Abstract
Fanconi anemia (FA) is a recessive genetic disease characterized by congenital abnormalities, chromosome instability, progressive bone marrow failure (BMF), and a strong predisposition to cancer. Twenty FA genes have been identified, and the FANC proteins they encode cooperate in a common pathway that regulates DNA crosslink repair and replication fork stability. We identified a child with severe BMF who harbored biallelic inactivating mutations of the translesion DNA synthesis (TLS) gene REV7 (also known as MAD2L2), which encodes the mutant REV7 protein REV7-V85E. Patient-derived cells demonstrated an extended FA phenotype, which included increased chromosome breaks and G2/M accumulation upon exposure to DNA crosslinking agents, γH2AX and 53BP1 foci accumulation, and enhanced p53/p21 activation relative to cells derived from healthy patients. Expression of WT REV7 restored normal cellular and functional phenotypes in the patient’s cells, and CRISPR/Cas9 inactivation of REV7 in a non-FA human cell line produced an FA phenotype. Finally, silencing Rev7 in primary hematopoietic cells impaired progenitor function, suggesting that the DNA repair defect underlies the development of BMF in FA. Taken together, our genetic and functional analyses identified REV7 as a previously undescribed FA gene, which we term FANCV.
Introduction
Fanconi anemia (FA) is the most frequent cause of inherited bone marrow failure (IBMF) syndromes (1, 2). Twenty FA genes have been identified, including FANCA, FANCB, FANCC, FANCD1 (also known as BRCA2), FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ (also known as BRIP1 or BACH1), FANCL, FANCM, FANCN (also known as PALB2), FANCO (also known as RAD51C), FANCP (also known as SLX4), FANCQ (also known as XPF or ERCC4), FANCR (also known as RAD51), FANCS (also known as BRCA1), FANCT (also known as UBE2T), and FANCU (also known as XRCC2) (3–5). The products of these genes cooperate in a unique FA/BRCA pathway, regulating the response to physiological stress or exposure to genotoxic agents and maintaining genome integrity (3–5). Most FA patients develop a progressive bone marrow failure (BMF) during childhood due to the depletion or impairment of the hematopoietic stem cell (HSC) pool (1, 2, 6–8).
Here, we identified biallelic mutations in REV7 (also known as MAD2L2) in a child with a classic presentation of FA. Interestingly, REV7 has several cellular functions including translesion DNA synthesis (TLS) (9, 10), mitotic checkpoint regulation (11, 12), and DNA repair pathway choice (13, 14). Which if any of these functions of REV7 is required for suppressing the FA cellular and developmental phenotypes is unknown.
Results and Discussion
A child with a clinical and cellular FA phenotype and a constitutive REV7 mutation.
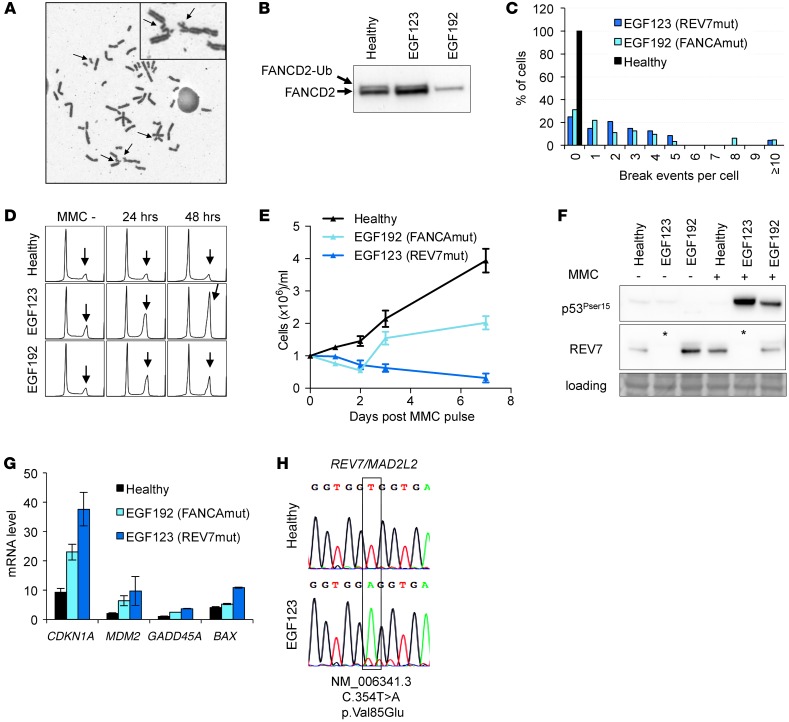
Patient EGF123, an 8-year-old girl, presented with severe BMF involving all 3 lineages (hemoglobin, 8.0 g/dl; neutrophil count, 0.43 × 109/l; and platelets, 10 × 109/l). She exhibited FA physical signs (short size at less than tenth percentile, microcephaly, and abnormal facial features), a renal tubulopathy, elevated serum α-fetoprotein, and a positive mitomycine C (MMC) chromosome breakage test of blood lymphocytes (15), establishing a definitive diagnosis of FA (Figure 1A). Monoubiquitination of the FANCD2 protein in the patient cells suggested an abnormality downstream or independent of the FA core complex (Figure 1B) (16). Functional analysis of the patient’s skin fibroblasts and EBV-transformed lymphoid cells confirmed the FA phenotype (i.e., a hypersensitivity to the interstrand-crosslinking [ICL] agent MMC), with increased chromosome radials (Figure 1C), arrest at the G2 phase of the cell cycle, and growth inhibition (Figure 1, D and E). Consistent with previous studies with FA cells (7, 17), MMC exposure strongly activated Ser15 phosphorylation of p53 and CDKN1A (also known as p21) transcriptional induction (Figure 1, F and G).
Figure 1. Genetic and cellular phenotype of the REV7-mutated patient EGF123.
(A) Metaphase EGF123 lymphoid cells upon MMC exposure; arrows show chromosome breaks. Original magnification, ×630. (B) Immunoblot analysis of FANCD2 monoubiquitination of the REV7-mutated (EGF123), FANCA-mutated (EGF192), and healthy fibroblasts. (C) Quantification of the MMC-induced breaks per cell in REV7-mutated (EGF123) and FANCA-mutated (EGF192) EBV-transformed cells and cells of a healthy subject. (D) Cell-cycle analysis after MMC pulse; arrows show the late S/G2 arrest. (E) Proliferation curves after MMC pulse. (F) Protein immunoblot analysis before (–) and 24 hours after (+) MMC pulse; asterisks underline the absence of REV7 in EGF123 protein extracts. (G) Transcript expression levels of a set of DNA damage response genes analyzed by quantitative reverse-transcriptase PCR (qRT-PCR) relative to HPRT (primers are indicated in Supplemental Table 1). Experiments shown in panels E–H were performed using EBV-transformed cells. (H) Partial REV7 exon5 sequence in gDNA from primary fibroblasts.
Sequencing of known FANC genes failed to identify a deleterious nucleotide variant or deletion. Whole exome sequencing (WES) on genomic DNA from the EGF123 proband identified a homozygous REV7 variant, c.354T>A. Sanger resequencing confirmed the homozygous variant in EFG123 skin fibroblast and EBV-transformed cells (Figure 1H). Rare regions of germline homozygosity (which included the REV7 gene) were observed in this patient, consistent with distant consanguinity (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI88010DS1). The c.354 C>T REV7 is a variant based on a survey of publicly accessible variant databases. The mutation affects an amino acid at a highly conserved position, resulting in an amino acid substitution, p.V85E, predicted to be pathogenic by different tools. The substitution was located in the highly conserved HORMA domain of the protein (Supplemental Figure 2), known to mediate the REV7 interaction with REV1 and REV3 (10). Moreover, the absence of detectable levels of REV7 protein in several tissues, despite normal transcript levels, suggested that protein destabilization may result from the mutation (Figure 1F).
Lentiviral complementation of REV7 restores cellular and functional phenotype in patient cells.
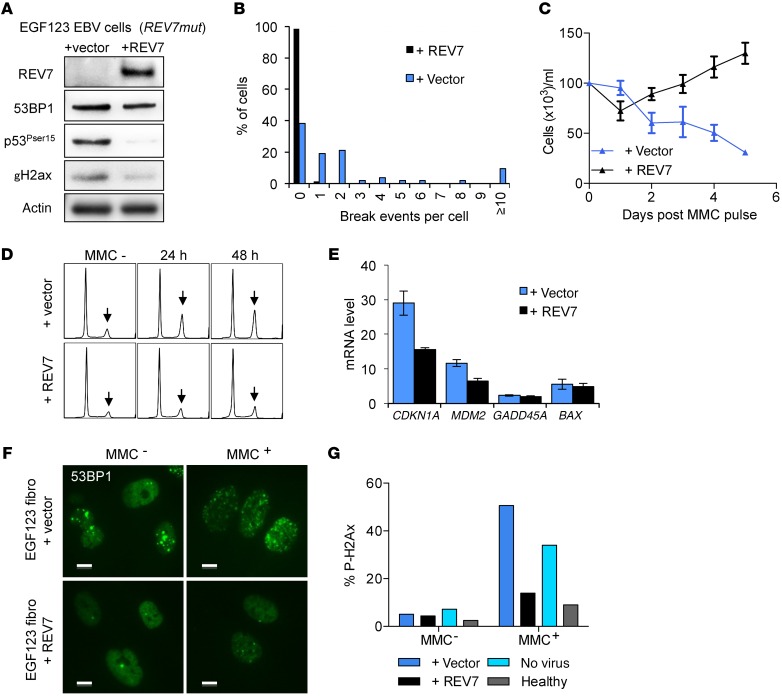
To provide additional evidence for the disease causality of the REV7 mutation, we lentivirally reexpressed a WT REV7 cDNA in patient fibroblasts and EBV-transformed cells. Western blot analysis of whole cell lysates revealed that REV7 expression was restored in REV7-transduced cells (Figure 2A). Reexpression of the WT REV7 fully rescued chromosome breakage, cell cycle arrest, and cell proliferation defects (Figure 2, B–D and Supplemental Figure 3). Ser15 phosphorylation of p53 and phosphorylated γH2ax levels was also decreased in the REV7 complemented cells, and transcript analysis revealed a decrease of CDKN1A (Figure 2, A and E). Moreover, EGF123 cells exhibited an excess of γH2AX and 53BP1 foci, suggesting unresolved DNA damage at steady state and upon MMC exposure, and these defects were resolved by REV7 reexpression (Figure 2, A, F, and G, and Supplemental Figure 4).
Figure 2. Correction of the extended phenotype of the EGF123 cells by WT REV7 expression.
(A) Protein immunoblot analysis 24 hours after MMC pulse of EBV-transformed cells from the patient transduced with REV7 or the empty vector. (B) Quantification of MMC-induced breaks per cell. (C) Proliferation curves after MMC pulse. (D) Cell-cycle analysis after MMC pulse; arrows show late S/G2 arrests. (E) Transcript expression levels of a set of DNA damage response genes analyzed by qRT-PCR relative to HPRT. (F) 53BP1 immunofluorescence analysis of EGF123 fibroblasts transduced with REV7 or empty vector, with or without MMC exposure. Scale bars: 10 μm. (G) Percentage of γH2AX-positive cells by flow cytometry analysis with or without MMC exposure. Experiments shown in panels B–F were performed using EBV-transformed cells.
CRISPR/Cas9-mediated knockout of REV7 recapitulates an FA phenotype.
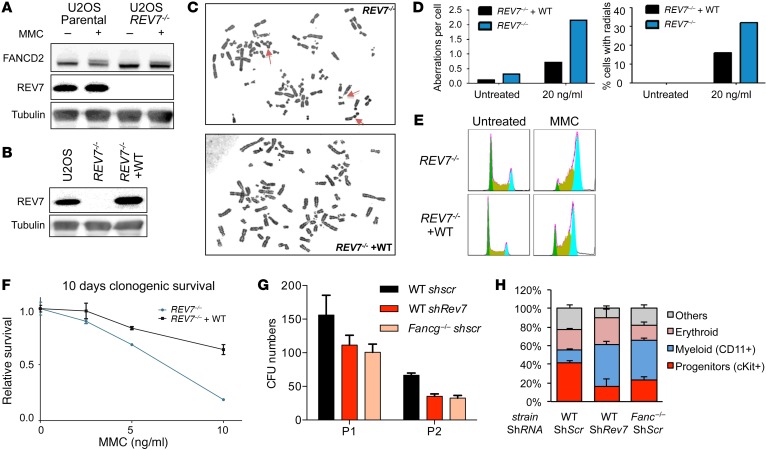
To confirm that loss of REV7 directly causes the FA phenotype, we used CRISPR/Cas9-mediated gene editing to generate a homozygous knockout in U2OS cells (denoted REV7–/–) (Figure 3). Western blotting confirmed the absence of detectable REV7 protein in the REV7–/– line, whereas REV7 was readily detectable in the parental cell line (Figure 3A). Consistent with the patient-derived EGF123 cells, FANCD2 monoubiquitination was largely unaffected in REV7–/– cells. As expected, REV7–/– cells exhibited the hallmark FA phenotypes, including an increase in chromosome breaks and radials upon MMC treatment, cellular hypersensitivity to MMC, and a pronounced G2/M arrest (Figure 3, C–F). All of these phenotypes were corrected by retroviral transduction with WT REV7 cDNA (Figure 3, B–F).
Figure 3. CRISPR/Cas9 knockout of REV7 recapitulates the FA phenotype.
(A) Western blot of whole cell lysates shows no detectable REV7 protein in the REV7–/– line. FANCD2 is still efficiently monoubiquitylated in response to 100 ng/ml MMC. (B) Western blot showing reconstitution of WT REV7 expression in REV7–/– cells. (C–D) Increase in chromosome aberrations (left graph) and radials (right) after 48-hour treatment with 20 ng/ml MMC as compared with cells complemented with WT REV7 cDNA. Original magnification, ×1,000. (E) Increase in G2/M arrest with and without MMC treatment (20 ng/ml). (F) Impaired clonogenic capacity of REV7–/– cells over 10-day treatment with MMC. (G) REV7 is required for normal hematopoiesis; CFU scoring of bone marrow Lin– cells after lentiviral silencing using REV7 or scramble (scr) shRNAs. Fancg-/- cells are used as FA cell control. CFUs were numbered after 7 days on methylcellulose; 2 passages (P1 and P2) were performed. (H) Cell population percentages in CFUs after 7 days of culture.
REV7 deficiency directly impairs hematopoietic cells.
Previous studies have indicated that FA proteins are required for normal hematopoiesis and hematopoietic progenitor cell (HPC) survival (6–8, 18). To evaluate whether REV7 deficiency can affect hematopoiesis, we silenced this gene in mouse HPCs. Rev7 knockdown impaired the ability of HPCs to form in vitro CFU in short-term methylcellulose culture, similar to the defect observed in Fancg–/– HPCs (Figure 3G). In addition, colonies had a decreased proportion of c-Kit+/progenitor cells and an increased proportion of CD11b+/differentiated cells (Figure 3H). Together, these data suggest that the Rev7 defect directly alters HPCs, leading to increased lineage engagement, consistent with a DNA damage–mediated mechanism of BMF.
FA cells are hypersensitive to DNA ICL agents and exhibit DNA repair and replication fork defects. How a deficiency in REV7 contributes to these cellular phenotypes remains unknown. Previous studies have demonstrated that the REV7 protein has several normal cellular functions. First, REV7 is a subunit of the DNA polymerase ζ complex (POL ζ), a TLS error-prone polymerase complex containing REV1, REV7, REV3, and 2 additional accessory subunits, POLD2 and POLD3 (9, 10, 19). POL ζ acts downstream of the FA core proteins in the repair of DNA ICL lesions (3–5, 20). In this role, REV7 acts as an adaptor protein between REV1 and REV3, the catalytic subunit of the POL ζ complex. A deficiency in REV7 could account for the well-known hypomutability of FA cells, resulting from a defect in TLS repair (20–23). Second, REV7 plays a role in mitosis by preventing premature activation of the anaphase promoting complex/cyclosome (APC/C) by the activation of CDH1 (11, 12). Accordingly, a deficiency in REV7 could account for the mitotic defects and cytokinesis failure observed in FA cells (24–26). Third, REV7 participates in the cellular choice of DNA repair pathways during DNA double-strand break repair. In this role, REV7 functions downstream of 53BP1 and RIF1 and inhibits the 5′ DNA end resection, thereby promoting nonhomologous end joining (NHEJ) and suppressing homologous recombination (HR) (13, 14). A deficiency in REV7 could therefore account, at least in part, for the dysregulated levels of NHEJ repair and HR repair observed in FA cells (27, 28).
Previous work in chicken DT40 cells had demonstrated that inactivation of Rev7 confers a cell hypersensitivity to ICL agents, suggesting that REV7 may be an FA gene (29). Interestingly, 2 Rev7 deficient mouse models have been generated, Rev7–/– and Rev7C70R mice (30, 31). In one model, Rev7–/– mice exhibited growth retardation and a partial embryonic lethal phenotype, and those mice that survived to adulthood were infertile and showed germ cell aplasia in the testes and ovaries (30). In the second model, a missense mutation in Rev7C70R disrupted Pol ζ assembly, thereby impairing mouse development and the repair of genotoxic agent–induced DNA lesions (31). Rev7C70R mutant cells also showed decreased proliferation, increased apoptosis, and arrest in the S phase with extensive γH2AX foci in nuclei, indicating accumulation of DNA damage (31). The Rev7-deficient mice, therefore, have a very similar defect in development, embryonic lethality, infertility, and DNA damage accumulation, which is also observed in other Fanc–/– mouse models (32).
In conclusion, these human genetic and functional data establish REV7 as a new bona fide FA gene, FANCV.
Methods
Further details can be found in the Supplemental Methods.
Samples and cell lines.
This study was based on a cohort of 268 consecutive FA patients referred for medical diagnosis at the French Reference Center for Constitutional Bone Marrow Failure, Saint-Louis and Robert Debré Hospitals. All patients had an FA diagnosis based on FA tests, including the chromosomal breakage test. Primary fibroblasts were established and cells were analyzed at early passage. Genetic analyses of the FANC gene mutations were performed in fibroblast genomic DNA. EBV-transformed cell lines were produced for functional analyses (7).
Exome sequencing.
All sequencing data were deposited in ArrayExpress (E-MTAB-4817).
CRISPR-Cas9 knockout.
Guide RNA sequences were cloned into the pSpCas9(BB)-2A-GFP (PX458) vector, a gift from Feng Zhang (Addgene plasmid no. 48138; Massachusetts Institute of Technology, Cambridge, Massachusetts, USA.) . The genomic sequence targeted for CRISPR-Cas9 disruption in REV7 was GAGGTCTTGTCGTGTGAGCG. U2OS cells were transfected using Lipofectamine 2000 (Invitrogen catalog 11668). Twenty-four hours after transfection, GFP+ cells were selected and single cells were seeded using a BD FACSAria II cell sorter. Single cells were grown for approximately 3 weeks. Putative REV7 knockouts were identified using Western blotting. For reconstitution, WT REV7 cDNA was cloned into the pBabe-puro retroviral vector.
Statistics.
Results were evaluated by performing unpaired 2-tailed t tests using GraphPadPrism version 5.0 software (GraphPad Software). Results are presented as mean ± SEM.
Study approval.
Informed consent for medical diagnosis and research was obtained from the patients and their relatives. The study was approved by the Institutional Review Board of Institut Universitaire d’Hématologie (IUH) at Saint-Louis Hospital (project IUH2012-12-078). All experiments in mice were performed in accordance with a protocol approved by the Committee on the Ethics of Animal Experiments Paris-Nord (C2EA-121), project No. 2014-IUH013.
Author contributions
DB, JMP, CC, AR, RC, CDE, WC, and SG performed experiments. All authors contributed to the research. DB, JMP, ADD, and JS designed the study and wrote the manuscript. All authors approved the final manuscript.
Supplementary Material
Acknowledgments
This work was supported by the European Research Council (ERC) Grant Consolidator 311660, an INCa grant (TRANSLA-12-011), and the Saint-Louis Institute program (ANR-10-IBHU-0002) (to JS). Saint-Louis/Robert Debré Hospital is supported by the French Government (Direction de l’Hospitalisation et de l’Organisation des Soins) as Centre de Référence Maladies Rares “Aplasies Médullaires Constitutionnelles” (to TL, GS, AB, JS). AR was supported by a fellowship from the Fondation ARC. ADD was supported by NIH grant DK43889. We thank the FA patients and their families, the Association Française de la Maladie de Fanconi (AFMF) for their support, and the physicians and nurses from French pediatric, genetic, and/or hematological centers who have taken care of the patients.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(9):3580–3584. doi:10.1172/JCI88010.
Contributor Information
Dominique Bluteau, Email: dominique.bluteau@inserm.fr.
Julien Masliah-Planchon, Email: julien.masliahplanchon@curie.fr.
Connor Clairmont, Email: clairmont.connor@gmail.com.
Alix Rousseau, Email: alixr6@gmail.com.
Raphael Ceccaldi, Email: Raphael_Ceccaldi@DFCI.HARVARD.EDU.
Olivier Bluteau, Email: olivier.bluteau@inserm.fr.
Wendy Cuccuini, Email: wendy.cuccuini@sls.aphp.fr.
Stéphanie Gachet, Email: stephanie.gachet@hotmail.fr.
Régis Peffault de Latour, Email: regis.peffaultdelatour@sls.aphp.fr.
Thierry Leblanc, Email: thierry.leblanc@rdb.aphp.fr.
Dominique Stoppa-Lyonnet, Email: dominique.stoppa-lyonnet@curie.net.
Jean Soulier, Email: jean.soulier@sls.aphp.fr.
References
- 1.Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24(3):101–122. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soulier J. Fanconi anemia. Hematology Am Soc Hematol Educ Program. 2011;2011:492–497. doi: 10.1182/asheducation-2011.1.492. [DOI] [PubMed] [Google Scholar]
- 3.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493(7432):356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogliolo M, Surrallés J. Fanconi anemia: a model disease for studies on human genetics and advanced therapeutics. Curr Opin Genet Dev. 2015;33:32–40. doi: 10.1016/j.gde.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17(6):337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 6.Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489(7417):571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 7.Ceccaldi R, et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11(1):36–49. doi: 10.1016/j.stem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter D, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520(7548):549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- 9.Sharma S, Helchowski CM, Canman CE. The roles of DNA polymerase ζ and the Y family DNA polymerases in promoting or preventing genome instability. Mutat Res. 2013;743–744:97–110. doi: 10.1016/j.mrfmmm.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomida J, et al. REV7 is essential for DNA damage tolerance via two REV3L binding sites in mammalian DNA polymerase ζ. Nucleic Acids Res. 2015;43(2):1000–1011. doi: 10.1093/nar/gku1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sale JE. REV7/MAD2L2: the multitasking maestro emerges as a barrier to recombination. EMBO J. 2015;34(12):1609–1611. doi: 10.15252/embj.201591697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Listovsky T, Sale JE. Sequestration of CDH1 by MAD2L2 prevents premature APC/C activation prior to anaphase onset. J Cell Biol. 2013;203(1):87–100. doi: 10.1083/jcb.201302060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu G, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521(7553):541–544. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boersma V, et al. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5’ end resection. Nature. 2015;521(7553):537–540. doi: 10.1038/nature14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auerbach AD. Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp Hematol. 1993;21(6):731–733. [PubMed] [Google Scholar]
- 16.Shimamura A, et al. A novel diagnostic screen for defects in the Fanconi anemia pathway. Blood. 2002;100(13):4649–4654. doi: 10.1182/blood-2002-05-1399. [DOI] [PubMed] [Google Scholar]
- 17.Freie B, et al. Fanconi anemia type C and p53 cooperate in apoptosis and tumorigenesis. Blood. 2003;102(12):4146–4152. doi: 10.1182/blood-2003-03-0971. [DOI] [PubMed] [Google Scholar]
- 18.Yung SK, et al. Brief report: human pluripotent stem cell models of fanconi anemia deficiency reveal an important role for fanconi anemia proteins in cellular reprogramming and survival of hematopoietic progenitors. Stem Cells. 2013;31(5):1022–1029. doi: 10.1002/stem.1308. [DOI] [PubMed] [Google Scholar]
- 19.Lee YS, Gregory MT, Yang W. Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc Natl Acad Sci U S A. 2014;111(8):2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knipscheer P, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326(5960):1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H, Yang K, Dejsuphong D, D’Andrea AD. Regulation of Rev1 by the Fanconi anemia core complex. Nat Struct Mol Biol. 2012;19(2):164–170. doi: 10.1038/nsmb.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirchandani KD, McCaffrey RM, D’Andrea AD. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amst) 2008;7(6):902–911. doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papadopoulo D, Guillouf C, Mohrenweiser H, Moustacchi E. Hypomutability in Fanconi anemia cells is associated with increased deletion frequency at the HPRT locus. Proc Natl Acad Sci U S A. 1990;87(21):8383–8387. doi: 10.1073/pnas.87.21.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11(6):753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 25.Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol. 2009;11(6):761–768. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- 26.Vinciguerra P, Godinho SA, Parmar K, Pellman D, D’Andrea AD. Cytokinesis failure occurs in Fanconi anemia pathway-deficient murine and human bone marrow hematopoietic cells. J Clin Invest. 2010;120(11):3834–3842. doi: 10.1172/JCI43391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adamo A, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39(1):25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 28.Pace P, Mosedale G, Hodskinson MR, Rosado, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329(5988):219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 29.Okada T, et al. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol Cell Biol. 2005;25(14):6103–6111. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe N, et al. The REV7 subunit of DNA polymerase ζ is essential for primordial germ cell maintenance in the mouse. J Biol Chem. 2013;288(15):10459–10471. doi: 10.1074/jbc.M112.421966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khalaj M, et al. A missense mutation in Rev7 disrupts formation of Polζ, impairing mouse development and repair of genotoxic agent-induced DNA lesions. J Biol Chem. 2014;289(6):3811–3824. doi: 10.1074/jbc.M113.514752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parmar K, D’Andrea A, Niedernhofer LJ. Mouse models of Fanconi anemia. Mutat Res. 2009;668(1–2):133–140. doi: 10.1016/j.mrfmmm.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.