Abstract
The D-genome progenitor of wheat (Triticum aestivum), Aegilops tauschii, possesses numerous genes for resistance to abiotic stresses, including drought. Therefore, information on the genetic architecture of A. tauschii can aid the development of drought-resistant wheat varieties. Here, we evaluated 13 traits in 373 A. tauschii accessions grown under normal and polyethylene glycol-simulated drought stress conditions and performed a genome-wide association study using 7,185 single nucleotide polymorphism (SNP) markers. We identified 208 and 28 SNPs associated with all traits using the general linear model and mixed linear model, respectively, while both models detected 25 significant SNPs with genome-wide distribution. Public database searches revealed several candidate/flanking genes related to drought resistance that were grouped into three categories according to the type of encoded protein (enzyme, storage protein, and drought-induced protein). This study provided essential information for SNPs and genes related to drought resistance in A. tauschii and wheat, and represents a foundation for breeding drought-resistant wheat cultivars using marker-assisted selection.
Keywords: Aegilops tauschii, drought resistance, genome-wide association study, single nucleotide polymorphism, wheat
Introduction
The current global climate change is projected to have a significant impact on temperature and precipitation profiles, with consequent increases in drought incidence and severity. It is known that severe drought occurs in nearly half of the world's countries (Wilhite and Glantz, 1985). Since drought is probably the major abiotic factor limiting yields, the development of crops that are high yielding under environmentally stressful conditions is essential (Ergen and Budak, 2009; Fleury et al., 2010).
Wheat (Triticum spp.) is the leading human food source, accounting for more than half of the world's total food consumption (Ergen and Budak, 2009; Habash et al., 2009); therefore, it is a major target for the development of cultivars that are high-yielding under water-limited conditions. For drought-related research and the improvement of modern crop varieties, plants exhibiting high drought resistance are the most suitable targets and the most promising sources of drought-related genes and gene regions. Many wild species also retain superior genetic resources that have not yet been investigated. One such species is Aegilops tauschii, the diploid D-genome progenitor of hexaploid wheat (T. aestivum). A. tauschii is more drought resistant than T. aestivum and wild emmer wheat (T. dicoccoides) and harbors drought-resistance traits that were lost during the breeding processes (Ashraf et al., 2009). Breeders have increasingly focused on A. tauschii, since an understanding of the genetic basis of drought resistance in A. tauschii can contribute to the development of drought-resistant wheat cultivars.
Drought resistance is a quantitative trait with a complex phenotype affected by plant development stages (Budak et al., 2013). Linkage analysis is the most commonly used strategy for detecting quantitative trait loci (QTLs) in plants; however, linkage mapping using biparental crosses has some serious limitations. This method can only reveal information regarding two alleles at a given locus, or a few loci segregating in a studied population. In addition, the genetic resolution of detected QTLs is poor (Holland, 2007; Navakode et al., 2014). Furthermore, linkage analysis can only sample a small fraction of all possible alleles in the parental source population, while the development of mapping populations is costly and time-consuming.
Association mapping (AM), also known as linkage disequilibrium mapping, relies on existing natural populations or specially designed populations to overcome the constraints of linkage mapping (Pasam et al., 2012). This technique is a powerful tool to resolve complex trait variation and identify different loci and/or novel and superior alleles in natural populations (Zhu et al., 2008). In recent years, association studies have been extensively used to discover and validate QTLs or genes for important traits and to map candidate genes in many crop plants, including wheat. The benefit of this method over traditional bi-parental mapping approaches depends on the extent of linkage (Huang et al., 2010; Kump et al., 2011; Erena et al., 2013). In wheat, different association panels have been used in many AM studies to identify loci controlling agronomic (Breseghello and Sorrells, 2006; Crossa et al., 2007; Neumann et al., 2007; Bordes et al., 2013) and quality (Ravel et al., 2009; Bordes et al., 2011) traits.
Only a few genome-wide association studies have been carried out in A. tauschii for drought resistance traits. Here, we aimed to: 1) investigate marker-trait associations for drought resistance based on a genome-wide AM approach using single nucleotide polymorphism (SNP) markers in a core collection of 373 A. tauschii accessions of diverse origin; 2) identify SNPs highly associated with drought resistance traits; and 3) search for candidate genes controlling these traits. This study could provide important information for cloning genes related to drought-resistance in A. tauschii and develop resistant wheat cultivars using marker-assisted selection.
Material and Methods
Plant materials and phenotypic evaluation
The natural population used for the association analysis comprised of 373 A. tauschii accessions collected by the Triticeae Research Institute of Sichuan Agricultural University. A. tauschii plants were grown in a phytotron in Wenjiang, Sichuan Province, China, from September 2012 to March 2013 and evaluated under normal conditions (NC) and polyethylene glycol (PEG)-simulated drought-stress conditions (SC) in a completely randomized design with four replications per treatment. Hydroponic tanks were filled with standard Hoagland's nutrient solution (1 mM KH2PO4, 2 mM MgSO47H2O, 4 mM CaNO34H2O, 6 mM KNO3, 0.046 mM H3BO3, 0.76 μM ZnSO4, 0.32 μM CuSO45H2O, 9.146 μM MnCl2, 0.0161 μM (NH4)6 MoO44H2O, and 100 μM NaFeEDTA; Hoagland and Arnon, 1950) with or without PEG (19.2%) for SC and NC, respectively. Seedlings were grown at a temperature of 25/22 ± 1 °C day/night, relative humidity of 65/85% day/night, and a 16-h photoperiod with 500 mmolm-2s-1 photon flux density at the level of plant canopy.
Uniform seedlings were transferred to the phytotron 8 d after germination and evaluated 22 d later with a WinRHizo Pro 2008a image analysis system (Régent Instruments, Quebec, Canada) for the following traits: root length (RL), root diameter (RD), the number of root tips (RT), and the number of roots with a diameter of 0.000-0.500 mm (TNOR). The plants were then separated into shoots and roots for measuring total fresh weight (TFW), root fresh weight (RFW), shoot fresh weight (SFW), and shoot height (SH). To determine total dry weight (TDW), root dry weight (RDW), and shoot dry weight (SDW), shoots and roots were stored in paper bags, heated at 105 °C for 30 min to kill the cells, and dried at 75 °C until a constant mass was obtained.
Descriptive statistics, correlation analysis, analysis of variance, principal component analysis and multiple linear stepwise regressions were conducted for all traits using IBM SPSS Statistics for Windows 20.0 (IBM Corp., Chicago, IL, USA). Heritability was calculated as follows (Smith et al., 1998):
where VG and VE represent estimates of genetic and environmental variances, respectively.
In order to eliminate individual variation resulting from inherent genetic differences unrelated to drought resistance, the drought resistance index (DI) was used as a standardizing measure across A. tauschii accessions and calculated as follows (Bouslama and Schapaugh, 1950):
where TSC and TNC are the traits measured for each plant under SC and NC, respectively.
We also calculated the weighted comprehensive evaluation value (D value) for each genotype as follows (Xie, 1993; Zhou et al., 2003):
where Wj is the weighting variable calculated as:
with Pj being the percent of variance and u(Xj) the membership function value calculated as:
10K Infinium iSelect SNP array and SNP genotyping
The construction of the A. tauschii 10K SNP array was described previously by Luo et al. (2014). A total of 7,185 SNP markers was mapped to an A. tauschii genetic map and a physical map built by bacterial artificial chromosome clones (Luo et al., 2014). SNPs were assayed according to the manufacturer's protocol (Illumina, San Diego, CA, USA) at the Genome Center, University of California, Davis, CA, USA. Normalized Cy3 and Cy5 fluorescence for each DNA sample was graphed using Genome Studio (Illumina, San Diego, CA, USA), resulting in genotype clustering for each SNP marker. SNP genotyping was carried out as described previously by Wang et al. (2013).
Population structure
Population structure was estimated with a set of 7,185 SNP markers mapped to the A. tauschii genetic map using STRUCTURE 2.3.3, which implements a model-based Bayesian cluster analysis (Pritchard et al., 2000; Wang et al., 2013). The linkage ancestry model and the allele frequency-correlated model were used. A total of 100 burn-in iterations followed by 100 Markov chain Monte Carlo iterations for K = 1 to 10 clusters were used to identify the optimal range of K. Five runs were performed separately for each value of K, and the optimal K-value was determined using the delta K method (Evanno et al., 2005). Using K = 4 (Wang et al., 2013), the population was divided into Subp1, Subp2, Subp3, Subp4, and mixed individuals.
Genome-wide association study
Marker-trait associations were calculated in Tassel 2.1 (Bradbury et al., 2007) using both the general linear model (GLM) and the mixed linear model (MLM). Both models used 6,905 SNP markers with a minor allele frequency threshold (> 0.05). To correct the population structure, the GLM incorporated a Q-matrix and the MLM incorporated Q- and K-matrices. The Bonferroni-corrected threshold at α = 1 (Yang et al., 2014) was used as the cutoff value, which was 144.823 × 10-6 with a corresponding -log p-value of 3.839. Significant markers were visualized with a Manhattan plot drawn in R 3.03 (http://www.r-project.org/). Important p-value distributions (observed vs. cumulative p-values on a -log10 scale) were displayed in a quantile-quantile plot drawn in R. To find candidate genes, flanking genes, and trait-related proteins, we performed a Basic Local Alignment Search Tool (BLAST) search against the International Wheat Genome Sequencing Consortium database (IWGSC; http://www.wheatgenome.org/) using SNP sequences. The IWGSC BLAST results were used to perform a BLAST search of the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/) and then a direct BLASTx search of the NCBI database.
Results
Phenotypic evaluation
Significant phenotypic variation was observed for all traits, and the means were significantly different between NC and SC (Table 1). The mean values of the root to shoot ratio of fresh weight (FRS), the root to shoot ratio of dry weight (DRS), RT, and RL were higher under SC, whereas RFW, SFW, RDW, SDW, SH, TFW, TDW, RD, and TNOR were lower under SC compared with those under NC (Table 1). Significant differences between NC and SC were observed for all traits, except for RFW, FRS, TFW, and TDW, indicating that most of the tested traits were significantly affected by drought. Medium to high heritability estimates were obtained for most of the traits, and heritability was higher for five traits under NC and seven traits under SC. Heritability ranged from 0.333 to 0.971 under NC and 0.331 to 0.983 under SC (Table 1). Pearson correlations were calculated among all traits, and we found 56 and 50 significant correlation coefficients (P < 0.05) under NC and SC, respectively (Table S1 (47.9KB, pdf) ).
Table 1. Phenotypic variation in 13 traits in 373 Aegilops tauschii accessions under the normal condition (NC) and the PEG-induced, simulated drought-stress condition (SC).
| Trait | Condition | Mean ± s.d. | CV(%) | F-value | h B(%)a |
|---|---|---|---|---|---|
| RDW | NC | 0.016 ± 0.009 | 55.983 | 48.191** | 0.431 |
| SC | 0.013 ± 0.009 | 70.672 | 0.440 | ||
| SDW | NC | 0.041 ± 0.020 | 49.342 | 21.498** | 0.552 |
| SC | 0.022 ± 0.011 | 49.682 | 0.552 | ||
| DRS | NC | 0.419 ± 0.285 | 67.962 | 37.497** | 0.719 |
| SC | 0.987 ± 1.792 | 181.476 | 0.822 | ||
| RFW | NC | 0.276 ± 0.130 | 47.209 | 0.287ns | 0.964 |
| SC | 0.108 ± 0.048 | 43.921 | 0.958 | ||
| SFW | NC | 0.278 ± 0.145 | 52.219 | 1.335** | 0.924 |
| SC | 0.073 ± 0.034 | 46.294 | 0.920 | ||
| FRS | NC | 1.073 ± 0.649 | 60.544 | 0.142ns | 0.971 |
| SC | 1.572 ± 0.556 | 35.415 | 0.983 | ||
| SH | NC | 17.267 ± 3.998 | 23.155 | 6.833** | 0.333 |
| SC | 13.785 ± 3.196 | 23.185 | 0.337 | ||
| RL | NC | 246.692 ± 129.523 | 52.504 | 20.049** | 0.341 |
| SC | 340.228 ± 415.846 | 122.226 | 0.331 | ||
| RD | NC | 7.749 ± 33.842 | 436.727 | 10.66** | 0.475 |
| SC | 3.481 ± 10.981 | 315.422 | 0.440 | ||
| TDW | NC | 0.057 ± 0.025 | 44.074 | 1.521ns | 0.862 |
| SC | 0.035 ± 0.014 | 39.802 | 0.902 | ||
| TFW | NC | 0.554 ± 0.264 | 47.622 | 0.592ns | 0.666 |
| SC | 0.182 ± 0.075 | 41.300 | 0.927 | ||
| RT | NC | 1229.254 ± 912.330 | 74.218 | 58.931** | 0.343 |
| SC | 2180.079 ± 3181.680 | 145.943 | 0.334 | ||
| TNOR | NC | 2148.141 ± 864.048 | 74.578 | 58.574** | 0.342 |
| SC | 1158.575 ± 3163.958 | 147.288 | 0.355 |
RFW: root fresh weight; SFW: shoot fresh weight; FRS: root to shoot ratio of fresh weight; RDW: root dry weight; SFW: shoot dry weight; FRS: root to shoot ratio of dry weight; SH: shoot height; TFW: total fresh weight; TDW: total dry weight; TRL: total root length; RD: root diameter; RT: number of root tips; TNOR: the number of root in diameter 0.000 to 0.500.
Broad-sense heritability of the tested traits.
significant at p < 0.01;
ns: not significant.
Principal component analysis (PCA) and multiple linear stepwise regressions
PCA were performed for all traits using DI (Table 2) that were highly correlated according to the Bartlett's test of sphericity (χ2 = 5056.738; P < 0.001). To establish selection indices involving multiple drought-resistance traits, a series of linear regressions were performed for all traits. We built the regression to explain TDW and chose our predictive variables through stepwise regression (Table 3). The final stepwise model explained 93.9% and 65.3% of the phenotypic variation in TDW under NC and SC, respectively. The model contained nine traits for NC (RFW, RDW, FRS, DRS, TFW, RD, RL, RT, and TNOR) and seven traits for SC (RFW, RDW, FRS, DRS, TFW, RL, and TNOR).
Table 2. Principal component analysis (PCA). For trait abbreviations see Table 1.
| Trait | PC 1 | PC 2 | PC 3 | PC 4 | |
|---|---|---|---|---|---|
| RFW | 0.655 | -0.082 | 0.618 | 0.238 | |
| SFW | 0.584 | -0.179 | -0.144 | -0.264 | |
| FRS | -0.050 | 0.084 | 0.831 | 0.469 | |
| RDW | 0.734 | -0.348 | -0.210 | 0.350 | |
| SDW | 0.365 | 0.244 | 0.365 | -0.677 | |
| DRS | 0.483 | -0.411 | -0.400 | 0.495 | |
| Characteristic vector | SH | 0.608 | -0.042 | -0.132 | -0.282 |
| TFW | 0.865 | -0.166 | 0.086 | 0.024 | |
| TDW | 0.815 | -0.014 | 0.094 | -0.265 | |
| RL | 0.278 | 0.765 | -0.111 | 0.173 | |
| RD | 0.083 | -0.362 | -0.065 | -0.005 | |
| RT | 0.294 | 0.891 | -0.170 | 0.157 | |
| TNOR | 0.295 | 0.891 | -0.167 | 0.154 | |
| Eigenvalues | 3.720 | 2.731 | 1.538 | 1.400 | |
| Contribution % | 28.614 | 21.005 | 11.831 | 10.766 | |
| Cumulative contribution % | 28.614 | 49.618 | 61.449 | 72.215 |
Table 3. Multiple linear stepwise regression to explain total dry weight (TDW) from root traits built with Aegilops tauschii genotypes means. For trait abbreviations see Table 1.
| Treatment | Final stepwise model | R2 | P value |
|---|---|---|---|
| NC | TDW = 0.011 – 0.08RFW + 2.014RDW + 0.02FRS – 0.032DRS + 0.089TFW + 0.00005817RD – 0.000002274RL -0.000001614RT + 0.000008294TNOR | 0.939 | < 0.001 |
| SC | TDW = 0.011 – 0.033RFW + 0.92RDW – 0.001FRS – 0.003DRS – 0.105TFW + 0.000002321RL + 0.000002292TNOR | 0.653 | < 0.001 |
We performed a comprehensive evaluation of drought resistance in A. tauschii using D values and DI ( Table S2 (49.5KB, pdf) ). Among the 373 A. tauschii accessions, AS623213 that had the highest D value and AS623095 that had the lowest D value were selected as extremely resistant and susceptible genotypes, respectively. Overall, we identified six genotypes (1.6%) with high resistance (D ≥ 0.5), 262 (70.2%) with moderate resistance (0.30 ≤ D < 0.5), and 105 (28.2%) with low resistance (D < 0.30). Next, we observed that A. tauschii accessions with a higher D value also had a higher DI (Table S2 (49.5KB, pdf) ), which suggested that the two selection indicators were effective for screening A. tauschii under SC.
Marker-trait association analysis
The Bonferroni-corrected threshold (-log p > 3.839, α = 1) was used as the cutoff value for identifying marker-trait associations (Yang et al., 2014). A total of 208 and 28 SNPs were detected by the GLM and MLM, respectively, while 25 significant SNPs with genome-wide distribution (chromosomes [Chr.] 1D-7D) markers were detected by both models (Table 4; Figure S1 (1.5MB, pdf) and Table S3 (16.2KB, xlsx) ).
Table 4. Genome-wide association of 13 tested traits under the normal condition (NC) and the PEG-induced, simulated drought-stress condition (SC) detected using general linear (GLM) and mixed linear (MLM) models. For trait abbreviations see Table 1.
| Trait | GLM | MLM | No. Sharec | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No.siga | Average -log(P) | Range -log(P) | Average R2 (%)b | Range R2 (%) b | No.siga | Average -log(P) | Range -log(P) | Average R2 (%) b | Range R2 (%)b | |||
| NC | FRS | 31 | 4.476 | 3.843-5.522 | 4.958 | 4.183-6.240 | 1 | 3.970 | 4.732 | 1 | ||
| RD | 9 | 4.055 | 3.884-4.334 | 4.367 | 4.160-4.702 | |||||||
| RDW | 1 | 4.314 | 4.891 | |||||||||
| RFW | 28 | 4.555 | 3.873-6.217 | 5.087 | 4.243-7.128 | |||||||
| RL | 16 | 4.734 | 3.866-7.607 | 4.912 | 3.896-8.144 | |||||||
| RT | 12 | 4.635 | 3.858-5.551 | 4.674 | 3.866-6.016 | 1 | 3.980 | 4.805 | ||||
| SDW | 5 | 4.703 | 3.855-6.332 | 4.983 | 3.983-6.860 | 1 | 4.040 | 4.803 | 1 | |||
| SFW | 7 | 4.564 | 3.878-6.596 | 4.883 | 4.074-7.277 | 2 | 4.122 | 4.109-4.136 | 4.932 | 4.912-4.951 | 2 | |
| SH | 1 | 3.932 | 4.410 | |||||||||
| TDW | 9 | 4.567 | 3.901-6.883 | 4.826 | 4.044-7.508 | 1 | 4.217 | 5.033 | 1 | |||
| TFW | 21 | 4.763 | 3.875-6.930 | 5.116 | 4.062-7.653 | 2 | 3.893 | 3.857-3.930 | 4.566 | 4.516-4.616 | 2 | |
| TNOR | 11 | 4.701 | 3.873-5.462 | 4.728 | 3.780-5.896 | 1 | 3.945 | 4.760 | 1 | |||
| SC | DRS | 1 | 4.238 | 7.197 | ||||||||
| FRS | 1 | 4.242 | 4.588 | |||||||||
| RD | 8 | 5.628 | 3.875-7.932 | 6.569 | 4.319-9.367 | 6 | 5.793 | 3.844-6.505 | 8.140 | 4.995-9.211 | 5 | |
| RDW | 6 | 4.184 | 3.959-5.076 | 4.404 | 4.129-5.395 | |||||||
| RT | 1 | 3.967 | 4.460 | |||||||||
| SFW | 1 | 3.991 | 4.339 | |||||||||
| TDW | 8 | 4.561 | 4.006-5.631 | 4.898 | 4.238-6.162 | 2 | 4.087 | 3.992-4.183 | 4.857 | 4.725-4.989 | 2 | |
| TFW | 6 | 4.447 | 3.868-5.290 | 4.792 | 4.112-5.796 | 3 | 4.678 | 4.089-4.973 | 5.637 | 4.813-6.049 | 3 | |
| TNOR | 1 | 4.148 | 4.708 | |||||||||
| DI | DRS | 1 | 4.639 | 5.288 | 1 | 5.286 | 9.930 | 1 | ||||
| FRS | 7 | 4.264 | 3.868-5.330 | 4.965 | 4.370-6.229 | |||||||
| RD | 3 | 4.432 | 4.432-4.432 | 5.154 | 5.154-5.154 | 3 | 4.225 | 4.225-4.225 | 5.133 | 5.133-5.133 | 3 | |
| RL | 1 | 4.425 | 4.979 | 1 | 3.848 | 4.513 | 1 | |||||
| RT | 3 | 4.323 | 3.872-4.906 | 5.228 | 4.415-6.447 | 1 | 4.401 | 5.838 | 1 | |||
| SDW | 5 | 4.850 | 4.421-5.085 | 5.625 | 5.064-5.907 | |||||||
| TDW | 2 | 4.274 | 4.059-4.490 | 4.902 | 4.604-5.199 | |||||||
| TNOR | 3 | 4.366 | 3.982-4.872 | 5.280 | 4.554-6.395 | 1 | 4.396 | 5.818 | 1 | |||
| Total | 208 | 28 | 25 | |||||||||
Total number of significantly associated SNPs detected by GLM and MLM at the threshold of -log10 p = 3.839
R2 value showing the percentage of explained phenotypic variation
Number of significant SNPs detected by both models
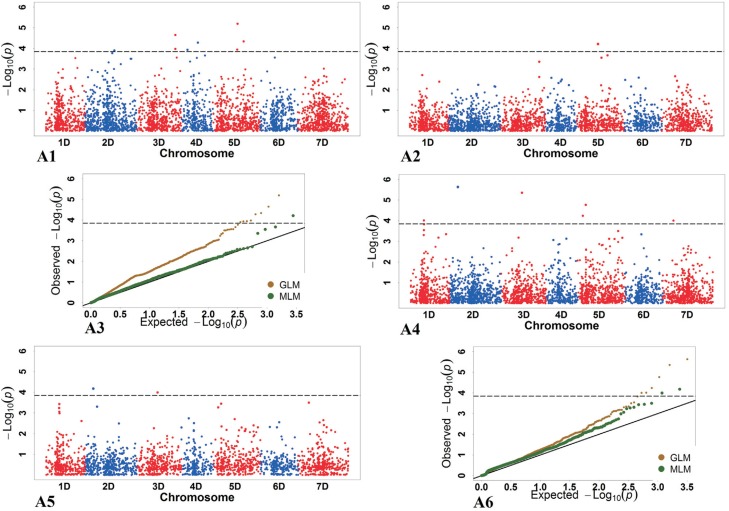
Under NC, significant markers were detected by both the GLM and MLM for FRS, RT, SDW, SFW, TDW, TFW, and TNOR (Table 4), and by the GLM for RD, RDW, RFW, RL, and SH (partly shown in Figure 1). No significant markers were detected for FRS by any of the two models.
Figure 1. The p values of the SNPs and quantile-quantile (Q-Q) plots of p values for total dry weight (TDW) under the normal condition (NC) and the PEG-induced, simulated drought-stress condition (SC). Summary of GWAS results for TDW. A1 and A2) GLM and MLM results for association under NC condition. A3) Q-Q plots of GLM and MLM under NC condition. A4 and A5) GLM and MLM results for association under SC condition. A6) Q-Q plots of GLM and MLM under SC condition.
Under SC, significant markers were detected by both the GLM and MLM for RD, TDW, and TFW, and by the GLM for FRS, RDW, RT, SFW, and TNOR (partly shown in Figure 1). No significant markers were detected for RFW, RT, SH, and SDW by any of the two models.
Numerous SNPs were significantly associated with the DI in both the GLM and MLM, and a relatively large amount of phenotypic variation in DI was explained by the studied markers (Table 4).
We performed a BLAST search against the IWGSC using the SNP sequences, and we found that their chromosomal locations were different from those of the best hits returned from IWGSC. For example, the SNP markers contig10767_892 and contig50332_70 located on Chr. 7D and 6D, respectively, on the genetic map of Luo et al. (2014) were located on Chr. 5DL and 6BL, respectively, according to the IWGSC BLAST results.
QTLs and putative candidate genes associated with significant loci
To compare the identified regions between the 373 A. tauschii accessions, markers separated by less than 5 cM were considered to be part of the same QTL (Massman et al., 2011). The results revealed three QTLs that were related to RD-SC, RD-DI, and TFW-SC. To find candidate genes, flanking genes, and trait-related proteins, we performed a BLAST search of the NCBI database using the IWGSC BLAST results and then a direct BLASTX search of the NCBI database. Putative and flanking genes associated with significant loci are listed in Table S3 (16.2KB, xlsx) . We identified several candidate genes that were associated with different traits. Examples include Rht-A that was associated with TFW-SC, RD-SC, TNOR-NC, SDW-NC, SFW-NC, TDW-NC, and TFW-NC; Rht-B associated with TFW-SC; Glo-2 associated with TFW-SC and TDW-NC; WM1.7 associated with RD-SC and RD-DI; and Acc-2 associated with RD-SC, RD-DI, TDW-SC, TNOR-NC, and FRS-DI. We also found two candidate vernalization-requirement genes, VRN2 and VRN-B1, suggesting that vernalization might be related to drought resistance.
We also identified a few putative candidate genes associated with phenotypic traits. These genes could be roughly divided into three groups: the first group included genes encoding enzymes, such as RUBISCO, CKX2.5, Acc-1 and Acc-2, suggesting that many biochemical pathways were activated under SC; the second group included genes encoding storage proteins, such as Glo-2, WM1.12, and WM1.7, which might be activated in response to drought stress; and the final group included genes encoding drought-induced proteins, such as Hotr1, Rht-A, Rht-B, VRN-B1, and VRN2, that might play a crucial role in the drought-resistance reaction of A. tauschii.
Discussion
Importance of the wheat wild relative A. tauschii
A. tauschii possesses numerous traits of high agronomic interest, such as yield, insect resistance, disease resistance, and drought resistance (Cox, 1994; Ma et al., 1995; Assefa, 2000; Aghaee-Sarbarzeh et al., 2002), and its genes can be incorporated into the wheat genome via intergenic crossing (Valkoun et al., 1990; Cox et al., 1992; Li et al., 2006; Zhang and Ma, 2008). Many agronomically useful traits have been already incorporated into wheat (Raupp et al., 1993; Cox and Hatchett, 1994; Friebe et al., 1996). In addition, artificial hybridization between tetraploid wheat and A. tauschii has resulted in allohexaploid wheat lines, known as 'resynthesized' or 'synthetic hexaploid' wheat (SW) (Mujeeb-Kazi et al., 1996), i.e. 'Chuanmai 42' (CM42), which is derived from a cross between Triticum durum and A. tauschii and is resistant to Chinese new stripe rust races (Li et al., 2006).
Based on the results of this study, we believe that drought resistance is another A. tauschii trait that could be incorporated into the wheat breeding programs. We identified A. tauschii accessions with high drought resistance that could be used as germplasm resources to widen the genetic diversity of cultivated wheat and, thus, to reduce the time required to breed for drought resistance.
Loci controlling drought resistance traits
Here, we reported the outcome of a genome-wide association study for the identification of genomic regions in A. tauschii responding to NC and SC. AM involved 7,185 SNP markers genotyped in a core collection of 373 A. tauschii accessions. Linkage mapping using different segregation populations tested in different environments could be also applied to detect QTLs, but there are only a few reports on QTL mapping related to drought-resistance traits in A. tauschii, compared with the high number of such studies in wheat using linkage mapping.
Landjeva et al. (2008) detected QTLs for RL on Chr. 1A, 6D, and 7D under SC, while Zhang et al. (2013) found two QTLs for RL associated with drought resistance on Chr. 6D in two F8:9 recombinant inbred line populations (Weimai 8 x Yannong 19 and Weimai 8 x Luohan 2). In our study, we also identified a significant locus (contig03437_336) on Chr. 6D (28.073 cM) that was associated with RL-DI, and we also found two loci related to RD-SC and RD-DI on Chr. 7D. However, Liu et al. (2013) found QTLs for RL on Chr. 2D and 5D under two different water conditions. Quarrie et al. (2005) mapped QTLs for drought resistance in hexaploid wheat on Chr. 2D and 3D, and found that three yield QTL clusters were coincident with Vrn-A1 on Chr. 5AL and Vrn-D1 on Chr. 5DL. By comparison, we identified seven significant loci on Chr. 2D and one significant locus on Chr. 2D. Furthermore, we found a candidate VRN2 at the significant loci GCE8AKX01BMYMJ_66 and GDEEGVY01D8PT5_76 located on Chr. 5D and associated with RD-SC and RD-DI. These results indicated that vernalization-required genes probably affect drought resistance in wheat. These findings further suggested the importance of exploring the relationship between drought resistance and vernalization-required genes.
Significant genome-wide loci were detected by both the GLM and MLM. Some traits were associated with multiple chromosomes, including RD-DI associated with SNPs on Chr. 1D and 6D, TFW-NC associated with SNPs on Chr. 1D and 5D, and RD-NC associated with SNPs on Chr. 4D, 5D, and 7D. Massman et al. (2011) stated that significant SNP markers separated by less than 5 cM could be considered as a single QTL. Accordingly, GCE8AKX02IHJOC_389, contig37658_165, and GA8KES402HD74L_87 (Chr. 1D) separated by less than 1 cM were considered as a single QTL related to TFW-SC. Similarly, GCE8AKX01BMYMJ_66 and GDEEGVY01D8PT5_76 (Chr. 5D) also separated by less than 1 cM were considered as a single QTL related to RD-DI and RD-SC (Table S3 (16.2KB, xlsx) ).
Until the wheat genome map is complete, loci identified in this study as associated with drought resistance traits cannot be directly compared with QTLs reported by previous studies in wheat. In addition, since the genome of A. tauschii is not equivalent to the D-genome of wheat, only approximate chromosomal locations that control drought resistance traits can be inferred. For example, contig10767_892 located on Chr. 7D in A. tauschii was found on Chr. 5DL in hexaploid wheat. Similarly, contig50332_70 located on Chr. 6D in A. tauschii was found on Chr. 6BL in wheat. One possible reason for these differences could be the translocation of chromosomal regions during the hexaploidization of common wheat, in which A. tauschii was involved.
Analysis of putative candidate and flanking genes
Drought resistance is a complex trait resulting from the interaction of root and shoot traits. In response to drought stress, wheat has developed highly specialized morphological, physiological and biochemical mechanisms to increase the efficiency of nutrient and water acquisition from soil (Ludlow and Muchow 1990; Richards et al., 2002; Nicotra and Davidson, 2010). These mechanisms are closely associated with genes controlling drought resistance and apparently responsive traits under drought conditions. Previous studies have reported many genes related to drought resistance in wheat, such as DREB that plays a central role in plant stress response (Agarwal et al., 2006; Mizoi et al., 2012) and TaAIDFa that encodes a C-repeat/dehydration-responsive element-binding factor responsive to drought (Xu et al., 2008). In addition, the silencing of TaBTF3 impairs resistance to drought stress, suggesting that it may be involved in abiotic stress response in higher plants (Kang et al., 2013). Jiang et al. (2014) isolated a strongly drought-induced C3H zinc finger gene, AetTZF1, in A. tauschii. Uga et al. (2013) characterized the DRO1 gene that controls root growth angle in rice, which was the first root QTL that cloned in a crop species. Rice OsTZF1 confers increased stress resistance to drought by regulating stress-related genes (Jan et al., 2013).
In this study, we identified several putative candidate genes associated with phenotypic traits related to drought resistance. These genes could be broadly divided into three groups: (1) genes related to various enzymes, suggesting that many biochemical pathways are activated under drought conditions; (2) genes related to storage proteins that may be synthesized in response to drought stress; and (3) genes related to drought-induced proteins that probably play a crucial role in drought resistance. These findings reflected the complexity of drought-resistance mechanisms and the large number of genes involved in these mechanisms. Information on SNPs and genes related to drought-resistance might provide a genetic basis for gene cloning and marker-assisted selection in the wheat breeding programs.
Conclusion
We performed a genome-wide association study for drought resistance traits in a population of 373 A. tauschii accessions using 7,185 SNP markers and we detected 25 significant markers using GLM and MLM analysis. Furthermore, we identified candidate genes at significant loci and their flanking regions that might control drought resistance traits, including genes encoding enzymes, storage proteins, and drought-induced proteins. The results provided essential information on SNPs and genes related to drought resistance in A. tauschii that could be used for breeding drought-resistant wheat cultivars.
Supplementary Material
The following online material is available for this article:
This material is available as part of the online article from http://www.scielo.br/gmb
Footnotes
Associate Editor: Everaldo Gonçalves de Barros
References
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25:1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Aghaee-Sarbarzeh M, Ferrahi M, Singh S, Singh H, Friebe B, Gill BS, Dhaliwal HS. Transfer of leaf and stripe rust-resistance genes from Aegilops triuncialis and Ae. Geniculata to bread wheat. Euphytica. 2002;127:377–382. [Google Scholar]
- Ashraf M, Ozturk M, Athar HR. Salinity and Water Stress: Improving Crop Efficiency. Springer; Berlin: 2009. pp. 1–243. [Google Scholar]
- Assefa S. Resistance to wheat leaf rust in Aegilops tauschii Coss and inheritance of resistance in hexaploid wheat. Genet Resour Crop Evol. 2000;47:135–140. [Google Scholar]
- Bordes J, Ravel C, Le Gouis J, Charmet G, Balfourier F. Use of global wheat core collection for association analysis of flour and dough quality traits. J Cereal Sci. 2011;54:137–147. [Google Scholar]
- Bordes J, Ravel C, Jaubertie JP, Duperrier B, Gardet O, Heumez E, Pissavy AL, Charmet G, Le Gouis J, Balfourier F. Genomic regions associated with the nitrogen limitation response revealed in a global wheat core collection. Theor Appl Genet. 2013;126:805–822. doi: 10.1007/s00122-012-2019-z. [DOI] [PubMed] [Google Scholar]
- Bouslama M, Schapaugh WT. Stress tolerance in soybeans. I. Evaluation of three screening techniques for heat and drought tolerance. Crop Sci. 1950;24:933–937. [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Breseghello F, Sorrells ME. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics. 2006;172:1165–1177. doi: 10.1534/genetics.105.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budak H, Kantar M, Yucebilgili Kurtoglu K. Drought tolerance in modern and wild wheat. Sci World J. 2013;2013 doi: 10.1155/2013/548246. 548246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox TS. Leaf rust-resistance genes Lr41, Lr42, and Lr43 transferred from Triticum tauschii to common wheat. Crop Sci. 1994;34:39–43. [Google Scholar]
- Cox TS, Hatchett JH. Hessian fly resistance gene H26 transferred from Triticum tauschii to common wheat. Crop Sci. 1994;34:958–960. [Google Scholar]
- Cox TS, Raupp WJ, Wilson DL, Gill BS, Leath S, Bockus WW. Resistance to foliar diseases in a collection of Triticum tauschii germplasm. Plant Dis. 1992;76:1061–1064. [Google Scholar]
- Crossa J, Burgueno J, Dreisickacker S, Vargas M, Herrera-Foessel SA, Lillemo M, Singh RP, Trethowan R, Warburton M, Franco J, et al. Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics. 2007;177:1889–1913. doi: 10.1534/genetics.107.078659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erena EA, Patrick PF, Byrne SD, Marta MS, Matthew MP. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor Appl Genet. 2013;4:791–807. doi: 10.1007/s00122-013-2257-8. [DOI] [PubMed] [Google Scholar]
- Ergen NZ, Budak H. Sequencing over 13,000 expressed sequence tags from six subtractive cDNA libraries of wild and modern wheats following slow drought stress. Plant Cell Environ. 2009;32:220–236. doi: 10.1111/j.1365-3040.2008.01915.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Fleury D, Jefferies S, Kuchel H, Langridge P. Genetic and genomic tools to improve drought tolerance in wheat. J Exp Bot. 2010;61:3211–3222. doi: 10.1093/jxb/erq152. [DOI] [PubMed] [Google Scholar]
- Friebe B, Jiang J, Raupp WJ, McIntSCh RA, Gill BS. Characterization of wheat alien translocations conferring resistance to diseases and pests: Current status. Euphytica. 1996;71:59–83. [Google Scholar]
- Habash DZ, Kehel Z, Nachit M. Genomic approaches for designing durum wheat ready for climate change with a focus on drought. J Exp Bot. 2009;60:2805–2815. doi: 10.1093/jxb/erp211. [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon IR. The water-culture method for growing plants without soils. Circ Calif Agric Exp Stn. 1950;347:4–32. [Google Scholar]
- Holland JB. Genetic architecture of complex traits in plants. Curr Opin Plant Biol. 2007;10:156–161. doi: 10.1016/j.pbi.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li CY, Zhu CR, Lu TT, Zhang ZW, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nature Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- Jan A, Maruyama K, Todaka D, Kidokoro S, Abo M, Yoshimura E, Shinozaki K, Nakashima K, Yamaguchi-Shinozaki K. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013;161:1202–1216. doi: 10.1104/pp.112.205385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang AL, Xu ZS, Zhao GY, Cui XY, Chen M, Li LC, Ma YZ. Genome-Wide Analysis of the C3H Zinc Finger Transcription Factor Family and Drought Responses of Members in Aegilops tauschii . Plant Mol Biol. 2014;6:1241–1256. [Google Scholar]
- Kang GZ, Ma HZ, Liu GQ, Han QX, Li CW, Guo TC. Silencing of TaBTF3 gene impairs tolerance to freezing and drought stresses in wheat. Mol Genet Genomics. 2013;11:591–599. doi: 10.1007/s00438-013-0773-5. [DOI] [PubMed] [Google Scholar]
- Kump K, Bradbury PJ, Wisser RJ, Buckler ES, Belcher AR, Oropeza-Rosas MA, Zwonitzer JC, Kresovich S, McMullen MD, Ware D, et al. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat Genet. 2011;43:163–168. doi: 10.1038/ng.747. [DOI] [PubMed] [Google Scholar]
- Landjeva S, Neumann K, Lohwasser U. Molecular mapping of genomic regions associated with wheat seedling growth under osmotic stress. Biol Plant. 2008;2:259–266. [Google Scholar]
- Li GQ, Li ZF, Yang WY, Zhang Y, He ZH, Xu SC, Singh RP, Qu YY, Xia XC. Molecular mapping of stripe rust resistance gene YrCH42 in Chinese wheat cultivar Chuanmai 42 and its allelism with Yr 24 and Yr26. Theor Appl Genet. 2006;112:1434–1440. doi: 10.1007/s00122-006-0245-y. [DOI] [PubMed] [Google Scholar]
- Liu XL, Li RZ, Chang XP, Jing RL. Mapping QTLs for seedling root traits in a doubled haploid wheat population under different water regimes. Euphytica. 2013;189:51–66. [Google Scholar]
- Ludlow MM, Muchow RC. A critical evaluation of traits for improving crop yields in water-limited environments. Advan Agron. 1990;43:107–153. [Google Scholar]
- Luo MC, Gu YQ, You FM, Deal KR, Ma Y, Hu Y, Huo N, Wang Y, Wang J, Chen S, et al. A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc Natl Acad Sci U S A. 2014;110:7940–7945. doi: 10.1073/pnas.1219082110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Singll RP, Muieeb-kazi A. Resistance to stripe rust in Triticum turgidum, T. tauschii and their synthetic hexaploids. Euphytica. 1995;82:117–120. [Google Scholar]
- Massman J, Cooper B, Horsley R, Neate S, Dill-Macky R, Chao S, Dong Y, Schwarz P, Muehlbauer GJ, Smith KP. Genome-wide association mapping of Fusarium head blight resistance in contemporary barley breeding germplasm. Mol Breeding. 2011;27:439–454. [Google Scholar]
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. Review AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819:86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Mujeeb-Kazi A, Rosas V, Roldan S. Conservation of the genetic variation of Triticum tauschii in synthetic hexaploid wheats and its potential utilization for wheat improvement. Genet Resour Crop Evol. 1996;43:129–134. [Google Scholar]
- Navakode S, Neumann K, Kobiljski B, Lohwasser U, Börner A. Genome wide association mapping to identify aluminium tolerance loci in bread wheat. Euphytica. 2014;198:401–411. [Google Scholar]
- Neumann K, Kobiljski B, Dencie S, Varshney RK, Borner A. Genome-wide association mapping: A case study in bread wheat (Triticum aestivum L.) Mol Breed. 2007;27:37–58. [Google Scholar]
- Nicotra AB, Davidson A. Adaptive phenotypic and plant water use. Funct Plant Biol. 2010;37:117–127. [Google Scholar]
- Pasam RK, Sharma R, Malosetti M, van Eeuwijk FA, Haseneyer G, Kilian B, Graner A. Genome-wide association studies for agronomical traits in a worldwide spring barley collection. BMC Plant Biol. 2012;12:16–37. doi: 10.1186/1471-2229-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;55:945–995. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrie SA, Steed A, Calestani C, Semikhodskii A, Lebreton C, Chinoy C, Steele ND, Pljevljakusi CD, Waterman E, Weyen J, et al. A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring x SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor Appl Genet. 2005;110:865–880. doi: 10.1007/s00122-004-1902-7. [DOI] [PubMed] [Google Scholar]
- Raupp WJ, Amri A, Hatchett JH, Gill BS, Wilson DL, Cox TS. Chromosomal location of Hessian fly-resistance genes H22, H23 and H24 derived from Triticum tauschii in the D genome of wheat. J Hered. 1993;84:142–145. [Google Scholar]
- Ravel C, Martre P, Romeuf I, Dardevet M, El-Malki R, Bordes J, Duchateau N, Brunel D, Balfourier F, Charmet G. Nucleotide polymorphism in the wheat transcriptional activator Spa influences its pattern of expression and has pleiotropic effects on grain protein composition, dough viscoelasticity and grain hardness. Plant Physiol. 2009;151:33–44. doi: 10.1104/pp.109.146076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RA, Rebetzke GJ, Condon AG, van Herwaarden AF. Breeding opportunities for increasing the efficiency of water use and crop yield in temperate cereals. Crop Sci. 2002;42:111–121. doi: 10.2135/cropsci2002.1110. [DOI] [PubMed] [Google Scholar]
- Smith SE, Kuehl RO, Ray IM, Hui R, Soleri D. Evaluation of simple methods for estimating broad-sense heritability in stands of randomly planted genotypes. Crop Sci. 1998;38:1125–1129. [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet. 2013;45:1097–1102. doi: 10.1038/ng.2725. [DOI] [PubMed] [Google Scholar]
- Valkoun J, Dostal J, Kucerova D. Wheat. Springer; Berlin: 1990. Triticum x Aegilops hybrids through embryo culture; pp. 152–166. [Google Scholar]
- Wang JR, Luo MC, Chen ZX, You FM, Wei YM, Zheng YL, Dvorak J. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013;198:925–937. doi: 10.1111/nph.12164. [DOI] [PubMed] [Google Scholar]
- Wilhite DA, Glantz MH. Understanding the drought phenomenon: The role of definitions. Water Int. 1985;10:111–120. [Google Scholar]
- Xie JJ. Agricultural Science and the Method of Fuzzy Mathematics. Huazhong University of Science Press; Wuhan: 1993. pp. 99–193. [Google Scholar]
- Xu ZS, Ni ZY, Liu L, Nie LN, Li LC, Chen M, Ma YZ. Characterization of the TaAIDF a gene encoding a CRT/DRE-binding factor responsive to drought, high-salt, and cold stress in wheat. Mol Genet Genomics. 2008;6:497–508. doi: 10.1007/s00438-008-0382-x. [DOI] [PubMed] [Google Scholar]
- Yang N, Lu YL, Yang XH, Huang J, Zhou Y, Ali FH, Wen WW, Liu J, Li JS, Yan JB. Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genet. 2014;10: doi: 10.1371/journal.pgen.1004573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Cui F, Wang L, Li J, Ding AM, Zhao CH, Bao YG, Yang QP, Wang H. Conditional and unconditional QTL mapping of drought-tolerance-related traits of wheat seedling using two related RIL populations. J Genet. 2013;2:213–231. doi: 10.1007/s12041-013-0253-z. [DOI] [PubMed] [Google Scholar]
- Zhang HQ, Ma SQ. Transfer of resistant genes from Aegilops tauschii L. to Triticum aestivum L. and their mapping by SSR. Zhongguo Nong Ye Da Xue Xue Bao. 2008;13:5–11. [Google Scholar]
- Zhou GS, Mei FZ, Zhou QZ, Zhu XT. Different wheat varieties during physiological index comprehensive evaluation and prediction. Zhongguo Nong Ye Ke Xue. 2003;36:1378–1382. [Google Scholar]
- Zhu C, Gore M, Buckler ES, Yu J. Status and prospects of association mapping in plants. Int J Plant Genomics. 2008;1:5–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.