Abstract
Senescence-associated proteolysis in plants is a complex and controlled process, essential for mobilization of nutrients from old or stressed tissues, mainly leaves, to growing or sink organs. Protein breakdown in senescing leaves involves many plastidial and nuclear proteases, regulators, different subcellular locations and dynamic protein traffic to ensure the complete transformation of proteins of high molecular weight into transportable and useful hydrolysed products. Protease activities are strictly regulated by specific inhibitors and through the activation of zymogens to develop their proteolytic activity at the right place and at the proper time. All these events associated with senescence have deep effects on the relocation of nutrients and as a consequence, on grain quality and crop yield. Thus, it can be considered that nutrient recycling is the common destiny of two processes, plant senescence and, proteolysis. This review article covers the most recent findings about leaf senescence features mediated by abiotic and biotic stresses as well as the participants and steps required in this physiological process, paying special attention to C1A cysteine proteases, their specific inhibitors, known as cystatins, and their potential targets, particularly the chloroplastic proteins as source for nitrogen recycling.
Keywords: barley, cysteine-proteases, leaf senescence, protein traffic, protein recycling, proteolysis
Proteolysis is associated with leaf senescence
Leaf senescence is a physiological process critical for plant survival. It is characterized by the dismantling of cellular structures, massive degradation of macromolecules and efficient relocation of nutrients from senescing leaves to growing tissues or sink organs (Gregersen et al., 2008, Krupinska et al., 2012, Avice and Etiene, 2014, Diaz-Mendoza et al., 2014). This coordinated sequence of events associated with senescence is triggered by the reprograming of thousands of genes, down- or up-regulated, in response to specific senescence-promoting factors. Accordingly, many hydrolytic enzymes targeted to degrade proteins, lipids, nucleic acids and pigments are activated. At the same time, basic metabolic activities are maintained to ensure the processing of high molecular weight molecules and the subsequent mobilization of the hydrolyzed products to the phloem (Gregersen et al., 2008, Yang and Ohlrogge, 2009, Roberts et al., 2012, Avila-Ospina et al., 2014, Sakamoto and Takami, 2014).
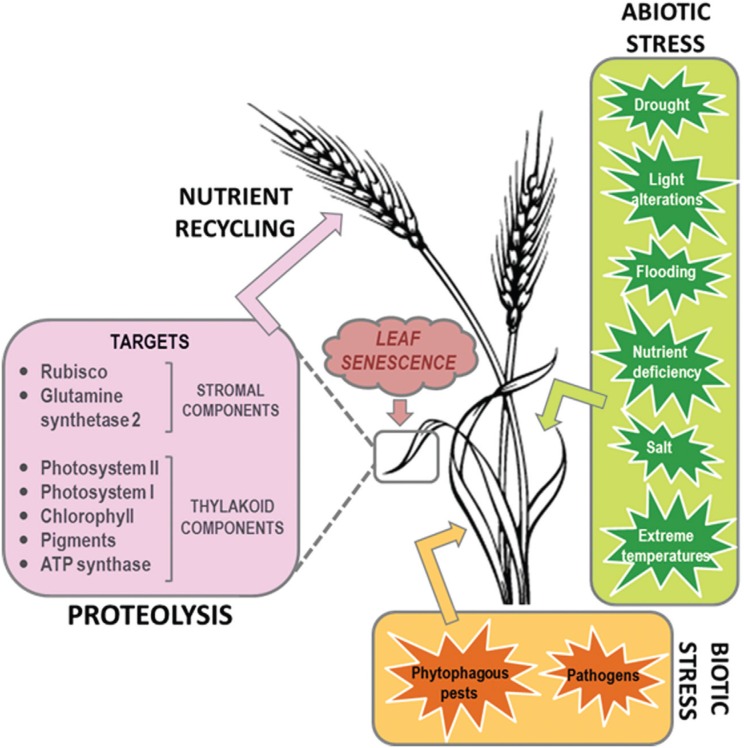
Protein breakdown is one of the most important catabolic processes associated with leaf senescence with an essential role in nutrient recycling, especially nitrogen. Changes in the temporal expression pattern of proteases take place not only in nuclei but also in chloroplasts and mitochondria to cooperatively ensure protein degradation into amino acids, amides and ammonium (Diaz-Mendoza et al., 2014, Roberts et al., 2012). As a result, a complex traffic of proteins, peptides and amino acids takes place among cell compartments involving chloroplasts, cytosol, special vesicles and lytic vacuoles (Roberts et al., 2012, Carrion et al., 2013, Avila-Ospina et al., 2014, Diaz-Mendoza et al., 2014). Finally, the major part of the nitrogen is released as ammonium after being re-assimilated into amino acids to be exported via the phloem to developing grains, fruits and tubers. In consequence, the timing of leaf senescence is of pivotal importance for yield in crop species (Gregersen et al., 2013). Figure 1 summarizes the whole set of events related to the proteolytic processes during leaf senescence.
Figure 1. Physiological events involved in induced-senescence mediated by biotic/abiotic stresses. Multiple biotic and abiotic stresses induce leaf senescence characterized by a dismantling of organelles and proteolysis, mainly from chloroplastic proteins. In consequence, protein breakdown and mobilization from stressed tissues to growing and sink organs are the major metabolic features essential for nutrient recycling.
Leaf senescence is induced by abiotic and biotic stresses
Leaf senescence is a natural developmental process but it is also closely linked to abiotic and biotic stresses. This physiological set of events can be modulated by endogenous and exogenous factors such as plant growth regulators (abscisic acid, cytokinin, ethylene, salicylic acid), sucrose starvation, dark, cold, heat, drought, salt, or wound. Moreover, pathogen infection (bacteria, fungi, viruses) and phytophagous arthropod infestation can also promote or alter senescence (Figure 1). There are numerous reports demonstrating how abiotic stresses trigger leaf senescence by reprograming specific subsets of senescence-associated genes (SAGs) that are differentially expressed in distinct tissues (Roberts et al., 2012, Diaz-Mendoza et al., 2014). This has been reviewed in recent special issues published: J Exp Bot vol. 65 and J Plant Growth Reg vol. 33 in 2014, and Plants vol. 4 in 2015, as well as in other reviews from previous years (Quirino et al., 2000; Yoshida, 2003; Guo and Gan, 2005; Gregersen et al., 2008; Martinez et al., 2008b). In contrast, information about the interplay between leaf senescence and biotic stresses is more limited, particularly with respect to leaf senescence linked to phytophagous pests. Regarding this interaction between senescence and biotic stress, it is sometimes difficult to elucidate which event comes first. Pathogen and pest lifestyles determine the developmental program of the host, and on the other side, the developmental status of the host may affect the outcome of the host-pathogen/pest interactions (Haffner et al., 2015). Pathogen infection and herbivore infestation influence leaf senescence via modulation of the plant metabolite status directly affecting primary metabolism or by regulating levels of plant hormones (Masclaux-Daubresse et al., 2010; Machado et al., 2013; Seifi et al., 2013, Fagard et al., 2014).
There are a wealth of data analysing the relationship between pathogens and plants. Likewise, induced-senescence genes have been detected during the hypersensitive response (HR) against incompatible bacteria and fungi as well as interactions with viruses (Pontier et al., 1999, Schenk et al., 2005, Espinoza et al., 2007, Fernandez-Calvino et al., 2015). The same SAGs were overexpressed during HR produced by fungal, bacterial and viral infection (Fagard et al., 2014). In Arabidopsis and grapevine, transcripts coding for aspartyl- and cysteine-protease (CysProt) increased during senescence and as a part of plant responses during compatible viral interactions (Espinoza et al., 2007). In tobacco, expression of the CysProt SAG12 was also induced during the HR against viruses and bacteria (Pontier et al., 1999). Down-regulation of OsSAG12-1 in rice brings about early senescence and enhances cell death when inoculated with Xanthomonas oryzae (Singh et al., 2013). Biotic stresses mediated by pathogens induce N mobilization in Arabidopsis (Masclaux-Daubresse et al., 2010, Fagard et al., 2014). However, references about proteolysis in leaf senescence upon arthropod feeding are occasional. Very recently, Kempema et al. (2015) have demonstrated that three SAGs, one of them SAG12, were induced in Arabidopsis plants by infestation of the hemipteran Bemisia tabaci. The green peach aphid, Myzus persicae, when fed on Arabidopsis, induces the expression of SAG13, SAG21 and SAG27 genes, cell death alongside chlorophyll degradation (Pegadaraju et al., 2005). Petrova and Smith (2015) demonstrated that the application of salivary secretions of the planthopper Nilaparvata lugens to rice induced host mRNAs associated with nutrient mobilization.
Plant responses to confluent abiotic and biotic stresses are not only the addition of the responses to independent stress. Abiotic stress factors alter not only plant defence responses but also their susceptibility to biotic interactions (Prasch and Sonnewald, 2013). The presence of an abiotic stress may reduce or enhance susceptibility to a biotic pest or pathogen and vice versa (Atkinson and Urwin, 2012). Thus, dark-induced senescence in potato promoted feeding and nymph development of the aphid Myzus persicae probably due to amino acid mobilization and phloem sap loading (Machado-Assefh et al., 2014). Similarly, nitrogen deficiency in barley seedlings induced molecular and metabolic adjustments that trigger aphid resistance. This is because N-deficient leaves were enriched in amino acids and sugars providing a more nutritive diet to phloem-feeding insects (Comadira et al., 2015), The metabolic plant profiles demonstrated that plants were adapted to low N availability by reducing photosynthesis but not respiration or protein turnover. The significance of this overlap and the precise roles of biotic- and senescence-responsive pathways remain still unknown.
Interplay between proteases, protease inhibitors and target proteins
Proteases
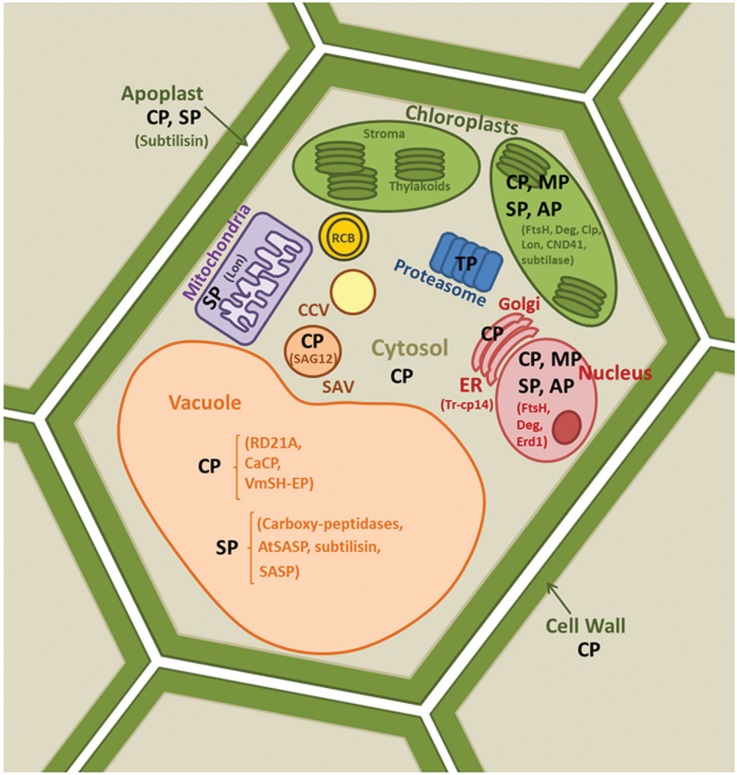
Among the more than 800 proteases identified in plant genomes (Rawlings et al., 2016), serine-proteases and CysProt have been described as the most abundant enzymes associated with leaf senescence in different plant species (Roberts et al., 2012, Diaz and Martinez, 2013, Bhalerao et al., 2003, Diaz-Mendoza et al., 2014, Kidric et al., 2014a). Aspartic-, threonine- and metallo-proteases also participate in this physiological process but their role has been less documented (Graham et al., 1991, Roberts et al., 2012). Expression studies have shown changes in the temporal patterns and subcellular location of proteases during senescence, which is consistent with alterations in proteolytic activities (Breeze et al., 2011, Roberts et al., 2012, Kidric et al., 2014a). Plant proteases have been detected in different cellular compartments such as nuclei, chloroplasts, cytosol, endoplasmic reticulum (ER), vacuoles, mitochondria, apoplast, cell wall or special vesicles (Figure 2), where they fulfil specific functions.
Figure 2. Location of plant proteases in different cellular compartments involved in plant senescence. Different families of proteases are represented: Cysteine-Proteases (CP), Serine-Proteases (SP), Metallo-Proteases (MP), Threonine-Proteases (TP) and Aspartic-Proteases (AP). Subcellular localization of specific proteases, indicated in brackets, are in chloroplast (FtsH-, Clp-, Lon-, DegP- CND41- and subtilase-like proteases), vacuole (RD21A, CaCP, VmSh-EP, AtSAP, subtilase-, SASP-like proteases), mitochondria (Lon-like protease), SAV (SAG12-like protease), ER (Tr-cp-14-like protease) and apoplast (subtilisin). Endoplasmic Reticulum (ER), Rubisco Containing Body (RCB), Senescence Associated Vesicle (SAV) and Chloroplast Containing Vesicles (CCV).
The main proteolytic system in the cytosol is the ubiquitin/26S proteasome pathway, a complex structure involving several proteolytic activities as well as a large set of enzymes needed for covalent binding of targeted proteins to ubiquitin for degradation (Vierstra, 2009). Organelles such as mitochondria, peroxisomes and chloroplasts possess their own conserved proteolytic machinery. In particular, the degradation of the chloroplastic proteins associated with senescence is mediated by the combination of their own proteases and the action of nuclear encoded proteases. These nuclear genes encode precursor proteins with N-terminal extensions known as signal peptides that redirect the processed protein to specific cell locations (Teixeira and Glaser, 2013). Intra-plastidial proteolysis takes place mainly by the action of different forms of FtsH metallo-proteases, Clp serine-proteases, and Lon-like ATP-dependent proteases and DegP serine-proteases, ATP-independent proteases. Members of the DegP, Clp and FtsH proteases are up-regulated in senescing leaves and participate in the degradation of plastidial photosystem II (Roberts et al., 2012). Kato et al. (2004) described the proteolytic action of the chloroplast CND41 aspartic-protease on Rubisco (Ribulose 1,5- bisphosphate carboxylase-oxygenase) breakdown during senescence as well as its implication in nitrogen translocation. The over-expression of CND41 reduced Rubisco in senescent tobacco leaves whereas CND41-silenced lines delayed senescence and maintained higher levels of Rubisco in old leaves (Kato et al., 2005). Besides, activities of other chloroplast proteases have been shown to increase under senescence, as in the case of an alkaline serine-protease (subtilase) induced in leaves of spinach under salinity stress and in desiccated leaves of Ramonda serbica (Srivastava et al., 2009, Kidric et al., 2014b).
Plastidial proteolysis proceeds outside the organelle through the action of proteases in the cytosol, apoplast or vacuoles (Figure 2). Subunits of the proteasome system are up-regulated by abiotic stress-induced senescence in leaves of tomato and seedlings of Arabidopsis thaliana under iron or potassium deficiency (Kidric et al., 2014a). Another example is the Tr-cp14 CysProt of Trifolium repens, localized in the ER and associated with senescence in leaves (Mulisch et al., 2013). Proteolytic activities have been also detected in cell walls and inter-cellular spaces (Brzin and Kidric, 1995). Extra-cellular proteases that catalyze the hydrolysis of proteins into peptides and amino acids for subsequent incorporation into the cell constitute a very important step in nitrogen metabolism at this level (Vierstra, 1996, Lopez-Otin and Bond, 2008, Kidric et al., 2014a). Proteolysis during senescence is completed in the acidic environment of the vacuole, which mainly contains C1A CysProt with acidic pH optima, among other enzymes (Thoenen et al., 2007, Ishida et al., 2008, van Doorn et al., 2011). One example is CysProt RD21A, a major protease activity in Arabidopsis leaf extracts and responsible for inducing proteome degradation in the vacuoles of senescing leaves (Yamada et al., 2001, Gu et al., 2012). The silencing of the CaCP gene encoding the vacuolar CysProt CaCP of Capsicum annuum L. delays salt- and osmotic-induced leaf senescence (Xiao et al., 2014). Although less abundant, carboxy-proteases belonging to serine-protease are also present in vacuoles (van der Hoorn, 2008). For instance, the Arabidopsis AtSASP subtilisin serine-protease has been detected in the proteome of central vacuoles isolated from vegetative leaves (Carter et al., 2004), as well as other two subtilisins, also termed Senescence-Associated Subtilisin Proteases (SASP) with increased proteolytic activity in senescing leaves of this model plant species (Martinez et al., 2015). Recently, Distelfeld et al. (2014) have listed several direct and indirect lines of evidence demonstrating the importance of the vacuolar proteases for the complete plastidial protein degradation.
This extra-plastidial pathway of degradation is dependent on ATG genes which contribute at different levels in the autophagy pathway and requires a complex trafficking of proteins from the chloroplast to the central vacuole. A recent review published by Carrion et al. (2014) has characterized the Senescence-Associated Vacuoles (SAVs) as specific lytic compartments for degradation of chloroplastic proteins. SAVs coexist with the central vacuole in senescent leaves and they are part of the vesicular transport system where proteolysis may continue due to the presence of active CysProt (Otegui et al., 2005, Martinez et al., 2007, 2008a). The detection of the large subunit of Rubisco and glutamine synthetase 2 (GS2) in a SAV- enriched fraction purified from senescent leaves, and the presence of the CysProt SAG12-GFP, confirms the co-location of both plastidial proteins and CysProt to SAVs during senescence (Martinez et al., 2008a, Carrion et al., 2013). Martinez et al. (2008a) also detected a small fraction of chlorophyll a using HPLC technology in these acidic vesicles suggesting that pigment disassembly may be carried out through this transport pathway under certain conditions. Moreover, in vivo inhibition of CysProt completely abolished Rubisco degradation in isolated SAVs (Carrion et al., 2013, 2014). In addition to SAVs, autophagic bodies named Rubisco-Containing Bodies (RCBs), double membrane bounded vesicles of small size derived from chloroplasts, have been detected in the cytosol of senescent leaves and are also redirected to the central vacuole (Chiba et al., 2003, Prins et al., 2008,). These vesicles carry stromal proteins or their hydrolytic products but not thylakoid proteins, with no evidences of any protease activity within them (Ishida et al., 2008, Carrion et al., 2013). Wang and Blumwald (2014) described a third pathway for the degradation of chloroplastic proteins in stress-induced senescing Arabidopsis based on chloroplast vesiculation (CV), independent of RCBs or SAVs. CCVs (Chloroplast-Containing Vesicles) carry stromal proteins as thylakoid membrane protein (FtsH1), luminal protein (PsbO1) and inner envelope membrane protein (Tic20-II), are released from chloroplasts and redirected to the central vacuole for proteolysis. Further studies are needed to identify partners and protein trafficking routes to senescence in which plastids, vesicles, vacuoles and cytosol are inter-connected for an efficient protein degradation and the subsequent relocation of nutrients.
Protease activity regulation
Senescence-associated proteolysis in plants is a controlled process where protease activities can be regulated by controlling the protease transcript content through transcriptional regulation, or by control of the activity itself by post-translational processing (Cambra et al., 2012b, Christiansen and Gregersen, 2014, Hollmann et al., 2014). Additionally, enzyme activity is regulated by specific inhibitors and cofactors and through the activation of zymogens. In particular, members of the papain-like subfamily C1A CysProt, probably the most widely studied among plant proteases, are synthesized as inactive or little active precursors to prevent spatio-temporal inappropriate proteolysis. To become active, the C1A proteases are either self-processed or require the aid of other enzymes. Activation takes places by limited intra- or inter-molecular proteolysis cleaving off the inhibitory pro-peptide (Wiederanders, 2003). Therefore, the pro-sequences play important roles as modulators of the protease activity to guarantee that the mature enzyme is formed in the right place and/or at the right time. Pro-peptides are not only able to inhibit their cognate enzymes but also other related proteases in trans (Cambra et al., 2012a). Until now, no information has been published about the pro-peptide function as modulators of leaf senescence, but there are some examples demonstrating their regulatory role of such peptides in barley grain germination (Cambra et al., 2012a, 2012b).
Although protease inhibitors act as modulators of the protease activities to control protein turnover, plant protease-inhibitor interactions in response to abiotic stresses are still poorly documented apart from the CysProt and their specific inhibitors known as phytocystatins (Kidric et al., 2014a, Kunert et al., 2015). Cystatins and proteases not only co-localize to the ER and to the Golgi complex, but also interact in these compartments (Martinez et al., 2009). Evidence of in vivo interactions has been obtained from BiFC (Bimolecular Fluorescent Complementation) assays using barley cystatins and cathepsin L-like CysProt fused to moieties of the green fluorescent protein marker (GFP). The formation of a CysProt-cystatin complex has also been reported in senescent spinach leaves (Tajima et al., 2011). In a recent publication, Kunert et al. (2015) have comented on a current study that they are doing on the interaction between purified recombinant cystatins and CysProt that are expressed during drought using in vitro assay systems. These data, in combination with immuno-histochemistry assays will allow them to analyze the intra-cellular localization under optimal and stress conditions. These studies can be essential to demonstrate the specificity of any protease-inhibitor interaction. The CysProt-cystatin interaction has also been indirectly explored in transgenic plants. Prins et al. (2008) found an increase of immunogold-labelled Rubisco in chloroplasts as well as in RCBs of tobacco plants overexpressing the rice cystatin OC-I in comparison to the non-transformed controls, whereas OC-I in the cytosol, vacuole, and chloroplasts of these transgenic plants (Prins et al., 2008). Expression of this rice cystatin in soybean and Arabidopsis plants leads to enhanced drought stress tolerance through effects on strigolactone pathways and can also result in improved seed traits (Quain et al., 2014). Overexpression of the broccoli BoCPI-1 cystatin leads to a decrease in total protease activity and delays chlorophyll degradation and, in consequence, the onset of senescence in broccoli florets after harvest (Eason et al., 2014). Je et al. (2014) have shown that DREB2 (Dehydration-Responsive Element-Binding factor) acts as transcriptional activator of the thermotolerance-related cystatin 4 gene from Arabidopsis, reducing CysProt activity. These findings demonstrate that cystatins can be applied as important regulatory proteins of senescence in biotechnological systems. Other classes of protease inhibitors, mainly targeting serine-proteases, also enhance tolerance to abiotic stress conditions (Shan et al., 2008, Srinivasan et al., 2009), but there is still little information about their importance for leaf senescence.
Protease targets
Leaf senescence is characterized by a yellowish phenotype parallel to strong plastid disorganization while the rest of the cell organelles remain practically intact until the end of the senescence period. This main hallmark of senescence, leaf yellowing, is caused by the preferential degradation of chlorophyll over carotenoids (Matile, 1992). Chlorophyll degradation occurs by a specific catalytic route termed PAO (from pheophorbide a oxygenase). It includes the formation of a primary fluorescent catabolite in the plastid, followed by isomerization to produce non-fluorescent catabolites in the central vacuole (Hörtensteiner, 2013, Christ and Hörtensteiner, 2014). Our own results have demonstrated that a retarded chlorophyll loss parallel to higher protein content is produced in knock-down HvPap-1 CysProt barley lines grown under darkness (unpublished data). Proteolysis associated with senescence provides free peptides or amino acids and redistributes them within the plant. Degradation of plastidial proteins represents the main source of nitrogen remobilization, and most studies have been focused in this organelle rather than in the other cell compartments (Schiltz et al., 2004, Keech et al., 2007, Avila-Ospina et al., 2014). Most evidence suggests that degradation of stromal proteins, mainly Rubisco and GS2, occurs earlier than degradation of chlorophyll and thylakoidal proteins proteins such as D1, LHCII of the PSII reaction centre and PSII antenna (Krupinska, 2007). However, variability in protein degradation depending on species/cultivars and environmental conditions has also been found (Simova-Stoilova et al., 2010; Krupinska et al., 2012). Degradation of thylakoidal proteins, in particular these from PSII, represents the second largest pool of remobilizable nitrogen from chloroplast during leaf senescence, about 30% of the total chloroplast protein (Matile, 1992; Simova-Stoilova et al., 2010). A faster decline in PSII vs PSI was detected during heat stress-promoted leaf senescence in wheat (Hortensteiner and Matile, 2004). Apart from membrane disassembling, photosynthetic proteins both PSI and PSII as well as ATP synthase are hydrolyzed as observed in ultrastructural studies (Ghosh et al., 2001, Guiamet et al., 2002, Krupinska et al., 2012). Immunoblot and ultrastructural results have shown a preferential degradation of granal over stromal proteins, resulting in an unexpected increase in the chlorophyll a/b ratio, meaning that chlorophyll b is degraded faster (Desimone et al., 1996).
Regarding stromal proteins, Rubisco and Rubisco activase seem to be principal targets for CysProt during leaf senescence in C3 plants (Prins et al., 2008). For this reason, understanding the mechanisms of Rubisco degradation has become a key purpose (Desimone et al., 1996, Schiltz et al., 2004, Irving and Robinson, 2006). Rubisco fragmentation has been detected in intact isolated chloroplasts of pea and wheat incubated under continuous light or dark conditions (Mitsuhashi and Feller, 192, Zhang et al., 2007). Likewise, Desimone et al. (1996) studied the nature of Rubisco degradation under oxidative stress in isolated chloroplasts of barley. Several hypotheses put forward that Reactive Oxygen Species (ROS) might be involved in the initial denaturation of Rubisco, by oxydazing certain cysteine residues and thus rendering the protein as a more susceptible target for protease cleavage (Garcia-Ferris and Moreno, 1994). Nonetheless, this ROS prompted degradation does not seem to be sufficient for complete degradation (Desimone et al., 1996). It remains to be elucidated if denaturation and breakdown events are sequential or if both occur at the same time.
GS2 is also susceptible to proteolysis as shown in isolated tobacco chloroplasts. This enzyme is almost lost during early stages of senescence in cereal leaves but the cytosolic GS1, the key enzyme for ammonia assimilation and de novo synthesis of amino acids from released nutrients, is maintained (Mitsuhashi and Feller, 1992, Khanna-Chopra, 2012). There are some in vitro approaches showing that GS2 is degraded before other enzymes involved in carbon assimilation, such as Rubisco (Thoenen and Feller, 1998). Proteolysis of GS2 seems to be initiated through oxidative carbonylation of histidine residues (Palatnik et al., 1999, Ishida et al., 2002). The presence of only stromal proteins (Rubisco and GS2) within specialized SAVs enriched in CysProt activity, suggests that these enzymes are responsible for the degradation of this stromal protein fraction (Martinez et al., 2008a, Carrion et al., 2013, 2014). Fischer and Feller (1994) examined a broad set of other stromal enzymes in young winter wheat leaves, detecting different degradation rates and discussed how proteolysis within the same organelle can be selectively regulated.
Influence of leaf senescence on cereal grain yield and quality
Up to 90% of the nitrogen in the grain of several cereals like barley, wheat and rice comes from senescing vegetative tissues. Thomas and Stoddart (1980) postulated that delayed senescence, which means an extended period of maximal photosynthetic activity, should lead to higher yields. However, grain yield is a complex trait that involves different physiological processes. For example, in cereal crops, it is determined by both the source tissues and the sink organs. Fischer (2008) proposed that in small-grain cereals such as wheat, the main limiting factor for grain yield is the strength of the seed, suggesting that the physiological events in the period around seed setting are crucial for determining yield levels. Conversely, there are numerous examples for a positive correlation between delayed senescence and yield levels in both small-grain cereals such as wheat and barley and large-grain cereals such as maize (Gregersen et al., 2013). Furthermore, Egli (2011) stated that enhanced seed yields are obtained when the grain filling period is longer. Thus, manipulation of senescence events could be a way to obtain higher grain yield and quality.
Yield is not the only grain feature affected by plant senescence. Grain quality parameters such as protein and micronutrient (Fe, Zn) concentrations are also affected by alterations in the senescence process (Distelfeld et al., 2014). Grain yield and protein content are negatively correlated in wheat and barley (Thoenen and Feller, 1998, Palatnik et al., 1999, Gregersen et al., 2008). While delayed senescence may lead to higher grain yield but it also provokes inefficient nitrogen remobilization and lower harvest index (Gong et al., 2005), acceleration of senescence confers efficient nitrogen remobilization and high protein content, but also a lower total grain yield. Distelfeld et al. (2014) indicated two possible justifications for this effect. The first explanation suggests that in delayed senescence the prolonged accumulation of carbohydrates dilutes stored proteins and micronutrients leading to increased grain weight and yield but lower grain quality (Slafer et al., 1990, Gregersen et al., 2008). The second explanation is based on the fact that the synthesis of storage proteins requires more carbon consumption than the synthesis of starch, which enhances the accumulation of carbohydrates (Munier-Jolain and Salon, 2005).
To cope with this dilemma it is important to balance the agronomical or economic consequences with the quality of the end-product. For barley grains, low protein content is desirable in malting processes to obtain beer whereas high protein content is desirable to be used for animal feeding. Moreover, slow grain filling associated with delayed senescence implies an increased probability of damage by environmental factors such as heat and drought stress and drought during the later stages of crop development (Mi et al., 2002, Yang and Zhang, 2006). Manipulation of the proteolytic machinery is a potential way to enhance grain yield and quality. Using this approach, novel breeding strategies that consider the complexity of the feature are promising tools to achieve a higher grain yield and quality.
Concluding remarks
Strong indications from multiple studies indicate that prevention of premature senescence induced by biotic/abiotic stresses may be the key in engineering stress tolerance. This review compiled the current knowledge on different aspects related to degradation of proteins as part of the leaf senescence mediated by different stresses, and their potential effects on crop yields. Future research should be done to acquire more precise information about protease action and protease targets and to increase knowledge concerning the traffic of hydrolysed proteins from their original subcellular locations via specialized vesicles to the central lytic vacuole. Clarifying how protease activity is regulated by a specific inhibitor may contribute to understand the balance between recovery from stress and excessive protein degradation resulting in cell death. Overall, research is needed to encompass the complete set of information in order to understand why, how, where and when leaf senescence is produced. The final goal will be to control the impact of senescence in agriculture keeping in mind the current and future effects of extreme weather events related to climate changes.
Acknowledgments
This work was supported by the Ministerio de Economia y Competitividad of Spain (projects BIO2014-53508-R and 618105 FACCE Era Net Plus) and by the Juan de la Ciera 2012 subprogram.
Footnotes
Associate Editor: Carlos F. M. Menck
References
- Atkinson NJ, Urwin PE. The interaction of biotic and abiotic stresses: From genes to the field. J Exp Bot. 2012;63:3523–3543. doi: 10.1093/jxb/ers100. [DOI] [PubMed] [Google Scholar]
- Avice JC, Etiene P. Leaf senescence and nitrogen remobilization efficiency in oilseed rape (Brassica napus L.) J Exp Bot. 2014;65:3813–3824. doi: 10.1093/jxb/eru177. [DOI] [PubMed] [Google Scholar]
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C. Autophagy, plant senescence, and nutrient recycling. J Exp Bot. 2014;65:3799–3811. doi: 10.1093/jxb/eru039. [DOI] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F, Erlandsson R, Björkbacka H, Birve SJ, Karlsson J, Gardeström P, Gustafsson P, Lundeberg J, et al. Gene expression in autumn leaves. Plant Physiol. 2003;131:430–442. doi: 10.1104/pp.012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell. 2011;23:873–894. doi: 10.1105/tpc.111.083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzin J, Kidric M. Proteinases and their inhibitors in plants: Role in normal growth and in response to various stress conditions. Biotech Genetic Eng Rev. 1995;13:420–467. [Google Scholar]
- Cambra I, Hernandez D, Diaz I, Martinez M. Structural basis for specificity of propeptide-enzyme interaction in barley C1A cysteine peptidase. PLoS One. 2012a;7: doi: 10.1371/journal.pone.0037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambra I, Martinez M, Dader B, Gonzalez-Melendi P, Gandullo J, Santamaria ME, Diaz I. A cathepsin F-like peptidase involved in barley grain protein mobilization, HvPap-1 is modulated by its own propeptide and by cystatins. J Exp Bot. 2012b;63:4615–4629. doi: 10.1093/jxb/ers137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion CA, Costa ML, Martinez DE, Mohr C, Humbeck K, Guiamet JJ. In vivo inhibition of cysteine proteases provides evidence for the involvement of 'senescence-associated vacuoles' in chloroplast protein degradation during dark-induced senescence of tobacco leaves. J Exp Bot. 2013;64:4967–4980. doi: 10.1093/jxb/ert285. [DOI] [PubMed] [Google Scholar]
- Carrion CA, Martinez DE, Costa ML, Guiamet JJ. Senescence-associated vacuoles, a specific lytic compartment for degradation of chloroplastic proteins? Plants. 2014;3:498–512. doi: 10.3390/plants3040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell. 2004;16:3285–3303. doi: 10.1105/tpc.104.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba A, Ishida H, Nishizawa NK, Makino A, Mae T. Exclusion of ribulose-1,5-bisphosphate carboxylase/oxygenase from chloroplasts by specific bodies in naturally senescing leaves of wheat. Plant Cell Physiol. 2003;44:914–921. doi: 10.1093/pcp/pcg118. [DOI] [PubMed] [Google Scholar]
- Christ B, Hörtensteiner S. Mechanism and significance of chlorophyll breakdown. J Plant Growth Regul. 2014;33:4–20. [Google Scholar]
- Christiansen MW, Gregersen PL. Members of the barley NAC transcription factor gene family show differential co-regulation with senescence-associated genes during senescence of flag leaves. J Exp Bot. 2014;65:4009–4022. doi: 10.1093/jxb/eru046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comadira G, Rasool B, Kaprinska B, Morris J, Verrall SR, Hedley PE, Foyer CH. Nitrogen deficiency in barley (Hordeum vulgare) seedling induces molecular and metabolic adjustments that trigger aphis resistance. J Exp Bot. 2015;66:3639–3655. doi: 10.1093/jxb/erv276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desimone M, Henke A, Wagner E. Oxidative stress induces partial degradation of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase in isolated chloroplasts of barley. Plant Physiol. 1996;111:789–796. doi: 10.1104/pp.111.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz I, Martinez M. Plant C1A cysteine peptidases in germination and senescence. In: Rawlings ND, Salvesen G, editors. Handbook of Proteolytic Enzymes. Elsevier; Amsterdam: 2013. pp. 1853–1858. [Google Scholar]
- Diaz-Mendoza M, Arroyo-Velasco B, Gonzalez-Melendi P, Martinez M, Diaz I. C1A cysteine protease-cystatin interactions in leaf senescence. J Exp Bot. 2014;65:3825–3833. doi: 10.1093/jxb/eru043. [DOI] [PubMed] [Google Scholar]
- Distelfeld A, Avni R, Fischer AM. Senescence, nutrient remobilization, and yield in wheat and barley. J Exp Bot. 2014;65:3783–3798. doi: 10.1093/jxb/ert477. [DOI] [PubMed] [Google Scholar]
- Eason JR, West PJ, Brummell DA, Watson LM, Somerfield SD, McLachlan ARG. Overexpression of the protease inhibitor BoCPI-1 in broccoli delays chlorophyll loss after harvest and causes down-regulation of cysteine protease gene expression. Postharv Biol Technol. 2014;97:23–31. [Google Scholar]
- Egli DB. Time and the productivity of agronomic crops and cropping systems. Agron J. 2011;103:743–750. [Google Scholar]
- Espinoza C, Medina C, Somerville S, Arce-Johnson P. Senescence-associated genes induced during compatible viral interactions with grapevine and Arabidopsis. J Exp Bot. 2007;58:3197–3212. doi: 10.1093/jxb/erm165. [DOI] [PubMed] [Google Scholar]
- Fagard M, Launay A, Clement G, Courtial J, Dellagi A, Farjad M, Krapp A, Soulie MC, Masclaux-Daubresse C. Nitrogen metabolism meets phytopathology. J Exp Bot. 2014;65:5643–5656. doi: 10.1093/jxb/eru323. [DOI] [PubMed] [Google Scholar]
- Fernandez-Calvino L, Guzman-Benito I, Toro FJ, Donaire L, Castro-Sanz AB, Ruiz-Ferrer V, Llave C. Activation of senescence-associated Dark-inducible (DIN) genes during infection contributes to enhanced susceptibility to plant viruses. Mol Plant Pathol. 2015;17:3–15. doi: 10.1111/mpp.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RA. The importance of grain and kernel number in wheat: A reply to Sinclair and Jamieson. Fields Crops Res. 2008;105:15–21. [Google Scholar]
- Fischer A, Feller U. Senescence and protein degradation in leaf segments of young winter wheat - Influence of lead age. J Exp Bot. 1994;45:103–109. [Google Scholar]
- Garcia-Ferris C, Moreno J. Oxidative modification and breakdown of ribulose-1,5-bisphosphate carboxylase oxygenase induced in Euglena gracilis by nitrogen starvation. Planta. 1994;193:208–215. [Google Scholar]
- Gong Y, Zhang J, Gao J, Lu L, Wang J. Slow export of photoassimilate from stay-green leaves during late grain-filling stage in hybrid winter wheat (Triticum aestivum L.) J Agron Crop Sci. 2005;191:292–299. [Google Scholar]
- Graham JS, Xiong J, Gillikin JW. Purification and development analysis of a metallo-endoproteinase from the leaves of Glycine max . Plant Physiol. 1991;97:786–792. doi: 10.1104/pp.97.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen PL, Culetic A, Boschian L, Krupinska K. Plant senescence and crop productivity. Plant Mol Biol. 2013;82:603–622. doi: 10.1007/s11103-013-0013-8. [DOI] [PubMed] [Google Scholar]
- Gregersen PL, Holm PB, Krupinska K. Leaf senescence and nutrient remobilization in barley and wheat. Plant Biol. 2008;10:37–49. doi: 10.1111/j.1438-8677.2008.00114.x. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Mahoney SR, Penterman JN, Peirson D, Dumbroff EB. Ultrastructural and biochemical changes in chloroplasts during Brassica napus senescence. Plant Physiol Biochem. 2001;39:777–784. [Google Scholar]
- Gu C, Shabab M, Strasser R, Wolters PJ, Shindo T, Niemer M, Kaschani F, Mach L, van der Hoorn RA. Post-translational regulation and trafficking of the granulin-containing protease RD21 of Arabidopsis thaliana . PLoS One. 2012;7: doi: 10.1371/journal.pone.0032422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Gan S. Leaf senescence: Signals, execution, and regulation. Current Top Dev Biol. 2005;71:83–112. doi: 10.1016/S0070-2153(05)71003-6. [DOI] [PubMed] [Google Scholar]
- Guiamet JJ, Tyystjärvi E, Tyystjärvi T, John I, Kairavuo M, Pichersky P, Noodén LD. Photoinhibition and loss of photosystem II reaction center proteins during senescence of soybean leaves. Enhancement of photoinhibition by the "stay- green" mutation cytG. Physiol Plant. 2002;115:468–478. doi: 10.1034/j.1399-3054.2002.1150317.x. [DOI] [PubMed] [Google Scholar]
- Haffner E, Konietzki P, Diederichsen E. Keeping control: The role of senescence and development in plant pathogenesis and defense. Plants. 2015;4:449–488. doi: 10.3390/plants4030449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann J, Gregersen PL, Krupinska K. Identification of predominant genes involved in regulation and execution of senescence-associated nitrogen remobilization in flag leaves of filed grown barley. J Exp Bot. 2014;65:3963–3973. doi: 10.1093/jxb/eru094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. Update on the biochemistry of chlorophyll breakdown. Plant Mol Biol. 2013;82:505–517. doi: 10.1007/s11103-012-9940-z. [DOI] [PubMed] [Google Scholar]
- Hortensteiner S, Matile P. How leaves turn yellow: Catabolism of chlorophyll. In: Nooden LD, editor. Plant Cell Death Processes. Elsevier Academic Press; San Diego: 2004. pp. 189–202. [Google Scholar]
- Irving LJ, Robinson D. A dynamic model of Rubisco turnover in cereal leaves. New Phytol. 2006;169:493–504. doi: 10.1111/j.1469-8137.2005.01584.x. [DOI] [PubMed] [Google Scholar]
- Ishida H, Anzawa D, Kokubun N, Makino A, Mae T. Direct evidence for non-enzymatic fragmentation of chloroplastic glutamine synthetase by a reactive oxygen species. Plant Cell Environ. 2002;25:625–631. [Google Scholar]
- Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T. Mobilization of Rubisco and stroma-localized fluorescent proteins of chloroplast to the vacuole by ATG gene-dependent autophagic process. Plant Physiol. 2008;148:142–155. doi: 10.1104/pp.108.122770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je J, Song C, Hwang JE, Chyng WS, Lim CO. DREB2C acts a transcriptional activator of the thermo tolerance-related phytocystatin 4 (AtCYS4) gene. Transgenic Res. 2014;23:10–123. doi: 10.1007/s11248-013-9735-2. [DOI] [PubMed] [Google Scholar]
- Kato Y, Murakami S, Yamamoto Y, Chantani H, Kondo Y, Nakano T, Yokota A, Sato F. The DNA-binding protease, CND41, and the degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase in senescent leaves of tobacco. Planta. 2004;220:97–104. doi: 10.1007/s00425-004-1328-0. [DOI] [PubMed] [Google Scholar]
- Kato Y, Yamamoto Y, Murakami S, Sato F. Post-translational regulation of CND41 protease activity in senescent tobacco leaves. Planta. 2005;222:643–651. doi: 10.1007/s00425-005-0011-4. [DOI] [PubMed] [Google Scholar]
- Keech O, Pesquet E, Ahad A, Askne A, Nordvall D, Vodnala SM, Tuominen H, Hurry V, Dizengremel P, Gardeström P. The different fates of mitochondria and chloroplasts during dark-induced senescence in Arabidopsis leaves. Plant Cell Environ. 2007;30:1523–1534. doi: 10.1111/j.1365-3040.2007.01724.x. [DOI] [PubMed] [Google Scholar]
- Kempema LA, Cui X, Holzer FM, Walling LL. Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol. 2015;143:849–865. doi: 10.1104/pp.106.090662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna-Chopra R. Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma. 2012;249:469–481. doi: 10.1007/s00709-011-0308-z. [DOI] [PubMed] [Google Scholar]
- Kidric M, Kos J, Sabotic J. Protease and their endogenous inhibitors in the plant response to abiotic stress. Bot Serb. 2014a;38:139–158. [Google Scholar]
- Kidric M, Sabotic J, Stevanovic B. Desiccation tolerance of resurrection plant Ramonda serbica is associated with dehydration dependent changes in levels of proteolytic activities. J Plant Physiol. 2014b;171:998–1002. doi: 10.1016/j.jplph.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Krupinska K. Fate and activities of plastids during leaf senescence. In: Wise RR, Hoober JK, editors. The Structure and Function of Plastids. Springer; Dordrecht: 2007. pp. 403–449. [Google Scholar]
- Krupinska K, Mulisch M, Hollmann J, Tokarz K, Zschiesche W, Kage H, Humbeck K, Biler W. An alternative strategy of dismantling of the chloroplast during leaf senescence observed in a high-yield variety of barley. Physiol Plant. 2012;144:189–200. doi: 10.1111/j.1399-3054.2011.01545.x. [DOI] [PubMed] [Google Scholar]
- Kunert KJ, van Wyk SG, Cullis CA, Vorster BJ, Foyer CH. Potential use of phytocystatins in crop improvement, with a particular focus on legumes. J Exp Bot. 2015;66:3559–35570. doi: 10.1093/jxb/erv211. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Bond JS. Proteases: Multifunctional enzymes in life and disease. J Biol Chem. 2008;283:30433–30437. doi: 10.1074/jbc.R800035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado RA, Ferrieri AP, Robert CA, Glauser G, Kallenbach M, Baldwin IT, Erb M. Leaf-herbivore attack reduces carbon reserves and regrowth from the roots via jasmonate and auxin signalling. New Phytol. 2013;200:124–1246. doi: 10.1111/nph.12438. [DOI] [PubMed] [Google Scholar]
- Machado-Assefh CR, Lucatti AF, Alvarez AE. Induced senescence promotes the feeding activities and nymph development of Myzus persicae (Hemiptera: Aphididae) on potato plants. J Insect Sci. 2014;14:155–155. doi: 10.1093/jisesa/ieu017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez DE, Bartoli CG, Grbic V, Guiamet JJ. Vacuolar cysteine proteases of wheat (Triticum aestivum L) are common to leaf senescence induced by different factors. J Exp Bot. 2007;58:1099–1107. doi: 10.1093/jxb/erl270. [DOI] [PubMed] [Google Scholar]
- Martinez DE, Borniego ML, Battchikova N, Aro EM, Tyystjärvi E, Guiamet JJ. SASP, a Senescence-Associated Subtilisin Protease, is involved in reproductive development and determination of silique number in Arabidopsis. J Exp Bot. 2015;66:161–174. doi: 10.1093/jxb/eru409. [DOI] [PubMed] [Google Scholar]
- Martinez DE, Costa ML, Guiamet JJ. Senescence-associated degradation of chloroplast proteins inside and outside the organelle. Plant Biol. 2008a;10(1):15–22. doi: 10.1111/j.1438-8677.2008.00089.x. [DOI] [PubMed] [Google Scholar]
- Martinez DE, Costa ML, Gomez FM, Otegui MS, Guiamet JJ. "Senescence-associated vacuoles" are involved in the degradation of chloroplasts proteins in tobacco leaves. Plant J. 2008b;56:196–206. doi: 10.1111/j.1365-313X.2008.03585.x. [DOI] [PubMed] [Google Scholar]
- Martinez M, Cambra I, Carrillo L, Diaz-Mendoza M, Diaz I. Characterization of the entire cystatin gene family in barley and their target cathepsin L-Like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiol. 2009;151:1531–1545. doi: 10.1104/pp.109.146019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. Nitrogen uptake, assimilation and remobilization in plants: Challengues for sustainable and reproductive agriculture. Ann Bot. 2010;105:1141–1157. doi: 10.1093/aob/mcq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P. Chloroplast senescence. In: Baker NR, Thomas H, editors. Crop Photosynthesis: Spatial and Temporal Determinants. Elsevier; Amsterdam: 1992. pp. 413–440. [Google Scholar]
- Mi G, Tang L, Zhang F, Zhang J. Carbohydrate storage and utilization during grain filling as regulated by nitrogen application in two wheat cultivars. J Plant Nutr. 2002;25:213–229. [Google Scholar]
- Mitsuhashi W, Feller U. Effects of light and external solutes on the catabolism of nuclear-encoded stromal proteins in intact chloroplasts isolated from pea leaves. Plant Physiol. 1992;100:2100–2105. doi: 10.1104/pp.100.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulisch M, Asp T, Krupinska K, Hollmann J, Holm PB. The Tr-cp 14 cysteine protease in white clover (Trifolium repens) is localized to the endoplasmic reticulum and is associated with programmed cell death during development of tracheary elements. Protoplasma. 2013;250:623–629. doi: 10.1007/s00709-012-0427-1. [DOI] [PubMed] [Google Scholar]
- Munier-Jolain NG, Salon C. Are the carbon costs of seed production related to the quantitative and qualitative performance? An appraisal for legumes and other crops. Plant Cell Environ. 2005;28:1388–1395. [Google Scholar]
- Otegui MS, Noh Y-S, Martinez DE, Petroff AGV, Staehelin LA, Amasino RM, Guiamet JJ. Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 2005;41:831–844. doi: 10.1111/j.1365-313X.2005.02346.x. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Carillo N, Valle EM. The role of photosynthetic electron transport in the oxidative degradation of chloroplastic glutamine synthetase. Plant Physiol. 1999;121:471–478. doi: 10.1104/pp.121.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegadaraju V, Knepper C, Reese J, Shah J. Premature leaf senescence modulated by the Arabidopsis PHYTOALEXIN DEFICIENT4 gene is associated with defense against the phloem-feeding green peach aphid. Plant Physiol. 2005;139:1927–1934. doi: 10.1104/pp.105.070433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova A, Smith CM. Application of brown planthopper salivary gland extract to rice plants induces systemic host mRNA patterns associated with nutrient remobilization. PLoS One. 2015;7: doi: 10.1371/journal.pone.0141769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Gan S, Amasino RM, Roby D, Lam E. Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol Biol. 1999;39:1243–1255. doi: 10.1023/a:1006133311402. [DOI] [PubMed] [Google Scholar]
- Prasch C, Sonnewald U. Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol. 2013;162:1849–1866. doi: 10.1104/pp.113.221044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins A, van Heerden PDR, Olmos E, Kunert KJ, Foyer C. Cysteine proteinases regulate chloroplast protein content and composition in tobacco leaves: A model for dynamic interactions with ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) vesicular bodies. J Exp Bot. 2008;59:1935–1950. doi: 10.1093/jxb/ern086. [DOI] [PubMed] [Google Scholar]
- Quain MD, Makgopa ME, Marquez-Garcia B, Comadira G, Fernandez-Garcia N, Olmos E, Schnaubelt D, Kunert KJ, Foyer CH. Ectopic phytocystatin expression leads to enhances drought stress tolerance in soybean (Glycine max) and Arabidopsis thaliana through effects on strigolactone pathways and can result in improved seed traits. Plant Biotechnol J. 2014;12:903–913. doi: 10.1111/pbi.12193. [DOI] [PubMed] [Google Scholar]
- Quirino BF, Noh Y-S, Himelblau E, Amasino RM. Molecualr aspects of leaf senescence. Trends Plant Sci. 2000;5:278–282. doi: 10.1016/s1360-1385(00)01655-1. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barret AJ, Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016;44:D343–D350. doi: 10.1093/nar/gkv1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts IN, Caputo C, Criado MV, Funk C. Senescence-associated proteases in plants. Physiol Plant. 2012;145:130–139. doi: 10.1111/j.1399-3054.2012.01574.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Takami T. Nucleases and higher plants and their possible involvement in DNA degradation during senescence. J Exp Bot. 2014;65:3835–3843. doi: 10.1093/jxb/eru091. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Rusu AG, Manners JM, Maclean DJ. The SEN1 gene of Arabidopsis is regulated by signals that link plant defence responses and senescence. Plant Physiol Biochem. 2005;43:997–1005. doi: 10.1016/j.plaphy.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Schiltz S, Gallardo K, Huart M, Negroni L, Sommerer N, Burstin J. Proteome reference maps of vegetative tissues in pea. An investigation of nitrogen mobilization from leaves during seed filling. Plant Physiol. 2004;135:2241–2260. doi: 10.1104/pp.104.041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifi HS, Van Bockhaven J, Angenon G, Höfte M. Glutamate metabolism in plant disease and defense: Friend or foe? Mol Plant Microbe-Interact. 2013;26:475–485. doi: 10.1094/MPMI-07-12-0176-CR. [DOI] [PubMed] [Google Scholar]
- Shan L, Li C, Chen F, Zhao S, Xia G. A Bowman-Birk type protease inhibitor is involved in the tolerance to salt stress in wheat. Plant Cell Environ. 2008;31:1128–1137. doi: 10.1111/j.1365-3040.2008.01825.x. [DOI] [PubMed] [Google Scholar]
- Simova-Stoilova L, Vaseva I, Grigorova B, Demirevska K, Feller U. Proteolytic activity and cysteine protease expression in wheat leaves under severe soil drought and recovery. Plant Physiol Biochem. 2010;48:200–206. doi: 10.1016/j.plaphy.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Singh S, Giri MK, Singh PK, Siddiqui A, Nandi AK. Down-regulation of OsSAG12-1 results in enhanced senescence and pathogen-induced cell death in transgenic rice plants. J Biosci. 2013;38:583–592. doi: 10.1007/s12038-013-9334-7. [DOI] [PubMed] [Google Scholar]
- Slafer GA, Andrade FH, Feingold SE. Genetic improvement of bread wheat (Triticum aestivum L.) in Argentina: Relationships between nitrogen and dry matter. Euphytica. 1990;50:63–71. [Google Scholar]
- Srinivasan T, Kumar KRR, Kirti PB. Constitutive expression of a trypsin protease inhibitor confers multiple stress tolerance in transgenic tobacco. Plant Cell Physiol. 2009;50:541–553. doi: 10.1093/pcp/pcp014. [DOI] [PubMed] [Google Scholar]
- Srivastava AK, Nair JS, Bendigeri D, Vijaykumar A, Ramaswamy NK, D'Souza SF. Purification and characterization of a salinity induced alkaline protease from isolated spinach chloroplasts. Acta Physiol Plant. 2009;31:187–197. [Google Scholar]
- Tajima T, Yamaguchi A, Matsushima S, Satoh M, Hayasaka S, Yoshimatsu K, Shioi Y. Biochemical and molecular characterization of senescence-related cysteine protease-cystatin complex from spinach leaf. Physiol Plant. 2011;141:97–116. doi: 10.1111/j.1399-3054.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- Teixeira PF, Glaser E. Processing peptidases in mitochondria and chloroplasts. Biochim Biophys Acta. 2013;1833:360–370. doi: 10.1016/j.bbamcr.2012.03.012. [DOI] [PubMed] [Google Scholar]
- Thoenen M, Feller U. Degradation of glutamine synthetase in intact chloroplasts isolated from pea (Pisum sativum) leaves. Aust J Plant Physiol. 1998;25:279–286. [Google Scholar]
- Thoenen M, Herrmann B, Feller U. Senescence in wheat leaves: Is a cysteine endopeptidase involved in the degradation of the large subunit of Rubisco? Acta Physiol Plant. 2007;29:339–350. [Google Scholar]
- Thomas H, Stoddart J. Leaf senescence. Annu Rev Plant Physiol. 1980;31:83–111. [Google Scholar]
- van der Hoorn RA. Plant proteases: From phenotypes to molecular mechanisms. Annu Rev Plant Biol. 2008;59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Beers EP, Dangl JL, Franklin-Tong VE, Gallois P, Hara-Nishimura I, Jones AM, Kawai-Yamada M, Lam E, Mundy J, et al. Morphological classification of plant cell deaths. Cell Death Diff. 2011;18:1241–1246. doi: 10.1038/cdd.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. Proteolysis in plants: Mechanisms and functions. Plant Mol Biol. 1996;32:275–302. doi: 10.1007/BF00039386. [DOI] [PubMed] [Google Scholar]
- Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- Wang S, Blumwald E. Stress-induced chloroplast degradation in Arabidopsis is regulated via a process independent of autophagy and senescence-associated vacuoles. Plant Cell. 2014;26:4875–4888. doi: 10.1105/tpc.114.133116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederanders B. Structure-function relationship in class CA1 cysteine peptidases. Acta Biochim Pol. 2003;50:691–713. [PubMed] [Google Scholar]
- Xiao HJ, Yin YX, Chai WG, Gong ZH. Silencing of the CaCP gene delays salt- and osmotic-induced leaf senescence in Capsicum annuum L. Int J Mol Sci. 2014;15:8316–8334. doi: 10.3390/ijms15058316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Matsushima R, Nishimura M, Hara-Nishimura I. A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol. 2001;127:1626–1634. [PMC free article] [PubMed] [Google Scholar]
- Yang ZL, Ohlrogge JB. Turnover of fatty acids during natural senescence of Arabidopsis Brachypodium and switchgrass and in Arabidopsis beta-oxidation mutants. Plant Physiol. 2009;150:1981–1989. doi: 10.1104/pp.109.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang J. Grain filling of cereals under soil drying. New Phytol. 2006;169:223–236. doi: 10.1111/j.1469-8137.2005.01597.x. [DOI] [PubMed] [Google Scholar]
- Yoshida S. Molecular regulation of leaf senescence. Current Opin Plant Biol. 2003;6:79–84. doi: 10.1016/s1369526602000092. [DOI] [PubMed] [Google Scholar]
- Zhang LF, Rui Q, Zhang P, Wang XY, Xu LL. A novel 51-kDa fragment of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase formed in the stroma of chloroplasts in dark-induced senescing wheat leaves. Physiol Plant. 2007;131:64–71. doi: 10.1111/j.1399-3054.2007.00928.x. [DOI] [PubMed] [Google Scholar]