the kinases overexpressed in M compared to N or T tissue were pyridoxal 5'-phosphate salvage, salvage pathways of pyrimidine ribonucleotides, NF-kB signaling, NGF signaling, and cell cycle control of chromosomal replication (Table 4).
Table 2. Top kinase genes overexpressed in all metastases compared to all primary ccRCC tumors in 35 patients from the discovery dataset.
| Gene name | Metastatic tumor mean | Primary tumor mean | Fold change | P VALUE (Metastatic vs primary tumor) |
|---|---|---|---|---|
| EPHB2 | 15.06 | 5.63 | 2.67 | 0.011 |
| AURKA | 24.79 | 14.11 | 1.75 | 0.000 |
| GUCY2C* | 2.22 | 1.27 | 1.75 | 0.023 |
| GSG2 | 3.75 | 2.34 | 1.60 | 0.006 |
| IKBKE | 66.02 | 41.29 | 1.59 | 0.025 |
| MELK | 57.23 | 35.94 | 1.59 | 0.005 |
| CSK | 382.90 | 241.12 | 1.59 | 0.031 |
| CHEK2 | 47.63 | 30.43 | 1.56 | 0.001 |
| CDC7 | 19.58 | 12.51 | 1.56 | 0.031 |
| MAP3K8 | 144.50 | 95.63 | 1.51 | 0.011 |
Table 3. Top kinase genes overexpressed >1.7 fold in metastases in > a third of patients in all metastases compared to all primary ccRCC tumors in 35 patients from the discovery dataset.
| No. of patients (%) | Genes |
|---|---|
| 18 (51.4%) | ADCK1, DYRK4, PDGFRA, PRKCA, ROR2, TYRO3 |
| 17 (48.5%) | TEC, PLK4, EPHB3 |
| 16 (45.7%) | PLK1, NEK2, EPHB2, EPHA3, CKD1 |
| 15 (42.8%) | AURKA, ROS1, PHMYT1, PASK, MAPK11, Kit, BUB1B |
| 14 (40%) | IRAK3, CHEK1, AATK |
| 13 (37.1%) | PRKCE, CAMK4, BUB1 |
| 12 (34.2%) | PRKCQ, MAPK12, KALRN, IKBKE, FES, CDC7, BTK, BLK |
Table 4. Top signaling pathways overexpressed in all metastases compared to all primary ccRCC tumors in 35 patients from the discovery dataset.
| Ingenuity Canonical Pathways | Kinases |
|---|---|
| Pyridoxical 5’-Phosphate Salvage Pathway | EIF2AK2, IRAK1, MAP3K8, LIMK1 |
| Salvage Pathways of Pyrimidine Ribonucleotides | EIF2AK2, IRAK1, MAP3K8, LIMK1 |
| NK-kB signaling | EIF2AK2, IRAK1, MAP3K8, INSR |
| NGF signaling | MAP3K8, TRIO, IKBKE |
| Cell cycle control of chromosomal replication | CDC7, CHEK2 |
Kinase gene overexpression in primary tumors from patients in the TCGA dataset with metastatic vs. non-metastatic ccRCC
To examine our findings in an independent cohort, we analyzed RNA-seq data from the TCGA ccRCC cohort, which includes kinase gene expression data in both primary tumor (n = 497) and adjacent benign (n = 72) kidney tissue. First we compared primary tumor kinase gene expression in patients with metastasis at baseline (n = 79) to patients without metastasis for at least 2 years (n = 187). Nine of the 10 significant kinase genes identified in our discovery study were significantly overexpressed in the primary tumors of patients with metastasis at baseline compared to primary tumors in patients without metastases for at least two years (adjusted p<0.001). Although GUCY2C was not significantly differentially expressed between these groups, a closer examination reveals low levels of expression in both the discovery and validation cohorts, which may explain our inability to validate it in the second cohort (Table 5). When comparing patients with localized disease at baseline who subsequently developed metastasis (n = 28) to patients who did not develop metastasis for at least 2 years (n = 187), 8 of the 10 kinase genes were significantly over-expressed in primary tumors from metastatic patients (adjusted p <0.05), with CSK and MAP3k8 being the exceptions. When comparing primary tumors in patients with no metastasis at baseline (n = 418) to primary tumors with metastasis at baseline (n = 79), again, with the exception of GUCY2C, the other 9 kinase genes were significantly overexpressed (adjusted p<0.05). These 9 kinases were examined for alterations in DNA sequence or copy number and no significant alterations were identified (data not shown). Alterations at the protein level of these kinases could not be examined since they were not analyzed by TCGA.
Table 5. Expression of kinase genes in primary tumors from metastatic vs. non-metastatic patients in the validation dataset*.
| Genes | baseMean | p-value | adjusted p-value |
|---|---|---|---|
| MELK.9833 | 199.5619451 | 8.74E-18 | 2.80E-15 |
| GSG2.83903 | 46.98529174 | 1.87E-15 | 3.30E-13 |
| CHEK2.11200 | 307.6903398 | 3.99E-11 | 2.87E-09 |
| AURKA.6790 | 353.7879497 | 3.39E-10 | 1.85E-08 |
| IKBKE.9641 | 496.7497848 | 3.98E-10 | 2.12E-08 |
| EPHB2.2048 | 389.0931124 | 1.69E-09 | 7.51E-08 |
| CDC7.8317 | 251.725834 | 2.29E-05 | 0.000202497 |
| MAP3K8.1326 | 480.1769944 | 0.000472431 | 0.002394953 |
| CSK.1445 | 3162.71356 | 0.001042731 | 0.004538433 |
| GUCY2C.2984 | 7.891334454 | 0.691714419 | 0.772671281 |
*comparing expression of top 10 discovery dataset kinase genes in TCGA patients who had metastasis at baseline (n = 79) vs. those who did not develop metastasis for at least 2 years (n = 187); baseMean (DESeq2 generated normalized mean count); *GUCY2C was the only kinase gene that was not externally validated in the TCGA dataset, but GUCY2C.2984 had a very low mean count in the TCGA dataset (8.6), as well as in the discovery dataset (2.22).
We also evaluated the TCGA dataset for gene expression changes of the 33 kinase genes with >1.7 fold increased expression in M vs. T in at least a third of patients in our discovery dataset. We found when comparing primary tumors in patients with metastasis at baseline (n = 79) to primary tumors in patients who did not develop metastasis for at least 2 years (n = 187), 21 of 33 kinase genes were differentially expressed (adjusted p<0.05) (S3 Table). When comparing primary tumors who developed metastasis (n = 28) vs. those who did not develop metastasis for at least 2 years (n = 187) (S4 Table), or when comparing those who had no metastasis at baseline (n = 418) vs. those that did (n = 79), 18 and 19, respectively, of the 33 kinase genes were significantly over-expressed in metastatic patients (adjusted p<0.05) (S5 Table).
Discussion
Molecular profiling of metastatic tumor tissue may be critical for making further advances in the therapy of metastatic ccRCC. Given that most studies have focused on profiling primary renal tumors and because kinases may be therapeutically actionable, we analyzed FFPE tissue from the intra-patient primary tumor, benign adjacent normal kidney and metastases to explore whether kinase genes are differentially expressed. Of the 10 kinase genes overexpressed in metastases compared to primary tumor in the discovery cohort, we demonstrated 9 genes were also differentially expressed in TCGA primary tumors with metastasis at baseline (n = 79) compared to primary tumors without metastasis for at least 2 years (n = 187). These 9 kinases are not currently readily recognized as therapeutic targets in ccRCC and include EPHB2, AURKA, GSG2, IKBKE, MELK, CSK, CHEK2, CDC7 and MAP3K8. The study also highlights the substantial molecular heterogeneity of this disease, as no single kinase gene appeared dominant [14, 20, 21]. The targeted assay for gene expression made possible accurate quantification of kinases from small blocks of tissue with high sensitivity and accuracy.
Many of the validated 9 kinases have previously been implicated data in the metastatic progression of other malignancies. Moreover, inhibitors of some of these kinases are already in early phase I trials. EphB2 is important in cell motility, epithelial mesenchymal transition (EMT), adhesion and angiogenesis, and appears prognostic in multiple other malignancies, especially colorectal cancers [31–37]. AURKA plays a role in chromosome segregation during mitosis and is correlated with mTOR pathway overexpression in sarcomatoid RCC [38]. GSG2 is required for mitotic histone phosphorylation and metaphase chromosome alignment [39]. Additionally, a preclinical study reported a potential role for GSG2 inhibitors in multiple malignancies [40]. IKBKE is a member of the nuclear factor-kappaB (NF-κB) signaling pathway and expression predicted poor/reduced survival in one study of ccRCC patients [41]. Another meta-analysis suggested that NF-κB may represent a therapeutic target in ccRCC [42]. MELK is thought to bind and activate transcription factors c-JUN and FOXM1 and play a role in maintaining tumor initiating cells [43–46]. Over-expression of CSK-binding protein appeared to contribute to renal cell carcinogenesis in a preclinical study [47]. CHEK2 is activated in response to DNA damage, and is involved in cell cycle arrest, but its role in ccRCC has not been studied [48]. Similarly, CDC7 (essential for the G1/S transition and initiation of DNA replication) and MAP3K8 (which activates MAPK and JNK) have not been validated as therapeutic targets in ccRCC and warrant further study. Notably, widely targeted kinases in other malignancies including EGFR/HER2 family and PI3K/mTOR pathway kinases were among those measured, but not found to be differentially expressed. However, when examining the 9 validated genes and other genes with >1.7-fold increase in M in at least a third of patients, agents are either approved for other indications (e.g. BTK, MAPK and ROS1 inhibitors) or undergoing investigation in phase I trials (e.g. AURKA, CHEK1, CHEK2 and PLK1 inhibitors), suggesting that clinical validation of their potential actionability may be feasible. Although our study focused on kinases, pathway analysis showed significance of salvage pathways of pyridoxal 5'-phosphate and pyrimidine ribonucleotides, which accords with the importance of a metabolic shift demonstrated by TCGA [14].
Although our study is limited by the size of the discovery dataset (n = 35), using paired intra-patient T, N and M samples is a strength. Additionally, because fresh frozen tumors are not readily available, we used FFPE tumor tissue. RNA extracted from FFPE material is typically highly degraded, but the nCounterTM technology largely overcomes this obstacle [49]. Although genomic material may undergo degradation with age, all tumors in this study were obtained from biopsies performed after the year 2000. The TCGA validation cohort differs from our discovery cohort in two important ways. First, the discovery cohort employed NanoString, while the validation dataset used RNA-seq. However, these different but robust platforms assess gene expression through independent techniques and yielded highly concordant results in different patient cohorts. Second, while the TCGA validation cohort did not directly measure metastatic tissues, kinases were overexpressed in primary tumors with metastases compared to primary tumors without metastases. Future studies can further corroborate our results in a larger dataset of metastatic FFPE tumors. A limitation is that tumor heterogeneity leads to the presence of different clonal populations within the same primary or metastatic mass displaying differing kinase gene expressions, although our sample size of 35 patients may be expected to partly overcome this confounding factor and discover broadly important overexpressed genes in metastases. Another important future direction is to further investigate the activity and expression of the corresponding kinase proteins. While kinase gene expression may correlate with corresponding kinase protein levels and activity, many kinases are regulated at the translational or post-translational modification levels. Immunohistochemistry (IHC) could be performed to assay protein expression, but will likely require fresh frozen tissues because this platform is characterized by methodological and analytical limitations when performed on FFPE tissue. Inadequate tissue precluded performing quantitative polymerase chain reaction (PCR) to validate gene expression using a different platform and IHC to confirm expression of the corresponding proteins.
Nevertheless, our study has identified altered gene expression in kinase transcripts measured by two different platforms in two independent cohorts. These transcripts have broad deterministic ability to drive metastatic ccRCC as opposed to stochastic alterations important in smaller subsets of tumors. It is also possible that the 9 validated overexpressed kinase genes may be ‘shared’ among metastatic sites across patients, as opposed to being ‘privately’ overexpressed in small subsets of patients or metastatic sites. Furthermore, the expression of these genes may assist in improving the prognostic stratification of patients with localized ccRCC following surgery. Notably, other molecular panels such as the recurrence score and MET gene polymorphisms have been previously reported to improve prognostication of localized ccRCC [50, 51]. We could not evaluate the prognostic impact of kinase gene expression on outcomes with systemic therapy for metastatic disease because information on systemic therapy was unavailable. Moreover, this objective is beyond the scope of the study since the sample size is modest and such an analysis would be under-powered.
The only kinase gene expression change that was not externally validated in TCGA was GUCY2C, which had low absolute expression in both the discovery cohort and the TCGA cohort. Nevertheless, GUCY2C gene expression by microarray was associated with increasing metastatic potential in a study of primary ccRCC tumors, suggesting that further study of this gene in a larger dataset of metastatic tumor tissue is warranted [52]. Examining genes where more than a third of patients had >1.7 fold increase in expression between M vs. T sites, we observed a significant kinase gene expression increase in TCGA primary tumors with metastases at baseline compared to primary tumors in patients who did not develop metastasis for at least 2 years in 21 of the 33 genes identified in the discovery dataset. While differential gene expression changes in the TCGA cohort are not as significant in this set of kinases as the panel of 10 kinases overexpressed in all M compared to T tumors, this is not surprising given that this group of genes had a modest expression change in only a subset of patients.
Conclusions
This study is the first to examine intra-patient kinase gene expression in ccRCC metastatic and primary tumor tissues and identifies nine over-expressed kinase genes (EPHB2, AURKA, GSG2, IKBKE, MELK, CSK, CHEK2, CDC7 and MAP3K8; p<0.001) in metastatic ccRCC tumor tissue, which may represent drivers of disease and novel therapeutic targets. Our results remain hypothesis-generating and require further validation to confirm functional significance of findings. Preclinical activity of inhibitors of the identified kinases needs to be demonstrated to support clinical trials. Collaboration between institutions to comprehensively study a large dataset of metastatic ccRCC tissue will enable better understanding of this disease and inform both rational drug development and patient selection for targeted agents.
Supporting Information
(XLS)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The results published/shown here are in part based upon data generated by the TCGA Research network: http://cancergenome.nih.gov/.
Funding: Funded by genitourinary oncology research support grant provided to Guru Sonpavde by the UAB Department of Medicine. Additionally, Ryne Ramaker was supported by the NIH-National Institute of General Medical Sciences Medical Scientist Training Program (5T32GM008361-21) and Brittany Lasseigne and Richard Myers were supported by a cancer research fund from the Alabama Cancer Research Consortium.
Data Availability
All relevant data are within the paper and its Supporting Information files. The RNASeq data employed for validation is freely available from TCGA and data accession is described in the Methods section of the manuscript.
Funding Statement
This work was funded by genitourinary oncology research support grant provided to Guru Sonpavde by the UAB Department of Medicine. Additionally, Ryne Ramaker was supported by the NIH-National Institute of General Medical Sciences Medical Scientist Training Program (5T32GM008361-21) and Brittany Lasseigne and Richard Myers were supported by a cancer research fund from the state of Alabama. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353(23):2477–90. Epub 2005/12/13. doi: 353/23/2477 [pii] 10.1056/NEJMra043172 . [DOI] [PubMed] [Google Scholar]
- 2.McDermott DF, Regan MM, Clark JI, Flaherty LE, Weiss GR, Logan TF, et al. Randomized phase III trial of high-dose interleukin-2 versus subcutaneous interleukin-2 and interferon in patients with metastatic renal cell carcinoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23(1):133–41. Epub 2004/12/31. doi: 23/1/133 [pii] 10.1200/JCO.2005.03.206 . [DOI] [PubMed] [Google Scholar]
- 3.Fyfe G, Fisher RI, Rosenberg SA, Sznol M, Parkinson DR, Louie AC. Results of treatment of 255 patients with metastatic renal cell carcinoma who received high-dose recombinant interleukin-2 therapy. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1995;13(3):688–96. Epub 1995/03/01. . [DOI] [PubMed] [Google Scholar]
- 4.Vlahovic G, Crawford J. Activation of tyrosine kinases in cancer. Oncologist. 2003;8(6):531–8. . [DOI] [PubMed] [Google Scholar]
- 5.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. The New England journal of medicine. 2007;356(2):115–24. Epub 2007/01/12. doi: 356/2/115 [pii] 10.1056/NEJMoa065044 . [DOI] [PubMed] [Google Scholar]
- 6.Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. The New England journal of medicine. 2007;356(2):125–34. 10.1056/NEJMoa060655 . [DOI] [PubMed] [Google Scholar]
- 7.Rini BI, Escudier B, Tomczak P, Kaprin A, Szczylik C, Hutson TE, et al. Comparative effectiveness of axitinib versus sorafenib in advanced renal cell carcinoma (AXIS): a randomised phase 3 trial. Lancet. 378(9807):1931–9. Epub 2011/11/08. S0140-6736(11)61613-9 [pii] 10.1016/S0140-6736(11)61613-9 . [DOI] [PubMed] [Google Scholar]
- 8.Sternberg CN, Davis ID, Mardiak J, Szczylik C, Lee E, Wagstaff J, et al. Pazopanib in locally advanced or metastatic renal cell carcinoma: results of a randomized phase III trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28(6):1061–8. 10.1200/JCO.2009.23.9764 . [DOI] [PubMed] [Google Scholar]
- 9.Hudes G, Carducci M, Tomczak P, Dutcher J, Figlin R, Kapoor A, et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. The New England journal of medicine. 2007;356(22):2271–81. Epub 2007/06/01. 356/22/2271 [pii] 10.1056/NEJMoa066838 . [DOI] [PubMed] [Google Scholar]
- 10.Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372(9637):449–56. 10.1016/S0140-6736(08)61039-9 . [DOI] [PubMed] [Google Scholar]
- 11.Choueiri TK, Escudier B, Powles T, Mainwaring PN, Rini BI, Donskov F, et al. Cabozantinib versus Everolimus in Advanced Renal-Cell Carcinoma. The New England journal of medicine. 2015;373(19):1814–23. 10.1056/NEJMoa1510016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. The New England journal of medicine. 2015;373(19):1803–13. 10.1056/NEJMoa1510665 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motzer RJ, Hutson TE, Glen H, Michaelson MD, Molina A, Eisen T, et al. Lenvatinib, everolimus, and the combination in patients with metastatic renal cell carcinoma: a randomised, phase 2, open-label, multicentre trial. The lancet oncology. 2015;16(15):1473–82. 10.1016/S1470-2045(15)00290-9 . [DOI] [PubMed] [Google Scholar]
- 14.Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9. Epub 2013/06/25. 10.1038/nature12222 ; PubMed Central PMCID: PMCPmc3771322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brugarolas J. Molecular genetics of clear-cell renal cell carcinoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2014;32(18):1968–76. 10.1200/JCO.2012.45.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behbahani TE, Thierse C, Baumann C, Holl D, Bastian PJ, von Ruecker A, et al. Tyrosine kinase expression profile in clear cell renal cell carcinoma. World J Urol. 2012;30(4):559–65. 10.1007/s00345-011-0767-z . [DOI] [PubMed] [Google Scholar]
- 17.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nature genetics. 2012;44(7):751–9. 10.1038/ng.2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlo MI, Molina AM, Lakhman Y, Patil S, Woo K, DeLuca J, et al. A Phase Ib Study of BEZ235, a Dual Inhibitor of Phosphatidylinositol 3-Kinase (PI3K) and Mammalian Target of Rapamycin (mTOR), in Patients With Advanced Renal Cell Carcinoma. The oncologist. 2016. 10.1634/theoncologist.2016-0145 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powles T, Lackner MR, Oudard S, Escudier B, Ralph C, Brown JE, et al. Randomized Open-Label Phase II Trial of Apitolisib (GDC-0980), a Novel Inhibitor of the PI3K/Mammalian Target of Rapamycin Pathway, Versus Everolimus in Patients With Metastatic Renal Cell Carcinoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2016;34(14):1660–8. 10.1200/JCO.2015.64.8808 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 366(10):883–92. Epub 2012/03/09. 10.1056/NEJMoa1113205 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nature genetics. 2014;46(3):225–33. 10.1038/ng.2891 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu P, Nielsen TE, Clausen MH. FDA-approved small-molecule kinase inhibitors. Trends Pharmacol Sci. 2015;36(7):422–39. 10.1016/j.tips.2015.04.005 . [DOI] [PubMed] [Google Scholar]
- 23.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature biotechnology. 2008;26(3):317–25. 10.1038/nbt1385 . [DOI] [PubMed] [Google Scholar]
- 24.Beard RE, Abate-Daga D, Rosati SF, Zheng Z, Wunderlich JR, Rosenberg SA, et al. Gene expression profiling using nanostring digital RNA counting to identify potential target antigens for melanoma immunotherapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19(18):4941–50. 10.1158/1078-0432.CCR-13-1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sestak I, Dowsett M, Zabaglo L, Lopez-Knowles E, Ferree S, Cowens JW, et al. Factors predicting late recurrence for estrogen receptor-positive breast cancer. Journal of the National Cancer Institute. 2013;105(19):1504–11. 10.1093/jnci/djt244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dowsett M, Sestak I, Lopez-Knowles E, Sidhu K, Dunbier AK, Cowens JW, et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol. 2013;31(22):2783–90. 10.1200/JCO.2012.46.1558 . [DOI] [PubMed] [Google Scholar]
- 27.Lohavanichbutr P, Mendez E, Holsinger FC, Rue TC, Zhang Y, Houck J, et al. A 13-gene signature prognostic of HPV-negative OSCC: discovery and external validation. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19(5):1197–203. 10.1158/1078-0432.CCR-12-2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulkarni MM. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol. 2011;Chapter 25:Unit25B.10. 10.1002/0471142727.mb25b10s94 . [DOI] [PubMed] [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Statistics in medicine. 1990;9(7):811–8. . [DOI] [PubMed] [Google Scholar]
- 31.Mosch B, Reissenweber B, Neuber C, Pietzsch J. Eph receptors and ephrin ligands: important players in angiogenesis and tumor angiogenesis. J Oncol. 2010;2010:135285 10.1155/2010/135285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khansaard W, Techasen A, Namwat N, Yongvanit P, Khuntikeo N, Puapairoj A, et al. Increased EphB2 expression predicts cholangiocarcinoma metastasis. Tumour Biol. 2014;35(10):10031–41. 10.1007/s13277-014-2295-0 . [DOI] [PubMed] [Google Scholar]
- 33.Gao Q, Liu W, Cai J, Li M, Gao Y, Lin W, et al. EphB2 promotes cervical cancer progression by inducing epithelial-mesenchymal transition. Hum Pathol. 2014;45(2):372–81. 10.1016/j.humpath.2013.10.001 . [DOI] [PubMed] [Google Scholar]
- 34.Jubb AM, Zhong F, Bheddah S, Grabsch HI, Frantz GD, Mueller W, et al. EphB2 is a prognostic factor in colorectal cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11(14):5181–7. 10.1158/1078-0432.CCR-05-0143 . [DOI] [PubMed] [Google Scholar]
- 35.Kandouz M, Haidara K, Zhao J, Brisson ML, Batist G. The EphB2 tumor suppressor induces autophagic cell death via concomitant activation of the ERK1/2 and PI3K pathways. Cell Cycle. 2010;9(2):398–407. . [DOI] [PubMed] [Google Scholar]
- 36.Lugli A, Spichtin H, Maurer R, Mirlacher M, Kiefer J, Huusko P, et al. EphB2 expression across 138 human tumor types in a tissue microarray: high levels of expression in gastrointestinal cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11(18):6450–8. 10.1158/1078-0432.CCR-04-2458 . [DOI] [PubMed] [Google Scholar]
- 37.Mao W, Luis E, Ross S, Silva J, Tan C, Crowley C, et al. EphB2 as a therapeutic antibody drug target for the treatment of colorectal cancer. Cancer research. 2004;64(3):781–8. . [DOI] [PubMed] [Google Scholar]
- 38.Pal SK, He M, Tong T, Wu H, Liu X, Lau C, et al. RNA-seq Reveals Aurora Kinase Driven-mTOR Pathway Activation in Patients with Sarcomatoid Metastatic Renal Cell Carcinoma. Mol Cancer Res. 2014. 10.1158/1541-7786.MCR-14-0352 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai J, Sultan S, Taylor SS, Higgins JM. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19(4):472–88. 10.1101/gad.1267105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huertas D, Soler M, Moreto J, Villanueva A, Martinez A, Vidal A, et al. Antitumor activity of a small-molecule inhibitor of the histone kinase Haspin. Oncogene. 2012;31(11):1408–18. 10.1038/onc.2011.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hildebrandt MA, Tan W, Tamboli P, Huang M, Ye Y, Lin J, et al. Kinome expression profiling identifies IKBKE as a predictor of overall survival in clear cell renal cell carcinoma patients. Carcinogenesis. 2012;33(4):799–803. 10.1093/carcin/bgs018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peri S, Devarajan K, Yang DH, Knudson AG, Balachandran S. Meta-analysis identifies NF-kappaB as a therapeutic target in renal cancer. PLoS One. 2013;8(10):e76746 10.1371/journal.pone.0076746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung S, Suzuki H, Miyamoto T, Takamatsu N, Tatsuguchi A, Ueda K, et al. Development of an orally-administrative MELK-targeting inhibitor that suppresses the growth of various types of human cancer. Oncotarget. 2012;3(12):1629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joshi K, Banasavadi-Siddegowda Y, Mo X, Kim SH, Mao P, Kig C, et al. MELK-dependent FOXM1 phosphorylation is essential for proliferation of glioma stem cells. Stem Cells. 2013;31(6):1051–63. 10.1002/stem.1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu C, Banasavadi-Siddegowda YK, Joshi K, Nakamura Y, Kurt H, Gupta S, et al. Tumor-specific activation of the C-JUN/MELK pathway regulates glioma stem cell growth in a p53-dependent manner. Stem Cells. 2013;31(5):870–81. 10.1002/stem.1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minata M, Gu C, Joshi K, Nakano-Okuno M, Hong C, Nguyen CH, et al. Multi-kinase inhibitor C1 triggers mitotic catastrophe of glioma stem cells mainly through MELK kinase inhibition. PLoS One. 2014;9(4):e92546 10.1371/journal.pone.0092546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng X, Lu X, Man X, Zhou W, Jiang LQ, Knyazev P, et al. Overexpression of Csk-binding protein contributes to renal cell carcinogenesis. Oncogene. 2009;28(37):3320–31. 10.1038/onc.2009.185 . [DOI] [PubMed] [Google Scholar]
- 48.Cybulski C, Gorski B, Huzarski T, Masojc B, Mierzejewski M, Debniak T, et al. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet. 2004;75(6):1131–5. 10.1086/426403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stricker TP, Morales La Madrid A, Chlenski A, Guerrero L, Salwen HR, Gosiengfiao Y, et al. Validation of a prognostic multi-gene signature in high-risk neuroblastoma using the high throughput digital NanoString nCounter™ system. Mol Oncol. 2014;8(3):669–78. 10.1016/j.molonc.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rini B, Goddard A, Knezevic D, Maddala T, Zhou M, Aydin H, et al. A 16-gene assay to predict recurrence after surgery in localised renal cell carcinoma: development and validation studies. The lancet oncology. 2015;16(6):676–85. 10.1016/S1470-2045(15)70167-1 . [DOI] [PubMed] [Google Scholar]
- 51.Schutz FA, Pomerantz MM, Gray KP, Atkins MB, Rosenberg JE, Hirsch MS, et al. Single nucleotide polymorphisms and risk of recurrence of renal-cell carcinoma: a cohort study. The lancet oncology. 2013;14(1):81–7. 10.1016/S1470-2045(12)70517-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sültmann H, von Heydebreck A, Huber W, Kuner R, Buness A, Vogt M, et al. Gene expression in kidney cancer is associated with cytogenetic abnormalities, metastasis formation, and patient survival. Clin Cancer Res. 2005;11(2 Pt 1):646–55. . [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. The RNASeq data employed for validation is freely available from TCGA and data accession is described in the Methods section of the manuscript.