Abstract
Precise gene expression ensures proper stem and progenitor cell differentiation, lineage commitment and organogenesis during mammalian development. ATP-dependent chromatin-remodeling complexes utilize the energy from ATP hydrolysis to reorganize chromatin and, hence, regulate gene expression. These complexes contain diverse subunits that together provide a multitude of functions, from early embryogenesis through cell differentiation and development into various adult tissues. Here, we review the functions of chromatin remodelers and their different subunits during mammalian development. We discuss the mechanisms by which chromatin remodelers function and highlight their specificities during mammalian cell differentiation and organogenesis.
KEY WORDS: Gene expression, Chromatin remodeling, Chromatin remodeler, Transcription, Differentiation
Summary: This Review summarizes the mechanisms by which chromatin remodelers function and highlights their specificities during mammalian cell differentiation and organogenesis.
Introduction
Organogenesis involves the development, differentiation, migration and maturation of a variety of cell types, many of which are defined by specific gene expression programs (Young, 2011). Gene expression programs operate within the confines of chromatin and hence chromatin organization influences key aspects of mammalian development. Major factors that affect chromatin organization are: (1) DNA methylation; (2) covalent modification of histones; and (3) ATP-dependent alterations to chromatin. ATP-dependent chromatin alteration is accomplished by several multi-subunit chromatin-remodeling complexes that utilize energy derived from ATP hydrolysis to alter nucleosome structure or conformation and, thereby, regulate the access of transcription factors to their cognate DNA binding sites. These chromatin-remodeling complexes are primarily made up of a single ATPase and multiple associated subunits. The ATPase subunit binds and hydrolyzes ATP, while the associated subunits modulate the catalytic activity of the ATPase subunit and provide specificity to genome binding. Thus, combinatorial assembly of different ATPase and associated subunits gives rise to a diverse set of chromatin-remodeling complexes with cell- and tissue-specific functions (Ho and Crabtree, 2010; Wang et al., 1996; Wu et al., 2009).
Recent advances in mass spectrometry and high-throughput sequencing have identified and characterized several ATP-dependent chromatin-remodeling complexes in mammals. One conclusion of these studies is that chromatin-remodeling complexes facilitate the transcription of a group of genes while simultaneously inhibiting inappropriate expression of other genes to establish a specific cell identity. Here, we review four major classes of ATP-dependent chromatin-remodeling enzymes and their subunits, and discuss how they function during cell differentiation and development in mammals. We summarize evidence suggesting that tissue-specific chromatin-remodeler subunits often form a unique complex that collaborates with lineage-specific transcription factors for recruitment to regulatory sites in the genome, where they remodel chromatin to favor transcription factor binding and regulate lineage-specific gene transcription.
Types of ATP-dependent chromatin-remodeling complexes
All ATP-dependent chromatin-remodeling complexes contain an ATPase subunit belonging to the SNF2 family of DNA helicases and one or more associated subunits. SNF2 family proteins can be divided into four major subfamilies based on sequence similarity between their ATPase domains, with each subfamily being distinguished by the presence of distinct domains (Fig. 1). These four major subfamilies are SWI/SNF (switch/sucrose non-fermentable), ISWI (imitation SWI), CHD (chromodomain helicase DNA-binding) and INO80 [SWI2/SNF2 related (SWR)]. Each of these subfamilies consists of several protein(s) or protein complexes that contains up to 16 different subunits. While the ATPase subunits perform the basic catalytic activities of the complex, the associated subunits have important roles in modulating the catalytic activity. For example, the associated subunit IES2 in the INO80 complex activates the ATPase activity of the catalytic INO80 subunit, whereas two other subunits – IES6 and ARP5 – facilitate binding of the INO80 complex to nucleosomes (Chen et al., 2013). Similar modulation of the ATPase domain activity by other domains and associated subunits has been shown for SWI/SNF (Cairns et al., 1996; Sen et al., 2013) and ISWI (Clapier and Cairns, 2012; Hota et al., 2013) complexes. Furthermore, the assembly of tissue-specific isoforms of associated subunits into these complexes confers unique properties that are particularly suited to the tissue type and help in recruiting these complexes to tissue-specific regulatory genomic loci. Indeed, various complexes with different subunit compositions have been identified in specific cells and tissues during development. For example, the BAF60C isoform of the associated BAF60 subunit in the mammalian SWI/SNF complex has important roles in regulating heart and muscle gene transcription, whereas a SWI/SNF complex containing BAF60A has minimal role in these tissue types (Forcales et al., 2012; Lickert et al., 2004). As more and more tissue-specific chromatin-remodeler subunits are identified, it will be crucial to understand the subunit composition of these remodelers and the mechanisms through which they regulate tissue-specific gene expression.
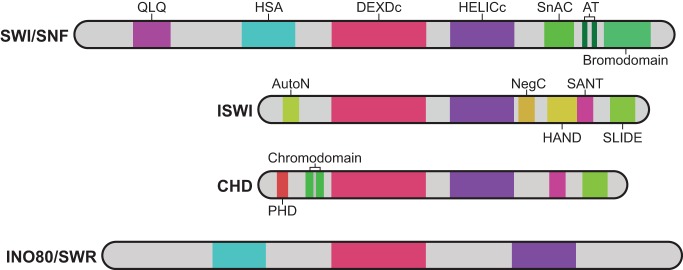
Fig. 1.
The domain structures of SNF2 family proteins. The domain organization of the catalytic subunits of SWI/SNF, ISWI, CHD and INO80/SWR subfamilies of chromatin remodelers are shown. All of these subunits are SNF2 family proteins. They all contain an ATPase domain, which consists of DEXDc and HELICc domains, with each subfamily possessing additional domains. SWI/SNF proteins, for example, are defined by the presence of an N-terminal helicase-SANT (HSA) domain and a C-terminal bromodomain. These proteins also contain QLQ and SNF2 ATP-coupling (SnAC) domains, as well as two A-T hook motifs. By contrast, ISWI proteins harbor a C-terminal SANT domain as well as SANT-like ISWI (SLIDE) and HAND domains. They also contain AutoN and NegC regulatory domains. CHD proteins are defined by the presence of tandem N-terminal chromodomains, with some family members containing N-terminal plant homeodomain (PHD) domains. INO80R/SWR proteins notably contain a split ATPase domain, with a spacer between the DEXDc and HELICc domains.
Developmental roles of SWI/SNF-like complexes
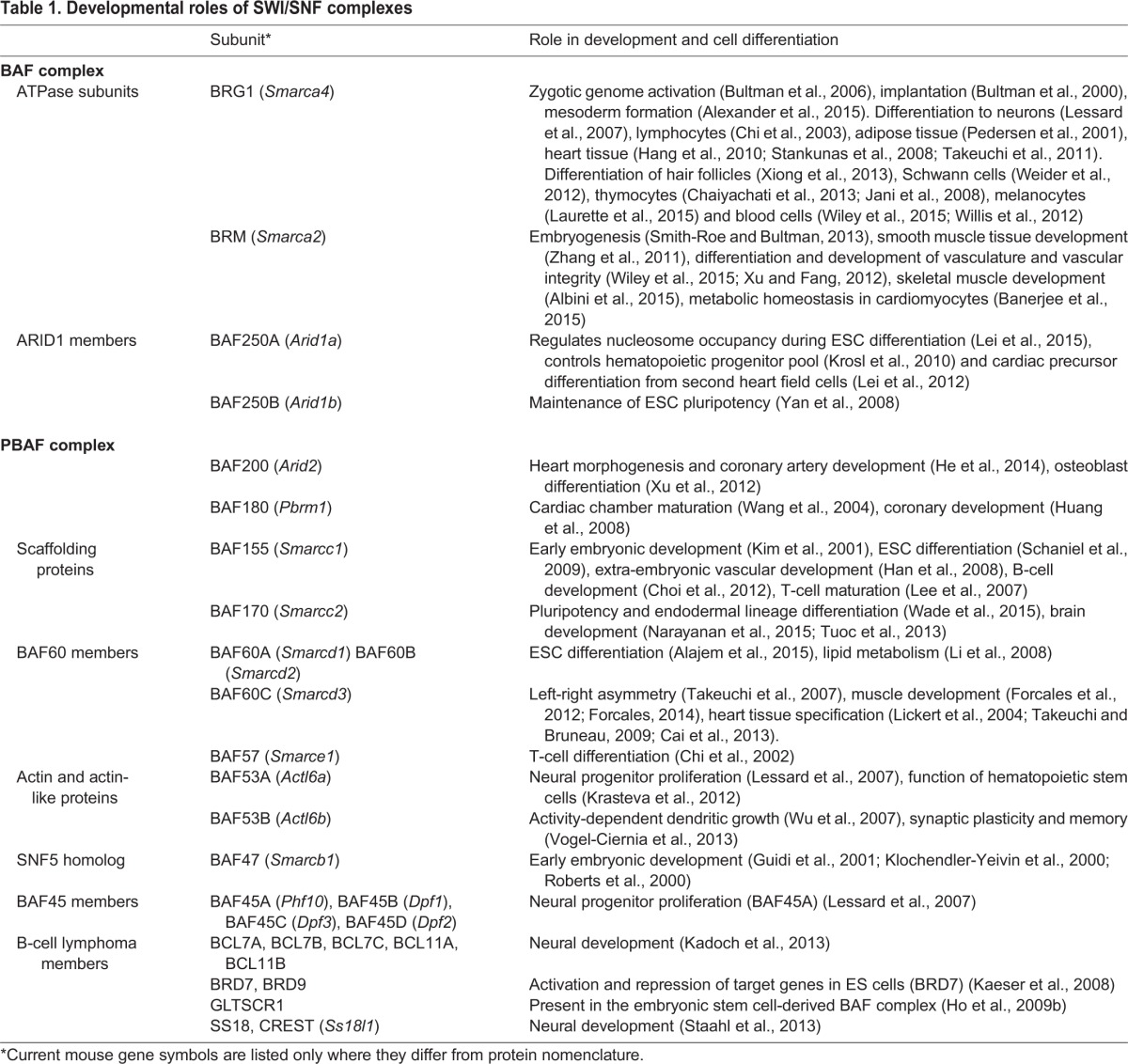
SWI/SNF complexes, which contain SWI2 or SNF2 ATPase subunits, were initially identified in Saccharomyces cerevisiae, with brahma (brm) in Drosophila and Brm and Brahma-related gene 1 (Brg1) in mammals subsequently being identified as homologs. This led to the identification of various complexes containing these subunits in mammalian cells. BRM/BRG1-associated factor (BAF) complexes are large ∼1.5 MDa complexes (Fig. 2A) and consist of at least 15 different subunits, including either BRM or BRG1 as the ATPase subunit (Table 1). Depending upon the ATPase and associated subunits that they contain, these complexes can be classed into BAF250A-containing BAF-A complexes or BAF250B-containing BAF-B complexes. Furthermore, BAF200-, BAF180 (polybromo)- and BRD7-containing complexes can associate with the BRG1 ATPase subunit, but not BRM, to form a polybromo-associated BAF (PBAF) complex (Yan et al., 2008). BAF complexes are similar to yeast SWI/SNF complexes, but are more diverse in their subunit composition and have roles both in activation and repression of genes, performing key functions during development (Ho and Crabtree, 2010; Wang et al., 1996). Below, we describe some of the key functions of BAF complexes and subunits at different stages of mammalian development and discuss their mechanisms of action.
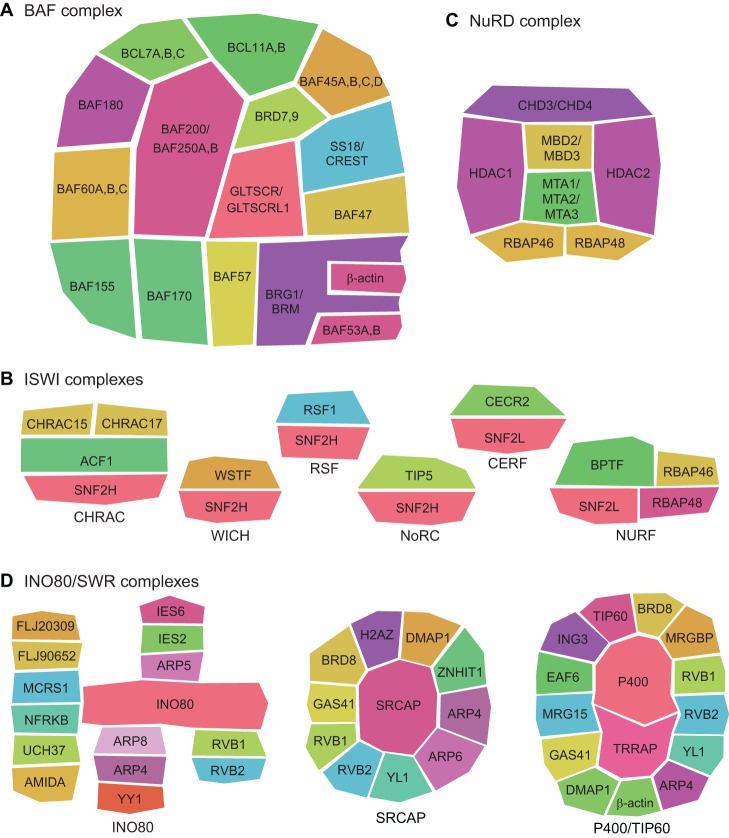
Fig. 2.
The composition of chromatin-remodeling complexes. The subunit composition of some mammalian chromatin-remodeling complexes is shown: (A) BAF complex, (B) ISWI complexes, (C) the CHD-containing NuRD complex and (D) INO80/SWR complexes.
Table 1.
Developmental roles of SWI/SNF complexes
BAF complex function in preimplantation development and embryonic stem cells
BAF complex components have varied roles throughout early development in mammals (Fig. 3). After fertilization, the transcriptionally inert zygotic genome is activated to initiate high transcriptional activity, a process known as zygotic genome activation (ZGA). BRG1 has essential roles in ZGA as it regulates global dimethyl H3K4 levels to control zygotic transcription (Bultman et al., 2006). After ZGA, blastomeres rapidly divide to form a compact blastula of 16-40 cells. By embryonic day 3 (E3), blastula cells polarize to form cells of the inner cell mass (ICM) and the outer layer of trophectoderm (TE).
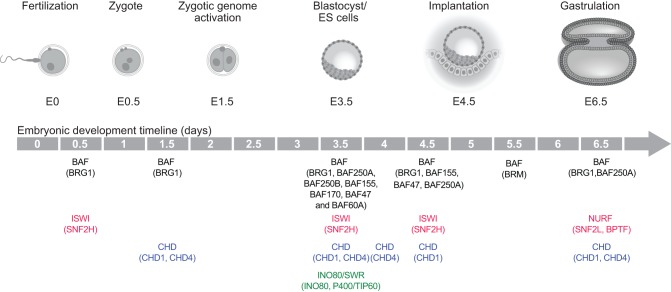
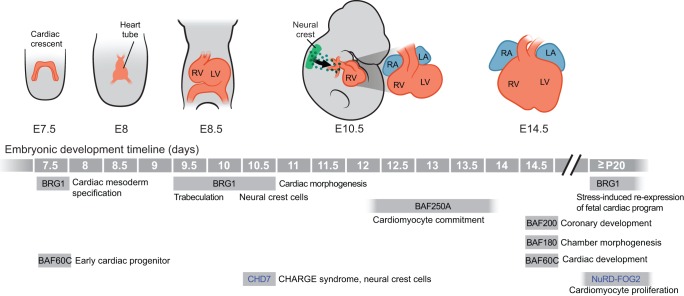
Fig. 3.
Roles of chromatin remodelers in early embryogenesis. Stages of early mammalian development are pictured, with mouse embryonic day (E) indicated below each pictured stage. Key demonstrated roles for specific chromatin-remodeling factors are positioned at relevant stages below the developmental timeline. Roles for BAF complex subunits are in black; ISWI/NURF subunits are in red; CHD proteins are in blue; INO80/SWR factors are in green.
Embryonic stem cells (ESCs) are derived from the ICM of blastocysts and are characterized by their properties of self-renewal and pluripotency. A network of core transcription factors (OCT4, SOX2 and NANOG) regulates gene expression to maintain pluripotency in ESCs and Polycomb group (PcG) proteins modify chromatin to prevent differentiation (Jaenisch and Young, 2008; Young, 2011). Several components of BAF chromatin-remodeling complexes have been implicated in this self-renewal and pluripotency transcriptional network (Table 1). For example, BRG1 is required to maintain ESC self-renewal and pluripotency (Fazzio et al., 2008; Ho et al., 2009b; Kidder et al., 2009), by activating the expression of core pluripotency transcription factors while repressing differentiation-specific genes. BRG1 binds to the promoter regions of key pluripotency genes, PcG genes and some of their target genes (Ho et al., 2009a; Kidder et al., 2009), and appears to act by modulating LIF/STAT3 signaling and Polycomb function (Ho et al., 2011).
In mouse ESCs, this specialized role of BRG1 is mediated by the formation of an ESC-specific BAF complex called esBAF, which contains BRG1, BAF155 and BAF60A but lacks BRM, BAF170 and BAF60C (Ho et al., 2009b). In human ESCs, however, esBAF complex composition is different; human esBAF contains both BAF155 and BAF170, and BAF170 is required to maintain human ESC pluripotency (Wade et al., 2015; Zhang et al., 2014). Accordingly, BAF170 depletion causes loss of pluripotency and its overexpression in human ESCs causes impaired differentiation to mesoderm and endoderm lineages. This is not due to the primed state of human ESCs versus the naive state of mouse ESCs but could be due to the inherent difference between mouse and human and might mark rodent-human divergence in BAF complex function. In addition to its role in maintaining pluripotency, BRG1 is essential during trophoblast development, where it represses Nanog via HDAC1-mediated H3K9/14 deacetylation (Carey et al., 2015) and Oct4 by a Cdx2-mediated mechanism (Wang et al., 2010). Thus, BRG1 appears to support self-renewal and pluripotency by chromatin remodeling and conditioning the genome for proper signaling while simultaneously repressing core pluripotency factors in non-ESC lineages.
Several other subunits of the BAF complex have important roles in maintaining pluripotency, largely via reorganizing chromatin and redistributing histone marks. Both BAF250A and BAF250B have distinct roles in early stages of development, regulating embryonic stem cell pluripotency, differentiation and cell lineage decisions (Gao et al., 2008; Yan et al., 2008). BAF250A mutant ESCs differentiate into ectoderm lineages but are defective in differentiation towards cardiomyocyte or hepatocytes lineages. BAF250A appears to regulate lineage decisions by modulating esBAF and PcG activities and nucleosome occupancy at poised chromatin regions (Lei et al., 2015). The subunits BAF155 and BAF60A are also crucial for ESC differentiation. They act to repress the pluripotency genes Nanog and Klf4, respectively, via chromatin reorganization, compaction and heterochromatin formation upon initiation of differentiation (Alajem et al., 2015; Schaniel et al., 2009). Furthermore, BAF60A regulates transcription by binding to bivalent genes and re-distributing H3K4me3 and H3K27me3 marks to regulate gene expression.
The role of BAF complex subunits in maintaining pluripotency is also supported by their ability to induce the reprogramming of terminally differentiated cells. Knockdown of the ATPase subunit BRM or the core subunit BAF170, both of which are mostly expressed in terminally differentiated cells and absent in ESCs, gives rise to better reprogramming outcomes (Jiang et al., 2015). BRG1 and BAF155 facilitate OCT4-SOX2-KLF4-MYC-mediated reprogramming by facilitating OCT4 binding and increasing the euchromatin content of the cells (Singhal et al., 2014, 2010). BAF complexes are thus important regulators of the core pluripotency transcriptional network that function to establish and maintain pluripotency in ESCs, and reorganize chromatin to allow differentiation.
From implantation through to gastrulation
A number of BAF subunits are involved in implantation and early development of the embryo. BRG1 is essential during peri-implantation development, and Brg1 homozygous mutants die from failure to develop both the ICM and the trophectoderm (Bultman et al., 2000). Unlike BRG1, BRM is not essential for early embryonic development, and Brm-null mice are viable (Reyes et al., 1998), although a recent study revealed that Brm-knockout mice express functional transcripts, suggesting incomplete knockout (Thompson et al., 2015). BRM has non-redundant roles with BRG1 during some, but not all, developmental processes in embryogenesis (Smith-Roe and Bultman, 2013). The associated subunits, BAF155, BAF47 and BAF250A are also required for peri-implantation development (Gao et al., 2008; Guidi et al., 2001; Han et al., 2008; Kim et al., 2001; Klochendler-Yeivin et al., 2000; Roberts et al., 2000), further establishing the crucial role of BAF subunits during early embryonic development.
It is also evident that several BAF complex subunits have active roles as embryos undergo gastrulation and form the three germ layers. BRG1 and BAF250A, for example, are required for mesoderm formation. During in vitro directed differentiation of ESCs to cardiomyocytes, loss of Brg1 (Alexander et al., 2015) impairs mesoderm induction, leading to a loss of mesoderm-derived cardiomyocytes. BRG1 colocalizes with and activates H3K27ac-enriched distal enhancers for mesodermal gene induction and represses non-mesodermal developmental genes via a PcG-mediated mechanism. In agreement with these observations, BRG1 binds distal regulatory elements in multiple developing organs during embryogenesis, and BRG1 occupancy correlates with active and repressive histone marks and gene expression (Attanasio et al., 2014). The loss of BAF250A also results in defective mesoderm formation, with a reduction in mesoderm-derived cardiomyocytes and adipocytes (Gao et al., 2008). Thus, BAF subunits collaborate with different histone modifications to drive gene expression during mesoderm lineage commitment.
BAF complexes in neural development
Many BAF complex subunits are involved in cell differentiation and development throughout neural development (Fig. 4). Both Brg1 and Baf155 heterozygous mutants exhibit neural tube closure defects and exencephaly, suggesting that BAF complexes have an important and dosage-sensitive role in neural differentiation and development (Bultman et al., 2000; Kim et al., 2001). In agreement with these observations, BRG1 is required during both neuronal and glial differentiation (Marathe et al., 2013; Matsumoto et al., 2006; Weider et al., 2012; Yu et al., 2013). In the central nervous system, the differentiation of glial progenitor cells to form first immature and subsequently mature myelinating oligodendrocytes requires BRG1 (Yu et al., 2013). In this context, the oligodendrocyte-specific transcription factor OLIG2 coats the H3K27ac-containing enhancer regions of myelination-specific genes and recruits BRG1 to these sites. This combination of transcription factor, chromatin remodeler and activating histone mark drives oligodendrocyte differentiation and myelination. In the peripheral nervous system, BAF complexes are essential for the differentiation of Schwann cells (Weider et al., 2012). Here, SOX10, a Schwann cell- and melanocyte-specific marker, interacts with BAF60A and recruits the BAF complex to enhancer regions of genes encoding the Schwann cell differentiation-specific transcription factors OCT6 and KROX20 to drive differentiation and maturation. However, a recent study found that the role of BRG1 is less significant during oligodendrocyte differentiation than it is during the differentiation of Schwann cells (Bischof et al., 2015).
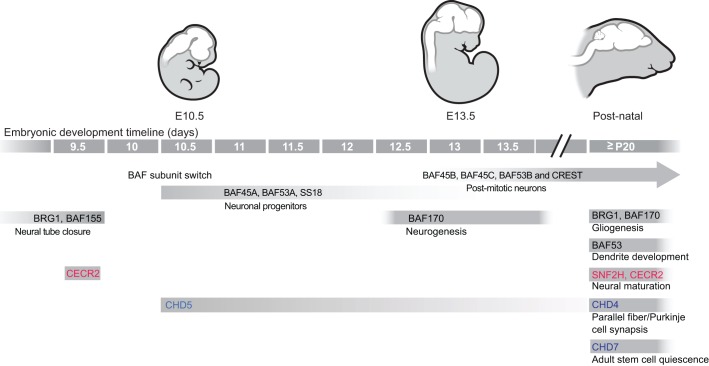
Fig. 4.
Chromatin remodeler functions and transitions in neural development. Stages of mammalian neural development are pictured, with mouse embryonic day (E) indicated below each pictured stage. Key demonstrated roles for specific chromatin remodeling factors are positioned at relevant stages below the developmental timeline. Roles for BAF complex subunits are in black; ISWI/NURF subunits are in red; CHD proteins are in blue.
BAF complexes also play essential roles in neural progenitors. An important switch in BAF complex composition occurs as neural progenitors differentiate into mature neurons, whereby the neural progenitor-specific BAF45A (Phf10) and BAF53A (Actl6a) subunits are replaced by mature neuron-specific BAF45B (Dpf1), BAF45C (Dpf3) and BAF53B (Actl6b) isoforms (Lessard et al., 2007; Wu et al., 2007). In line with this, short hairpin-mediated knockdown of Baf45a and/or Baf53a causes reduced proliferation, whereas overexpression of Baf45a increases the mitotic activity of neural progenitors (Lessard et al., 2007). The neural progenitor-specific BAF complex modulates both Notch and Sonic hedgehog (Shh) signaling to support proliferation and keep cells in a state poised for differentiation to post-mitotic neurons (Lessard et al., 2007). In post-mitotic neurons, BAF53B is essential for activity-dependent dendritic growth and branching. A neuron-specific BAF complex containing BAF53B recruits the Ca2+-regulated dendritic regulator CREST to the promoter regions of genes involved in dendritic growth and directly regulates their expression (Wu et al., 2007). In agreement with these observations, BAF53B was found to be essential for long-term memory and synaptic plasticity (Vogel-Ciernia et al., 2013). The switch from neural progenitor-specific BAF53A to neuron-specific BAF53B is mediated by repression of BAF53A by the microRNAs (miRNAs) miR-9* and miR-124, which are highly expressed in post-mitotic neurons. In neuronal progenitors, by contrast, miR-9* and miR-124 are repressed by the transcriptional repressor REST. As REST activity declines in post-mitotic neurons, miR-9* and miR-124 bind to the 3′ untranslated region of BAF53A, facilitating its repression and the expression of BAF53B (Yoo et al., 2009).
These miRNAs are also crucial for the direct reprogramming of human fibroblasts to neurons (Yoo et al., 2011); miR-9/9* and miR-124 can reprogram fibroblasts to mature neurons, facilitated by NEUROD2. Reprogramming efficiency is increased in the presence of the neurogenic transcription factors ASCL1 and MYT1L; however, these transcription factors alone (i.e. without miR-9/9* and miR-124) cannot reprogram fibroblasts to neurons. Furthermore, the switch in BAF subunit composition involves the removal of SS18, a target of oncogenic translocation in synovial sarcoma that is present in the BAF complex at the progenitor stage, and its replacement by CREST, a calcium-responsive transactivator and SS18-like protein that is found in mature neuron-specific BAF complexes (Kadoch and Crabtree, 2013; Staahl et al., 2013).
The BAF170 subunit also has significant roles in neurogenesis. During neurogenesis, radial glia act as neuronal progenitors that undergo direct neurogenesis to produce deep layer neurons or indirect neurogenesis to produce intermediate progenitors that differentiate into neurons to populate the upper layers of the developing cerebral cortex. BAF170-containing BAF complexes facilitate direct neurogenesis and inhibit indirect neurogenesis to regulate cortex size (Tuoc et al., 2013). They do so by regulating the chromatin landscape and inhibiting the binding of REST to promoters of PAX6 target genes, thereby blocking the proliferation of intermediate and late progenitors during indirect neurogenesis. It appears that BAF170 competes with BAF155 to regulate BAF complex composition and activity. The conditional deletion of both BAF155 and BAF170 subunits revealed that BAF complexes containing these essential subunits are essential for forebrain development (Narayanan et al., 2015). In this setting, global H3K9ac histone acetylation levels were reduced, and repressive H3K27me2/3 levels were increased, resulting in the downregulation of several genes essential for brain development. Furthermore, BAF complexes regulate H3K27 methylation by interacting with and increasing the activity of the histone lysine demethylases JMJD3 (Kdm6b) and UTX (Kdm6a), regulating their access to histone H3. Together, these findings suggest that distinct BAF complexes undergo compositional changes, interact with specific transcription factors and chromatin regulators, and reorganize the global chromatin landscape to regulate gene expression during brain development.
BAF complex functions during hair follicle and skin development
BAF complex components have been implicated in the development and regeneration of various cell types within the epidermis. A molecular circuitry between BRG1 and Sonic Hedgehog (SHH) signaling, for example, regulates hair follicle regeneration (Xiong et al., 2013). In bulge cells – the multipotent stem cells found in the hair follicle – SHH signaling through GLI transcription factors (GLI1 and GLI2) activates Brg1 (Smarca4) to initiate hair regeneration. However, in matrix cells, which contribute to the bulk of the hair shaft, NFκβ recruitment by BRG1 activates Shh to maintain hair growth. The loss of Brg1 or inhibition of SHH signaling disrupts this molecular circuitry, resulting in failure in hair regeneration and tissue repair (Wang et al., 2000; Xiong et al., 2013). Thus co-operation between chromatin remodelers, signaling pathways and transcription factors regulates mammalian hair development.
BAF chromatin remodelers are also involved in keratinocyte and melanocyte differentiation. In keratinocytes, both BRG1 and, to a lesser extent, BRM are required for terminal epidermal differentiation (Indra et al., 2005). Brg1 is expressed at late stages of keratinocyte differentiation, and its conditional deletion results in skin permeability barrier defects, which are slightly accentuated in a Brm (Smarca2) mutant background. Expression of the actin-related protein subunit of the BAF complex, BAF53A, is essential to maintain the epidermal progenitor state and is reduced during terminal keratinocyte differentiation (Bao et al., 2013). In the absence of BAF53A, the BAF complex binds to epidermal differentiation genes to facilitate their expression. BAF complexes also coordinate with the epidermal differentiation transcription factor P63 to maintain an open chromatin state at P63 binding sites, facilitating the expression of epidermal differentiation-specific genes in keratinocytes (Bao et al., 2015). BRG1 also has an important role during melanocyte development (Laurette et al., 2015). Microphthalmia-associated transcription factor (MITF), an important regulator of the melanocyte lineage, physically interacts with several subunits of the PBAF complex, including BRG1 and the chromo and helicase DNA-binding domain protein CHD7, to regulate melanocyte differentiation. At melanocyte-specific active enhancers, BRG1 is recruited by MITF and SOX2, which in turn remodel chromatin and recruit other transcription factors, such as YY1 and TFAP2A, to regulate gene expression during melanocyte development. Overall, these studies highlight that the cell type-specific roles of BAF subunits are mediated by their interaction with lineage-specific transcription factors during hair and skin development.
Roles for BAF complexes during cardiovascular development
BAF complex components are essential for multiple stages of cell differentiation during heart development, and for heart formation and post-natal heart function (Fig. 5). BRG1, in particular, has crucial roles in multiple transcriptional regulatory pathways. For instance, it is required for the formation and differentiation of neural crest cells, which are essential for the formation of pharyngeal arch arteries and the cardiac outflow tract. BRG1 maintains a neural crest cell progenitor pool by promoting mitosis and inhibiting apoptosis, facilitates neural crest cell maturation to vascular smooth muscle cells and regulates neural crest cell migration to the outflow tract (Li et al., 2013). BRG1 is also a crucial regulator of cardiomyocyte gene expression and differentiation, and its dosage is essential during mouse and zebrafish heart development (Takeuchi et al., 2011). Deletion of Brg1 in the developing mouse heart leads to severe anomalies in cardiac morphogenesis and dysregulation of cardiac gene expression programs. Brg1 is haploinsufficient in the mouse heart, as heterozygous loss of function results in dilated and disorganized ventricles, ventricular septal defects and a double outlet right ventricle, anomalies that resemble human congenital heart defects. Brg1 genetically interacts with Tbx5, Tbx20 and Nkx2-5, and Brg1 dosage regulates transcription factor occupancy at cardiac promoters. The zebrafish brahma mutant (Gregg et al., 2003) also exhibits severe but specific defects in heart chamber cell allocation (Takeuchi et al., 2011). In addition, BRG1 is crucial during the development of cardiac trabeculae (Stankunas et al., 2008), the ridge-like outgrowths that project into the ventricle. In the endocardium, which overlies developing trabeculae, BRG1 directly binds to the promoter region and represses the expression of Adamts1, which encodes a metalloproteinase that degrades cardiac jelly, to facilitate trabeculation. Accordingly, the endocardial-specific loss of Brg1 derepresses Adamts1 and prevents trabeculation, whereas inhibition of metalloproteinase activity rescues the trabeculation defects observed in Brg1 mutants.
Fig. 5.
Roles for chromatin remodelers in heart development. Stages of mammalian heart development are pictured, with mouse embryonic day (E) indicated below each pictured stage. Key demonstrated roles for specific chromatin-remodeling factors are positioned at relevant stages below the developmental timeline. Roles for BAF complex subunits are in black; CHD proteins in blue. LA, left atrium; LV, left ventricle; RA, right atrium; RV, right ventricle.
BRG1 also functions in the adult heart. The transition from embryonic to adult heart involves a switch in myosin heavy chain (MHC) composition. In mice, this is due to a postnatal change in MHC expression, from predominant Myh7 (β-MHC) expression to Myh6 (α-MHC) expression. This switch occurs at the same time as a reduction in BRG1 expression is seen (Hang et al., 2010). In addition, Brg1 is reactivated in adult ventricular cardiomyocytes upon cardiac stress, and initiates a gene expression program that switches expression from Myh6 back to Myh7. Brg1 appears to be important for the maladaptive response to stress, as this can be reversed by preventing Brg1 re-expression in the stressed heart.
Several other subunits of the BAF complex have important tissue-specific roles in cardiac development. BAF60C, for example, is highly expressed in the developing heart and is required for some BAF complex functions. After gastrulation, cardiac mesoderm cells become rapidly specialized into cardiac progenitors that are marked by the expression of Smarcd3 (also known as Baf60c) (Devine et al., 2014). The RNAi-mediated knockdown of Baf60c in mouse embryos results in severe defects in heart development and defects in cardiac and skeletal muscle differentiation (Lickert et al., 2004). The function of BAF60C is ascribed to its ability to potentiate interactions between cardiac transcription factors and a BRG1-containing BAF complex to promote cardiac gene expression. Indeed, BAF60C along with the cardiac transcription factors TBX5 and GATA4 function together to reprogram mouse mesoderm to beating cardiomyocytes (Takeuchi and Bruneau, 2009). BAF60C is also an essential factor that cooperates with other cardiac transcription factors to reprogram fibroblasts into cardiovascular precursors, further highlighting its importance in cardiac differentiation (Lalit et al., 2016). Expression of Baf60c (Smarcd3), along with Tbx5 and Gata4, is induced by the Nodal and BMP4 antagonist CER1, resulting in recruitment of BAF60C to an enhancer of Nkx2-5 during cardiomyocyte differentiation (Cai et al., 2013).
Another BAF subunit, BAF250A, has crucial functions during the commitment of cardiac progenitor cells to beating cardiomyocytes or pacemaker cells (Lei et al., 2012; Wu et al., 2014). In the second heart field, Baf250a (Arid1a) deletion is embryonic lethal at E13, giving rise to a range of structural heart defects. BAF250A mediates BRG1 recruitment and the remodeling of promoter chromatin to regulate Mef2c, Nkx2-5 and Bmp10 expression as cardiac progenitor cells differentiate into beating cardiomyocytes (Lei et al., 2012). In sinoatrial node cells, in contrast, BAF250A along with TBX3 and the histone deacetylase HDAC3 repress Nkx2-5 expression to prevent differentiation to a ‘working’ cardiomyocyte fate (Wu et al., 2014).
The PBAF complex subunits BAF200 and BAF180 are implicated in cardiac chamber morphogenesis and coronary artery development (He et al., 2014; Huang et al., 2008; Wang et al., 2004). Baf180 (Pbrm1)-null mice die at E14.5, exhibiting severely hypoplastic ventricles due to failure in ventricular cell growth. BAF180 is also required for placental trophoblast growth, although the heart phenotype appears independent of placental defects. BAF180 directly regulates retinoic acid (RA) signaling by binding to the promoter region of Rarb2 (which encodes an RA receptor) and Crabp2 (which encodes an RA-binding protein) to regulate heart development (Wang et al., 2004). BAF180 is also involved in epithelial to mesenchymal transition (EMT), epicardial cell maturation and ventricular coronary vessel formation, and it positively regulates FGF, TGFβ and VEGF signaling during coronary development (Huang et al., 2008). BAF200, on the other hand, has a specific role in coronary artery development, where it is required for the conversion of venous cells to arterial endothelial cells (He et al., 2014). Thus, it appears that BAF complex components integrate signaling pathways and transcription factors occupancy to regulate different aspects of cardiac development.
BAF complexes in skeletal muscle development
The BAF complex subunits BRG1, BRM and BAF60C play distinct roles during different stages of muscle development, acting to regulate the expression of four myogenic regulatory factors – Myf5, Myod1, Myf6 and Myog – that are key for muscle progenitor commitment and differentiation. BRG1 is recruited to the Myog promoter in a MYOD1-dependent manner to regulate myogenic miRNA expression and muscle gene transcription (de la Serna et al., 2005; Mallappa et al., 2010). BRG1 is also responsible for the initiation of a muscle-specific transcriptional program (Albini et al., 2015). The other BAF complex catalytic subunit, BRM, represses the cell-cycle gene Ccnd1 to facilitate cell cycle exit upon differentiation, before the initiation of myogenic transcription. Separately, BRM also activates the transcription of late muscle-specific genes (Albini et al., 2015). BAF60C, however, interacts with MYOD1 and facilitates its binding at MYOD1 target sites, before muscle differentiation. In response to differentiation, BAF60C is phosphorylated by P38α, promoting its incorporation into a BRG1-containing BAF complex, which participates in initiating muscle-specific gene expression (Forcales et al., 2012). In addition, the forced expression of BAF60C and MYOD1 can directly reprogram human ESCs to a committed muscle progenitor fate – bypassing the mesoderm stage – and produce skeletal myofibers upon differentiation, further demonstrating the instructive role of BAF60C in myogenesis (Albini et al., 2013). While MYOD1 is responsible for repressing ESC-specific genes, it is not sufficient to initiate myogenic transcription, and thus, BAF60C is additionally required to induce muscle-specific genes.
There are three isoforms of BAF60 protein in mammals: BAF60A, BAF60B and BAF60C. How the expression of BAF60C is selected over that of the other two isoforms during myogenesis has recently been uncovered (Goljanek-Whysall et al., 2014; Saccone et al., 2014). These studies showed that, during early somite differentiation, the muscle-specific miRNAs miR-133 and miR1/206 repress the translation of BAF60A and BAF60B, resulting in a switch from primarily a BAF60A-containing complex to a BAF60C-containing complex. Thus, a change in the subunit composition of the BAF complex, via microRNA-mediated repression of alternative BAF60 subunit isoforms, appears to facilitate the formation of a muscle-specific BAF complex that is capable of MYOD1 interaction and muscle-specific gene expression.
Roles for BAF complexes during immune cell development
BAF complexes influence the development of various immune cell types. For example, BAF complex subunits have specific and important roles in maintaining hematopoietic stem cells (HSCs), the stem cells that give rise to all cellular components of blood (Krasteva et al., 2012). They also control the differentiation of HSCs to different lineages (Bultman et al., 2005; Griffin et al., 2008; Vradii et al., 2006) and, subsequently, different stages of B- and T-cell development.
The BAF complex subunits BRG1 and BAF155 regulate early stages of B-cell development (Choi et al., 2012). In the absence of BAF155, HSCs show defects in common lymphoid progenitor formation, whereas either Baf155 (Smarcc1) inactivation or loss of Brg1 affects pro-B cell development and survival by directly affecting the expression of the pro-B-cell-specific genes Ebf1 and Il7ra. Compared with its role in B-cell development, BRG1 has a far greater role during B-cell activation and activation-dependent cell proliferation, as evaluated by BRG1-mediated gene expression in naive and activated B cells (Holley et al., 2014). However, it is unknown if the specific assembly of BAF complex subunits into a BRG1-containing complex mediate the role of BRG1 in B-cell development and activation.
BAF complexes also play essential roles in various stages of T-cell development, where they remodel chromatin in response to different signals and regulate transcription (Chi et al., 2003; Gebuhr et al., 2003). In particular, they control the expression of T-cell co-receptors on the surface of T cells. Mature T cells express either CD4 or CD8 co-receptors, and BRG1 regulates CD8 activation, whereas both BRG1 and BAF57 are involved in silencing of CD4 (Chi et al., 2002). Accordingly, CD8+ cells are greatly reduced in Brg1+/− mutants, and both Brg1 and Baf57 (Smarce1) mutants prematurely produce CD4+ cells. CD4 repression depends on a 434 bp transcriptional silencer region, and both BRG1 and BAF57 increase chromatin accessibility by reducing H1 linker histone content and facilitating the binding of RUNX1, a transcriptional repressor, to this silencer region (Wan et al., 2009). BRG1 is also involved in CD4 activation and regulatory T-cell activation, which partially require its chromatin-remodeling ability and physical interactions between BRG1 and the Cd4 locus (Chaiyachati et al., 2013; Jani et al., 2008).
During erythropoiesis, the protein products of both α- and β-globin genes are combined to form αβ heterotetrameric proteins that ultimately form hemoglobin, the central component of erythrocytes. BRG1 regulates expression of both α- and β-globin genes by modifying nucleosome architecture near the promoter of these genes and by promoting the interaction between the promoter and a major regulatory element present far upstream. Furthermore, it modifies global histone H3 acetylation and H3K4me2 levels at these sites. Consistently, loss of BRG1 function leads to reduced expression of both α- and β-globin genes (Kim et al., 2009a,b). Thus, BAF complexes promote long-range interactions between enhancer and promoter regions to facilitate gene expression during erythropoiesis.
In summary, BAF complexes are involved in almost every stage of cell proliferation and differentiation during mammalian development. Distinct subunits form specialized BAF complexes and recruit them to specific genomic loci to provide cell type-specific functions. Far from being general transcriptional facilitators, BAF complexes have surprisingly specialized functions across the genome. A central function for BAF complexes appears to be their role downstream of signaling cascades, leading to the regulation of transcription by modulating transcription factor dosage. BAF complex-mediated chromatin reorganization by virtue of nucleosome redistribution appears to play a central role in regulating the transcriptional outcomes in different tissue types, with tissue specificity attributed to temporal expression and association of specific subunits with specialized BAF complexes. Furthermore, BAF complexes also function via non-coding RNAs (Flynn et al., 2016; Han et al., 2014; Prensner et al., 2013) and might mediate long-range chromosomal interactions to regulate gene expression. As new subunits are identified (for example the B-cell lymphoma family of proteins, such as BCL7A-C, BCL11A,B and bromodomain-containing proteins BRD7,9) and found to be part of stable BAF complexes (Kadoch et al., 2013; Kaeser et al., 2008), analyzing the function of these subunits in in vitro biochemical assays and via in vivo genetic perturbations may provide new insights into the role of these specialized BAF complexes.
Developmental roles of ISWI complexes
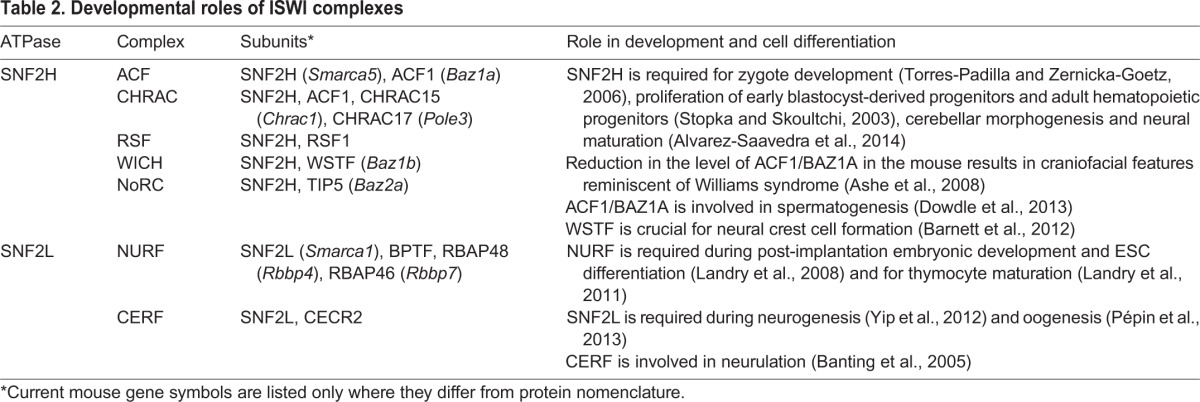
Similar to BAF complexes, ISWI complexes (Fig. 2B) contain one of two catalytic subunits with conserved ATPase and helicase domains, and 2-4 accessory subunits (Table 2). The two ISWI ATPase subunits found in mammals are SNF2H and SNF2L, both of which show distinct expression profiles (Lazzaro and Picketts, 2001). Snf2h (Smarca5) is expressed ubiquitously across a variety of tissue types, and is required for early embryonic development, whereas Snf2l (Smarca1) appears to be restricted to post-natal reproductive tissues and the brain (Lazzaro and Picketts, 2001; Stopka and Skoultchi, 2003). Consistent with these observations, Snf2h but not Snf2l is essential for survival, and Snf2l-knockout mice survive and reproduce normally (Yip et al., 2012). Below, we describe the developmental roles of different ISWI complexes.
Table 2.
Developmental roles of ISWI complexes
A number of studies highlight key roles for ISWI complex components during early mouse embryo development (Fig. 3). SNF2H, as well as the BAF complex component BRG1, colocalizes with and regulates the function of transcription intermediary factor 1α (TIF1α), which mediates the initial wave of transcription in the one-cell zygote before ZGA occurs. Both SNF2H and BRG1 are mislocalized in the absence of TIF1α and inhibition of Snf2h levels results in misregulation of a subset of TIF1α-regulated genes, indicating a crucial role for SNF2H in zygotic transcription regulation (Torres-Padilla and Zernicka-Goetz, 2006). Consistent with this role, SNF2H was shown to be essential for early mouse development; homozygous Snf2h mutants display peri-implantation lethality because of a failure to proliferate both trophectoderm and ICM (Stopka and Skoultchi, 2003). A SNF2L-containing NURF remodeling complex (Barak et al., 2003) is required shortly after embryo implantation (Landry et al., 2008). Mutants for Bptf (bromodomain PHD finger transcription factor), which encodes the largest subunit of NURF, fail to differentiate any mesoderm, endoderm or ectoderm tissue lineages, indicating a crucial role for BPTF in germ layer formation. Furthermore, the inability of Bptf mutants to form distal ventral endoderm and the dependence of SMAD-responsive gene expression on BPTF suggest a crucial role for NURF as a cofactor for SMAD transcription factors (Landry et al., 2008). Thus, both BAF and ISWI complexes have similar roles during development in early embryonic growth and implantation.
Both SNF2H- and SNF2L-containing complexes are also essential during the development of ectoderm-derived lineages (Alvarez-Saavedra et al., 2014; Barak et al., 2003; Koludrovic et al., 2015; Yip et al., 2012). During nervous system development (Fig. 4), SNF2H has a role in the proliferation of neural progenitors that is partially compensated by SNF2L. Conditional Snf2h mutants show impaired proliferation of granule neuron progenitors and Purkinje cells, and increased cell death leading to postnatal neural maturation defects. SNF2H seems to act on the C-terminal tail of histone H2A to block H1 linker histone loading to chromatin and, thus, maintains a permissive environment for progenitor proliferation and neural gene expression during cerebellum development (Alvarez-Saavedra et al., 2014). SNF2L, on the other hand, is involved in repressing the proliferation of neural progenitors to regulate proper brain size. Conditional deletion of Snf2l increases neural progenitor proliferation and self-renewal, and expression of the transcription factor FOXG1. SNF2L directly represses Foxg1 expression by occupying its promoter region, hence maintaining a balance between neural progenitor proliferation and subsequent differentiation during brain development (Yip et al., 2012). Another SNF2L-containing complex comprising SNF2L and CECR2, called the CERF complex, is involved in neurulation. Homozygous gene-trap Cecr2 mutations cause neural tube defects leading to perinatal lethality (Banting et al., 2005). Thus, both ISWI ATPase subunits (i.e. SNF2H and SNF2L) have crucial functions and appear to function antagonistically in regulating transcription during brain development.
ISWI complexes are also involved in the differentiation of mesoderm-derived lineages. The SNF2L-containing NURF complex has essential roles in thymocyte development and erythropoiesis (Landry et al., 2011; Stopka and Skoultchi, 2003). During thymocyte development, the NURF subunit BPTF is essential for the differentiation of CD4 or CD8 single-positive cells to mature T cells and functions by regulating the expression of several thymocyte maturation genes in a TCR-signaling dependent manner. BPTF regulates DNaseI hypersensitivity and interacts with the transcription factor SRF to regulate NURF recruitment at Egr1, a thymocyte maturation-specific gene (Landry et al., 2011). SNF2H, by contrast, is required for the proliferation of early stage hematopoietic progenitor cells during erythropoiesis (Stopka and Skoultchi, 2003). SNF2H- and SNF2L-containing complexes thus have distinct roles during the early and late stages of hematopoiesis, respectively.
SNF2L is also essential for the proliferation and differentiation of granulosa cells during folliculogenesis (Lazzaro et al., 2006; Pépin et al., 2013). Mice mutant for Snf2l respond differently to gonadotropin induction, produce very few eggs and have fewer secondary follicles. It appears that Snf2l regulates the transcription of Fgl2, which encodes a prothrombinase required for normal mouse reproduction, to regulate folliculogenesis.
In summary, ISWI proteins form specific complexes containing either one of the catalytic subunits SNF2H or SNF2L, and they appear to have very specific roles in progenitor proliferation and in cell differentiation and maturation, respectively. SNF2H-containing complexes are mostly involved in early embryonic development and in progenitor cells, whereas SNF2L-containing complexes have roles during differentiation and maturation. It seems that these complexes regulate chromatin structure in distinct ways; while SNF2H helps to maintain an open and permissive chromatin structure, SNF2L complexes appear to promote a closed chromatin structure consistent with differentiation and maturation processes. Compared with BAF complexes, the role of ISWI complexes is limited; however, ISWI complexes seem to non-redundantly supplement the function of BAF complexes in both early mammalian development and organogenesis.
Developmental roles of CHD complexes
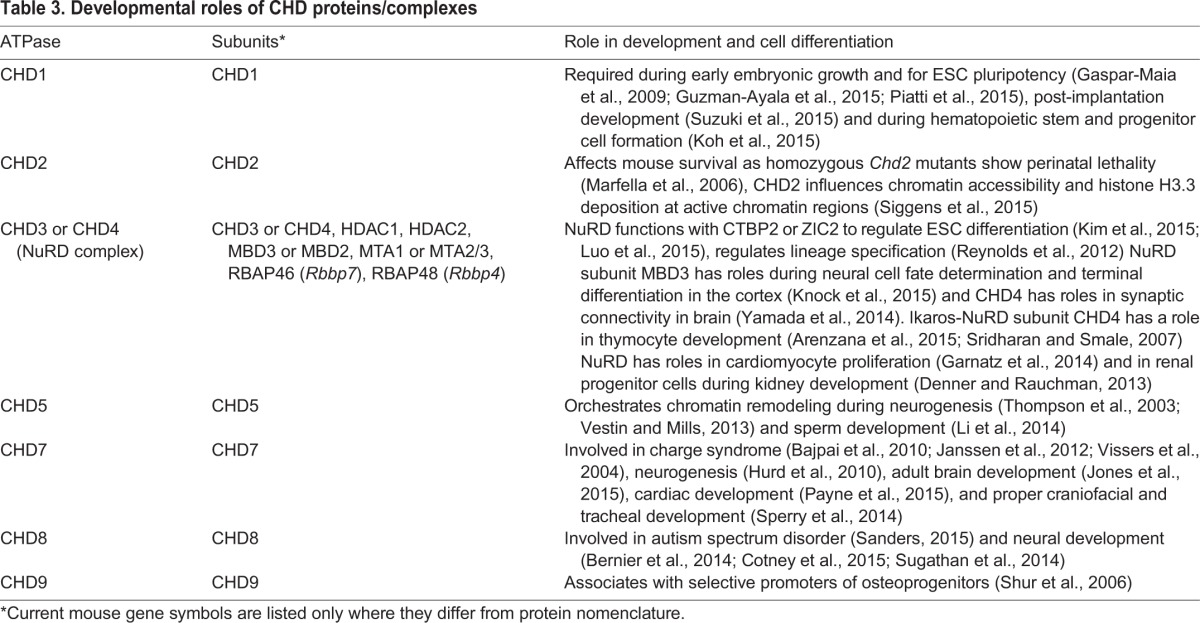
There are nine different chromodomain helicase DNA-binding (CHD) proteins in mammals (CHD1-9) that either act alone or form complexes with other proteins (Fig. 2C). As we discuss below, CHD proteins carry out diverse roles (Table 3) during early embryonic development (Fig. 3) and in terminally differentiated cell lineages.
Table 3.
Developmental roles of CHD proteins/complexes
CHD1 is essential during pre-implantation embryo development. Genetic deletion or RNAi-mediated knockdown of Chd1 at ZGA results in post-implantation lethality. CHD1 controls the differentiation of ICM and TE by regulating the expression of transcription factor genes such as Oct4, Nanog and Cdx2 during ZGA. Regulation of ICM and TE gene expression by CHD1 is thought to be mediated by its activation of high-mobility group protein transcription factor HMGPI (Suzuki et al., 2015). CHD4 also has a redundant role during the differentiation of ICM and TE as it facilitates the expression of appropriate genes and restricts expression of inappropriate genes in the ICM and TE (O'Shaughnessy-Kirwan et al., 2015). In ESCs and epiblast cells, CHD1 maintains optimal transcriptional output by engaging with RNA polymerases I and II to regulate both mRNA and rRNA transcription (Guzman-Ayala et al., 2015). CHD1 was also found to be associated with euchromatin in ESCs and is essential for maintaining their self-renewal and pluripotency (Gaspar-Maia et al., 2009). In line with this, the RNAi-mediated knockdown of Chd1 in ESCs results in self-renewal and pluripotency defects, the loss of differentiation potential to primitive endoderm and an increase in heterochromatic foci. A more recent study suggests that a serine-rich region in the N-terminus region of CHD1 is important for its function in maintaining pluripotency (Piatti et al., 2015).
CHD-containing NuRD (nucleosome remodeling deacetylase) complexes have also been identified in ESCs and are involved in regulating proper ESC differentiation (Kim et al., 2015). In ESCs, C-terminal binding protein 2 (CTBP2) colocalizes with pluripotency factors and H3K27ac marks on active genes. However, upon differentiation, CTBP2 associates with CHD4-containing NuRD complexes to deacetylate H3K27 and recruit PRC2 to facilitate H3K27me3-mediated repression. A NuRD complex containing CHD4 and MBD3 also regulates the differentiation of ESCs, interacting with zinc finger protein of the cerebellum 2 (Zic2) at ESC enhancers (Luo et al., 2015). In agreement with the above findings, CHD proteins are crucial for reprogramming somatic cells to a pluripotent state, and both the NuRD complex and CHD1 improve reprogramming efficiency (Gaspar-Maia et al., 2009; Ozmadenci et al., 2015). Thus, CHD1 and CHD-containing NuRD complexes appear to have roles similar to those of BAF complexes during pre-implantation development and ES cell pluripotency.
CHD proteins are also associated with lineage commitment decisions and terminal differentiation processes. In the developing brain, for example, CHD family members (CHD4-8) transcriptionally regulate various stages of nervous system development (Fig. 4). A CHD4-containing NuRD complex is involved in synapse formation (Yamada et al., 2014), where it represses a number of developmentally downregulated genes in presynaptic granule neurons to drive synaptogenesis. This NuRD-mediated repression involves deacetylation of histone 3 at K9/14/27 and subsequent demethylation of H3K4me3 at the promoters of these genes. In contrast, CHD5, which is highly expressed in both fetal and adult brain, is required during neuronal differentiation to repress genes of non-neuronal lineages (Egan et al., 2013; Thompson et al., 2003; Vestin and Mills, 2013). During adult neurogenesis, CHD7 is required for keeping neural stem cells in a quiescent state. It represses several cell cycle activators and induces Notch signaling to maintain quiescence (Jones et al., 2015). Accordingly, the conditional loss of Chd7 in mice results in loss of the neural stem cell pool, premature neural progenitor formation and impaired neuronal maturation. In humans, CHD7 is mutated in patients with CHARGE syndrome, which is characterized by neural, craniofacial, ear, eye and heart defects (Janssen et al., 2012; Vissers et al., 2004). The morpholino-mediated downregulation of Chd7 in Xenopus embryos recapitulates most CHARGE syndrome symptoms (Bajpai et al., 2010) and the conditional deletion of Chd7 in mice also recapitulates these symptoms (Sperry et al., 2014). CHD7 is also required for the formation and migration of neural crest cells (Bajpai et al., 2010), which contribute to the development of craniofacial, peripheral nervous system and heart (Fig. 5). Specifically, CHD7 interacts with PBAF complex subunits and regulates genes involved in neural crest specific transcription, such as Sox9, Twist and Slug (Bajpai et al., 2010). Furthermore, CHD7 is required for neurogenesis during inner ear morphogenesis (Hurd et al., 2010). CHD8, in contrast, is linked to autism spectrum disorder (ASD); reducing the Chd8 dose by half in neural progenitors downregulates genes involved in neural development, including those associated with ASD (Sugathan et al., 2014).
CHD proteins are also involved during heart development. A CHD3/4-containing NuRD complex supports cardiomyocyte proliferation through its interaction with the transcription cofactor FOG2 (Garnatz et al., 2014). FOG2 interacts with GATA4 and components of the NuRD complex to repress GATA4-mediated transcription. Disruption of the FOG2-NuRD interaction results in perinatal lethality due to atrial and ventricular septal defects and a thin ventricular myocardium. At the molecular level, the FOG2-NuRD interaction inhibits the cell cycle inhibitor gene Cdkn1a to maintain cardiomyocyte proliferation in the myocardium; the FOG2-NuRD disruption phenotype can, thus, be rescued via ablation of Cdkn1a. CHD7, by contrast, transcriptionally regulates multiple processes during heart development and Chd7 mutants, as mentioned above, recapitulate the cardiac aspects of CHARGE syndrome (Liu et al., 2014; Payne et al., 2015) and mutations in CHD7 are found in sporadic cases of human congenital heart defects (Zaidi et al., 2013).
CHD proteins have also been implicated in several other cell differentiation events during development. For example, CHD1 is specifically required for endothelial to hematopoietic transition (EHT), the process by which definitive hematopoietic stem and progenitor cells arise from endothelial cells of various organs. In this context, CHD1 supports the high transcriptional output of hematopoietic progenitors in a very specific time window and is dispensable before and after formation of hematopoietic stem and progenitor cells (Koh et al., 2015). CHD5 has crucial roles during spermatogenesis, in particular during the process of spermiogenesis, in which haploid round spermatids mature into elongated spermatozoa. This event involves extensive sperm chromatin compaction whereby histone proteins are first replaced by transition proteins and then protamines (Govin et al., 2004). Homozygous Chd5-null mutants exhibit fertility defects owing to abnormal histone H4 acetylation, altered expression of variant histones, delayed nucleosome removal, impaired double-strand break repair, and increased transition protein and protamine expression in both round and differentiated spermatids (Li et al., 2014).
In summary, CHD proteins act to regulate the transcription of specific sets of genes and restrict inappropriate expression of a different set of genes, with the common goal of promoting context-dependent cell proliferation, pluripotency and differentiation. The mechanism of transcriptional regulation is mediated by the interaction of CHD proteins with different lineage-specific transcription factors and histone modifiers, and by their own ability to recognize specific histone marks. Cooperation between CHD proteins and other chromatin remodelers such as members of the PBAF complex has important consequences in neural crest cell proliferation and migration. While basic chromatin recognition and remodeling function is provided by the CHD proteins, recruitment of these complexes to appropriate genomic loci may involve other subunits of the complex they reside in or collaboration with a cell type-specific transcription factor.
Developmental roles of INO80/SWR complexes
The fourth major subfamily of ATP-dependent chromatin remodelers comprises the INO80 and SWR complexes. The ATPase subunits of both of these complexes are characterized by the presence of a conserved insertion in their ATPase/helicase domain (Fig. 1) that is responsible for association of the RVB1/RVB2 helicase with these complexes (Mizuguchi et al., 2004). In mammals, the INO80 subfamily consists of the INO80 complex (Chen et al., 2013; Jin et al., 2005), whereas the SWR subfamily consists of the SRCAP (Ruhl et al., 2006) and P400/TIP60 (Cai et al., 2005) complexes, which share subunits both with each other and with the INO80 complex (Table 4, Fig. 2D). Chromatin remodeling by the INO80 complex involves ATP-dependent nucleosome mobilization and exchange of histone variant H2A.Z (Papamichos-Chronakis et al., 2011), whereas SWR complexes are primarily involved in the deposition of H2A.Z into H2A-containing nucleosomes (Krogan et al., 2003; Mizuguchi et al., 2004). As is the case for other chromatin remodelers, these complexes play varied roles during mammalian development.
Table 4.
Developmental roles of INO80/SWR complexes

The INO80 complex is required for ESC self-renewal and pluripotency. The INO80 subunit of the complex binds to the promoter region of ESC master transcription factor genes and facilitates recruitment of the mediator complex and RNA polymerase II (Wang et al., 2014). It maintains an open chromatin region at these sites to facilitate ESC-specific gene transcription. Consistent with its role in regulating pluripotency, INO80 facilitates the reprogramming of somatic cells to induced pluripotent stem cells (Wang et al., 2014). The SWR-related P400/TIP60 complex is also a critical regulator of ESC self-renewal and pluripotency (Fazzio et al., 2008). The crucial role of INO80 complexes (both INO80 and SWR) that manage the exchange and deposition of histone variant H2A.Z to canonical H2A-containing nucleosomes is emphasized by the finding that H2A.Z variants occupy both active and poised promoters in ESCs and are required for proper ESC differentiation (Creyghton et al., 2008; Subramanian et al., 2013).
SRCAP and INO80 complexes also play roles later in development. The SRCAP subunit P18Hamlet (ZNHIT1), for instance, is involved in muscle differentiation (Cuadrado et al., 2010). At the promoter region of the muscle-specific transcription factor gene Myog, P18Hamlet is phosphorylated by P38 MAP kinase (MAPK14); phosphorylated P18Hamlet/SRCAP then recruits H2A.Z (H2AFZ), leading to a chromatin structure that is essential for MYOD1 binding and the transcription of Myog. The SWR complex subunit P400 (EP400) has crucial functions during embryonic hematopoiesis (Fujii et al., 2010; Ueda et al., 2007). It regulates the expression of several embryonic globin genes and deregulates HOX gene expression. In addition, the conditional deletion of P400 in bone marrow cells results in loss of the hematopoietic stem and progenitor cell pool due to defects in cell cycle progression (Fujii et al., 2010). Finally, INO80 is required for meiotic recombination during spermatogenesis (Serber et al., 2015). Conditional spermatogonial mutation of Ino80 before meiosis results in impaired synapse formation and double-strand break defects, consistent with its role in DNA replication and DNA damage repair processes (Kato et al., 2012; Vassileva et al., 2014).
In summary, INO80 complexes are involved in many developmental processes, exerting roles in the regulation of transcription. They share similarities with BAF complexes with regards to maintaining ESC pluripotency, by regulating the expression of master ESC regulators; however, the mechanisms by which they act could be different and may involve their ability to alter chromatin structure by histone variant exchange.
Conclusions and perspectives
As we have highlighted above, ATP-dependent chromatin-remodeling complexes catalyze crucial functions during the development of diverse tissue types in mammals. A close inspection of these various activities suggests that a combination of factors contributes to the specificity of chromatin-remodeler function. First, it is clear that chromatin remodelers condition chromatin in an ATP-dependent manner to facilitate or impede the binding of transcriptional regulators. However, in a non-ATP-dependent manner, they also interact with different transcription factors to regulate transcription. Importantly, chromatin remodelers bring specificity to transcription by incorporating tissue-specific subunits into the complex. Often these subunits are permissive to signaling cues and recruit transcription factors to pre-coat chromatin before assembling into a specialized remodeling enzyme with specific chromatin-remodeling activities on the target genes. Second, the combinatorial assembly of multiple subunits creates chromatin remodelers with very specific functions. Often a particular combination of subunits has a function in a progenitor cell, whereas another combination is required for chromatin remodeling in differentiated cells. How such subunit switches occur in response to differentiation signals is incompletely understood at present, although miRNA-based post-transcriptional regulation appears to be important. Third, it is becoming evident that chromatin remodelers collaborate with histone-modifying complexes to install specific histone marks at regulatory sites, which in turn regulate transcription. Depending upon the collaboration (e.g. activating or repressing histone modifications), chromatin remodelers can exert their specificity in different cell types or on the same cell type but on different transcriptional programs. Finally, a number of studies have revealed that chromatin-remodeler specificity is determined by several other mechanisms, including regulation by non-coding RNAs, chromatin architecture, transcriptional memory and the covalent modification status of histone and DNA.
Our understanding of the roles of ATP-dependent chromatin-remodeling complexes is slowly increasing but could be improved, for example, by employing new techniques and approaches in analyzing their biochemical, cellular and developmental functions during mammalian development. The isolation of chromatin-remodeling complexes from different tissue types together with a comparison of their remodeling activities in vitro will shed light on the types of reaction they each catalyze. Knowing whether a tissue-specific remodeler assembles, dissembles or slides nucleosomes, or whether it exchanges one histone for a variant, might help in understanding its tissue-specific function. Understanding the relative abundance of different remodeler subunits and their alterations during differentiation will also be important for understanding the biochemical nature of tissue-specific complexes. This is more achievable now with the advent of sensitive mass spectrometers and peptide quantification methods. In addition, determining how chromatin remodelers collaborate with transcription factors and other chromatin regulators in specific tissues will be important and tissue-specific chromatin or DNA capture combined with sensitive mass spectrometry would provide such information. Finally, cell type-specific, whole-genome RNA analyses, and the examination of patterns of protein binding and histone and DNA modification will be essential to understand how chromatin remodelers function in diverse cell types. Such an improved understanding of chromatin remodelers, their activities and their roles during development will provide opportunities for designing therapeutic strategies for regeneration and for the treatment of developmental disorders.
Acknowledgements
We thank Gary Howard for editorial assistance and Giovanni Maki for graphics. Deposited in PMC for release after 12 months.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
B.G.B. was funded by the National Institutes of Health National Heart, Lung, and Blood Institute (NHLBI) [Bench to Bassinet Program UM1 HL098179; P01 HL089707] and by William H. Younger, Jr. S.K.H. was supported by the Tobacco-Related Disease Research Program [22FT-0079]. Deposited in PMC for release after 12 months.
References
- Alajem A., Biran A., Harikumar A., Sailaja B. S., Aaronson Y., Livyatan I., Nissim-Rafinia M., Sommer A. G., Mostoslavsky G., Gerbasi V. R. et al. (2015). Differential association of chromatin proteins identifies BAF60a/SMARCD1 as a regulator of embryonic stem cell differentiation. Cell Rep. 10, 2019-2031. 10.1016/j.celrep.2015.02.064 [DOI] [PubMed] [Google Scholar]
- Albini S., Coutinho P., Malecova B., Giordani L., Savchenko A., Forcales S.-V. and Puri P. L. (2013). Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Rep. 3, 661-670. 10.1016/j.celrep.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini S., Coutinho Toto P., Dall'Agnese A., Malecova B., Cenciarelli C., Felsani A., Caruso M., Bultman S. J. and Puri P. L. (2015). Brahma is required for cell cycle arrest and late muscle gene expression during skeletal myogenesis. EMBO Rep. 16, 1037-1050. 10.15252/embr.201540159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J. M., Hota S. K., He D., Thomas S., Ho L., Pennacchio L. A. and Bruneau B. G. (2015). Brg1 modulates enhancer activation in mesoderm lineage commitment. Development 142, 1418-1430. 10.1242/dev.109496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra M., De Repentigny Y., Lagali P. S., Raghu Ram E. V. S., Yan K., Hashem E., Ivanochko D., Huh M. S., Yang D., Mears A. J. et al. (2014). Snf2h-mediated chromatin organization and histone H1 dynamics govern cerebellar morphogenesis and neural maturation. Nat. Commun. 5, 4181 10.1038/ncomms5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenzana T. L., Schjerven H. and Smale S. T. (2015). Regulation of gene expression dynamics during developmental transitions by the Ikaros transcription factor. Genes Dev. 29, 1801-1816. 10.1101/gad.266999.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A., Morgan D. K., Whitelaw N. C., Bruxner T. J., Vickaryous N. K., Cox L. L., Butterfield N. C., Wicking C., Blewitt M. E., Wilkins S. J. et al. (2008). A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development. Genome Biol. 9, R182 10.1186/gb-2008-9-12-r182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio C., Nord A. S., Zhu Y., Blow M. J., Biddie S. C., Mendenhall E. M., Dixon J., Wright C., Hosseini R., Akiyama J. A. et al. (2014). Tissue-specific SMARCA4 binding at active and repressed regulatory elements during embryogenesis. Genome Res. 24, 920-929. 10.1101/gr.168930.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai R., Chen D. A., Rada-Iglesias A., Zhang J., Xiong Y., Helms J., Chang C.-P., Zhao Y., Swigut T. and Wysocka J. (2010). CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463, 958-962. 10.1038/nature08733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R., Bultman S. J., Holley D., Hillhouse C., Bain J. R., Newgard C. B., Muehlbauer M. J. and Willis M. S. (2015). Non-targeted metabolomics of Brg1/Brm double-mutant cardiomyocytes reveals a novel role for SWI/SNF complexes in metabolic homeostasis. Metabolomics 11, 1287-1301. 10.1007/s11306-015-0786-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banting G. S., Barak O., Ames T. M., Burnham A. C., Kardel M. D., Cooch N. S., Davidson C. E., Godbout R., McDermid H. E. and Shiekhattar R. (2005). CECR2, a protein involved in neurulation, forms a novel chromatin remodeling complex with SNF2L. Hum. Mol. Genet. 14, 513-524. 10.1093/hmg/ddi048 [DOI] [PubMed] [Google Scholar]
- Bao X., Tang J., Lopez-Pajares V., Tao S., Qu K., Crabtree G. R. and Khavari P. A. (2013). ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell 12, 193-203. 10.1016/j.stem.2012.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Rubin A. J., Qu K., Zhang J., Giresi P. G., Chang H. Y. and Khavari P. A. (2015). A novel ATAC-seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol. 16, 284 10.1186/s13059-015-0840-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak O., Lazzaro M. A., Lane W. S., Speicher D. W., Picketts D. J. and Shiekhattar R. (2003). Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J. 22, 6089-6100. 10.1093/emboj/cdg582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett C., Yazgan O., Kuo H.-C., Malakar S., Thomas T., Fitzgerald A., Harbour W., Henry J. J. and Krebs J. E. (2012). Williams Syndrome Transcription Factor is critical for neural crest cell function in Xenopus laevis. Mech. Dev. 129, 324-338. 10.1016/j.mod.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier R., Golzio C., Xiong B., Stessman H. A., Coe B. P., Penn O., Witherspoon K., Gerdts J., Baker C., Vulto-van Silfhout A. T. et al. (2014). Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263-276. 10.1016/j.cell.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof M., Weider M., Küspert M., Nave K.-A. and Wegner M. (2015). Brg1-dependent chromatin remodelling is not essentially required during oligodendroglial differentiation. J. Neurosci. 35, 21-35. 10.1523/JNEUROSCI.1468-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S., Gebuhr T., Yee D., La Mantia C., Nicholson J., Gilliam A., Randazzo F., Metzger D., Chambon P., Crabtree G. et al. (2000). A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6, 1287-1295. 10.1016/S1097-2765(00)00127-1 [DOI] [PubMed] [Google Scholar]
- Bultman S. J., Gebuhr T. C. and Magnuson T. (2005). A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes Dev. 19, 2849-2861. 10.1101/gad.1364105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S. J., Gebuhr T. C., Pan H., Svoboda P., Schultz R. M. and Magnuson T. (2006). Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 20, 1744-1754. 10.1101/gad.1435106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Jin J., Florens L., Swanson S. K., Kusch T., Li B., Workman J. L., Washburn M. P., Conaway R. C. and Conaway J. W. (2005). The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 280, 13665-13670. 10.1074/jbc.M500001200 [DOI] [PubMed] [Google Scholar]
- Cai W., Albini S., Wei K., Willems E., Guzzo R. M., Tsuda M., Giordani L., Spiering S., Kurian L., Yeo G. W. et al. (2013). Coordinate Nodal and BMP inhibition directs Baf60c-dependent cardiomyocyte commitment. Genes Dev. 27, 2332-2344. 10.1101/gad.225144.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B. R., Levinson R. S., Yamamoto K. R. and Kornberg R. D. (1996). Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 10, 2131-2144. 10.1101/gad.10.17.2131 [DOI] [PubMed] [Google Scholar]
- Carey T. S., Cao Z., Choi I., Ganguly A., Wilson C. A., Paul S. and Knott J. G. (2015). BRG1 Governs Nanog Transcription in Early Mouse Embryos and Embryonic Stem Cells via Antagonism of Histone H3 Lysine 9/14 Acetylation. Mol. Cell. Biol. 35, 4158-4169. 10.1128/MCB.00546-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiyachati B. H., Jani A., Wan Y., Huang H., Flavell R. and Chi T. (2013). BRG1-mediated immune tolerance: facilitation of Treg activation and partial independence of chromatin remodelling. EMBO J. 32, 395-408. 10.1038/emboj.2012.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Conaway R. C. and Conaway J. W. (2013). Multiple modes of regulation of the human Ino80 SNF2 ATPase by subunits of the INO80 chromatin-remodeling complex. Proc. Natl. Acad. Sci. USA 110, 20497-20502. 10.1073/pnas.1317092110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi T. H., Wan M., Zhao K., Taniuchi I., Chen L., Littman D. R. and Crabtree G. R. (2002). Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418, 195-199. 10.1038/nature00876 [DOI] [PubMed] [Google Scholar]
- Chi T. H., Wan M., Lee P. P., Akashi K., Metzger D., Chambon P., Wilson C. B. and Crabtree G. R. (2003). Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity 19, 169-182. 10.1016/S1074-7613(03)00199-7 [DOI] [PubMed] [Google Scholar]
- Choi J., Ko M., Jeon S., Jeon Y., Park K., Lee C., Lee H. and Seong R. H. (2012). The SWI/SNF-like BAF complex is essential for early B cell development. J. Immunol. 188, 3791-3803. 10.4049/jimmunol.1103390 [DOI] [PubMed] [Google Scholar]
- Clapier C. R. and Cairns B. R. (2012). Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature 492, 280-284. 10.1038/nature11625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J., Muhle R. A., Sanders S. J., Liu L., Willsey A. J., Niu W., Liu W., Klei L., Lei J., Yin J. et al. (2015). The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 6, 6404 10.1038/ncomms7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M. P., Markoulaki S., Levine S. S., Hanna J., Lodato M. A., Sha K., Young R. A., Jaenisch R. and Boyer L. A. (2008). H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135, 649-661. 10.1016/j.cell.2008.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A., Corrado N., Perdiguero E., Lafarga V., Muñoz-Canoves P. and Nebreda A. R. (2010). Essential role of p18Hamlet/SRCAP-mediated histone H2A.Z chromatin incorporation in muscle differentiation. EMBO J. 29, 2014-2025. 10.1038/emboj.2010.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna I. L., Ohkawa Y., Berkes C. A., Bergstrom D. A., Dacwag C. S., Tapscott S. J. and Imbalzano A. N. (2005). MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25, 3997-4009. 10.1128/MCB.25.10.3997-4009.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denner D. R. and Rauchman M. (2013). Mi-2/NuRD is required in renal progenitor cells during embryonic kidney development. Dev. Biol. 375, 105-116. 10.1016/j.ydbio.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine W. P., Wythe J. D., George M., Koshiba-Takeuchi K. and Bruneau B. G. (2014). Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife 3, 508 10.7554/eLife.03848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle J. A., Mehta M., Kass E. M., Vuong B. Q., Inagaki A., Egli D., Jasin M. and Keeney S. (2013). Mouse BAZ1A (ACF1) is dispensable for double-strand break repair but is essential for averting improper gene expression during spermatogenesis. PLoS Genet. 9, e1003945 10.1371/journal.pgen.1003945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan C. M., Nyman U., Skotte J., Streubel G., Turner S., O'Connell D. J., Rraklli V., Dolan M. J., Chadderton N., Hansen K. et al. (2013). CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev. Cell 26, 223-236. 10.1016/j.devcel.2013.07.008 [DOI] [PubMed] [Google Scholar]
- Fazzio T. G., Huff J. T. and Panning B. (2008). An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134, 162-174. 10.1016/j.cell.2008.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn R. A., Do B. T., Rubin A. J., Calo E., Lee B., Kuchelmeister H., Rale M., Chu C., Kool E. T., Wysocka J. et al. (2016). 7SK-BAF axis controls pervasive transcription at enhancers. Nat. Struct. Mol. Biol. 23, 231-238. 10.1038/nsmb.3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcales S.-V. (2014). The BAF60c-MyoD complex poises chromatin for rapid transcription. BioArchitecture 2, 104-109. 10.4161/bioa.20970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcales S. V., Albini S., Giordani L., Malecova B., Cignolo L., Chernov A., Coutinho P., Saccone V., Consalvi S., Williams R. et al. (2012). Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J. 31, 301-316. 10.1038/emboj.2011.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T., Ueda T., Nagata S. and Fukunaga R. (2010). Essential role of p400/mDomino chromatin-remodeling ATPase in bone marrow hematopoiesis and cell-cycle progression. J. Biol. Chem. 285, 30214-30223. 10.1074/jbc.M110.104513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Tate P., Hu P., Tjian R., Skarnes W. C. and Wang Z. (2008). ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl. Acad. Sci. USA 105, 6656-6661. 10.1073/pnas.0801802105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnatz A. S., Gao Z., Broman M., Martens S., Earley J. U. and Svensson E. C. (2014). FOG-2 mediated recruitment of the NuRD complex regulates cardiomyocyte proliferation during heart development. Dev. Biol. 395, 50-61. 10.1016/j.ydbio.2014.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M. J., Heidersbach A., Ramalho-Santos J., McManus M. T., Plath K., Meshorer E. et al. (2009). Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460, 863-868. 10.1038/nature08212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebuhr T. C., Kovalev G. I., Bultman S., Godfrey V., Su L. and Magnuson T. (2003). The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. J. Exp. Med. 198, 1937-1949. 10.1084/jem.20030714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goljanek-Whysall K., Mok G. F., Fahad Alrefaei A., Kennerley N., Wheeler G. N. and Münsterberg A. (2014). myomiR-dependent switching of BAF60 variant incorporation into Brg1 chromatin remodeling complexes during embryo myogenesis. Development 141, 3378-3387. 10.1242/dev.108787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J., Caron C., Lestrat C., Rousseaux S. and Khochbin S. (2004). The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur. J. Biochem. 271, 3459-3469. 10.1111/j.1432-1033.2004.04266.x [DOI] [PubMed] [Google Scholar]
- Gregg R. G., Willer G. B., Fadool J. M., Dowling J. E. and Link B. A. (2003). Positional cloning of the young mutation identifies an essential role for the Brahma chromatin remodeling complex in mediating retinal cell differentiation. Proc. Natl. Acad. Sci. USA 100, 6535-6540. 10.1073/pnas.0631813100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin C. T., Brennan J. and Magnuson T. (2008). The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development 135, 493-500. 10.1242/dev.010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi C. J., Sands A. T., Zambrowicz B. P., Turner T. K., Demers D. A., Webster W., Smith T. W., Imbalzano A. N. and Jones S. N. (2001). Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 21, 3598-3603. 10.1128/MCB.21.10.3598-3603.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Ayala M., Sachs M., Koh F. M., Onodera C., Bulut-Karslioglu A., Lin C.-J., Wong P., Nitta R., Song J. S. and Ramalho-Santos M. (2015). Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development 142, 118-127. 10.1242/dev.114843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Jeon S., Sohn D. H., Lee C., Ahn S., Kim W. K., Chung H. and Seong R. H. (2008). SRG3, a core component of mouse SWI/SNF complex, is essential for extra-embryonic vascular development. Dev. Biol. 315, 136-146. 10.1016/j.ydbio.2007.12.024 [DOI] [PubMed] [Google Scholar]
- Han P., Li W., Lin C.-H., Yang J., Shang C., Nurnberg S. T., Jin K. K., Xu W., Lin C.-Y., Lin C.-J. et al. (2014). A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102-106. 10.1038/nature13596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang C. T., Yang J., Han P., Cheng H.-L., Shang C., Ashley E., Zhou B. and Chang C.-P. (2010). Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 466, 62-67. 10.1038/nature09130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Tian X., Zhang H., Hu T., Huang X., Zhang L., Wang Z. and Zhou B. (2014). BAF200 is required for heart morphogenesis and coronary artery development. PLoS ONE 9, e109493 10.1371/journal.pone.0109493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L. and Crabtree G. R. (2010). Chromatin remodelling during development. Nature 463, 474-484. 10.1038/nature08911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Jothi R., Ronan J. L., Cui K., Zhao K. and Crabtree G. R. (2009a). An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. USA 106, 5187-5191. 10.1073/pnas.0812888106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Ronan J. L., Wu J., Staahl B. T., Chen L., Kuo A., Lessard J., Nesvizhskii A. I., Ranish J. and Crabtree G. R. (2009b). An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA 106, 5181-5186. 10.1073/pnas.0812889106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Miller E. L., Ronan J. L., Ho W. Q., Jothi R. and Crabtree G. R. (2011). esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 13, 903-913. 10.1038/ncb2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley D. W., Groh B. S., Wozniak G., Donohoe D. R., Sun W., Godfrey V. and Bultman S. J. (2014). The BRG1 chromatin remodeler regulates widespread changes in gene expression and cell proliferation during B cell activation. J. Cell. Physiol. 229, 44-52. 10.1002/jcp.24414 [DOI] [PubMed] [Google Scholar]
- Hota S. K., Bhardwaj S. K., Deindl S., Lin Y.-C., Zhuang X. and Bartholomew B. (2013). Nucleosome mobilization by ISW2 requires the concerted action of the ATPase and SLIDE domains. Nat. Struct. Mol. Biol. 20, 222-229. 10.1038/nsmb.2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Gao X., Diaz-Trelles R., Ruiz-Lozano P. and Wang Z. (2008). Coronary development is regulated by ATP-dependent SWI/SNF chromatin remodeling component BAF180. Dev. Biol. 319, 258-266. 10.1016/j.ydbio.2008.04.020 [DOI] [PubMed] [Google Scholar]
- Hurd E. A., Poucher H. K., Cheng K., Raphael Y. and Martin D. M. (2010). The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development 137, 3139-3150. 10.1242/dev.047894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra A. K., Dupé V., Bornert J.-M., Messaddeq N., Yaniv M., Mark M., Chambon P. and Metzger D. (2005). Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development 132, 4533-4544. 10.1242/dev.02019 [DOI] [PubMed] [Google Scholar]
- Jaenisch R. and Young R. (2008). Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132, 567-582. 10.1016/j.cell.2008.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani A., Wan M., Zhang J., Cui K., Wu J., Preston-Hurlburt P., Khatri R., Zhao K. and Chi T. (2008). A novel genetic strategy reveals unexpected roles of the Swi-Snf-like chromatin-remodeling BAF complex in thymocyte development. J. Exp. Med. 205, 2813-2825. 10.1084/jem.20080938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen N., Bergman J. E. H., Swertz M. A., Tranebjaerg L., Lodahl M., Schoots J., Hofstra R. M. W., van Ravenswaaij-Arts C. M. A. and Hoefsloot L. H. (2012). Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum. Mutat. 33, 1149-1160. 10.1002/humu.22086 [DOI] [PubMed] [Google Scholar]
- Jiang Z., Tang Y., Zhao X., Zhang M., Donovan D. M. and Tian X. C. (2015). Knockdown of Brm and Baf170, components of chromatin remodeling complex, facilitates reprogramming of somatic cells. Stem Cells Dev. 24, 2328-2336. 10.1089/scd.2015.0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Cai Y., Yao T., Gottschalk A. J., Florens L., Swanson S. K., Gutiérrez J. L., Coleman M. K., Workman J. L., Mushegian A. et al. (2005). A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 280, 41207-41212. 10.1074/jbc.M509128200 [DOI] [PubMed] [Google Scholar]
- Jones K. M., Sarić N., Russell J. P., Andoniadou C. L., Scambler P. J. and Basson M. A. (2015). CHD7 maintains neural stem cell quiescence and prevents premature stem cell depletion in the adult hippocampus. Stem Cells 33, 196-210. 10.1002/stem.1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C. and Crabtree G. R. (2013). Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 153, 71-85. 10.1016/j.cell.2013.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C., Hargreaves D. C., Hodges C., Elias L., Ho L., Ranish J. and Crabtree G. R. (2013). Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592-601. 10.1038/ng.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser M. D., Aslanian A., Dong M.-Q., Yates J. R. and Emerson B. M. (2008). BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 283, 32254-32263. 10.1074/jbc.M806061200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato D., Waki M., Umezawa M., Aoki Y., Utsugi T., Ohtsu M. and Murakami Y. (2012). Phosphorylation of human INO80 is involved in DNA damage tolerance. Biochem. Biophys. Res. Commun. 417, 433-438. 10.1016/j.bbrc.2011.11.134 [DOI] [PubMed] [Google Scholar]
- Kidder B. L., Palmer S. and Knott J. G. (2009). SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 27, 317-328. 10.1634/stemcells.2008-0710 [DOI] [PubMed] [Google Scholar]
- Kim J. K., Huh S.-O., Choi H., Lee K.-S., Shin D., Lee C., Nam J.-S., Kim H., Chung H., Lee H. W. et al. (2001). Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell. Biol. 21, 7787-7795. 10.1128/MCB.21.22.7787-7795.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-I., Bresnick E. H. and Bultman S. J. (2009a). BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription. Nucleic Acids Res. 37, 6019-6027. 10.1093/nar/gkp677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-I., Bultman S. J., Kiefer C. M., Dean A. and Bresnick E. H. (2009b). BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc. Natl. Acad. Sci. USA 106, 2259-2264. 10.1073/pnas.0806420106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. W., Kang B.-H., Jang H., Kwak S., Shin J., Kim H., Lee S.-E., Lee S.-M., Lee J.-H., Kim J.-H. et al. (2015). Ctbp2 Modulates NuRD-Mediated Deacetylation of H3K27 and Facilitates PRC2-Mediated H3K27me3 in Active Embryonic Stem Cell Genes During Exit from Pluripotency. Stem Cells 33, 2442-2455. 10.1002/stem.2046 [DOI] [PubMed] [Google Scholar]
- Klochendler-Yeivin A., Fiette L., Barra J., Muchardt C., Babinet C. and Yaniv M. (2000). The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1, 500-506. 10.1093/embo-reports/kvd129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knock E., Pereira J., Lombard P. D., Dimond A., Leaford D., Livesey F. J. and Hendrich B. (2015). The methyl binding domain 3/nucleosome remodelling and deacetylase complex regulates neural cell fate determination and terminal differentiation in the cerebral cortex. Neural Dev. 10, 13 10.1186/s13064-015-0040-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh F. M., Lizama C. O., Wong P., Hawkins J. S., Zovein A. C. and Ramalho-Santos M. (2015). Emergence of hematopoietic stem and progenitor cells involves a Chd1-dependent increase in total nascent transcription. Proc. Natl. Acad. Sci. USA 112, E1734-E1743. 10.1073/pnas.1424850112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koludrovic D., Laurette P., Strub T., Keime C., Le Coz M., Coassolo S., Mengus G., Larue L. and Davidson I. (2015). Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells. PLoS Genet. 11, e1005555 10.1371/journal.pgen.1005555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva V., Buscarlet M., Diaz-Tellez A., Bernard M.-A., Crabtree G. R. and Lessard J. A. (2012). The BAF53a subunit of SWI/SNF-like BAF complexes is essential for hemopoietic stem cell function. Blood 120, 4720-4732. 10.1182/blood-2012-04-427047 [DOI] [PubMed] [Google Scholar]
- Krogan N. J., Keogh M.-C., Datta N., Sawa C., Ryan O. W., Ding H., Haw R. A., Pootoolal J., Tong A., Canadien V. et al. (2003). A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12, 1565-1576. 10.1016/S1097-2765(03)00497-0 [DOI] [PubMed] [Google Scholar]
- Krosl J., Mamo A., Chagraoui J., Wilhelm B. T., Girard S., Louis I., Lessard J., Perreault C. and Sauvageau G. (2010). A mutant allele of the Swi/Snf member BAF250a determines the pool size of fetal liver hemopoietic stem cell populations. Blood 116, 1678-1684. 10.1182/blood-2010-03-273862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalit P. A., Salick M. R., Nelson D. O., Squirrell J. M., Shafer C. M., Patel N. G., Saeed I., Schmuck E. G., Markandeya Y. S., Wong R. et al. (2016). Lineage reprogramming of fibroblasts into proliferative induced cardiac progenitor cells by defined factors. Cell Stem Cell 18, 354-367. 10.1016/j.stem.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J., Sharov A. A., Piao Y., Sharova L. V., Xiao H., Southon E., Matta J., Tessarollo L., Zhang Y. E., Ko M. S. H. et al. (2008). Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 4, e1000241 10.1371/journal.pgen.1000241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J. W., Banerjee S., Taylor B., Aplan P. D., Singer A. and Wu C. (2011). Chromatin remodeling complex NURF regulates thymocyte maturation. Genes Dev. 25, 275-286. 10.1101/gad.2007311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurette P., Strub T., Koludrovic D., Keime C., Le Gras S., Seberg H., Van Otterloo E., Imrichova H., Siddaway R., Aerts S. et al. (2015). Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. Elife 4, M111.014233 10.7554/eLife.06857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro M. A. and Picketts D. J. (2001). Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J. Neurochem. 77, 1145-1156. 10.1046/j.1471-4159.2001.00324.x [DOI] [PubMed] [Google Scholar]
- Lazzaro M. A., Pépin D., Pescador N., Murphy B. D., Vanderhyden B. C. and Picketts D. J. (2006). The imitation switch protein SNF2L regulates steroidogenic acute regulatory protein expression during terminal differentiation of ovarian granulosa cells. Mol. Endocrinol. 20, 2406-2417. 10.1210/me.2005-0213 [DOI] [PubMed] [Google Scholar]
- Lee K. Y., Choi Y. I., Kim J., Choi J. W., Sohn D. H., Lee C., Jeon S. H. and Seong R. H. (2007). Down-regulation of the SWI/SNF chromatin remodeling activity by TCR signaling is required for proper thymocyte maturation. J. Immunol. 178, 7088-7096. 10.4049/jimmunol.178.11.7088 [DOI] [PubMed] [Google Scholar]
- Lei I., Gao X., Sham M. H. and Wang Z. (2012). SWI/SNF protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. J. Biol. Chem. 287, 24255-24262. 10.1074/jbc.M112.365080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei I., West J., Yan Z., Gao X., Fang P., Dennis J. H., Gnatovskiy L., Wang W., Kingston R. E. and Wang Z. (2015). BAF250a protein regulates nucleosome occupancy and histone modifications in priming embryonic stem cell differentiation. J. Biol. Chem. 290, 19343-19352. 10.1074/jbc.M115.637389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J., Wu J. I., Ranish J. A., Wan M., Winslow M. M., Staahl B. T., Wu H., Aebersold R., Graef I. A. and Crabtree G. R. (2007). An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55, 201-215. 10.1016/j.neuron.2007.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Liu C., Li N., Hao T., Han T., Hill D. E., Vidal M. and Lin J. D. (2008). Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 8, 105-117. 10.1016/j.cmet.2008.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Xiong Y., Shang C., Twu K. Y., Hang C. T., Yang J., Han P., Lin C.-Y., Lin C.-J., Tsai F.-C. et al. (2013). Brg1 governs distinct pathways to direct multiple aspects of mammalian neural crest cell development. Proc. Natl. Acad. Sci. USA 110, 1738-1743. 10.1073/pnas.1218072110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wu J., Kim S.-Y., Zhao M., Hearn S. A., Zhang M. Q., Meistrich M. L. and Mills A. A. (2014). Chd5 orchestrates chromatin remodelling during sperm development. Nat. Commun. 5, 3812 10.1038/ncomms4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H., Takeuchi J. K., Von Both I., Walls J. R., McAuliffe F., Adamson S. L., Henkelman R. M., Wrana J. L., Rossant J. and Bruneau B. G. (2004). Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432, 107-112. 10.1038/nature03071 [DOI] [PubMed] [Google Scholar]
- Liu Y., Harmelink C., Peng Y., Chen Y., Wang Q. and Jiao K. (2014). CHD7 interacts with BMP R-SMADs to epigenetically regulate cardiogenesis in mice. Hum. Mol. Genet. 23, 2145-2156. 10.1093/hmg/ddt610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Gao X., Lin C., Smith E. R., Marshall S. A., Swanson S. K., Florens L., Washburn M. P. and Shilatifard A. (2015). Zic2 is an enhancer-binding factor required for embryonic stem cell specification. Mol. Cell 57, 685-694. 10.1016/j.molcel.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallappa C., Nasipak B. T., Etheridge L., Androphy E. J., Jones S. N., Sagerström C. G., Ohkawa Y. and Imbalzano A. N. (2010). Myogenic microRNA expression requires ATP-dependent chromatin remodeling enzyme function. Mol. Cell. Biol. 30, 3176-3186. 10.1128/MCB.00214-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe H. G., Mehta G., Zhang X., Datar I., Mehrotra A., Yeung K. C. and de la Serna I. L. (2013). SWI/SNF enzymes promote SOX10- mediated activation of myelin gene expression. PLoS ONE 8, e69037 10.1371/journal.pone.0069037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella C. G. A., Ohkawa Y., Coles A. H., Garlick D. S., Jones S. N. and Imbalzano A. N. (2006). Mutation of the SNF2 family member Chd2 affects mouse development and survival. J. Cell. Physiol. 209, 162-171. 10.1002/jcp.20718 [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Banine F., Struve J., Xing R., Adams C., Liu Y., Metzger D., Chambon P., Rao M. S. and Sherman L. S. (2006). Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev. Biol. 289, 372-383. 10.1016/j.ydbio.2005.10.044 [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W.-H., Sen S. and Wu C. (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343-348. 10.1126/science.1090701 [DOI] [PubMed] [Google Scholar]
- Narayanan R., Pirouz M., Kerimoglu C., Pham L., Wagener R. J., Kiszka K. A., Rosenbusch J., Seong R. H., Kessel M., Fischer A. et al. (2015). Loss of BAF (mSWI/SNF) complexes causes global transcriptional and chromatin state changes in forebrain development. Cell Rep. 13, 1842-1854. 10.1016/j.celrep.2015.10.046 [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy-Kirwan A., Signolet J., Costello I., Gharbi S. and Hendrich B. (2015). Constraint of gene expression by the chromatin remodelling protein CHD4 facilitates lineage specification. Development 142, 2586-2597. 10.1242/dev.125450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozmadenci D., Féraud O., Markossian S., Kress E., Ducarouge B., Gibert B., Ge J., Durand I., Gadot N., Plateroti M. et al. (2015). Netrin-1 regulates somatic cell reprogramming and pluripotency maintenance. Nat. Commun. 6, 7398 10.1038/ncomms8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Watanabe S., Rando O. J. and Peterson C. L. (2011). Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144, 200-213. 10.1016/j.cell.2010.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne S., Burney M. J., McCue K., Popal N., Davidson S. M., Anderson R. H. and Scambler P. J. (2015). A critical role for the chromatin remodeller CHD7 in anterior mesoderm during cardiovascular development. Dev. Biol. 405, 82-95. 10.1016/j.ydbio.2015.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen T. A., Kowenz-Leutz E., Leutz A. and Nerlov C. (2001). Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 15, 3208-3216. 10.1101/gad.209901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pépin D., Paradis F., Perez-Iratxeta C., Picketts D. J. and Vanderhyden B. C. (2013). The imitation switch ATPase Snf2l is required for superovulation and regulates Fgl2 in differentiating mouse granulosa cells. Biol. Reprod. 88, 142-142 10.1095/biolreprod.112.105742 [DOI] [PubMed] [Google Scholar]
- Piatti P., Lim C. Y., Nat R., Villunger A., Geley S., Shue Y. T., Soratroi C., Moser M. and Lusser A. (2015). Embryonic stem cell differentiation requires full length Chd1. Sci. Rep. 5, 8007 10.1038/srep08007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner J. R., Iyer M. K., Sahu A., Asangani I. A., Cao Q., Patel L., Vergara I. A., Davicioni E., Erho N., Ghadessi M. et al. (2013). The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 45, 1392-1398. 10.1038/ng.2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J. C., Barra J., Muchardt C., Camus A., Babinet C. and Yaniv M. (1998). Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J. 17, 6979-6991. 10.1093/emboj/17.23.6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N., Salmon-Divon M., Dvinge H., Hynes-Allen A., Balasooriya G., Leaford D., Behrens A., Bertone P. and Hendrich B. (2012). NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 31, 593-605. 10.1038/emboj.2011.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C. W. M., Galusha S. A., McMenamin M. E., Fletcher C. D. M. and Orkin S. H. (2000). Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. USA 97, 13796-13800. 10.1073/pnas.250492697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl D. D., Jin J., Cai Y., Swanson S., Florens L., Washburn M. P., Conaway R. C., Conaway J. W. and Chrivia J. C. (2006). Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry 45, 5671-5677. 10.1021/bi060043d [DOI] [PubMed] [Google Scholar]
- Saccone V., Consalvi S., Giordani L., Mozzetta C., Barozzi I., Sandoná M., Ryan T., Rojas-Muñoz A., Madaro L., Fasanaro P. et al. (2014). HDAC-regulated myomiRs control BAF60 variant exchange and direct the functional phenotype of fibro-adipogenic progenitors in dystrophic muscles. Genes Dev. 28, 841-857. 10.1101/gad.234468.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S. J. (2015). First glimpses of the neurobiology of autism spectrum disorder. Curr. Opin. Genet. Dev. 33, 80-92. 10.1016/j.gde.2015.10.002 [DOI] [PubMed] [Google Scholar]
- Schaniel C., Ang Y.-S., Ratnakumar K., Cormier C., James T., Bernstein E., Lemischka I. R. and Paddison P. J. (2009). Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells 27, 2979-2991. 10.1002/stem.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen P., Vivas P., Dechassa M. L., Mooney A. M., Poirier M. G. and Bartholomew B. (2013). The SnAC domain of SWI/SNF is a histone anchor required for remodeling. Mol. Cell. Biol. 33, 360-370. 10.1128/MCB.00922-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serber D. W., Runge J. S., Menon D. U. and Magnuson T. (2015). The mouse INO80 chromatin remodeling complex is an essential meiotic factor for spermatogenesis. Biol. Reprod. 10.1095/biolreprod.115.135533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shur I., Socher R. and Benayahu D. (2006). In vivo association of CReMM/CHD9 with promoters in osteogenic cells. J. Cell. Physiol. 207, 374-378. 10.1002/jcp.20586 [DOI] [PubMed] [Google Scholar]
- Siggens L., Cordeddu L., Rönnerblad M., Lennartsson A. and Ekwall K. (2015). Transcription-coupled recruitment of human CHD1 and CHD2 influences chromatin accessibility and histone H3 and H3.3 occupancy at active chromatin regions. Epigenet. Chromatin 8, 4 10.1186/1756-8935-8-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N., Graumann J., Wu G., Araúzo-Bravo M. J., Han D. W., Greber B., Gentile L., Mann M. and Schöler H. R. (2010). Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141, 943-955. 10.1016/j.cell.2010.04.037 [DOI] [PubMed] [Google Scholar]
- Singhal N., Esch D., Stehling M. and Schöler H. R. (2014). BRG1 is required to maintain pluripotency of murine embryonic stem cells. Biores. Open Access 3, 1-8. 10.1089/biores.2013.0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Roe S. L. and Bultman S. J. (2013). Combined gene dosage requirement for SWI/SNF catalytic subunits during early mammalian development. Mamm. Genome 24, 21-29. 10.1007/s00335-012-9433-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry E. D., Hurd E. A., Durham M. A., Reamer E. N., Stein A. B. and Martin D. M. (2014). The chromatin remodeling protein CHD7, mutated in CHARGE syndrome, is necessary for proper craniofacial and tracheal development. Dev. Dyn. 243, 1055-1066. 10.1002/dvdy.24156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R. and Smale S. T. (2007). Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J. Biol. Chem. 282, 30227-30238. 10.1074/jbc.M702541200 [DOI] [PubMed] [Google Scholar]
- Staahl B. T., Tang J., Wu W., Sun A., Gitler A. D., Yoo A. S. and Crabtree G. R. (2013). Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J. Neurosci. 33, 10348-10361. 10.1523/JNEUROSCI.1258-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankunas K., Hang C. T., Tsun Z.-Y., Chen H., Lee N. V., Wu J. I., Shang C., Bayle J. H., Shou W., Iruela-Arispe M. L. et al. (2008). Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev. Cell 14, 298-311. 10.1016/j.devcel.2007.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka T. and Skoultchi A. I. (2003). The ISWI ATPase Snf2h is required for early mouse development. Proc. Natl. Acad. Sci. USA 100, 14097-14102. 10.1073/pnas.2336105100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V., Mazumder A., Surface L. E., Butty V. L., Fields P. A., Alwan A., Torrey L., Thai K. K., Levine S. S., Bathe M. et al. (2013). H2A.Z acidic patch couples chromatin dynamics to regulation of gene expression programs during ESC differentiation. PLoS Genet. 9, e1003725 10.1371/journal.pgen.1003725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugathan A., Biagioli M., Golzio C., Erdin S., Blumenthal I., Manavalan P., Ragavendran A., Brand H., Lucente D., Miles J. et al. (2014). CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl. Acad. Sci. USA 111, E4468-E4477. 10.1073/pnas.1405266111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Nozawa Y., Tsukamoto S., Kaneko T., Manabe I., Imai H. and Minami N. (2015). CHD1 acts via the Hmgpi pathway to regulate mouse early embryogenesis. Development 142, 2375-2384. 10.1242/dev.120493 [DOI] [PubMed] [Google Scholar]
- Takeuchi J. K. and Bruneau B. G. (2009). Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459, 708-711. 10.1038/nature08039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi J. K., Lickert H., Bisgrove B. W., Sun X., Yamamoto M., Chawengsaksophak K., Hamada H., Yost H. J., Rossant J. and Bruneau B. G. (2007). Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry. Proc. Natl. Acad. Sci. USA 104, 846-851. 10.1073/pnas.0608118104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi J. K., Lou X., Alexander J. M., Sugizaki H., Delgado-Olguín P., Holloway A. K., Mori A. D., Wylie J. N., Munson C., Zhu Y. et al. (2011). Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat. Commun. 2, 187-111 10.1038/ncomms1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P. M., Gotoh T., Kok M., White P. S. and Brodeur G. M. (2003). CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene 22, 1002-1011. 10.1038/sj.onc.1206211 [DOI] [PubMed] [Google Scholar]
- Thompson K. W., Marquez S. B., Lu L. and Reisman D. (2015). Induction of functional Brm protein from Brm knockout mice. Oncoscience 2, 349-361. 10.18632/oncoscience.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla M. E. and Zernicka-Goetz M. (2006). Role of TIF1alpha as a modulator of embryonic transcription in the mouse zygote. J. Cell Biol. 174, 329-338. 10.1083/jcb.200603146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuoc T. C., Boretius S., Sansom S. N., Pitulescu M.-E., Frahm J., Livesey F. J. and Stoykova A. (2013). Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev. Cell 25, 256-269. 10.1016/j.devcel.2013.04.005 [DOI] [PubMed] [Google Scholar]
- Ueda T., Watanabe-Fukunaga R., Ogawa H., Fukuyama H., Higashi Y., Nagata S. and Fukunaga R. (2007). Critical role of the p400/mDomino chromatin-remodeling ATPase in embryonic hematopoiesis. Genes Cells 12, 581-592. 10.1111/j.1365-2443.2007.01080.x [DOI] [PubMed] [Google Scholar]
- Vassileva I., Yanakieva I., Peycheva M., Gospodinov A. and Anachkova B. (2014). The mammalian INO80 chromatin remodeling complex is required for replication stress recovery. Nucleic Acids Res. 42, 9074-9086. 10.1093/nar/gku605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestin A. and Mills A. A. (2013). The tumor suppressor Chd5 is induced during neuronal differentiation in the developing mouse brain. Gene Expr. Patterns 13, 482-489. 10.1016/j.gep.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers L. E. L. M., van Ravenswaaij C. M. A., Admiraal R., Hurst J. A., de Vries B. B. A., Janssen I. M., van der Vliet W. A., Huys E. H. L. P. G., de Jong P. J., Hamel B. C. J. et al. (2004). Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 36, 955-957. 10.1038/ng1407 [DOI] [PubMed] [Google Scholar]
- Vogel-Ciernia A., Matheos D. P., Barrett R. M., Kramár E. A., Azzawi S., Chen Y., Magnan C. N., Zeller M., Sylvain A., Haettig J. et al. (2013). The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat. Neurosci. 16, 552-561. 10.1038/nn.3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vradii D., Wagner S., Doan D. N., Nickerson J. A., Montecino M., Lian J. B., Stein J. L., van Wijnen A. J., Imbalzano A. N. and Stein G. S. (2006). Brg1, the ATPase subunit of the SWI/SNF chromatin remodeling complex, is required for myeloid differentiation to granulocytes. J. Cell. Physiol. 206, 112-118. 10.1002/jcp.20432 [DOI] [PubMed] [Google Scholar]
- Wade S. L., Langer L. F., Ward J. M. and Archer T. K. (2015). MiRNA-mediated regulation of the SWI/SNF chromatin remodeling complex controls pluripotency and endodermal differentiation in human ESCs. Stem Cells 33, 2925-2935. 10.1002/stem.2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M., Zhang J., Lai D., Jani A., Prestone-Hurlburt P., Zhao L., Ramachandran A., Schnitzler G. R. and Chi T. (2009). Molecular basis of CD4 repression by the Swi/Snf-like BAF chromatin remodeling complex. Eur. J. Immunol. 39, 580-588. 10.1002/eji.200838909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xue Y., Zhou S., Kuo A., Cairns B. R. and Crabtree G. R. (1996). Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10, 2117-2130. 10.1101/gad.10.17.2117 [DOI] [PubMed] [Google Scholar]
- Wang L. C., Liu Z.-Y., Shapiro R., Yang J., Sizing I., Rayhorn P., Garber E. A., Benjamin C. D., Williams K. P., Taylor F. R., et al. (2000). Conditional disruption of hedgehog signaling pathway defines its critical role in hair development and regeneration. J. Invest. Dermatol. 114, 901-908. 10.1046/j.1523-1747.2000.00951.x [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhai W., Richardson J. A., Olson E. N., Meneses J. J., Firpo M. T., Kang C., Skarnes W. C. and Tjian R. (2004). Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 18, 3106-3116. 10.1101/gad.1238104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Sengupta S., Magnani L., Wilson C. A., Henry R. W. and Knott J. G. (2010). Brg1 is required for Cdx2-mediated repression of Oct4 expression in mouse blastocysts. PLoS ONE 5, e10622 10.1371/journal.pone.0010622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Du Y., Ward J. M., Shimbo T., Lackford B., Zheng X., Miao Y.-L., Zhou B., Han L., Fargo D. C. et al. (2014). INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 14, 575-591. 10.1016/j.stem.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weider M., Küspert M., Bischof M., Vogl M. R., Hornig J., Loy K., Kosian T., Müller J., Hillgärtner S., Tamm E. R. et al. (2012). Chromatin-remodeling factor Brg1 is required for Schwann cell differentiation and myelination. Dev. Cell 23, 193-201. 10.1016/j.devcel.2012.05.017 [DOI] [PubMed] [Google Scholar]
- Wiley M. M., Muthukumar V., Griffin T. M. and Griffin C. T. (2015). SWI/SNF chromatin-remodeling enzymes Brahma-related gene 1 (BRG1) and Brahma (BRM) are dispensable in multiple models of postnatal angiogenesis but are required for vascular integrity in infant mice. J. Am. Heart Assoc. 4, e001972-e001972 10.1161/JAHA.115.001972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis M. S., Homeister J. W., Rosson G. B., Annayev Y., Holley D., Holly S. P., Madden V. J., Godfrey V., Parise L. V. and Bultman S. J. (2012). Functional redundancy of SWI/SNF catalytic subunits in maintaining vascular endothelial cells in the adult heart. Circ. Res. 111, e111-e122. 10.1161/CIRCRESAHA.112.265587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. I., Lessard J., Olave I. A., Qiu Z., Ghosh A., Graef I. A. and Crabtree G. R. (2007). Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56, 94-108. 10.1016/j.neuron.2007.08.021 [DOI] [PubMed] [Google Scholar]
- Wu J. I., Lessard J. and Crabtree G. R. (2009). Understanding the words of chromatin regulation. Cell 136, 200-206. 10.1016/j.cell.2009.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Peng S., Yang J., Tu Z., Cai X., Cai C.-L., Wang Z. and Zhao Y. (2014). Baf250a orchestrates an epigenetic pathway to repress the Nkx2.5-directed contractile cardiomyocyte program in the sinoatrial node. Cell Res. 24, 1201-1213. 10.1038/cr.2014.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Li W., Shang C., Chen R. M., Han P., Yang J., Stankunas K., Wu B., Pan M., Zhou B. et al. (2013). Brg1 governs a positive feedback circuit in the hair follicle for tissue regeneration and repair. Dev. Cell 25, 169-181. 10.1016/j.devcel.2013.03.015 [DOI] [PubMed] [Google Scholar]
- Xu Y. and Fang F. (2012). Regulatory role of Brg1 and Brm in the vasculature: from organogenesis to stress-induced cardiovascular disease. Cardiovasc. Hematol. Disord. Drug Targets 12, 141-145. 10.2174/1871529X11202020141 [DOI] [PubMed] [Google Scholar]
- Xu F., Flowers S. and Moran E. (2012). Essential role of ARID2 protein-containing SWI/SNF complex in tissue-specific gene expression. J. Biol. Chem. 287, 5033-5041. 10.1074/jbc.M111.279968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Yang Y., Hemberg M., Yoshida T., Cho H. Y., Murphy J. P., Fioravante D., Regehr W. G., Gygi S. P., Georgopoulos K. et al. (2014). Promoter decommissioning by the NuRD chromatin remodeling complex triggers synaptic connectivity in the mammalian brain. Neuron 83, 122-134. 10.1016/j.neuron.2014.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Wang Z., Sharova L., Sharov A. A., Ling C., Piao Y., Aiba K., Matoba R., Wang W. and Ko M. S. H. (2008). BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells 26, 1155-1165. 10.1634/stemcells.2007-0846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip D. J., Corcoran C. P., Alvarez-Saavedra M., DeMaria A., Rennick S., Mears A. J., Rudnicki M. A., Messier C. and Picketts D. J. (2012). Snf2l regulates Foxg1-dependent progenitor cell expansion in the developing brain. Dev. Cell 22, 871-878. 10.1016/j.devcel.2012.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A. S., Staahl B. T., Chen L. and Crabtree G. R. (2009). MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642-646. 10.1038/nature08139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A. S., Sun A. X., Li L., Shcheglovitov A., Portmann T., Li Y., Lee-Messer C., Dolmetsch R. E., Tsien R. W. and Crabtree G. R. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228-231. 10.1038/nature10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A. (2011). Control of the embryonic stem cell state. Cell 144, 940-954. 10.1016/j.cell.2011.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Chen Y., Kim B., Wang H., Zhao C., He X., Liu L., Liu W., Wu L. M. N., Mao M. et al. (2013). Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152, 248-261. 10.1016/j.cell.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S., Choi M., Wakimoto H., Ma L., Jiang J., Overton J. D., Romano-Adesman A., Bjornson R. D., Breitbart R. E., Brown K. K. et al. (2013). De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220-223. 10.1038/nature12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Chen M., Kim J.-R., Zhou J., Jones R. E., Tune J. D., Kassab G. S., Metzger D., Ahlfeld S., Conway S. J. et al. (2011). SWI/SNF complexes containing Brahma or Brahma-related gene 1 play distinct roles in smooth muscle development. Mol. Cell. Biol. 31, 2618-2631. 10.1128/MCB.01338-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li B., Li W., Ma L., Zheng D., Li L., Yang W., Chu M., Chen W., Mailman R. B. et al. (2014). Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Rep. 3, 460-474. 10.1016/j.stemcr.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]