Abstract
Oral epithelia protect against constant challenges by bacteria, viruses, toxins and injury while also contributing to the formation of ectodermal appendages such as teeth, salivary glands and lingual papillae. Despite increasing evidence that differentiation pathway genes are frequently mutated in oral cancers, comparatively little is known about the mechanisms that regulate normal oral epithelial development. Here, we characterize oral epithelial stratification and describe multiple distinct functions for the mitotic spindle orientation gene LGN (Gpsm2) in promoting differentiation and tissue patterning in the mouse oral cavity. Similar to its function in epidermis, apically localized LGN directs perpendicular divisions that promote stratification of the palatal, buccogingival and ventral tongue epithelia. Surprisingly, however, in dorsal tongue LGN is predominantly localized basally, circumferentially or bilaterally and promotes planar divisions. Loss of LGN disrupts the organization and morphogenesis of filiform papillae but appears to be dispensable for embryonic hair follicle development. Thus, LGN has crucial tissue-specific functions in patterning surface ectoderm and its appendages by controlling division orientation.
KEY WORDS: LGN, Gpsm2, Papillae, Oral epithelia, Tongue, Placode, Oriented cell divisions, Hair follicle
Summary: In different regions of the mouse oral epithelium, differential subcellular localization of the spindle orientation protein LGN regulates distinct patterns of cell division.
INTRODUCTION
The epithelia that line the oral cavity are a diverse collection of stratified mucosal epithelia that include the buccogingiva (BG), palate and tongue. The mechanisms that regulate the development of these oral epithelia (OE) remain understudied compared with related tissues such as the epidermis (Fuchs, 2007; Arwert et al., 2012; Beck and Blanpain, 2012; Sumigray and Lechler, 2015), intestine (Gregorieff and Clevers, 2005; Wells and Spence, 2014), lung (Rock and Hogan, 2011; Herriges and Morrisey, 2014) and esophagus (DeWard et al., 2014; Rosekrans et al., 2015; Jones and Klein, 2013). Crucial epidermal differentiation genes such as TP63, NOTCH1-3, NFE2L2 and CASP8 (Mills et al., 1999; Yang et al., 1999; Nicolas et al., 2003; Blanpain et al., 2006; Dotto, 2009; Lee et al., 2009; Huebner et al., 2012; Kumar et al., 2015) are frequently altered in oral squamous cell carcinomas (Agrawal et al., 2011; Stransky et al., 2011; Cancer Genome Atlas Network, 2015), suggesting that the core differentiation programs targeted in human cancers are shared among stratified epithelia. Yet, it remains unknown whether OE development shares similar developmental programs with skin.
Recent work in the epidermis has shown that proper orientation of the mitotic spindle in basal epidermal progenitors is essential for skin development (Lechler and Fuchs, 2005; Poulson and Lechler, 2010; Williams et al., 2011, 2014; Niessen et al., 2013). In stem cells, spindle orientation can influence cell fate choices, often by directing polarized inheritance of fate determinants that promote asymmetric cell divisions (Siller and Doe, 2009; Morin and Bellaiche, 2011; Lu and Johnston, 2013; Williams and Fuchs, 2013). In developing epidermis, division orientation undergoes dynamic changes during stratification and has been linked to cell fate choices, such that planar divisions are thought to be symmetric whereas perpendicular divisions are asymmetric (Poulson and Lechler, 2010; Williams et al., 2011, 2014; Panousopoulou and Green, 2014). Initially, most basal cells divide with the mitotic spindle parallel to the basement membrane (BM), but as the epidermis begins to stratify oblique and perpendicular divisions become more numerous and by E16 ∼60% of divisions are perpendicular (Smart, 1970; Byrne et al., 1994; Lechler and Fuchs, 2005; Williams et al., 2014).
Spindle positioning is controlled by a highly conserved complex of proteins that includes the Par polarity complex (aPKC-Par3-Par6), the Gαi/Gαo family of heterotrimeric G-proteins, and mitotically recruited spindle orientation proteins including LGN (Leu-Gly-Asn-enriched protein; also known as Gpsm2), inscuteable (Insc) and NuMA (also known as Numa1) (Gonczy and Hyman, 1996; Du and Macara, 2004; Knoblich, 2008). LGN is an adaptor protein that contains N-terminal TPR repeats that mediate competitive binding between Insc and NuMA (Culurgioni et al., 2011; Yuzawa et al., 2011; Zhu et al., 2011), a flexible central linker that can be phosphorylated by aPKC and Aurora A (Johnston et al., 2009; Hao et al., 2010), and C-terminal G-protein regulatory (GPR)/GoLoco motifs that mediate interactions with Gαi/Gαo (Schaefer et al., 2001; Willard et al., 2004). LGN reorients the mitotic spindle to align with cortical polarity cues by facilitating capture of astral microtubules through its interactions with the microtubule-binding protein NuMA and the dynein/dynactin motor complex (Knoblich, 2008; Siller and Doe, 2009; Culurgioni and Mapelli, 2013; Seldin et al., 2016).
Here, we address how LGN functions in OE stratification and the morphogenesis of oral and epidermal appendages in mouse. As in skin, LGN localizes to the apical cell cortex in palatal, BG (cheek) and ventral tongue (VT) epithelium, and LGN knockdown leads to a bias toward planar and oblique divisions at the expense of perpendicular divisions, causing epithelial thinning and impaired differentiation. However, LGN shows a unique localization pattern in developing filiform papillae of the dorsal tongue (DT), where it is frequently found in lateral, bipolar domains or basal crescents. Using 2D and 3D analyses, we show that filiform papillae are built through a spatially and temporally coordinated program of oriented cell divisions mediated by LGN. Surprisingly, rather than promoting perpendicular divisions as in epidermis and other OE regions, in DT LGN drives two distinct classes of parallel divisions, namely ‘circumferential' and ‘vertical', which occur within and orthogonal to the epithelial plane, respectively. Upon loss of LGN, division orientation is randomized, causing profound morphological defects in filiform papilla patterning. By contrast, hair follicles of the epidermis do not require LGN to develop normally, at least through embryonic stages. This demonstrates that epithelial appendages utilize discrete mechanisms for morphogenesis, with LGN being required for the proper development of filiform papillae but dispensable for hair follicle growth.
RESULTS
Oral epithelial development is diverse and dynamic
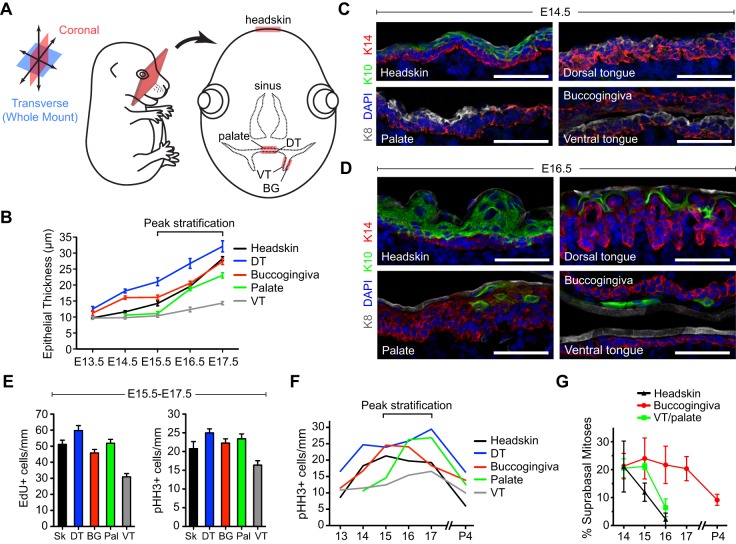
To investigate how OE development compares to that of epidermis, we analyzed coronal sections where both headskin and all major OE tissues can be visualized (schematized in Fig. 1A, see Fig. S3A for a composite image). As a first step, we measured epithelial layer thickness in the palate region, BG mucosa, VT, DT and headskin from E13.5 through E17.5 (Fig. 1B). At the earliest stages examined, headskin and VT consisted largely of a single layer of basal cells, whereas DT and BG epithelia were already two to three cell layers thick, in agreement with previous reports (Baratz and Farbman, 1975; Casey et al., 2006). Growth kinetics varied across tissue types, with DT and BG epithelia stratifying most rapidly and VT more slowly. Palatal epithelium initially appears thin and poorly organized between E13.5 and E15.0, but begins to rapidly stratify once the lateral palatal shelves fuse. Since peak OE stratification occurred between E15.5 and E17.5, we focused on ages just prior to (E14.5) and during (E16.5) this period to examine differentiation.
Fig. 1.
Oral epithelial stratification and proliferation are dynamic. (A) Visualizing oral epithelia (OE) in coronal sections. (B) E13.5-E17.5 OE/skin thickness. (C,D) Keratin expression in head and OE at E14.5 (C) and E16.5 (D), using basal (K14, red), suprabasal (K10, green), and periderm (K8, white) markers. (E) Proliferation rates during peak stratification, assessed by EdU incorporation (left) and levels of the G2/M marker pHH3 (right). (F) Proliferation rates during skin and OE development, representing aggregate pHH3+ cells/mm. All OE demonstrate higher proliferative activity postnatally compared with skin. (G) Suprabasal mitoses are transient during skin and ventral tongue (VT)/palate development but persist in buccogingival (BG) epithelia postnatally. (B,E,F) Data are derived from n=10-30 sections per tissue per age from n≥3 independent mouse embryos; (G) data are from 30-60 mitoses (from 3-6 embryos) per group. DT, dorsal tongue; Pal, palate; Sk, headskin. Scale bars: 50 µm.
As in epidermis, keratin 5 (K5, or Krt5) and K14 are co-expressed in OE (Fig. 1C,D, Fig. S1A,B). However, there are several notable differences between these tissues. First, by E14.5 K5/K14 become restricted to the basal layer in the epidermis while remaining more broadly expressed in OE, particularly in BG mucosa and palate. Second, the palate remains weakly K5/K14+ even at E14.5 (Fig. S1C,D). Third, prior to K5/K14, all OE broadly express K6, which becomes restricted suprabasally by E16.5. In epidermis, K6 expression is normally confined to the periderm – a transient embryonic barrier layer that also expresses K8 – but is also observed in pathological hyperproliferative stages (Mazzalupo and Coulombe, 2001). The periderm is well-defined on palatal and tongue epithelium from early stages (Fig. 1C, Fig. S1C), consistent with a known function of oral periderm in preventing palatal shelves from inappropriately fusing with other OE during palatogenesis (Casey et al., 2006; Richardson et al., 2009).
Differentiation, manifested by the expression of K1 and K10, is observed as early as E13.5 in dorsal backskin epidermal basal cells (Williams et al., 2014), but later K1/K10 become restricted to suprabasal layers (Kopan and Fuchs, 1989; Byrne et al., 1994). At E14.5, K10 labels suprabasal differentiating layers of headskin; however, differentiation is delayed in OE by at least 1 day (Fig. 1C,D). At E16.5, K10 remained absent from VT, whereas sparse K10+ cells were observed in BG and palatal epithelium, with coexpression of K10 and K14 evident in palate. Keratinization of DT filiform papillae was first observed at ∼E16, in agreement with observations in rat (Baratz and Farbman, 1975). Thus, stratification precedes differentiation to an even greater degree in OE than in epidermis (Byrne et al., 1994).
To assess proliferation, we performed 3-h pulses with 5-ethynyl-2′-deoxyuridine (EdU) to label cells in S-phase, and quantified G2/M cells by phospho-histone H3 (pHH3) (Fig. 1E,F). During peak stratification (E15.5-E17.5), proliferative activity, whether measured by EdU or pHH3, showed similar trends between tissues, with highest rates in DT and lowest in VT (Fig. 1E). Tissues with the highest (DT) and lowest (VT) mitotic rates also showed the fastest and slowest epithelial growth, respectively (Fig. 1B), demonstrating that proliferation is a driving force for stratification. Proliferation rates were highest between E15.5 and E17.5 for all tissues examined, although tongue and palate rates peaked later (E17.5) than BG and epidermis (E15.5), and all OE are more mitotically active postnatally than epidermis (Fig. 1F). Finally, suprabasal mitoses, which are thought to be an important transient driver of early stratification in skin (Byrne et al., 1994; Lechler and Fuchs, 2005), persist longer and are more frequent in BG (Fig. 1G). Together, these data demonstrate the diversity of the developing OE and implicate E15.5-E17.5 as a dynamic period for stratified and specialized OE development when proliferation rates are highest.
LGN localizes apically during peak stratification
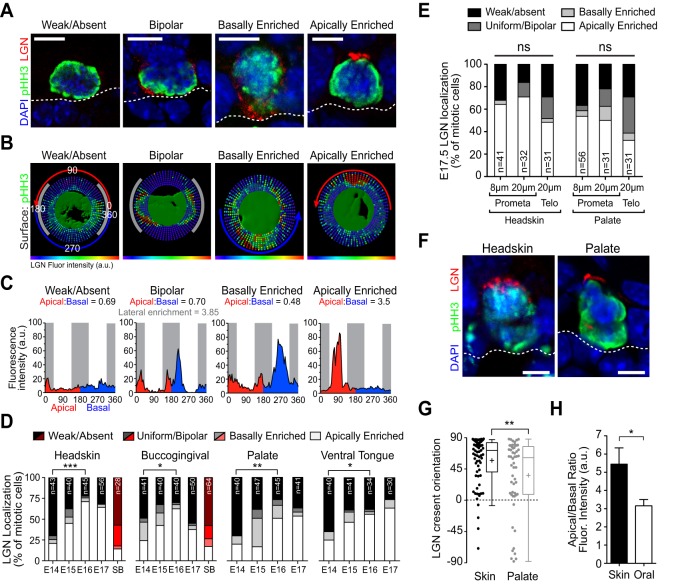
LGN rarely polarizes during mitosis in early epidermal development but is efficiently recruited to the apical cortex by E15.5-E16.5, promoting perpendicular differentiative divisions (Williams et al., 2011, 2014). In developing OE and headskin LGN is localized in four different patterns: weak/absent, uniform/bipolar, basally enriched, and apically enriched (Fig. 2A). LGN localization patterns were assessed quantitatively using a custom, open-source Python XTension that translates fluorescence intensity into ‘radial heat maps’ in Imaris software (Fig. 2B). Mean LGN fluorescence intensity at each radian around the perimeter of the cell cortex can be visualized relative to apical (red), basal (blue) and lateral (gray) domains, with integrated fluorescence intensity used to calculate apical:basal ratios (Fig. 2C). ‘Bipolar' cells could be further discriminated from ‘uniform' cells by measuring ‘lateral enrichment' (Fig. 2C), although for subsequent analyses these groups were combined. While applied to LGN here, this new computational tool allows for the unbiased radial quantification of any cortical protein (Fig. S6, Movie 3).
Fig. 2.
LGN is apically recruited during peak stratification. (A) Patterns of LGN localization, shown in mitotic (pHH3+) OE basal cells. Dashed line represents the basement membrane (BM). (B,C) Radial heat maps (B) and mean intensity plots (C) of LGN fluorescence intensity used to quantitatively categorize LGN localization patterns (shown in A), illustrating apical (red), basal (blue) and lateral domains (gray). (D,E) LGN localization patterns by category in different tissues during development. n values are indicated within bars. (D) Increasing proportions of mitotic basal cells polarize LGN apically by E16-E17, whereas suprabasal mitoses (red bars) rarely do. (E) LGN localization patterns in headskin (left) and palate (right) assessed by different methods. Cells were analyzed at early (pro/metaphase) or late (telophase) stages of mitosis, from single confocal planes in thin sections (8 μm) or from z-series confocal stacks in thick sections (20 μm). (F) Representative images from the same section of apical LGN in E16.5 headskin and palate. (G,H) Quantification of LGN crescent orientation (G) and apical:basal fluorescence intensity ratio (H) for headskin and palate (n=10 per group). *P<0.05, **P<0.01, ***P<0.0001, by χ2 (D,E) or Student's t-test (G,H). Scale bars: 5 µm.
As in headskin, by E16.5 LGN was apically enriched in most mitotic cells in each OE tissue, while tending to be weak/absent at earlier ages (Fig. 2D). When LGN did polarize, there was a significant shift from randomized orientation toward apical localization between early and late ages for all tissues (Fig. S2A). Interestingly, LGN failed to polarize in over 75% of suprabasal mitoses regardless of tissue or age, suggesting that suprabasal divisions do not generally utilize LGN (Fig. 2D, red bars). Postnatally, LGN continued to be expressed in most OE and epidermal mitoses, in remarkably similar patterns for all tissues examined (Fig. S2B). However, at P4, uniform/bipolar LGN was the predominant localization pattern in OE and headskin. This suggests that there might be a bias toward parallel divisions postnatally, as previously reported in backskin (Niessen et al., 2013) and tail epidermis (Clayton et al., 2007), although it has been reported that 85% of divisions in adult ear epidermis are perpendicular (Lechler and Fuchs, 2005). Similar patterns of LGN were observed at early (prometaphase) and late (telophase) mitosis, in thin (8 µm) or thick (20 µm) sections (Fig. 2E; no significant difference by chi-square test). Three-dimensional wholemount z-stack reconstructions of BG also revealed predominant patterns of apical enrichment (Fig. S2C). Interestingly, however, although LGN was frequently polarized apically in OE, this occurred less efficiently than in headskin (Fig. 2E), where apical crescents were less intense and more weakly polarized compared with headskin (Fig. 2F-H).
LGN is necessary for perpendicular division orientation in OE
We recently developed a technique to manipulate gene function in developing mouse embryos using ultrasound-guided in utero delivery of high-titer lentivirus into the amniotic fluid surrounding E9.5 embryos (Beronja et al., 2010). This leads to high-level transduction of surface epithelia including the epidermis and OE, where labeled cells express a stable nuclear histone H2B-mRFP1 (H2B-RFP) reporter (Fig. S3A). Using either a non-targeting scramble control or validated LGN shRNA (LGN 1617) (Williams et al., 2011, 2014), all OE were transduced at 60-90% efficiency (Fig. S3B-E). LGN knockdown efficiency was confirmed at the protein level in OE by quantitative analysis of fluorescence intensity in mitotic cells (Fig. S4, see also Fig. S8). In DT, LGN knockdown was highly efficient, reducing LGN accumulation in RFP+ mitotic cells to interphase levels, when LGN is normally not detectable by immunofluorescence. Moreover, interphase levels of LGN expression are essentially equivalent to background since no difference in LGN expression was observed between RFP+ and RFP− interphase cells in DT (Fig. S4E).
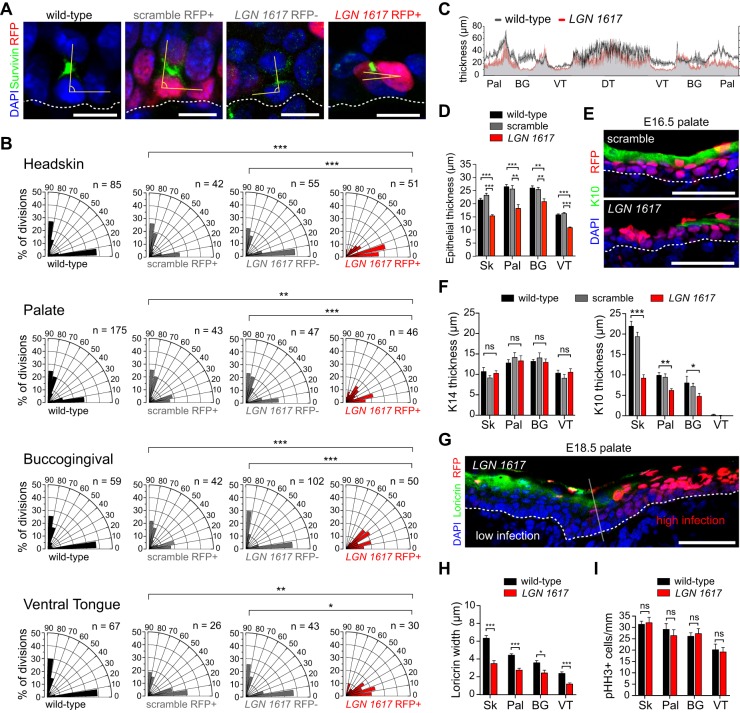
Since LGN can polarize apically during mitosis in both OE and epidermis, we asked whether LGN is required for proper division orientation in OE. Because spindle rotation can occur throughout metaphase, we used the cleavage furrow marker survivin (also known as Birc5) to identify late stage mitotic cells, and measured division orientation of these committed cells relative to the BM (Fig. 3A). Division orientation behavior of each population is depicted using radial histograms (Fig. 3B). The LGN shRNA experimental group (LGN 1617 RFP+) was compared with three controls: uninjected littermates (wild type), non-transduced cells from LGN 1617-injected embryos (LGN 1617 RFP−), and transduced cells from scramble-injected embryos (scramble RFP+). In E16.5 wild-type littermate headskin (Fig. 3B), as previously reported in backskin (Williams et al., 2011), division angles follow a bimodal distribution, with roughly equal numbers of perpendicular and parallel divisions, and very few oblique divisions. However, parallel divisions occur at a higher frequency in headskin (∼50%) than in backskin (∼40%), perhaps explaining why headskin stratification lags behind backskin. Bimodal patterns were also observed in wild-type palatal, BG and VT epithelia (Fig. 3B), with roughly equal proportions of parallel and perpendicular divisions. In headskin and OE, LGN loss recapitulated the phenotype of backskin (Williams et al., 2011), where perpendicular divisions are lost at the expense of oblique and parallel divisions (Fig. 3B). This phenotype was cell-autonomous and could not be attributed to non-specific effects of viral transduction or shRNA expression, since a wild-type distribution of division angles was observed in LGN 1617 RFP− and scramble groups, respectively.
Fig. 3.
Altered spindle orientation and differentiation upon LGN loss. (A) Representative wild-type littermate, scramble, LGN 1617 RFP− and LGN 1617 RFP+ telophase palatal cells indicating division orientation (yellow angle) relative to the BM (dashed line). (B) Radial histograms of division orientation for headskin, palatal epithelium, BG epithelium and VT at E16.5. Perpendicular and planar divisions are roughly equal in wild-type, LGN 1617 RFP− and scramble cells, whereas oblique and planar divisions are increased in LGN 1617 RFP+ mitoses. (C) Average OE thickness in wild-type and LGN knockdown embryos at E16.5, measured every 10 μm and averaged from three embryos per group. (D) Epithelial thickness for the indicated tissues in control groups (wild-type littermates and scramble) and LGN knockdowns (red). (E,F) Reduced immunolabeling for the spinous/granular marker K10 (green in E) in E16.5 LGN knockdowns, as quantified in F. Basal (K14) marker expression is unaffected. (G) Differentiation defects at E18.5 showing reduced loricrin immunolabeling specifically in RFP+ regions of LGN knockdown palatal epithelium. (H) Similar reductions in loricrin staining are observed across all OE. (I) LGN loss does not significantly impact proliferation rates. (D,F,H,I) Data are derived from n=10-30 sections per tissue per age from n≥4 independent embryos. *P<0.05, **P<0.01, ***P<0.0001, by χ2 test (B) or Student's t-test (D,F,H,I); ns, not significant. Scale bars: 10 µm in A; 50 µm in E,G.
We also observed stratification defects, manifested as thinning across all OE regions at E16.5 in LGN 1617-injected embryos compared with wild-type littermates (Fig. 3C,D). Differentiation, as measured by the spinous/granular layer marker K10 and the granular layer marker loricrin, was also specifically impaired in LGN 1617 RFP+ regions of palatal epithelium (Fig. 3E,F). At E16.5, the K10 layer was significantly thinner in headskin, palate and BG, with no observable alterations in the K14+ basal layer (Fig. 3F). At E18.5, loricrin expression was also significantly diminished (Fig. 3G,H). Importantly, mitotic rates were unaffected in OE of LGN 1617-injected embryos, demonstrating that delayed stratification and differentiation cannot be attributed to general proliferation defects (Fig. 3I, Fig. S2D). Collectively, these data present the first evidence that division orientation, mediated by apically localized LGN, is essential for OE differentiation.
LGN loss disrupts filiform papillae development
DT is populated by lingual papillae, which include both taste buds (fungiform, circumvallate and foliate papillae) and the more numerous hair-like, keratinized filiform papillae. These specialized appendages are continually renewed at one of the most rapid turnover rates in the body, displaying both capabilities for overgrowth (hairy tongue) and depapillation (atrophic glossitis) (Reamy et al., 2010; Gurvits and Tan, 2014). Analogous to the crypt-villus organization of the small intestine, progenitors are thought to lie in the interpapillary epithelium (IPE), where they contribute to the renewal of three to four neighboring papillae (Okubo et al., 2009; Tanaka et al., 2013). Although much is known about the mechanisms that regulate taste bud development (Thesleff and Mikkola, 2014; Barlow, 2015), comparatively little is known about how filiform papillae form.
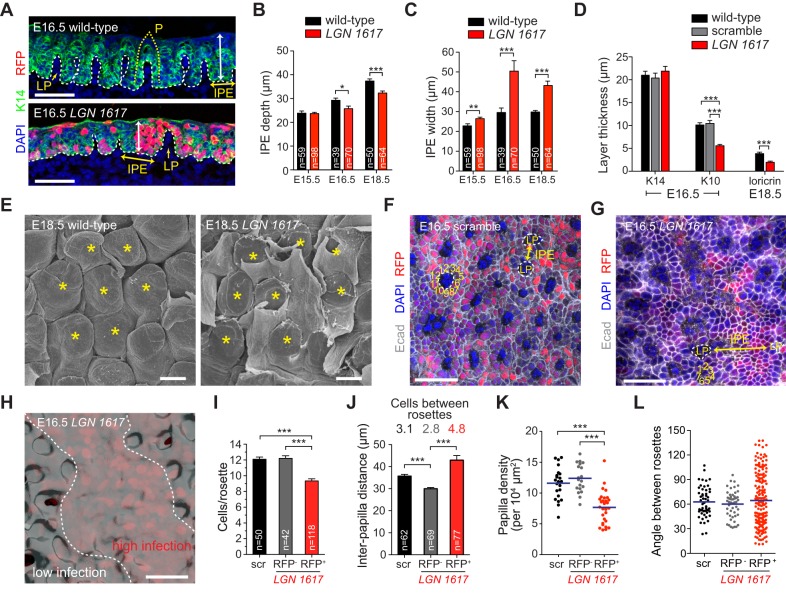
LGN loss caused striking defects in DT morphogenesis, causing the IPE region to become abnormally wide, with a concomitant decrease in the number of morphologically defined papillae (Fig. 4A-C, see also 4F-H). As with other OE, differentiation was impaired (Fig. 4D). Scanning electron microscopy (SEM) of E18.5 DT revealed delayed papillary eruption through the keratinized epithelial layer that covers the surface of the tongue upon LGN loss (Fig. 4E). SEM and confocal images of DT wholemounts showed a highly ordered hexagonal pattern of papilla that was visible at E16.5 in scramble controls but lost in LGN knockdowns (Fig. 4E-G). E-cadherin (Ecad, also known as Cdh1) surface plots rendered from confocal z-stacks showed stunted papilla growth in RFP+ regions of LGN 1617-transduced DT (Fig. 4H). Additional quantification of morphogenesis in LGN knockdowns revealed that (1) there are fewer papilla, (2) the distance and number of cells between papilla is increased and (3) the number of cells around the papilla circumference is decreased (Fig. 4I-K). Finally, whereas control papillae are arranged in an ordered hexagonal pattern in which placodes are separated by ∼60°, the distribution of angles observed in LGN 1617-transduced DT is significantly more variable (Fig. 4L).
Fig. 4.
LGN loss in DT causes defects in filiform papilla morphogenesis. (A-C) Coronal section of wild-type (A, top) DT epithelium showing ordered arrangement of papillary placodes. Future papillae (P, dotted yellow line) develop over evaginations of lamina propria (LP) and are separated by interpapillary epithelium (IPE). In LGN knockdown DT (A, bottom), fewer placodes develop, IPE width (yellow double arrowhead) increases, and depth (white double arrowhead) decreases, as quantified in B,C. (D) K14, K10 and loricrin layer thickness (derived from n=10-30 sections from n≥4 independent embryos per tissue per age) showing impaired differentiation in LGN knockdown DT. (E) SEM images of E18.5 wild-type (left) and LGN knockdown (right) central DT. Filiform papillae (asterisks) are arranged in regular hexagonal arrays in controls, whereas LGN knockdown papillae are disorganized and display delayed eruption through the keratinized outer epithelial layer. (F-H) Wholemount confocal images of E16.5 scramble (F) and LGN 1617 (G) transduced DT labeled for Ecad (gray). (H) Ecad 3D surface plot reconstructed from the region shown in G. Positions of LP and IPE are shown in F,G, with the number of cells in each papilla ‘rosette’ indicated. (I) Quantification of the number of cells per rosette. scr, scrambled. (J) Interpapilla distance (analogous to IPE width in C) is increased in RFP+ regions in LGN knockdowns, with compensatory decrease in RFP− regions. Values above bars indicate average number of cells between rosettes for each condition. (K) Reduction in papilla density in LGN 1617 RFP+ regions of DT. (L) Measurement of angles between papilla rosettes (blue bars indicate means) shows tight clustering around 60° in controls, but greater variation in LGN knockdowns (P<0.0001). *P<0.05, **P<0.01, ***P<0.0001, by two-tailed Student's t-tests except Mann–Whitney test (K) and F-test (L). Scale bars: 40 µm in A,F-H; 20 µm in E.
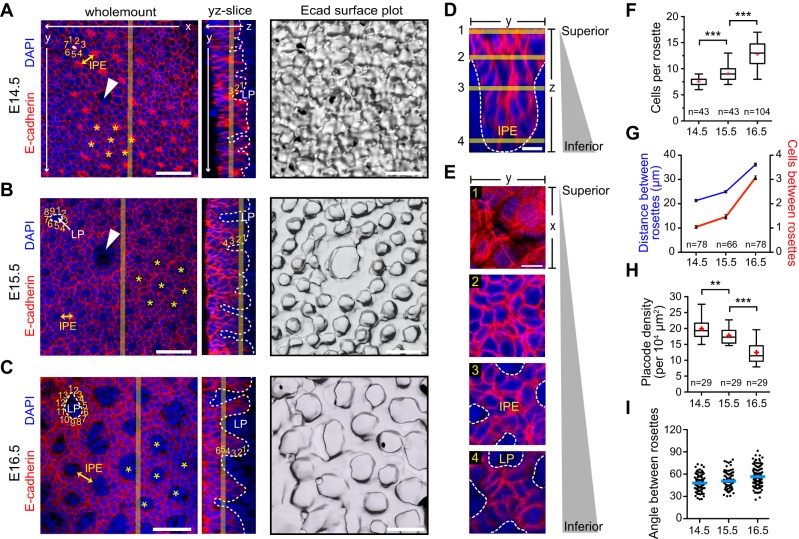
Since there are few studies of filiform papilla development (Baratz and Farbman, 1975; Hume and Potten, 1976), we next investigated wild-type papilla morphogenesis from E14.5-E16.5 using confocal microscopy of DT wholemounts. At E14.5, larger fungiform papillae were already well defined (arrowhead in Fig. 5A), while the remainder of the tongue epithelium was beginning to organize into hexagonally arranged ‘rosettes' of 6-7 cells with intense central stellate Ecad expression. By E15.5, mesenchymal cells of the lamina propria (LP) intercalated into the central region of these rosettes, surrounded by 7-9 epithelial cells. Ecad surface plots and z-slices clearly illustrate nascent papilla outgrowths from the DT surface, typically containing ∼4 cells per slope (Fig. 5B). By E16.5, papilla growth continued both centripetally and vertically, with LP now surrounded by ∼12 cells, ∼5-6 cells present along papilla slopes, and the distance between rosettes also increasing (Fig. 5C). A 3D reconstruction of the IPE region surrounded by four nascent papillae, with representative z-slices, is shown in Fig. 5D,E. Quantitative analyses show that the number of cells per rosette and the distance between rosettes increase over time, whereas placode density decreases in proportion with tongue growth (Fig. 5F-H). Importantly, placode positioning remains consistent between E14.5 and E16.5 (Fig. 5I), demonstrating that filiform papilla are pre-patterned prior to the vertical and circumferential growth phases that occur between E15.5 and E16.5.
Fig. 5.
Patterning and growth of papillary placodes. (A-C) Wholemount, yz slice and surface plot views of E14.5 (A), E15.5 (B) and E16.5 (C) central DT. Arrowheads indicate fungiform papillae. At E14.5, stellate regions of concentrated Ecad (asterisks) form a highly ordered heptagonal/hexagonal pattern representing early filiform papilla placodes. By E15.5, LP (dashed white lines) intercalates at the center of these rosettes, which increase in cell number. At E16.5, the LP area, rosette circumference and IPE width (yellow double arrows) increase. Surface plots reveal vertical growth of ordered papillary placodes beginning at E15.5. (D,E) High-magnification z-stack reconstruction of the IPE region between placodes (D), with representative z-slices illustrated (E). (F-I) Morphometric analyses of papilla development, showing increased circumferential growth around LP (F), increasing distance/cell number between rosettes (G), and decreasing placode density (H). Angles of rosettes relative to each other (blue bars indicate means) remain relatively consistent, indicating that filiform papilla position is pre-patterned by E14.5 (I). **P<0.01, ***P<0.0001, by Mann–Whitney test (F) or two-tailed Student's t-test (H). Scale bars: 50 µm in A-C; 10 µm in D,E.
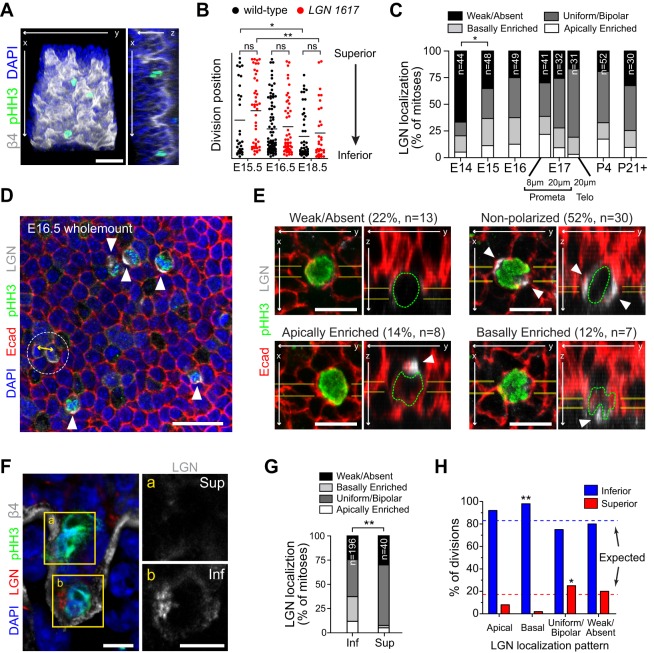
To investigate how LGN loss disrupts papilla development, we examined proliferation and LGN localization. Mitoses are generally confined to the base of the IPE between papilla, and LGN loss did not affect the position or frequency of mitotic cells throughout DT development (Fig. 6A,B), arguing that proliferation per se is not grossly affected. However, unlike other OE regions, where LGN shows a clear apical bias, LGN was rarely apically polarized in DT, instead showing bipolar/uniform or unilateral/basal polarization beginning around E15 and persisting into adulthood (Fig. 6C). As with other OE, our 2D analyses of LGN localization were consistent across sections of different thickness and at different stages of mitosis (Fig. 6C, E17 group). To further investigate LGN localization patterns in DT we utilized wholemounts analyzed by z-stack confocal microscopy, which enabled improved 3D resolution (Fig. 6D). Examples of localization patterns are shown in Fig. 6E and Movie 1, showing proportions similar to those observed in sections (Fig. 6C), with unpolarized (uniform or bipolar) distributions occurring most frequently. Interestingly, we noted regional differences within developing papilla, with mitotic cells showing different patterns of LGN localization in the IPE (‘inferior') compared with papilla slopes (‘superior') (Fig. 6F). A significant enrichment of uniform/bipolar patterns in superior cells (P=0.0013) and of basal localization in inferior cells (P=0.0445) was observed (Fig. 6G,H). These data suggest that spatially distinct DT populations might utilize LGN in different ways during papilla morphogenesis.
Fig. 6.
DT shows unique patterns of LGN localization. (A) 3D construction of E16.5 DT wholemount showing the position of mitotic cells (pHH3+, green) relative to BM (integrin β4, white). (B) Position (depth) of mitotic cells in DT from E15.5-E18.5 in wild-type and LGN knockdown (red) DT. Mitoses localize progressively deeper over time as papillae grow, and this is unchanged upon LGN loss. (C) LGN localization patterns from E14.5 to adulthood. LGN is initially undetectable, becoming primarily ‘basally enriched' or ‘uniform/bipolar' from E15.5 onward. E17 data were analyzed as in Fig. 2E. (D,E) Wholemount single confocal sections (D) and high-magnification xy/yz reconstructions from confocal stacks (E) of mitotic DT cells showing distinct patterns of LGN localization. Arrowheads indicate patterns of cortical LGN; observed percentages and n values are shown in E. Yellow double arrow in D indicates the division plane of telophase cell with uniformly distributed LGN. (F) Two mitotic cells positioned along papilla slope (a) or in deeper IPE (b). Insets show LGN, expressed weakly in superior cell and laterally in inferior cell. (G) Quantification of LGN localization patterns for inferior (papilla base/IPE) versus superior (papilla slope) mitoses in E14.5-E17.5 DT epithelium. (H) Data from G plotted by frequency of division location for each LGN localization pattern. Expected frequencies are indicated by dashed lines. Binomial tests indicate enrichment of basal polarization for inferior mitoses and of uniform/bipolar localization for superior mitoses. *P<0.05, **P<0.01, by χ2 test (C,G), Mann–Whitney test (B) or binomial test (H). Scale bars: 50 µm in A,D; 10 µm in E,F.
Papillary placodes develop through a coordinated pattern of oriented cell divisions (OCDs)
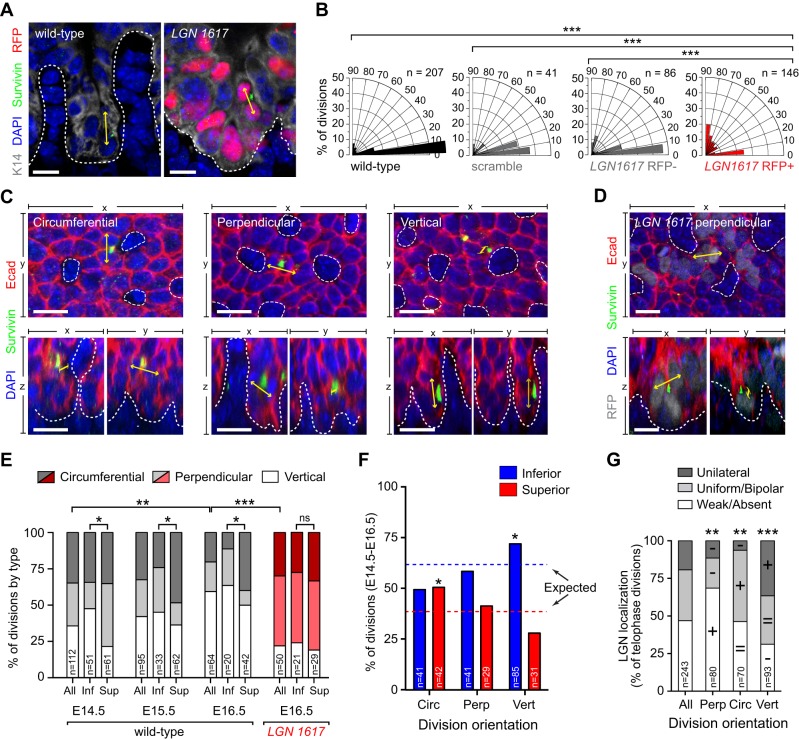
To understand how LGN loss alters papilla morphogenesis, we first examined division orientation in 2D coronal sections of E16.5 control and LGN 1617-transduced DT (Fig. 7A). The vast majority of divisions occurred parallel to the BM in wild-type, scramble and LGN 1617 RFP− cells, whereas a significant increase in perpendicular divisions was observed in LGN 1617 RFP+ cells (Fig. 7B). This strongly suggests that basal, bipolar and/or uniform LGN localization normally promote divisions parallel to the BM. Next, we conducted analyses of division orientation in 3D DT wholemounts, where three discrete categories of divisions were identified (Fig. 7C). Circumferential and perpendicular divisions occur in the plane of the epithelium, parallel and orthogonal to the BM, respectively. Vertical divisions occur in the z-axis, parallel to the BM. Effectively, circumferential divisions increase papilla girth, perpendicular divisions increase IPE width, and vertical divisions increase papilla height. In wild-type DT between E14.5 and E16.5 vertical divisions increased while circumferential divisions decreased, consistent with the developmental timing of initial radial papilla growth followed by later vertical growth (Fig. 5). Consistent with the 2D analyses, an increase in perpendicular divisions was observed in knockdowns with LGN 1617 (Fig. 7D,E), which is likely to explain the increased interpapillary distance in these mutants.
Fig. 7.
Spindle orientation is altered upon LGN loss in developing filiform papillae. DT division orientation in 2D coronal sections (A,B) and 3D wholemounts (C-G). (A) Wild-type and LGN knockdown telophase cells with division orientation indicated by yellow arrows. (B) Radial histograms of division orientation. Most control divisions occur parallel to the BM, whereas LGN knockdown divisions are frequently perpendicular. (C) xy (top) and xz/yz (bottom) views of telophase cells illustrating three division types occurring in papilla placodes. Circumferential (parallel to BM) and perpendicular (orthogonal to BM) divisions occupy the epithelial plane; vertical divisions occupy the z-plane parallel to the BM. (D) Example of abnormal perpendicular division in LGN knockdown DT. (E) Quantification of division type in wild-type and LGN knockdown DT, shown for all cells and also subdivided by inferior IPE (Inf) and superior papilla slope (Sup) positions. Vertical divisions increase and circumferential divisions decrease between E14.5 and E16.5, reflecting a switch from radial to vertical papilla growth. Upon LGN loss, division orientation is randomized and positional biases are eliminated. (F) Distribution of division types for inferior and superior mitoses in wild-type papillae. Expected frequencies are shown by dashed lines. (G) Circumferential divisions are enriched in the superior population, whereas vertical divisions are enriched in the inferior population. LGN localization patterns are marked as enriched (+), decreased (−) or equivalent (=) relative to the general population. *P<0.05, **P<0.01, ***P<0.0001, by χ2 test (B,E,G) or binomial test (F). Scale bars: 10 µm in A; 20 µm in C,D.
In agreement with our finding that LGN localization differed between IPE and papilla slopes (Fig. 6F-H), we observed distinct patterns of division orientation in inferior and superior regions (Fig. 7E,F). Specifically, we noted an overrepresentation of vertical divisions inferiorly, where polarized (generally basal) LGN is more common, and circumferential divisions superiorly, where bipolar/uniform LGN is more common (Fig. 7F). Notably, this positional bias was lost in LGN knockdowns and division orientation appeared randomized (Fig. 7E, Fig. S5A,B). Next, we sought to make a direct correlation between LGN localization and division orientation in late stage mitotic cells in DT wholemounts. This approach allowed us to visualize LGN inheritance in daughter cells in three dimensions (Fig. S5C,D, Movie 2). The majority of perpendicular divisions showed weak/absent LGN, whereas uniform/bipolar LGN patterns were most frequent in circumferential divisions, and polarized unilateral (apically or basally oriented) LGN localization patterns were enriched in vertical divisions (Fig. 7G).
Potential upstream regulators of differential LGN localization
We next sought to investigate how LGN could be differentially localized in distinct OE tissues. In the epidermis, both Insc and the heterotrimeric G-protein Gαi3 (also known as Gnai3) are required for the proper apical localization of LGN (Williams et al., 2014). Owing to a lack of suitable antibodies for Insc and because overexpression of tagged Insc can induce spindle orientation phenotypes (Poulson and Lechler, 2010; Williams et al., 2011, 2014), we focused on Gαi3. Three-dimensional cortical localization patterns and colocalization of LGN and Gαi3 were analyzed using our Imaris Python script (Fig. S6, Movie 3). In both epidermis and palate, LGN and Gαi3 were expressed in overlapping domains, with close alignment of their cortical crescents, even across multiple z-planes (Fig. S6A-G). However, in DT, the degree of colocalization between LGN and Gαi3 was more variable, with numerous instances in which Gαi3 was polarized but LGN was missing, or where LGN was polarized but Gαi3 was not (Fig. S6E,H). Thus, it seems likely that some as yet unidentified protein directs LGN subcellular localization in DT.
Hair follicle polarity and morphogenesis are unaffected by LGN loss
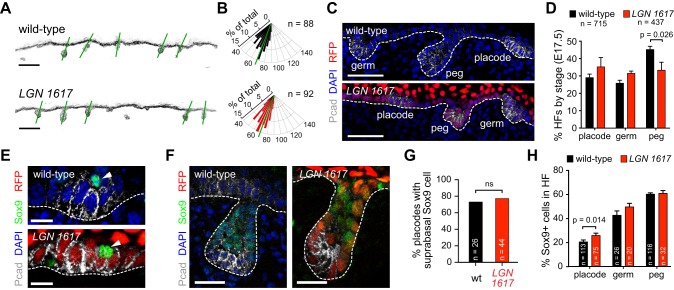
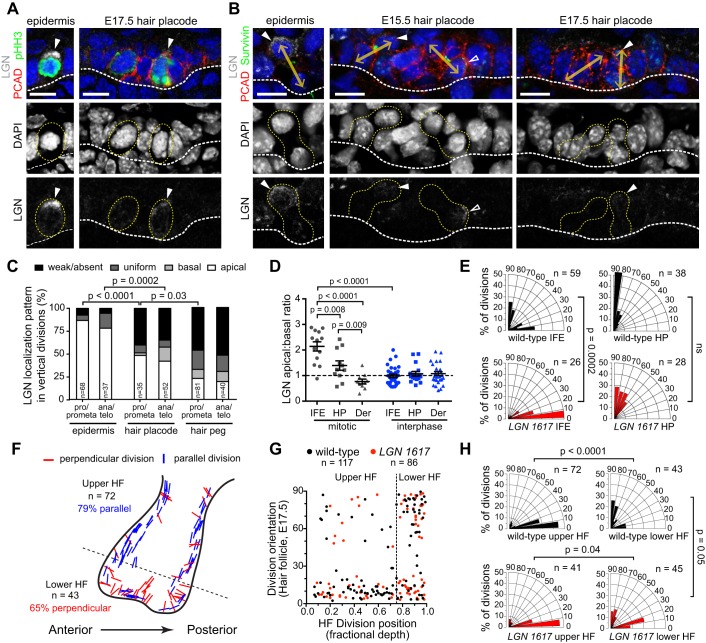
Given the crucial role of LGN in patterning filiform papilla, we wondered whether LGN directs the formation of another well-characterized epithelial appendage, the hair follicle (HF). Pelage HFs first appear as thickenings in the epidermis at ∼E14.5. These hair placodes (HPs) rapidly proliferate and grow downward into the dermis at the hair peg stage, eventually enveloping the specialized population of mesenchymal cells called the dermal papilla at the hair germ stage (Paus et al., 1999). Placodes are enriched for P-cadherin (Pcad, or Cdh3), which becomes restricted anteriorly at the germ stage, as HFs develop anteroposterior polarity (Tinkle et al., 2004; Devenport and Fuchs, 2008). Interestingly, LGN knockdown HFs appeared grossly normal, maintaining normal reciprocal expression of Pcad and Ecad (Fig. S7A) and proper angling along the anteroposterior axis (Fig. 8A,B). Because of the staggered development of different HF types, E17.5 is an ideal stage at which to examine HFs at each stage of morphogenesis. In wild-type backskin at this age, 29.1±2.0% of HFs are at the placode stage, 25.8±1.7% are at the germ stage, and 45.2±1.9% have reached the peg stage (Fig. 8C,D). Although knockdown with LGN 1617 caused a slight delay in HF differentiation (Fig. 8D), all three stages were observed with similar frequency, and HFs were present at normal density and appeared morphologically normal (Fig. S7B-F).
Fig. 8.
LGN loss does not affect hair follicle development. (A) Inverted images of K14 (basal marker) in wild-type and LGN knockdown E17.5 backskin, where green lines indicate hair follicle (HF) angling. (B) Radial histograms show HF orientation in wild type and LGN knockdowns (mean indicated with green lines; P=0.5307 by Mann–Whitney test). (C,D) Images (C) and quantification (D) of HFs at each stage in LGN knockdowns and wild-type littermates. (E-H) Characteristic suprabasal Sox9+ cells (arrowheads) in hair placodes (E) and in Pcad− regions of hair pegs (F), as quantified in G,H. P-values determined by two-tailed Student's t-test (D), binomial test (G) or Mann–Whitney test (H). Scale bars: 100 µm in A; 50 µm in C; 10 µm in E; 25 µm in F.
Sox9 is an early marker of hair follicle stem cells (HFSCs), first observed in sparse suprabasal cells in early HPs. This Sox9+ population increases in number as HFs develop and is found in a pattern complementary to that of Pcad, which labels a quiescent population of Wnthi/Shh+/Lhx2+ progenitors (Rhee et al., 2006; Nowak et al., 2008; Ouspenskaia et al., 2016). It was recently suggested that perpendicular asymmetric cell divisions in HPs help establish an asymmetry in Wnt and Shh responsiveness that specifies Sox9+ progenitors (Ouspenskaia et al., 2016). To address whether LGN is required for the specification of Sox9+ progenitors, we examined the position and number of Sox9+ cells at different stages of HF morphogenesis. In both early HPs and at more mature HF stages, we could find no evidence of any altered distribution of Sox9+ cells upon LGN loss (Fig. 8E-H). Similar numbers of HPs in LGN knockdowns and littermate controls (78% and 73%, respectively) contained a characteristic suprabasal Sox9+ cell (Fig. 8G). Moreover, the relative proportion of Sox9+ cells within developing HFs was similar at all stages, except for a slight increase in LGN knockdowns at the placode stage (Fig. 8H).
To understand how LGN loss might be tolerated in developing HFs, we examined LGN expression and division orientation. As a first step, we examined whether LGN knockdown was equally efficient in Pcadhi HPs compared with surrounding Pcadlo interfollicular epidermis (IFE). We confirmed significant and similar loss of LGN protein in both mitotic and interphase cells for both populations (Fig. S8A-C). Quantitative analyses of fluorescence intensity in wild-type mitotic cells revealed that although whole-cell LGN levels were similar in IFE and HPs, the apical and cortical LGN pools were significantly reduced in HPs (Fig. S8B,D,E). This suggested that LGN might not be polarized as efficiently in HPs, which was confirmed by examining LGN localization in both early and late stage mitotic cells of IFE and HPs (Fig. 9A,B). Whereas LGN was nearly always apically enriched in perpendicular divisions in the IFE, fewer than half of perpendicular divisions in both early placodes and more mature hair pegs displayed polarized apical LGN, and the degree of apical:basal enrichment was reduced (Fig. 9C,D). In agreement with Ouspenskaia et al. (2016), we observed that all HP divisions were oriented perpendicular to the BM, while a typical bimodal distribution was seen in the IFE (Fig. 9E). Surprisingly, however, whereas LGN loss eliminated perpendicular divisions in the IFE, all HP divisions remained perpendicular in the absence of LGN (Fig. 9E).
Fig. 9.
LGN is not required for oriented cell divisions in HFs. (A,B) Merged (top) and single-channel (middle for DAPI, bottom for LGN) images of early stage (pHH3+, A) and late stage (survivin+, B) mitotic cells in interfollicular epidermis (IFE, left) and hair placodes (HPs) (right). Yellow dashed outlines indicate mitotic cells; double arrows (B) indicate division orientation for telophase cells; solid arrowheads indicate apical LGN; open arrowheads indicate basal LGN. (C) Quantification of LGN localization patterns in perpendicular divisions in epidermis, HPs and hair pegs at both early and late stages of mitosis. LGN does not polarize apically as frequently in HFs at either stage compared with IFE. (D) LGN apical:basal ratio for epidermis (IFE), HP and dermis (Der) in prometaphase mitotic versus interphase cells. No polarization is observed in dermis or interphase cells, while stronger apical enrichment is observed in IFE compared with HPs. (E) Division orientation in IFE (left) and HP (right), showing LGN-dependent loss of perpendicular divisions in IFE but not HP. (F) Division orientation by position at hair peg stage. Most divisions in the upper HF are parallel, whereas most in the lower HF are perpendicular to the BM. No obvious bias in division orientation was observed between anterior and posterior regions. (G) Division orientation in E17.5 wild-type and LGN knockdown HFs plotted by position (depth). Divisions in the lower 25% are mostly perpendicular, in contrast to the upper HF where they are mostly parallel. (H) Radial histograms for upper and lower HF populations. LGN loss has no effect in the upper HF but causes a slight reduction in perpendicular divisions in the lower HF. P-values determined by χ2 test (C,E,H) or Mann–Whitney test (D). Scale bars: 10 µm.
We next examined division orientation in more mature peg stage HFs. Although most mitotic HF cells lacked detectable LGN (Fig. 9C), those cells that did polarize LGN were generally located in the lower HF. We observed a striking difference in division orientation between the upper (Sox9+/Pcad−) and lower (Sox9−/Pcad+) HF, with perpendicular divisions occurring almost exclusively in the lower HF (Fig. 9F,G). No significant differences in orientation preference were observed between the anterior and posterior faces (Fig. 9F). LGN loss did not alter division orientation in the upper HF, where most divisions remained parallel to the BM, but caused a mild reduction in perpendicular divisions in the lower HF (Fig. 9G,H). Collectively, these data reveal that even among similar epithelial appendages, the role of LGN in OCDs is context dependent. Although LGN plays an essential role in directing OCDs in the developing tongue, it appears to be dispensable for embryonic HF morphogenesis (Fig. 10).
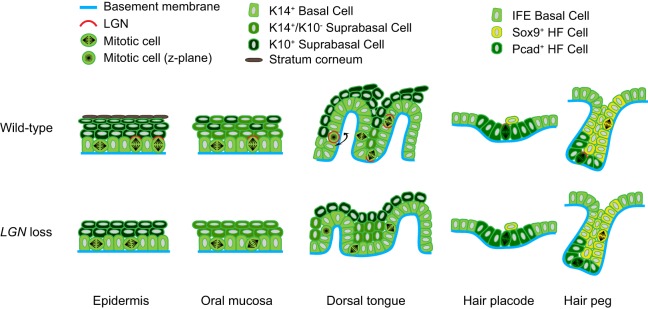
Fig. 10.
Model of LGN function in epithelia and placodes. Wild-type (top) and LGN knockdown (bottom) epithelial tissues at ∼E17.5. Representative oriented cell divisions (OCDs) are shown, with LGN localization patterns in red. Perpendicular OCDs with apical LGN occur in ∼60-65% of epidermal divisions and ∼50% of OE divisions. LGN loss biases toward parallel/oblique divisions, being more severe in epidermis than OE, and accompanied by impaired stratification (epithelial thinning) and differentiation (reduced or loss of K10 expression). Three types of OCD are observed in DT: circumferential, vertical and, more rarely, perpendicular. We propose that different patterns of LGN localization distinguish them (e.g. uniform is circumferential, absent is perpendicular, basal/bipolar is vertical). LGN loss causes randomization of division orientation, reducing papilla height and favoring IPE over papilla fate. In HPs, LGN is weakly and infrequently polarized apically, while in the upper (Sox9+) HF the mitotic cells do not express LGN and divide parallel to the BM. In the lower (Pcad+/Sox9−) HF divisions are mostly perpendicular to the BM. LGN loss randomizes divisions in the lower region, but has no effect on the upper region or in HPs.
DISCUSSION
Recent studies have shown that, in different contexts, LGN plays diverse roles in spindle orientation and cell fate choices. In both the spinal cord and cerebral cortex, as in the majority of DT divisions, LGN localizes to lateral domains, where LGN loss increases oblique/perpendicular divisions, suggesting that LGN normally promotes planar divisions (Morin et al., 2007; Konno et al., 2008; Peyre et al., 2011; Shitamukai et al., 2011). However, in epidermis, we and others have shown that LGN forms an apical complex with Insc, Gαi3, NuMA, dynactin and Par3 and that LGN loss leads to loss of perpendicular divisions, impaired Notch signaling and lethal defects in epidermal barrier formation and differentiation (Lechler and Fuchs, 2005; Poulson and Lechler, 2010; Williams et al., 2011, 2014). Similar roles for LGN in promoting perpendicular/asymmetric divisions have been reported in retina and mammary gland (Ballard et al., 2015; Chiu et al., 2016; Lacomme et al., 2016). The molecular mechanisms that regulate the differential subcellular localization of LGN in progenitors undergoing both symmetric and asymmetric cell divisions remain largely unknown. Here we have examined one candidate and show a weaker concordance between LGN and Gαi3 in DT than in other regions. Other potential regulators of LGN cortical localization include Insc, the LGN homolog Ags3 (Gpsm1), Sapcd2, afadin (Mllt4), 4.1G/R (Epb41l2/Epb41), 14-3-3 proteins, Par3 and aPKC (Prkcι) (Sanada and Tsai, 2005; Hao et al., 2010; Postiglione et al., 2011; Kiyomitsu and Cheeseman, 2013; Niessen et al., 2013; Seldin et al., 2013; Williams et al., 2014; Carminati et al., 2016; Chiu et al., 2016). Owing to the diverse patterns of LGN localization found in filiform papillae, we believe that the DT represents an ideal system with which to address this important question in future studies.
During embryogenesis, Lhx2 and Sox9 are among the earliest known transcription factors expressed in HPs, and are found in complementary patterns, with Lhx2 colocalizing with basal Pcad, and Sox9 positioned suprabasally (Rhee et al., 2006; Nowak et al., 2008). More recent studies have shown that the Pcad+/Lhx2+ population expresses Shh and high levels of Wnts, whereas the Sox9+ population is Wntlo (Ouspenskaia et al., 2016). Genetic and lineage-tracing studies have shown that Sox9+ HFSCs specified during embryogenesis later take up residence in the bulge niche where they contribute to all postnatal HF lineages (Nowak et al., 2008). Label-retention and lineage-tracing studies of HFSCs at the telogen to anagen transition have suggested that bulge stem cells most frequently divide symmetrically, whereas matrix transit amplifying cells divide asymmetrically (Zhang et al., 2009). Live imaging of postnatal HFs has shown that activated stem cells that have left the bulge and populate the outer root sheath divide centripetally (perpendicular to the axis of the HF), whereas in the matrix the division orientation is randomized (Rompolas et al., 2013). These studies demonstrate that different regions of the postnatal HF exhibit distinct patterns of OCDs.
The functional importance of OCDs in postnatal HF morphogenesis has recently been shown through analysis of mice lacking Prkcι and NuMA (Niessen et al., 2013; Seldin et al., 2016). In epidermis and HFs lacking Prkcι, a ∼20% increase in perpendicular divisions is observed, leading to prolonged anagen, progressive loss of quiescent bulge HFSCs and, ultimately, hair loss (Niessen et al., 2013). It is presently unknown whether LGN directs these OCDs, although other proteins that colocalize with LGN, including Par3 and NuMA, appear to be expressed normally. Deletion of the microtubule-binding domain of NuMA (NuMAΔMTBD) in matrix transit amplifying cells leads to a decrease in perpendicular divisions, defects in hair shaft differentiation and hair loss (Seldin et al., 2016). Since NuMAΔMTBD retains its interaction domain with LGN and thus its ability to be recruited to the cell cortex, it is unclear whether LGN loss in the matrix would cause a similar phenotype. Since epidermal LGN knockdown in our hands leads to neonatal lethality (Williams et al., 2011), this will be difficult to address experimentally. However, our analyses show that LGN loss leads to randomization of division orientation specifically in the lower HF at late embryonic stages, suggesting that later defects could occur.
Interestingly, in early HPs it was reported that nearly all divisions are perpendicular to the BM and asymmetric, leading to the production of one Pcad+ basal daughter and one Sox9+ suprabasal daughter (Ouspenskaia et al., 2016). This study also reported that LGN is expressed in placode cells undergoing perpendicular divisions (Ouspenskaia et al., 2016), although our quantitative analyses (Fig. 9C) show that this appears to be a minority population. We also demonstrate that LGN is not required in the HP for perpendicular divisions to occur, or to maintain the proper balance of Sox9+ and Sox9− progenitors. Nonetheless, our results do agree with respect to the observed wild-type division orientations in both HPs and HFs. We both report that 100% of HP divisions are perpendicular, and that lower HF divisions [where the Wnthi/Pcad+/Sox9− cells described by Ouspenskaia et al. (2016) reside] are biased toward perpendicular/asymmetric divisions, whereas upper HF divisions (where Sox9+ cells reside) are mostly planar/symmetric. Although our interpretation of these results seems to be contradictory to that of Ouspenskaia et al. (2016), we propose that they can be unified by assuming that perpendicular divisions in the HP yield asymmetric fates but do not utilize LGN. This can be explained by the existence of either an active LGN-independent spindle orientation mechanism in HFs, or a passive mechanism whereby perpendicular divisions represent a default state. We favor the latter model and propose that this may be attributed to ‘Hertwig's rule', which stipulates that cells preferentially divide along their long axis (Hertwig, 1884). Recent studies have confirmed Hertwig's observation by utilizing micropatterned substrates to manipulate cell shape and tension to observe how they influence division orientation (Thery et al., 2007). We postulate that because early placode cells are unusually columnar compared with neighboring IFE, they might not require an active mechanism to orient their mitotic spindle. In addition, acquisition of asymmetric cell fates in the developing HF may not require OCDs, as they could be acquired by other means, as long as Wnthi and Wntlo cells remain juxtaposed (Ouspenskaia et al., 2016). Directed cell migration is one mechanism by which this asymmetry could be maintained irrespective of proliferation (Ahtiainen et al., 2014). Live imaging might be necessary to reveal the relative contributions of OCDs, migration, planar cell polarity and actomyosin dynamics to HF morphogenesis.
MATERIALS AND METHODS
Animals, RNAi and in utero delivery of lentivirus
Wild-type outbred CD1 mice (Charles River) were maintained in an AAALAC certified animal facility under IACUC approved protocols. Embryonic ages were determined using timed pregnancies, ultrasound and Theiler stages (Theiler, 1989). In utero lentiviral injection, LGN 1617 and scramble hairpins have been described previously (Beronja et al., 2010; Williams et al., 2011, 2014).
Immunohistochemistry, antibodies and imaging
Fresh frozen sections (8-10 µm, or 20 µm for z-series) were prepared with a Leica CM1950 cryostat, air dried for 20 min, fixed for 10 min with 4% paraformaldehyde, washed with PBS, and blocked for 1 h with gelatin block (5% normal donkey serum, 3% BSA, 8% gelatin, 0.05% Triton X-100 in PBS). Primary antibodies diluted in gelatin block were incubated overnight at 4°C, washed three times with PBS, incubated 2 h with secondary antibodies at room temperature, counterstained with DAPI for 5 min at room temperature, and mounted in ProLong Gold (Invitrogen). Wholemount tongues were fixed for 1 h, except for analyses of LGN localization (5 min), before immunostaining using a modified procedure with longer incubation times (2 days for primary and secondary antibodies). Images were acquired using LAS AF software on a Leica TCS SPE-II 4 laser confocal system on a DM5500 microscope with ACS Apochromat 20×/0.60 multi-immersion, ACS Apochromat 40×/1.15 oil, and ACS Apochromat 63×/1.30 oil objectives. Tile-scanned images were acquired at 20×, and stitching was completed automatically by LAS AF. For wholemount imaging, z-volume was determined using an epithelial marker, such as integrin β4 or Ecad, and optical slices were taken at system-optimized intervals determined by the LAS X software or manually manipulated for thicker slices to preserve the fluorescent signal. Detailed descriptions of antibodies, immunostaining procedures and image analysis are provided in the supplementary Materials and Methods.
Scanning electron microscopy
SEM of DT from wild-type and LGN 1617-transduced E18.5 mice was performed using a Zeiss Supra 25 as detailed in the supplementary Materials and Methods.
Graphs and statistics
All statistical analyses, graphs and radial histograms were generated using GraphPad Prism 5/6 and Origin 2015 (OriginLab). Error bars represent s.e.m. unless otherwise indicated. Statistical analyses of numerical data were performed by Mann–Whitney U-test (non-parametric) or Student's t-tests (parametric), depending on whether the distributed data passed D'Agostino-Pearson and Shapiro-Wilk normality tests. Statistical analyses of enrichment (Fig. 6H, Fig. 7F,G and Fig. 8G) utilized a binomial test to compare with expected (control) distributions. Categorical values were analyzed by chi-square tests (e.g. Fig. 2D, Fig. 6C,G, Fig. 7E and Fig. 9C). For Tukey box-and-whisker plots (Fig. 2G, Fig. 5F,H, Fig. S2A), the box represents the interquartile range (IQR, defined by 25th-75th percentiles) and the horizontal line represents the median. Whiskers represent 1.5×IQR unless this is greater than the minimum or maximum value. Figures were assembled using Adobe Photoshop and Illustrator CS6.
Acknowledgements
We thank the UNC Microscopy Services Laboratory staff (Robert Bagnell, Pablo Ariel, Victoria Madden and Kristen White) for assistance with scanning electron microscopy, and expertise and guidance in light microscopy and image analysis; Michelle Mac for technical assistance and management of the mouse colony; Meredith Price at Bitplane for training our lab on Imaris 7.6.5 and for generating the Python scripts used for this project; Ophir Klein, Susan Henning and members of the Antonio Amelio and S.E.W. labs for helpful comments and suggestions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
K.M.B. designed and conducted experiments and analyzed data, with assistance and supervision from S.E.W. K.J.L. and T.A.C. oversaw management of the mouse facility and performed lentiviral injections. K.J.L. assisted in making lentivirus, aided in experimental design and analysis, and conceptualized the application and analysis of the radial heat maps. J.H.P. assisted with performing experiments, image acquisition and data analysis. C.P.D. performed analysis of LGN fluorescence intensity. S.E.W. and K.M.B. wrote the manuscript, with input from all authors.
Funding
K.M.B. is supported by a National Institutes of Health T90 Training Grant [NIH/NIDCR 5T90DE021986-05] and NIH Loan Repayment Plan [NOT-OD-15-122]. S.E.W. is supported by a Kimmel Scholar Award [SKF-15-065] from the Sidney Kimmel Foundation for Cancer Research and an IBM Junior Faculty Development Award. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.136010.supplemental
References
- Agrawal N., Frederick M. J., Pickering C. R., Bettegowda C., Chang K., Li R. J., Fakhry C., Xie T.-X., Zhang J., Wang J. et al. (2011). Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333, 1154-1157. 10.1126/science.1206923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahtiainen L., Lefebvre S., Lindfors P. H., Renvoisé E., Shirokova V., Vartiainen M. K., Thesleff I. and Mikkola M. L. (2014). Directional cell migration, but not proliferation, drives hair placode morphogenesis. Dev. Cell 28, 588-602. 10.1016/j.devcel.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Arwert E. N., Hoste E. and Watt F. M. (2012). Epithelial stem cells, wound healing and cancer. Nat. Rev. Cancer 12, 170-180. 10.1038/nrc3217 [DOI] [PubMed] [Google Scholar]
- Ballard M. S., Zhu A., Iwai N., Stensrud M., Mapps A., Postiglione M. P., Knoblich J. A. and Hinck L. (2015). Mammary stem cell self-renewal is regulated by Slit2/Robo1 signaling through SNAI1 and mINSC. Cell Rep. 13, 290-301. 10.1016/j.celrep.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratz R. S. and Farbman A. I. (1975). Morphogenesis of rat lingual filiform papillae. Am. J. Anat. 143, 283-301. 10.1002/aja.1001430303 [DOI] [PubMed] [Google Scholar]
- Barlow L. A. (2015). Progress and renewal in gustation: new insights into taste bud development. Development 142, 3620-3629. 10.1242/dev.120394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B. and Blanpain C. (2012). Mechanisms regulating epidermal stem cells. EMBO J. 31, 2067-2075. 10.1038/emboj.2012.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beronja S., Livshits G., Williams S. and Fuchs E. (2010). Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 16, 821-827. 10.1038/nm.2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., Lowry W. E., Pasolli H. A. and Fuchs E. (2006). Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 20, 3022-3035. 10.1101/gad.1477606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne C., Tainsky M. and Fuchs E. (1994). Programming gene expression in developing epidermis. Development 120, 2369-2383. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. (2015). Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576-582. 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati M., Gallini S., Pirovano L., Alfieri A., Bisi S. and Mapelli M. (2016). Concomitant binding of Afadin to LGN and F-actin directs planar spindle orientation. Nat. Struct. Mol. Biol. 23, 155-163. 10.1038/nsmb.3152 [DOI] [PubMed] [Google Scholar]
- Casey L. M., Lan Y., Cho E.-S., Maltby K. M., Gridley T. and Jiang R. (2006). Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev. Dyn. 235, 1830-1844. 10.1002/dvdy.20821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C. W. N., Monat C., Robitaille M., Lacomme M., Daulat A. M., Macleod G., McNeill H., Cayouette M. and Angers S. (2016). SAPCD2 controls spindle orientation and asymmetric divisions by negatively regulating the Galphai-LGN-NuMA ternary complex. Dev. Cell 36, 50-62. 10.1016/j.devcel.2015.12.016 [DOI] [PubMed] [Google Scholar]
- Clayton E., Doupé D. P., Klein A. M., Winton D. J., Simons B. D. and Jones P. H. (2007). A single type of progenitor cell maintains normal epidermis. Nature 446, 185-189. 10.1038/nature05574 [DOI] [PubMed] [Google Scholar]
- Culurgioni S. and Mapelli M. (2013). Going vertical: functional role and working principles of the protein Inscuteable in asymmetric cell divisions. Cell. Mol. Life Sci. 70, 4039-4046. 10.1007/s00018-013-1319-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culurgioni S., Alfieri A., Pendolino V., Laddomada F. and Mapelli M. (2011). Inscuteable and NuMA proteins bind competitively to Leu-Gly-Asn repeat-enriched protein (LGN) during asymmetric cell divisions. Proc. Natl. Acad. Sci. USA 108, 20998-21003. 10.1073/pnas.1113077108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenport D. and Fuchs E. (2008). Planar polarization in embryonic epidermis orchestrates global asymmetric morphogenesis of hair follicles. Nat. Cell Biol. 10, 1257-1268. 10.1038/ncb1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWard A. D., Cramer J. and Lagasse E. (2014). Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 9, 701-711. 10.1016/j.celrep.2014.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P. (2009). Crosstalk of Notch with p53 and p63 in cancer growth control. Nat. Rev. Cancer 9, 587-595. 10.1038/nrc2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q. and Macara I. G. (2004). Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119, 503-516. 10.1016/j.cell.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Fuchs E. (2007). Scratching the surface of skin development. Nature 445, 834-842. 10.1038/nature05659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P. and Hyman A. A. (1996). Cortical domains and the mechanisms of asymmetric cell division. Trends Cell Biol. 6, 382-387. 10.1016/0962-8924(96)10035-0 [DOI] [PubMed] [Google Scholar]
- Gregorieff A. and Clevers H. (2005). Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 19, 877-890. 10.1101/gad.1295405 [DOI] [PubMed] [Google Scholar]
- Gurvits G. E. and Tan A. (2014). Black hairy tongue syndrome. World J. Gastroenterol. 20, 10845-10850. 10.3748/wjg.v20.i31.10845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Du Q., Chen X., Zheng Z., Balsbaugh J. L., Maitra S., Shabanowitz J., Hunt D. F. and Macara I. G. (2010). Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr. Biol. 20, 1809-1818. 10.1016/j.cub.2010.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriges M. and Morrisey E. E. (2014). Lung development: orchestrating the generation and regeneration of a complex organ. Development 141, 502-513. 10.1242/dev.098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertwig O. (1884). Das Problem der Befruchtung und der Isotropie des Eies, eine Theorie der Vererbung. Jena: Verlag von Gustav Fischer. [Google Scholar]
- Huebner A. J., Dai D., Morasso M., Schmidt E. E., Schäfer M., Werner S. and Roop D. R. (2012). Amniotic fluid activates the nrf2/keap1 pathway to repair an epidermal barrier defect in utero. Dev. Cell 23, 1238-1246. 10.1016/j.devcel.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume W. J. and Potten C. S. (1976). The ordered columnar structure of mouse filiform papillae. J. Cell Sci. 22, 149-160. [DOI] [PubMed] [Google Scholar]
- Johnston C. A., Hirono K., Prehoda K. E. and Doe C. Q. (2009). Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell 138, 1150-1163. 10.1016/j.cell.2009.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. B. and Klein O. D. (2013). Oral epithelial stem cells in tissue maintenance and disease: the first steps in a long journey. Int. J. Oral Sci. 5, 121-129. 10.1038/ijos.2013.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T. and Cheeseman I. M. (2013). Cortical Dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell 154, 391-402. 10.1016/j.cell.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J. A. (2008). Mechanisms of asymmetric stem cell division. Cell 132, 583-597. 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Konno D., Shioi G., Shitamukai A., Mori A., Kiyonari H., Miyata T. and Matsuzaki F. (2008). Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat. Cell Biol. 10, 93-101. 10.1038/ncb1673 [DOI] [PubMed] [Google Scholar]
- Kopan R. and Fuchs E. (1989). A new look into an old problem: keratins as tools to investigate determination, morphogenesis, and differentiation in skin. Genes Dev. 3, 1-15. 10.1101/gad.3.1.1 [DOI] [PubMed] [Google Scholar]
- Kumar V., Bouameur J.-E., Bär J., Rice R. H., Hornig-Do H.-T., Roop D. R., Schwarz N., Brodesser S., Thiering S., Leube R. E. et al. (2015). A keratin scaffold regulates epidermal barrier formation, mitochondrial lipid composition, and activity. J. Cell Biol. 211, 1057-1075. 10.1083/jcb.201404147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme M., Tarchini B., Boudreau-Pinsonneault C., Monat C. and Cayouette M. (2016). The LGN protein promotes planar proliferative divisions in the neocortex but apicobasal asymmetric terminal divisions in the retina. Development 143, 575-581. 10.1242/dev.129783 [DOI] [PubMed] [Google Scholar]
- Lechler T. and Fuchs E. (2005). Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, 275-280. 10.1038/nature03922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P., Lee D.-J., Chan C., Chen S.-W., Ch'en I. and Jamora C. (2009). Dynamic expression of epidermal caspase 8 simulates a wound healing response. Nature 458, 519-523. 10.1038/nature07687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M. S. and Johnston C. A. (2013). Molecular pathways regulating mitotic spindle orientation in animal cells. Development 140, 1843-1856. 10.1242/dev.087627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzalupo S. and Coulombe P. A. (2001). A reporter transgene based on a human keratin 6 gene promoter is specifically expressed in the periderm of mouse embryos. Mech. Dev. 100, 65-69. 10.1016/S0925-4773(00)00489-5 [DOI] [PubMed] [Google Scholar]
- Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R. and Bradley A. (1999). p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708-713. 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- Morin X. and Bellaiche Y. (2011). Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell 21, 102-119. 10.1016/j.devcel.2011.06.012 [DOI] [PubMed] [Google Scholar]
- Morin X., Jaouen F. and Durbec P. (2007). Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat. Neurosci. 10, 1440-1448. 10.1038/nn1984 [DOI] [PubMed] [Google Scholar]
- Nicolas M., Wolfer A., Raj K., Kummer J. A., Mill P., van Noort M., Hui C.-C., Clevers H., Dotto G. P. and Radtke F. (2003). Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 33, 416-421. 10.1038/ng1099 [DOI] [PubMed] [Google Scholar]
- Niessen M. T., Scott J., Zielinski J. G., Vorhagen S., Sotiropoulou P. A., Blanpain C., Leitges M. and Niessen C. M. (2013). aPKClambda controls epidermal homeostasis and stem cell fate through regulation of division orientation. J. Cell Biol. 202, 887-900. 10.1083/jcb.201307001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak J. A., Polak L., Pasolli H. A. and Fuchs E. (2008). Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell 3, 33-43. 10.1016/j.stem.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo T., Clark C. and Hogan B. L. M. (2009). Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells 27, 442-450. 10.1634/stemcells.2008-0611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouspenskaia T., Matos I., Mertz A. F., Fiore V. F. and Fuchs E. (2016). WNT-SHH antagonism specifies and expands stem cells prior to niche formation. Cell 164, 156-169. 10.1016/j.cell.2015.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panousopoulou E. and Green J. B. A. (2014). Spindle orientation processes in epithelial growth and organisation. Semin. Cell Dev. Biol. 34, 124-132. 10.1016/j.semcdb.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Paus R., Muller-Rover S., Van Der Veen C., Maurer M., Eichmuller S., Ling G., Hofmann U., Foitzik K., Mecklenburg L. and Handjiski B. (1999). A comprehensive guide for the recognition and classification of distinct stages of hair follicle morphogenesis. J. Invest. Dermatol. 113, 523-532. 10.1046/j.1523-1747.1999.00740.x [DOI] [PubMed] [Google Scholar]
- Peyre E., Jaouen F., Saadaoui M., Haren L., Merdes A., Durbec P. and Morin X. (2011). A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J. Cell Biol. 193, 141-154. 10.1083/jcb.201101039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postiglione M. P., Jüschke C., Xie Y., Haas G. A., Charalambous C. and Knoblich J. A. (2011). Mouse inscuteable induces apical-Basal spindle orientation to facilitate intermediate progenitor generation in the developing neocortex. Neuron 72, 269-284. 10.1016/j.neuron.2011.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulson N. D. and Lechler T. (2010). Robust control of mitotic spindle orientation in the developing epidermis. J. Cell Biol. 191, 915-922. 10.1083/jcb.201008001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamy B. V., Derby R. and Bunt C. W. (2010). Common tongue conditions in primary care. Am. Fam. Phys. 81, 627-634. [PubMed] [Google Scholar]
- Rhee H., Polak L. and Fuchs E. (2006). Lhx2 maintains stem cell character in hair follicles. Science 312, 1946-1949. 10.1126/science.1128004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson R. J., Dixon J., Jiang R. and Dixon M. J. (2009). Integration of IRF6 and Jagged2 signalling is essential for controlling palatal adhesion and fusion competence. Hum. Mol. Genet. 18, 2632-2642. 10.1093/hmg/ddp201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J. R. and Hogan B. L. M. (2011). Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 27, 493-512. 10.1146/annurev-cellbio-100109-104040 [DOI] [PubMed] [Google Scholar]
- Rompolas P., Mesa K. R. and Greco V. (2013). Spatial organization within a niche as a determinant of stem-cell fate. Nature 502, 513-518. 10.1038/nature12602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosekrans S. L., Baan B., Muncan V. and van den Brink G. R. (2015). Esophageal development and epithelial homeostasis. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G216-G228. 10.1152/ajpgi.00088.2015 [DOI] [PubMed] [Google Scholar]
- Sanada K. and Tsai L.-H. (2005). G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell 122, 119-131. 10.1016/j.cell.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Schaefer M., Petronczki M., Dorner D., Forte M. and Knoblich J. A. (2001). Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell 107, 183-194. 10.1016/S0092-8674(01)00521-9 [DOI] [PubMed] [Google Scholar]
- Seldin L., Poulson N. D., Foote H. P. and Lechler T. (2013). NuMA localization, stability, and function in spindle orientation involve 4.1 and Cdk1 interactions. Mol. Biol. Cell 24, 3651-3662. 10.1091/mbc.E13-05-0277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin L., Muroyama A. and Lechler T. (2016). NuMA-microtubule interactions are critical for spindle orientation and the morphogenesis of diverse epidermal structures. Elife 5, e12504 10.7554/eLife.12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitamukai A., Konno D. and Matsuzaki F. (2011). Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J. Neurosci. 31, 3683-3695. 10.1523/JNEUROSCI.4773-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K. H. and Doe C. Q. (2009). Spindle orientation during asymmetric cell division. Nat. Cell Biol. 11, 365-374. 10.1038/ncb0409-365 [DOI] [PubMed] [Google Scholar]
- Smart I. H. M. (1970). Variation in the plane of cell cleavage during the process of stratification in the mouse epidermis. Br. J. Dermatol. 82, 276-282. 10.1111/j.1365-2133.1970.tb12437.x [DOI] [PubMed] [Google Scholar]
- Stransky N., Egloff A. M., Tward A. D., Kostic A. D., Cibulskis K., Sivachenko A., Kryukov G. V., Lawrence M. S., Sougnez C., McKenna A. et al. (2011). The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157-1160. 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumigray K. D. and Lechler T. (2015). Cell adhesion in epidermal development and barrier formation. Curr. Top. Dev. Biol. 112, 383-414. 10.1016/bs.ctdb.2014.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Komai Y., Tokuyama Y., Yanai H., Ohe S., Okazaki K. and Ueno H. (2013). Identification of stem cells that maintain and regenerate lingual keratinized epithelial cells. Nat. Cell Biol. 15, 511-518. 10.1038/ncb2719 [DOI] [PubMed] [Google Scholar]
- Theiler K. (1989). The House Mouse: Atlas of Embryonic Development. New York: Springer-Verlag. [Google Scholar]
- Thery M., Jimenez-Dalmaroni A., Racine V., Bornens M. and Julicher F. (2007). Experimental and theoretical study of mitotic spindle orientation. Nature 447, 493-496. 10.1038/nature05786 [DOI] [PubMed] [Google Scholar]
- Thesleff I. and Mikkola M. L. (2014). Development of ectodermal organs. Semin. Cell Dev. Biol. 25-26, 1-2. 10.1016/j.semcdb.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Tinkle C. L., Lechler T., Pasolli H. A. and Fuchs E. (2004). Conditional targeting of E-cadherin in skin: insights into hyperproliferative and degenerative responses. Proc. Natl. Acad. Sci. USA 101, 552-557. 10.1073/pnas.0307437100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. M. and Spence J. R. (2014). How to make an intestine. Development 141, 752-760. 10.1242/dev.097386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard F. S., Kimple R. J. and Siderovski D. P. (2004). Return of the GDI: the GoLoco motif in cell division. Annu. Rev. Biochem. 73, 925-951. 10.1146/annurev.biochem.73.011303.073756 [DOI] [PubMed] [Google Scholar]
- Williams S. E. and Fuchs E. (2013). Oriented divisions, fate decisions. Curr. Opin. Cell Biol. 25, 749-758. 10.1016/j.ceb.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. E., Beronja S., Pasolli H. A. and Fuchs E. (2011). Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature 470, 353-358. 10.1038/nature09793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. E., Ratliff L. A., Postiglione M. P., Knoblich J. A. and Fuchs E. (2014). Par3-mInsc and Galphai3 cooperate to promote oriented epidermal cell divisions through LGN. Nat. Cell Biol. 16, 758-769. 10.1038/ncb3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R. T., Tabin C., Sharpe A., Caput D., Crum C. et al. (1999). p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714-718. 10.1038/19539 [DOI] [PubMed] [Google Scholar]
- Yuzawa S., Kamakura S., Iwakiri Y., Hayase J. and Sumimoto H. (2011). Structural basis for interaction between the conserved cell polarity proteins Inscuteable and Leu-Gly-Asn repeat-enriched protein (LGN). Proc. Natl. Acad. Sci. USA 108, 19210-19215. 10.1073/pnas.1110951108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. V., Cheong J., Ciapurin N., McDermitt D. J. and Tumbar T. (2009). Distinct self-renewal and differentiation phases in the niche of infrequently dividing hair follicle stem cells. Cell Stem Cell 5, 267-278. 10.1016/j.stem.2009.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Wen W., Zheng Z., Shang Y., Wei Z., Xiao Z., Pan Z., Du Q., Wang W. and Zhang M. (2011). LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and Galphai/LGN/NuMA pathways. Mol. Cell 43, 418-431. 10.1016/j.molcel.2011.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]