ABSTRACT
Desmosomes are macromolecular junctions responsible for providing strong cell–cell adhesion. Because of their size and molecular complexity, the precise ultrastructural organization of desmosomes is challenging to study. Here, we used direct stochastic optical reconstruction microscopy (dSTORM) to resolve individual plaque pairs for inner and outer dense plaque proteins. Analysis methods based on desmosomal mirror symmetry were developed to measure plaque-to-plaque distances and create an integrated map. We quantified the organization of desmoglein 3, plakoglobin and desmoplakin (N-terminal, rod and C-terminal domains) in primary human keratinocytes. Longer desmosome lengths correlated with increasing plaque-to-plaque distance, suggesting that desmoplakin is arranged with its long axis at an angle within the plaque. We next examined whether plaque organization changed in different adhesive states. Plaque-to-plaque distance for the desmoplakin rod and C-terminal domains decreased in PKP-1-mediated hyperadhesive desmosomes, suggesting that protein reorganization correlates with function. Finally, in human epidermis we found a difference in plaque-to-plaque distance for the desmoplakin C-terminal domain, but not the desmoplakin rod domain or plakoglobin, between basal and suprabasal cells. Our data reveal the molecular organization of desmosomes in cultured keratinocytes and skin as defined by dSTORM.
KEY WORDS: Cell adhesion, Keratinocyte, Super-resolution, Fluorescence, Microscopy, Cadherin, dSTORM
Highlighted Article: The nanoscale organization of proteins in the desmosome, a structure mediating strong cell–cell adhesion, is elucidated by dSTORM super-resolution fluorescence microscopy.
INTRODUCTION
Desmosomes are intercellular junctions that are abundant in tissues that experience considerable mechanical stress, such as the skin and heart (Kowalczyk and Green, 2013). The importance of desmosome-mediated adhesion is evidenced by the numerous human diseases that result when desmosomes are compromised, manifesting in epidermal fragility, cardiomyopathies and cancers (Broussard et al., 2015; Stahley and Kowalczyk, 2015).
The desmosome is a macromolecular complex that forms a ‘spot-weld’ of strong intercellular adhesion and comprises transmembrane cadherins and cytoplasmic plaque proteins (Berika and Garrod, 2014; Saito et al., 2012). The desmosomal cadherins (desmogleins and desmocollins) link neighboring cells through extracellular adhesive interactions whereas the armadillo protein family members plakoglobin and plakophilin, and the plakin family member desmoplakin contribute to the intracellular plaque (Fig. 1A). Plaque ultrastructure is characterized by two electron-dense regions: the plasma-membrane-proximal outer dense plaque and the inner dense plaque (Desai et al., 2009; Farquhar and Palade, 1963; Stokes, 2007). The cadherin cytoplasmic tails bind to proteins in the outer dense plaque whereas the C-terminus of desmoplakin binds to intermediate filaments in the inner dense plaque. This tethers the desmosome to the intermediate filament cytoskeleton, establishing an integrated adhesive network (Bornslaeger et al., 1996; Harmon and Green, 2013).
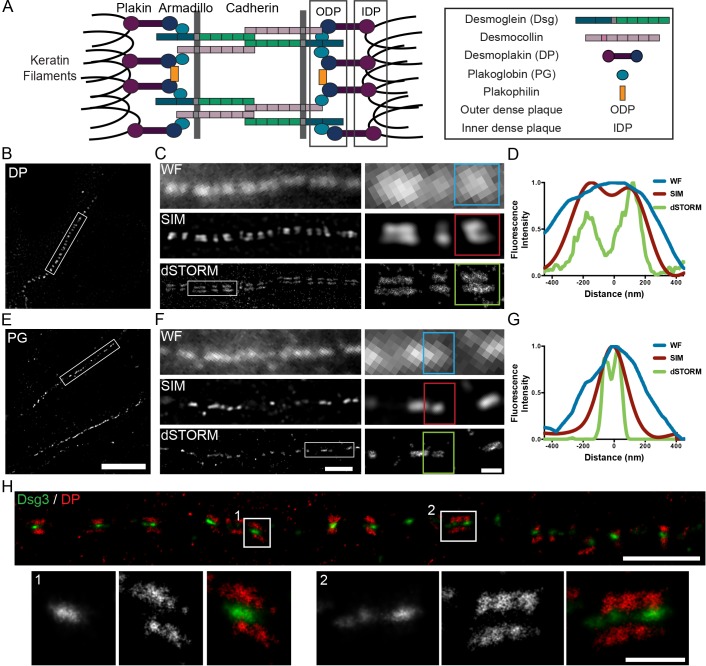
Fig. 1.
Super-resolution imaging of desmosomes. (A) Desmosome schematic. (B,C,E,F) Cultured human keratinocytes labeled for the desmoplakin (DP) C-terminal domain or plakoglobin (PG) N-terminal domain and imaged by widefield microscopy (WF), SIM and dSTORM. (B) SIM image of desmoplakin cell border. (C) Desmoplakin-labeled region of interest imaged by widefield microscopy, SIM and dSTORM. (D) Linescan across a single desmosome indicated by the colored boxes. (E) SIM image of plakoglobin cell border. (F) Plakoglobin-labeled region of interest imaged by widefield microscopy, SIM and dSTORM. (G) Linescan across a single desmosome indicated by the colored boxes. (H) dSTORM image of keratinocyte cell–cell border labeled for the Dsg3 N-terminal (Alexa Fluor 488, green) and desmoplakin C-terminal (Alexa Fluor 647, red) domains. Scale bars: 10 µm (B,E); 2 µm (C,F,H); 500 nm (C,F,H, insets).
Many desmosomal protein interactions have been characterized by biochemical studies (Bass-Zubek and Green, 2007; Green and Simpson, 2007; Thomason et al., 2010), and desmosomal ultrastructure has been studied by immunogold and tomographic electron microscopy (Al-Amoudi et al., 2011; Al-Amoudi and Frangakis, 2008; North et al., 1999; Owen et al., 2008; Stokes, 2007). Yet, studying desmosomes on a molecular level remains challenging due to their size, insolubility and molecular complexity. Measuring desmosomal protein organization requires a technique with high resolution coupled with minimal sample manipulation to minimize structural artifacts and preserve physiological relevance. Super-resolution microscopy has revealed sub-resolution protein organization in numerous macromolecular complexes including the nuclear pore, ciliary transition zone, kinetochore, neuronal synapse, focal adhesion and hemidesmosome (Chatel et al., 2012; Loschberger et al., 2014, 2012; Nahidiazar et al., 2015; Ribeiro et al., 2010; van Hoorn et al., 2014; Yang et al., 2015; Zhong, 2015). Here, we use direct stochastic optical reconstruction microscopy (dSTORM) (Rust et al., 2006) to elucidate protein organization within the desmosome at the molecular level both in vitro and in vivo.
RESULTS AND DISCUSSION
Desmosome plaques resolved by dSTORM
Two proteins, the inner dense plaque protein desmoplakin and the outer dense plaque protein plakoglobin, were utilized to establish dSTORM for studying desmosome organization. Primary human keratinocytes were pre-extracted prior to fixation and the desmosomal pool of proteins (Palka and Green, 1997) was immunostained with primary antibodies against the desmoplakin C-terminal or plakoglobin N-terminal domains. The same cell–cell border was then imaged by widefield, structured illumination microscopy (SIM) and dSTORM.
In widefield, desmoplakin fluorescence was punctate and pixilated with no discernable structural features. In contrast, two distinct desmoplakin plaques, one contributed by each cell, were resolved by SIM as previously shown (Fig. 1B,C) (Stahley et al., 2015). Likewise, two individual desmoplakin plaques were resolved by dSTORM, but with a more distinct separation (Fig. 1C). To analyze the difference in spatial resolution between the imaging techniques, fluorescence intensity was plotted as a function of distance along the desmosome axis, perpendicular to the plasma membrane. Although there is only a single intensity peak from the widefield image, two individual peaks, corresponding to the two plaques, are resolved from the SIM and dSTORM images (Fig. 1D).
Plakoglobin-labeled desmosomes imaged by widefield have a punctate appearance, similar to desmoplakin (Fig. 1E,F). Similarly, plakoglobin fluorescence by SIM is punctate with no evident internal structure. In contrast, dSTORM revealed two individual plakoglobin plaques (Fig. 1F). This is also shown in the plot of fluorescence intensity along the desmosome axis (Fig. 1G).
To confirm dSTORM resolves individual desmosomes consisting of plaques contributed by two different cells, keratinocytes were dual-labeled for the extracellular domain of desmoglein 3 (Dsg3) and the C-terminal domain of desmoplakin. dSTORM revealed a distinct separation, with the Dsg3 extracellular domain sandwiched between two desmoplakin plaques (Fig. 1H). In our experiments, the localization precision of Alexa Fluor 488 (∼19.5 nm) was less than that of Alexa Fluor 647 (∼5.8 nm), similar to that reported by others (Dempsey et al., 2011; Tam et al., 2014). Therefore, Alexa Fluor 647 was used for quantitative measurements unless otherwise indicated. Image resolution is also affected by the primary and secondary antibody labels, which increases the apparent size of structures (Bates et al., 2007).
In summary, these results demonstrate that dSTORM can resolve proteins in both the outer and inner dense plaques in individual desmosomes.
Molecular map of the desmosome by dSTORM
Next, we systematically quantified the molecular arrangement of proteins in the desmosome. Keratinocytes were labeled with antibodies against the Dsg3 N-terminal (Alexa Fluor 488), plakoglobin N-terminal, Dsg3 C-terminal, desmoplakin N-terminal, desmoplakin rod, and desmoplakin C-terminal domains (Fig. 2A; Table S1) and protein distributions along the desmosomal axis were determined using dSTORM. The mirror symmetry of desmosomal plaques allowed us to identify individual desmosomes, and only those with their axes in the x-y focal plane were analyzed (Fig. 2B; Fig. S1). A linescan of fluorescence intensity along the desmosome axis was created for each desmosome (Fig. 2C). The axial distribution of proteins was automatically measured as the distance between peaks from the linescans. This plaque-to-plaque distance was measured from multiple individual desmosomes labeled for each protein domain (Fig. 2D). There was a progressive increase in the average plaque-to-plaque distance with the plakoglobin N-terminal domain closest to the plasma membrane and the desmoplakin C-terminal domain furthest away (Table S2). All plaque-to-plaque distances were significantly different from one another with the exception of the plakoglobin N-terminal and Dsg3 C-terminal domains, and plakoglobin N-terminal and desmoplakin N-terminal domains (Fig. 2E). This is consistent with interactions between plakoglobin and both the cytoplasmic tail of Dsg3 and the N-terminal domain of desmoplakin (Delva et al., 2009).
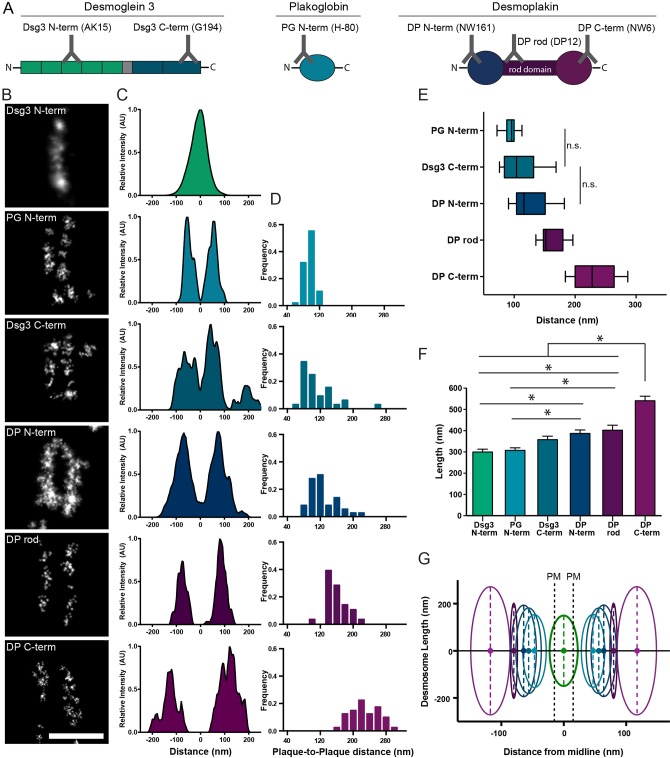
Fig. 2.
Molecular map of the desmosome as revealed by dSTORM. (A) Approximate binding locations of antibodies used for dSTORM in desmoglein 3 (Dsg3), desmoplakin (DP) or plakoglobin (PG). (B) Representative images of individual desmosomes labeled with the indicated antibodies, from 2–4 preparations. Scale bar: 500 nm. (C) Fluorescence intensity linescan along the desmosomal axis (horizontal through image as shown). (D) Histograms of plaque-to-plaque distance from all desmosomes analyzed for each protein domain. (E) Box-and-whisker plot of plaque-to-plaque distance. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the 10–90th percentiles. (F) Desmosome length. Mean±s.e.m. *P<0.05 (one-way ANOVA with Holm-Sidak's multiple comparisons test). For D–F, Dsg3 N-term, n=69; PG N-term, n=47; Dsg3 N-term, n=32; DP N-term, n=36; DP rod, n=28; DP C-term, n=40. (G) Composite dSTORM map where 0 is the midline, distances represent half the mean plaque-to-plaque distance, oval height represents desmosome length, and oval width represents the standard deviation of the peak width. PM, plasma membrane.
Desmosome length, parallel to the plasma membrane, was also determined for each protein domain (Fig. 2F). There was a trend of increasing length with increasing distance from the plasma membrane. This trend was also reflected at the protein domain level: the desmoplakin C-terminal domain length was significantly longer than that of either the desmoplakin N-terminal or desmoplakin rod domain length (Table S2). A composite molecular map including protein localizations and desmosome length, illustrates the possible arrangement of proteins splaying out from the membrane (Fig. 2G).
We next sought to determine whether the arrangement of proteins measured in primary human keratinocytes was conserved among the immortalized human keratinocyte cell line HaCaT and the human epidermoid carcinoma cell line A431. Individual desmoplakin rod and plakoglobin N-terminal-domain-labeled desmosomes were imaged in all cell types and there were no significant differences in plaque-to-plaque distances (Fig. S2). This suggests the general organization of proteins in desmosomes is conserved across primary keratinocytes and human cell lines derived from skin.
Desmosome protein organization has been previously studied with electron microscopy, measured as distance from the plasma membrane. For comparison, dSTORM plaque-to-plaque measurements were converted into a plasma membrane reference system by subtracting the width of the intercellular space (∼34 nm); (Al-Amoudi et al., 2004), subtracting two times the plasma membrane thickness (∼4-6 nm), and finally dividing by two (see Fig. 2G for reference). The dSTORM distributions of Dsg3 C-terminal and plakoglobin N-terminal domains overlap with the corresponding distributions of gold labels in immunoelectron microscopy studies (North et al., 1999), and the dSTORM plakoglobin distribution overlaps with the plakoglobin layer identified by cryoelectron tomography (Al-Amoudi et al., 2011). In contrast to immunogold studies, dSTORM indicates that desmoplakin is localized further from the plasma membrane.
Although desmosome length has been previously measured with electron microscopy, dSTORM allows for the analysis of specific protein domains. The corresponding increase in plaque length and plaque-to-plaque distance indicates that the desmosome splays out as it extends from the plasma membrane into the cell. Desmoplakin has two isoforms, desmoplakin I and desmoplakin II, each with globular N- and C-terminal domains separated by a central rod domain. The average N- to C-terminal length of desmoplakin I is 162 nm and desmoplakin II is 63 nm (O'Keefe et al., 1989). The distance between the desmoplakin N- and C-terminal domains measured perpendicular to the plasma membrane by dSTORM was 54±4 nm (mean±s.e.m; desmoplakin N-terminal n=36; C-terminal n=40), on par with previous measurements (North et al., 1999), whereas the length of the desmoplakin C-terminal plaque extends 77±13 nm beyond the desmoplakin N-terminal plaque. This suggests desmoplakin is oriented with its long axis at an angle in the plaque, not perpendicular to the plasma membrane. The minimum length that will fit this model is 94 nm, significantly less than the length of desmoplakin I. The desmoplakin II rod domain could span the 54 nm, but at a smaller angle. Although we cannot rule out an additional disorganized or kinked conformation of the rod, our data strongly suggests that desmoplakin is oriented at an angle in the plaque.
Reorganization of plaque proteins in PKP-1-mediated hyperadhesive desmosomes
Desmosomes can exist in a weaker Ca2+-dependent state and a stronger Ca2+-independent or hyperadhesive state (Garrod et al., 2005; Kimura et al., 2007). Plakophilin 1 (PKP-1) promotes desmosome formation by recruiting and clustering desmosomal proteins, and overexpression of PKP-1 has been shown to induce hyperadhesion in cultured keratinocytes (Bornslaeger et al., 2001; Hatzfeld et al., 2000; Tucker et al., 2014). To test whether plaque organization changes with adhesive state, adenoviral PKP-1–Myc overexpression was used to shift desmosomes to a hyperadhesive state. With PKP-1–Myc overexpression an approximate eightfold increase in PKP-1 protein level was observed compared to empty vector control (Fig. S3). Keratinocytes were labeled with antibodies against the plakoglobin N-terminal, Dsg3 C-terminal, desmoplakin N-terminal, desmoplakin rod and desmoplakin C-terminal domains (Fig. 3A). Cells were also labeled with anti-Myc antibody, and PKP-1–Myc expression was confirmed by widefield colocalization analysis. The plaque-to-plaque distance was measured from multiple individual desmosomes for each protein domain (Fig. 3B). A significant decrease in the plaque-to-plaque distance was measured for the Dsg3 C-terminal, desmoplakin rod and desmoplakin C-terminal domains. In contrast the distribution of plakoglobin and the desmoplakin N-terminal domain did not significantly change (Fig. 3C; Table S3). An increase in desmosome length was observed with PKP-1–Myc overexpression, similar to in previous studies (Fig. 3D; Table S4) (Tucker et al., 2014).
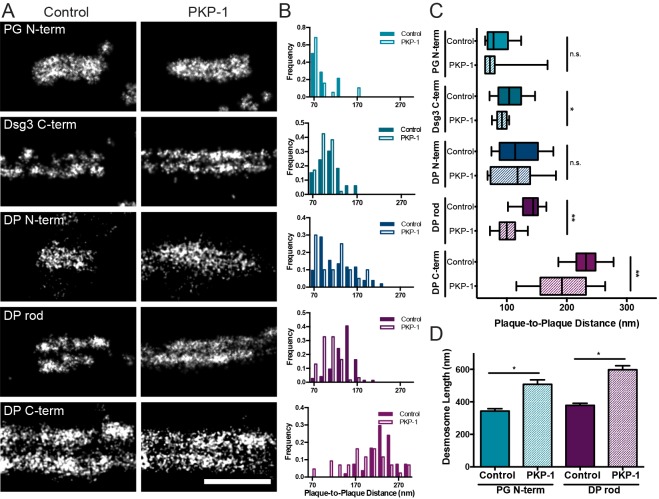
Fig. 3.
PKP-1-mediated hyperadhesion altered the molecular map of the desmosome. (A) dSTORM images of individual desmosomes from cultured primary keratinocytes transduced with control empty virus or PKP-1–Myc and labeled with indicated antibodies against plakoglobin (PG), Dsg3 or desmoplakin (DP). Images are representative of two experiments. Scale bar: 500 nm. (B) Histograms of the plaque-to-plaque distance measured in control (solid) and PKP-1–Myc (open). (C) Box-and-whisker plot of axial plaque-to-plaque distance of all protein domains analyzed in control (solid) and PKP-1–Myc overexpressing (striped) cells. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the 10–90th percentiles (for B,C, control: PG N-term, n=14; Dsg3 C-term, n=33; DP N-term, n=50; DP rod, n=74; DP C-term, n=54; for PKP-1–Myc: PG N-term, n=19; Dsg3 C-term n=47; DP N-term n=20; DP rod n=61; DP C-term n=43). *P<0.05, **P<0.0001; n.s., not significant (paired t-tests). (D) Desmosome length (mean±s.e.m., control: PG N-term, n=81; DP rod, n=116. PKP-1–Myc: PG N-term, n=90; DP rod, n=95) measured for the plakoglobin N-terminal and desmoplakin rod domains in control (solid) and PKP-1–Myc overexpressing (striped) cells. *P<0.05 (paired t-tests).
Our data show that the plaque-to-plaque distance for some, but not all, protein domains decreases in PKP-1-induced hyperadhesion. Notably, the desmoplakin N-terminal domain plaque-to-plaque distance does not change with PKP-1–Myc overexpression compared to control, whereas the plaque-to-plaque distance of both the desmoplakin rod and C-terminal domains decreased. The N-terminal plakin domain of desmoplakin has an ‘L’ conformation consisting of two perpendicular lobes separated by a flexible elbow, with a flexible linker connecting the N-terminal to the rod domain (Al-Jassar et al., 2011; Grum et al., 1999). Our data supports previously proposed models in which a conformational change within the plakin domain elbow or the flexible linker allows for motion of the rod and C-terminal domains within the plaque, whereas the N-terminal domain is stationary (Al-Jassar et al., 2011). PKP-1 has also been shown to interfere with plakoglobin-desmoplakin binding (Bornslaeger et al., 2001) and thus it is possible that the proposed conformational change also reflects a change in binding partner.
The difference in ultrastructure measured with dSTORM could contribute to the increased adhesive strength and mechanism of desmosome maturation. Interestingly, knockdown of PKP-3 in SCC9 cells has been shown to weaken cell adhesion and increase desmosome width (Todorovic et al., 2014). That result, along with those presented here, support the hypothesis that changes in protein organization within the desmosomal plaque correlate with changes in adhesive strength.
Desmosome molecular organization is conserved in skin
To investigate whether desmosomal plaques could be identified in vivo, human skin sections labeled with antibodies against the desmoplakin rod domain were imaged by dSTORM. The epidermis is a stratified epithelium, and imaging the desmoplakin rod domain revealed pairs of individual plaques in basal cells adjacent to the basement membrane and suprabasal cells in the spinous and granular layers (Fig. 4A).
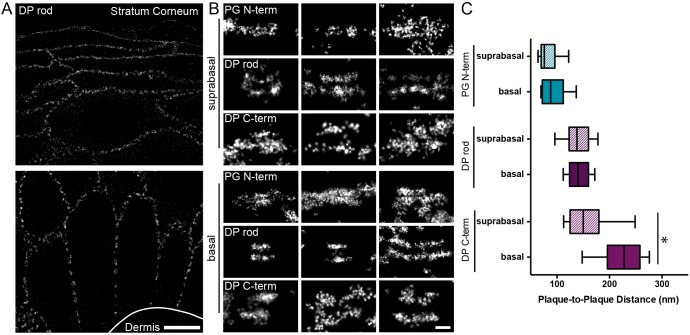
Fig. 4.
Molecular organization of desmosomes in human skin. (A) Human skin sections labeled for desmoplakin (DP, rod domain) with multiple desmosomes visible throughout the epidermis in basal (bottom) and suprabasal (top) cells. Scale bar: 5 µm. (B) Representative images of individual desmosomes labeled with the indicated antibodies from basal (bottom) and suprabasal (top) cells. PG, plakoglobin. Scale bar: 200 nm. (C) Box-and-whisker plot of plaque-to-plaque distance in basal (solid) and suprabasal (striped) cells. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the 10–90th percentiles (Basal: PG N-term, n=33; DP rod, n=80; DP C-term, n=29. Suprabasal: PG N-term, n=17; DP rod, n=34; DP C-term, n=12). *P<0.05 (paired t-tests).
To quantify the protein arrangement in skin, the plakoglobin N-terminal, desmoplakin rod or desmoplakin C-terminal domains were labeled and imaged by dSTORM. The plaque-to-plaque distances revealed that the overall axial arrangement of protein domains was similar in skin and cell culture. Next, we wanted to see whether the arrangement was the same in basal and suprabasal cells. Plaque-to-plaque distances measured in basal and suprabasal desmosomes for plakoglobin N-terminal or desmoplakin rod were not significantly different. However, for the desmoplakin C-terminal domain, the plaque-to-plaque distance decreased significantly by 65±11 nm (Fig. 4C; Table S5).
Expression levels of desmosomal proteins change with keratinocyte differentiation in the epidermis (Delva et al., 2009). PKP-1 expression is increased in suprabasal compared to basal cells, and the decrease in desmoplakin C-terminal plaque-to-plaque distance in these desmosome agrees with our PKP-1–Myc overexpression results in cell culture. However, the desmoplakin rod domain plaque-to-plaque distance was unchanged between basal and suprabasal desmosomes, which could reflect variations in plaque re-organization due to protein overexpression verses epithelial differentiation.
In summary, our results demonstrate the ability of dSTORM to investigate the nanoscale arrangement of proteins within intact desmosomal plaques, in cell culture and human tissue. Desmosomes are dynamic, exist in different adhesive states and can be disrupted by disease. Therefore, this resolution will likely prove powerful in understanding the relationship between molecular organization and desmosomal adhesive state (Garrod et al., 2005; Getsios et al., 2004; Green et al., 2010; Kitajima, 2014). The approach developed here is applicable to many aspects of desmosome biology including their interaction with intermediate filaments, the impact of intracellular signaling on plaque organization and disruption in disease. With relatively straightforward sample preparation, compatibility with cells and tissues, and single-desmosome resolution, this approach is poised to integrate functional and structural analysis of desmosomes.
MATERIALS AND METHODS
Cell culture
Human epidermal keratinocytes were isolated from neonatal foreskin as previously described (Calkins et al., 2006) and cultured in supplemented KBM-Gold (Lonza, Walkersville, MD). Keratinocytes, at passage two or three, were seeded onto eight-well no. 1.5 coverslip bottom microslides (Ibidi GmbH, Planegg-Martinsried, Germany). The Ca2+ concentration in the medium was increased to 550 µM at 16–20 h before fixation. Where indicated, keratinocytes were infected with empty or PKP-1–Myc adenovirus (Tucker et al., 2014) 36 h before fixation. HaCaT and A431 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Corning, Tewksbury, MA) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin.
Antibodies
Antibodies used were: anti-Dsg3 AK15 (Tsunoda et al., 2003) (gift from Dr Masayuki Amagai, Keio University, Tokyo, Japan); anti-Dsg3 G194 (Progen Biotechnik GmbH, Heidelberg); anti-γ-catenin (Santa Cruz Biotechnology); anti-desmoplakin I and II (Fitzgerald, Acton, MA); desmoplakin antibodies NW6 and NW161 (gift from Dr Kathleen Green, Northwestern University, Evanston, IL) and anti-Myc (Bethyl Labs, Montgomery, TX). Additional information is in Table S1. Alexa-Fluor-conjugated secondary antibodies (IgG H&L) were from Invitrogen (Grand Island, NY). For western blotting, anti-PKP-1 (Sobolik-Delmaire et al., 2006) (gift from Dr James Wahl III, University of Nebraska) and anti-Actin C2 (Santa Cruz Biotechnology) were used.
Immunofluorescence
Fixation and labeling protocols were adapted from Whelan and Bell (2015). Cells were pre-extracted (60 s) with 300 mM sucrose and 0.2% Triton-X 100 to remove non-desmosomal proteins. This was followed by fixation (12 min, 37°C) with 4% paraformaldehyde prepared fresh from 16% electron microscopy grade material (Electron Microscopy Sciences, Hatfield, PA). Samples were washed, blocked and permeabilized (30 min) in 3% bovine serum albumin (BSA) and 0.2% Triton-X 100. Samples were incubated with primary (3 h) and secondary antibodies (1 h), with multiple washes in-between. Samples were stored with protection from light (PBS+, 4°C) and imaged within 2 weeks.
Human tissue biopsy processing
Human skin biopsy use was approved by the Emory University Institutional Review Board. 5-µm-thick sections from OCT-embedded biopsies were mounted onto glass coverslips and processed for immunostaining. Tissue sections were labeled sequentially with primary and secondary antibodies (1 h) with triple PBS+ washes between incubations.
Immunoblotting
Cells were harvested and immunoblotting performed using standard procedures (Stahley et al., 2014).
Microscopy
SIM images were obtained and reconstructed with a Nikon N-SIM Eclipse Ti-E microscope system (Nikon Instruments, Melville, NY) equipped with a 100×1.49 NA oil immersion objective, 488 nm laser and EMCCD camera (DU-897, Andor Technology, Belfast, Northern Ireland). dSTORM images were obtained with a Nikon N-STORM microscope equipped with a 100×1.49 NA oil immersion objective, 488 and 647 nm lasers, and an iXon ultra EMCCD camera (Andor). Typically, 60,000–80,000 frames were collected with inclined sub-critical excitation. Images were reconstructed in Nikon Elements. Widefield images were obtained with low-intensity laser excitation on the N-STORM system. Photoswitching imaging buffer included glucose oxidase (Sigma, St Louis, Missouri), catalase (Roche, Penzberg, Germany), glucose (Sigma) and β-mercaptoethanol (Sigma) (Rust et al., 2006).
Image analysis
dSTORM images were exported at 4 nm/pixel and analyzed using MATLAB scripts (Mathworks, Natic, MA). Individual desmosomes were manually identified and automatically excised and aligned such that the desmosome axis was horizontal. Intensity was measured along the desmosome axis by averaging all pixels along the desmosome length. Linescans were normalized, smoothed and the peak finder function (40% intensity peak threshold) identified linescans with two peaks for quantification. For the Dsg3 N-terminal, linescans with one peak were selected. Custom MATLAB scripts are available upon request.
Acknowledgements
We thank Mattheyses and Kowalczyk laboratory members for discussion, Susan Summers for keratinocyte isolations and Sue Manos for aid with human tissue.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
S.N.S., E.I.B., A.P.K. and A.L.M. designed experiments; S.N.S., E.I.B. and A.L.M. performed the experiments, C.E.A. and A.L.M. wrote programs and analyzed data with S.N.S. S.N.S. and A.L.M. wrote the manuscript. All authors contributed intellectually and edited the manuscript.
Funding
This project was supported by the National Institutes of Health [grant numbers R21AR066920 to A.L.M., R01AR048266 to A.P.K.]; and National Science Foundation [grant number IDBR 1353939 to A.L.M.]. SIM was supported by the Integrated Cellular Imaging Core at Emory University. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.185785.supplemental
References
- Al-Amoudi A. and Frangakis A. S. (2008). Structural studies on desmosomes. Biochem. Soc. Trans. 36, 181-187. 10.1042/BST0360181 [DOI] [PubMed] [Google Scholar]
- Al-Amoudi A., Norlen L. P. O. and Dubochet J. (2004). Cryo-electron microscopy of vitreous sections of native biological cells and tissues. J. Struct. Biol. 148, 131-135. 10.1016/j.jsb.2004.03.010 [DOI] [PubMed] [Google Scholar]
- Al-Amoudi A., Castano-Diez D., Devos D. P., Russell R. B., Johnson G. T. and Frangakis A. S. (2011). The three-dimensional molecular structure of the desmosomal plaque. Proc. Natl. Acad. Sci. USA 108, 6480-6485. 10.1073/pnas.1019469108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Jassar C., Knowles T., Jeeves M., Kami K., Behr E., Bikker H., Overduin M. and Chidgey M. (2011). The nonlinear structure of the desmoplakin plakin domain and the effects of cardiomyopathy-linked mutations. J. Mol. Biol. 411, 1049-1061. 10.1016/j.jmb.2011.06.047 [DOI] [PubMed] [Google Scholar]
- Bass-Zubek A. E. and Green K. J. (2007). Biochemical characterization of the desmosome. J. Invest. Dermatol. 127, E4-E5. 10.1038/sj.skinbio.6250002 [DOI] [PubMed] [Google Scholar]
- Bates M., Huang B., Dempsey G. T. and Zhuang X. (2007). Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749-1753. 10.1126/science.1146598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berika M. and Garrod D. (2014). Desmosomal adhesion in vivo. Cell Commun. Adhes. 21, 65-75. 10.3109/15419061.2013.876018 [DOI] [PubMed] [Google Scholar]
- Bornslaeger E. A., Corcoran C. M., Stappenbeck T. S. and Green K. J. (1996). Breaking the connection: displacement of the desmosomal plaque protein desmoplakin from cell-cell interfaces disrupts anchorage of intermediate filament bundles and alters intercellular junction assembly. J. Cell Biol. 134, 985-1001. 10.1083/jcb.134.4.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornslaeger E. A., Godsel L. M., Corcoran C. M., Park J. K., Hatzfeld M., Kowalczyk A. P. and Green K. J. (2001). Plakophilin 1 interferes with plakoglobin binding to desmoplakin, yet together with plakoglobin promotes clustering of desmosomal plaque complexes at cell-cell borders. J. Cell. Sci. 114, 727-738. [DOI] [PubMed] [Google Scholar]
- Broussard J. A., Getsios S. and Green K. J. (2015). Desmosome regulation and signaling in disease. Cell Tissue Res. 360, 501-512. 10.1007/s00441-015-2136-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins C. C., Setzer S. V., Jennings J. M., Summers S., Tsunoda K., Amagai M. and Kowalczyk A. P. (2006). Desmoglein endocytosis and desmosome disassembly are coordinated responses to pemphigus autoantibodies. J. Biol. Chem. 281, 7623-7634. 10.1074/jbc.M512447200 [DOI] [PubMed] [Google Scholar]
- Chatel G., Desai S. H., Mattheyses A. L., Powers M. A. and Fahrenkrog B. (2012). Domain topology of nucleoporin Nup98 within the nuclear pore complex. J. Struct. Biol. 177, 81-89. 10.1016/j.jsb.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delva E., Tucker D. K. and Kowalczyk A. P. (2009). The desmosome. Cold Spring Harb. Perspect. Biol. 1, a002543 10.1101/cshperspect.a002543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey G. T., Vaughan J. C., Chen K. H., Bates M. and Zhuang X. (2011). Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 8, 1027-1036. 10.1038/nmeth.1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai B. V., Harmon R. M. and Green K. J. (2009). Desmosomes at a glance. J. Cell Sci. 122, 4401-4407. 10.1242/jcs.037457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar M. G. and Palade G. E. (1963). Junctional complexes in various epithelia. J. Cell Biol. 17, 375-412. 10.1083/jcb.17.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod D. R., Berika M. Y., Bardsley W. F., Holmes D. and Tabernero L. (2005). Hyper-adhesion in desmosomes: its regulation in wound healing and possible relationship to cadherin crystal structure. J. Cell Sci. 118, 5743-5754. 10.1242/jcs.02700 [DOI] [PubMed] [Google Scholar]
- Getsios S., Huen A. C. and Green K. J. (2004). Working out the strength and flexibility of desmosomes. Nat. Rev. Mol. Cell Biol. 5, 271-281. 10.1038/nrm1356 [DOI] [PubMed] [Google Scholar]
- Green K. J. and Simpson C. L. (2007). Desmosomes: new perspectives on a classic. J. Invest. Dermatol. 127, 2499-2515. 10.1038/sj.jid.5701015 [DOI] [PubMed] [Google Scholar]
- Green K. J., Getsios S., Troyanovsky S. and Godsel L. M. (2010). Intercellular junction assembly, dynamics, and homeostasis. Cold Spring Harb. Perspect. Biol. 2, a000125 10.1101/cshperspect.a000125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grum V. L., Li D., MacDonald R. I. and Mondragón A. (1999). Structures of two repeats of spectrin suggest models of flexibility. Cell 98, 523-535. 10.1016/S0092-8674(00)81980-7 [DOI] [PubMed] [Google Scholar]
- Harmon R. M. and Green K. J. (2013). Structural and functional diversity of desmosomes. Cell Commun. Adhes. 20, 171-187. 10.3109/15419061.2013.855204 [DOI] [PubMed] [Google Scholar]
- Hatzfeld M., Haffner C., Schulze K. and Vinzens U. (2000). The function of plakophilin 1 in desmosome assembly and actin filament organization. J. Cell Biol. 149, 209-222. 10.1083/jcb.149.1.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T. E., Merritt A. J. and Garrod D. R. (2007). Calcium-independent desmosomes of keratinocytes are hyper-adhesive. J. Invest. Dermatol. 127, 775-781. 10.1038/sj.jid.5700643 [DOI] [PubMed] [Google Scholar]
- Kitajima Y. (2014). 150th anniversary series: desmosomes and autoimmune disease, perspective of dynamic desmosome remodeling and its impairments in pemphigus. Cell Commun. Adhes. 21, 269-280. 10.3109/15419061.2014.943397 [DOI] [PubMed] [Google Scholar]
- Kowalczyk A. P. and Green K. J. (2013). Structure, function, and regulation of desmosomes. Prog. Mol. Biol. Transl. Sci. 116, 95-118. 10.1016/B978-0-12-394311-8.00005-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschberger A., van de Linde S., Dabauvalle M.-C., Rieger B., Heilemann M., Krohne G. and Sauer M. (2012). Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J. cell Sci. 125, 570-575. 10.1242/jcs.098822 [DOI] [PubMed] [Google Scholar]
- Loschberger A., Franke C., Krohne G., van de Linde S. and Sauer M. (2014). Correlative super-resolution fluorescence and electron microscopy of the nuclear pore complex with molecular resolution. J. cell Sci. 127, 4351-4355. 10.1242/jcs.156620 [DOI] [PubMed] [Google Scholar]
- Nahidiazar L., Kreft M., van den Broek B., Secades P., Manders E. M. M., Sonnenberg A. and Jalink K. (2015). The molecular architecture of hemidesmosomes, as revealed with super-resolution microscopy. J. cell Sci. 128, 3714-3719. 10.1242/jcs.171892 [DOI] [PubMed] [Google Scholar]
- North A. J., Bardsley W. G., Hyam J., Bornslaeger E. A., Cordingley H. C., Trinnaman B., Hatzfeld M., Green K. J., Magee A. I. and Garrod D. R. (1999). Molecular map of the desmosomal plaque. J. Cell Sci. 112, 4325-4336. [DOI] [PubMed] [Google Scholar]
- O'Keefe E. J., Erickson H. P. and Bennett V. (1989). Desmoplakin I and desmoplakin II. Purification and characterization. J. Biol. Chem. 264, 8310-8318. [PubMed] [Google Scholar]
- Owen G. R., Acehan D., Derr K. D., Rice W. J. and Stokes D. L. (2008). Cryoelectron tomography of isolated desmosomes. Biochem. Soc. Trans. 36, 173-179. 10.1042/BST0360173 [DOI] [PubMed] [Google Scholar]
- Palka H. L. and Green K. J. (1997). Roles of plakoglobin end domains in desmosome assembly. J. Cell Sci. 110, 2359-2371. [DOI] [PubMed] [Google Scholar]
- Ribeiro S. A., Vagnarelli P., Dong Y., Hori T., McEwen B. F., Fukagawa T., Flors C. and Earnshaw W. C. (2010). A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. USA 107, 10484-10489. 10.1073/pnas.1002325107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M. J., Bates M. and Zhuang X. (2006). Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793-795. 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Tucker D. K., Kohlhorst D., Niessen C. M. and Kowalczyk A. P. (2012). Classical and desmosomal cadherins at a glance. J. Cell Sci. 125, 2547-2552. 10.1242/jcs.066654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolik-Delmaire T., Katafiasz D. and Wahl J. K. III. (2006). Carboxyl terminus of Plakophilin-1 recruits it to plasma membrane, whereas amino terminus recruits desmoplakin and promotes desmosome assembly. J. Biol. Chem. 281, 16962-16970. 10.1074/jbc.M600570200 [DOI] [PubMed] [Google Scholar]
- Stahley S. N. and Kowalczyk A. P. (2015). Desmosomes in acquired disease. Cell Tissue Res. 360, 439-456. 10.1007/s00441-015-2155-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahley S. N., Saito M., Faundez V., Koval M., Mattheyses A. L. and Kowalczyk A. P. (2014). Desmosome assembly and disassembly are membrane raft-dependent. PLoS ONE 9, e87809 10.1371/journal.pone.0087809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahley S. N., Warren M. F., Feldman R. J., Swerlick R. A., Mattheyses A. L. and Kowalczyk A. P. (2015). Super-resolution microscopy reveals altered desmosomal protein organization in pemphigus vulgaris patient tissue. J. Invest. Dermatol. 136, 59-66. 10.1038/JID.2015.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes D. L. (2007). Desmosomes from a structural perspective. Curr. Opin. Cell Biol. 19, 565-571. 10.1016/j.ceb.2007.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J., Cordier G. A., Borbely J. S., Sandoval Álvarez A. and Lakadamyali M. (2014). Cross-talk-free multi-color STORM imaging using a single fluorophore. PLoS ONE 9, e101772 10.1371/journal.pone.0101772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason H. A., Scothern A., McHarg S. and Garrod D. R. (2010). Desmosomes: adhesive strength and signalling in health and disease. Biochem. J. 429, 419-433. 10.1042/BJ20100567 [DOI] [PubMed] [Google Scholar]
- Todorovic V., Koetsier J. L., Godsel L. M. and Green K. J. (2014). Plakophilin 3 mediates Rap1-dependent desmosome assembly and adherens junction maturation. Mol. Biol. Cell 25, 3749-3764. 10.1091/mbc.E14-05-0968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda K., Ota T., Aoki M., Yamada T., Nagai T., Nakagawa T., Koyasu S., Nishikawa T. and Amagai M. (2003). Induction of pemphigus phenotype by a mouse monoclonal antibody against the amino-terminal adhesive interface of desmoglein 3. J. Immunol. 170, 2170-2178. 10.4049/jimmunol.170.4.2170 [DOI] [PubMed] [Google Scholar]
- Tucker D. K., Stahley S. N. and Kowalczyk A. P. (2014). Plakophilin-1 protects keratinocytes from pemphigus vulgaris IgG by forming calcium-independent desmosomes. J. Invest. Dermatol. 134, 1033-1043. 10.1038/jid.2013.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoorn H., Harkes R., Spiesz E. M., Storm C., van Noort D., Ladoux B. and Schmidt T. (2014). The nanoscale architecture of force-bearing focal adhesions. Nano Lett. 14, 4257-4262. 10.1021/nl5008773 [DOI] [PubMed] [Google Scholar]
- Whelan D. R. and Bell T. D. (2015). Image artifacts in single molecule localization microscopy: why optimization of sample preparation protocols matters. Sci. Rep. 5, 7924 10.1038/srep07924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T. T., Su J., Wang W. J., Craige B., Witman G. B., Tsou M. F. and Liao J. C. (2015). Superresolution pattern recognition reveals the architectural map of the ciliary transition zone. Sci. Rep. 5, 14096 10.1038/srep14096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H. (2015). Applying superresolution localization-based microscopy to neurons. Synapse 69, 283-294. 10.1002/syn.21806 [DOI] [PMC free article] [PubMed] [Google Scholar]