Abstract
Myotubular myopathy (MTM) is a devastating pediatric neuromuscular disorder of phosphoinositide (PIP) metabolism resulting from mutations of the PIP phosphatase MTM1 for which there are no treatments. We have previously shown phosphatidylinositol-3-phosphate (PI3P) accumulation in animal models of MTM. Here, we tested the hypothesis that lowering PI3P levels may prevent or reverse the MTM disease process. To test this, we targeted class II and III PI3 kinases (PI3Ks) in an MTM1-deficient mouse model. Muscle-specific ablation of Pik3c2b, but not Pik3c3, resulted in complete prevention of the MTM phenotype, and postsymptomatic targeting promoted a striking rescue of disease. We confirmed this genetic interaction in zebrafish, and additionally showed that certain PI3K inhibitors prevented development of the zebrafish mtm phenotype. Finally, the PI3K inhibitor wortmannin improved motor function and prolonged lifespan of the Mtm1-deficient mice. In all, we have identified Pik3c2b as a genetic modifier of Mtm1 mutation and demonstrated that PIK3C2B inhibition is a potential treatment strategy for MTM. In addition, we set the groundwork for similar reciprocal inhibition approaches for treating other PIP metabolic disorders and highlight the importance of modifier gene pathways as therapeutic targets.
Introduction
Phosphoinositides (PIPs) are low-abundance phospholipids responsible for the regulation of myriad cellular processes, including membrane trafficking and intracellular signaling cascades (1, 2). The distribution and amount of PIPs are tightly controlled through the balancing actions of phosphatidylinositol (PI) kinases and PI phosphatases. The importance of these regulations is highlighted by a growing group of human neurogenetic diseases, including congenital brain malformations (3), amyotrophic lateral sclerosis (4), and Charcot-Marie-Tooth disease (5), that are associated with mutations in PIP-modifying enzymes. While class I PI3 kinase (PI3K) inhibitors have made an impact as anticancer agents (6), there are as yet no targeted therapeutics in the clinical arena for monogenetic disorders of PIP metabolism.
Myotubular myopathy (MTM) is an X-linked condition caused by loss-of-function mutations in the PI phosphatase MTM1 (7), and is one of the most devastating disorders of childhood. Approximately one-third of affected boys die in the first year of life, and survivors manifest significant morbidities, including chronic ventilator and wheelchair dependence, and drastically reduced lifespan (8). MTM1 functions to dephosphorylate phosphatidylinositol-3-phosphate (PI3P) and phosphatidylinositol-3,5-bisphosphate [PI(3,5)P2], and we have documented in vertebrate models that loss of MTM1 results in significantly increased PI3P levels (9–11).
Much progress has been made in understanding the pathogenesis of MTM (12). We and others have demonstrated that MTM1 mutations are associated with abnormalities in the triad (9, 13), a muscle structure critical for excitation-contraction coupling. Triad abnormalities are among the first changes identified in MTM and likely explain the profound weakness seen in MTM patients (14). However, an association between MTM1 mutation, altered PIP metabolism, and triad dysfunction has yet to be translated into corresponding therapies for MTM patients.
On the basis of our discovery of increased PI3P levels in MTM, and observations from cell culture (15) and invertebrate studies (16, 17) showing reciprocal interplay between MTM1 and PI3P-synthesizing kinases, we hypothesized that lowering PI3P levels may prevent MTM disease progression. We therefore examined the kinases responsible for generating PI3P (i.e., the class II and class III PI3Ks). We have previously shown that only PIK3C2B (a class II kinase) and PIK3C3 (the class III kinase) are appreciably expressed in skeletal muscle (18), the primary tissue affected in MTM, and have generated and characterized Pik3c3 muscle-specific KO mice. These animals have reduced levels of PI3P in skeletal muscle and manifest a severe cardiomyopathy, an autophagic vacuolar myopathy, and early lethality (18). The specific impact of Pik3c2b deletion on skeletal muscle has yet to be examined.
For this study, we generated muscle-specific Pik3c2b KOs, and studied the impact of reduced expression of either Pik3c2b or Pik3c3 on Mtm1 KO mice. We found that while Pik3c3 deletion exacerbates the Mtm1 KO phenotype, Pik3c2b ablation completely prevents its development. We additionally demonstrate using an inducible knockout that postsymptomatic targeting of Pik3c2b promotes complete rescue of the MTM phenotype. Furthermore, using a newly developed zebrafish model of MTM, we determine that chemical inhibitors with activity against PIK3C2B ameliorate the zebrafish phenotype. Lastly, we show that 1 of these inhibitors (wortmannin) can improve motor function and prolong lifespan of Mtm1 KO mice. We conclude that PIK3C2B is a potent second-site genetic modifier of Mtm1 mutation, and identify PIK3C2B inhibition as an exciting treatment strategy ideal for clinical translation for this devastating disorder.
Results
Pik3c2b is not required for the normal formation and function of murine skeletal muscle.
We generated muscle-specific Pik3c2b KO mice using a floxed Pik3c2b allele (Pik3c2bfl/fl) (19) and a muscle-specific Cre transgenic (Ckmm-Cre) (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI86841DS1). Muscle-specific reduction of RNA and protein expression was confirmed with quantitative RT-PCR and Western blot analysis, respectively (Supplemental Figure 1, C–E). Muscle-specific Pik3c2bfl/fl Ckmm-Cre (referred to here as Pik3c2b KOs) were born at mendelian ratios and were indistinguishable from WT siblings at all ages examined (Supplemental Figure 1F). Motor function testing did not reveal any differences between Pik3c2b KOs and their littermates (Supplemental Figure 1G), and muscle structure appeared normal (Supplemental Figure 1H). We thus conclude that Pik3c2b knockout does not appreciably alter the formation or maintenance of skeletal muscle structure and function.
Muscle-specific knockout of Pik3c2b prevents the phenotype associated with loss of Mtm1.
Mice with a deletion of Mtm1 on the X chromosome (Mtm1–/y mice, referred to here as Mtm1 KO mice) have a severe, progressive skeletal myopathy that accurately models human MTM (20). They become overtly symptomatic at approximately 21 days of age, with reduced body weight the first sign of disease. By 30 days, the animals become wasted and weak, exhibiting definitive changes in motor function (Figure 1). Histopathologic changes at this stage include myofiber hypotrophy, increased central nuclei, accumulation of subcellular organelles, and triad abnormalities (Figure 2). Mtm1 KOs continue to decline in health, with hindlimb paralysis manifesting by P35 (Supplemental Video 1) and with death occurring by P38 (Figure 1). Notably, the phenotypic presentation and survival of Mtm1 KO mice were consistent regardless of background strain (Supplemental Tables 1 and 2).
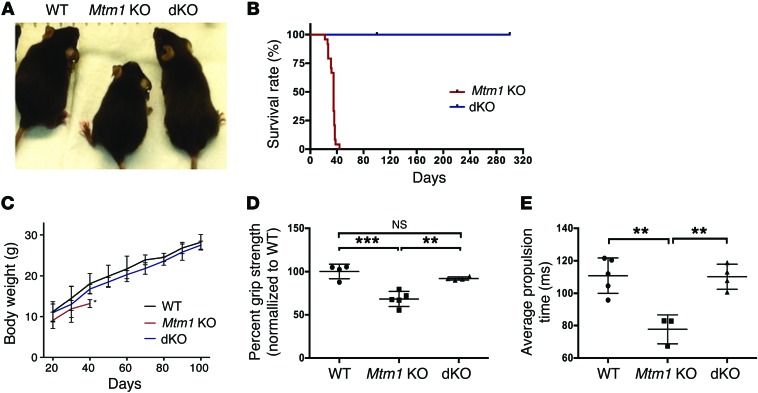
Figure 1. Muscle-specific knockout of Pik3c2b prevents the phenotype associated with Mtm1 knockout.
(A) Mtm1–/y (Mtm1 KO) mice have diminished size, hunched posture, and splayed hindlimbs (indicative of limb weakness), while Pik3c2b Mtm1 double KOs (dKOs) appear indistinguishable from WT. (B) Kaplan-Meier survival curve reveals that Pik3c2b knockout confers a survival benefit to the MTM mouse model (dKO median survival > 300 days, n = 11, vs. Mtm1 KO mice = 35 days, n = 24). (C) Plot of body weight versus postnatal age for WT, Mtm1 KO, and dKO mice. Mtm1 KO (n = 12) mice are significantly smaller than male WT siblings from 20 days onward, while dKO animals have weights indistinguishable from WT at all time points examined (n = 24, last age measured = 100 days). (D) dKO mice have restored muscle power. Mtm1 KO animal mean grip strength = 68% ± 4.8% compared with 100% ± 7.2% in the WT (***P < 0.001) and 92% ± 4.8% in dKOs (**P < 0.01 vs. Mtm1 KO). Values were normalized to the mean grip strength of WT littermates (n = 5 per condition). (E) dKO animals have normal motor function, as measured by treadmill analysis. The average propulsion time measurement at age 28 days for WT was 110 ± 9.7 ms (n = 4), for Mtm1 KO was 77 ± 6.9 ms (n = 5, **P < 0.01 compared with WT or dKO), and for dKO was 110 ± 7.2 ms (n = 4, P = 0.91 compared with WT). All comparisons in C–E done by 1-way ANOVA followed by Dunnett’s multicomparison test.
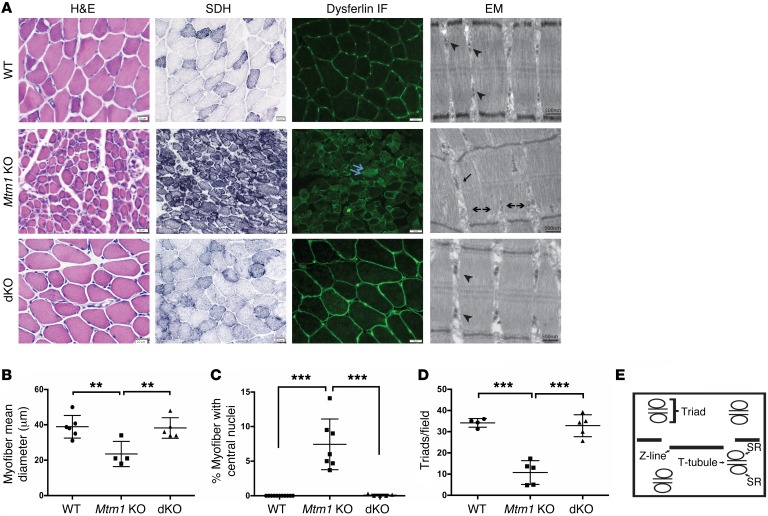
Figure 2. Restoration of normal muscle structure in Pik3c2b Mtm1 dKO mice.
(A) Cross sections from tibialis anterior muscle tissue of 35-day-old animals stained for H&E, SDH, and dysferlin. Mtm1 KO muscle has centrally located nuclei (arrow in H&E), mitochondrial aggregation and necklace myofibers on SDH staining (arrow), and abnormal distribution of dysferlin staining (double arrow). None of these changes were observed in muscle from dKO animals. Electron microscopic (EM) images reveal normal triad formation (arrowhead) in WT as well as dKO muscle but abnormal triads in Mtm1 KO with formation of longitudinal tubes (L-tubules, arrow) and absence of triads (double arrowhead). Scale bars: 20 μm; for EM: 500 nm. (B) dKO muscle has restored myofiber size, with average myofiber diameter for WT = 39 ± 5.8 μm (n = 6), for Mtm1 KO = 23 ± 4.1 μm (n = 4, **P < 0.01 vs. WT or dKO), and for dKO = 38 ± 4.3 μm (n = 5, P = 0.85 vs. WT). (C) Absence of central nuclei in the dKO. The average percent of central nuclei (per 100 fibers) in WT = 0% (n = 11), in Mtm1 KO = 7.4% ± 1.0% (n = 7, ***P < 0.001 vs. WT or dKO), and in dKO = 0.09% (n = 5, P = 0.2 vs. WT). (D) Quantification of number of triads per field. WT = 34 ± 3.1 (n = 4), Mtm1 KO = 10.7 ± 2.9 (n = 5, ***P < 0.001 vs. WT or dKO ), and dKO = 33 ± 4.6 (n = 5, P = 0.6 vs. WT). All comparisons in B–D done by 1-way ANOVA followed by Dunnett’s multicomparison test. (E) Schematic of the structure of triad with T-tubules, sarcoplasmic reticulum (SR), and the sarcomere ends (Z-line).
To test the ability of Pik3c2b to modify the Mtm1 KO phenotype, we generated Pik3c2b KO Mtm1 KO mice (Pik3c2b Mtm1 dKOs). Remarkably, these mice are indistinguishable from WT littermates at all ages examined (Supplemental Videos 2 and 3), with the oldest dKO animals alive and healthy at more than 18 months (Figure 1, A and B). At all ages tested, dKO animals have body weights and motor function similar to those of WT littermates (Figure 1, C–E). Histopathologic analyses reveal restoration of muscle structure, including normal myofiber size, an absence of central nuclei, and normal appearance and number of triads (Figure 2). In total, muscle-specific KO of Pik3c2b completely prevents the development of the murine MTM phenotype.
Muscle-specific KO of Pik3c2b restores PI3P levels in Mtm1 KO mice.
We next sought to measure PI3P levels in our dKO animals, first by using an ELISA technique previously described for Mtm1 KOs (10). Using this ELISA on muscle extracts from P35 mice (Figure 3A), we again found that PI3P levels were significantly increased in Mtm1 KOs. In contrast, Pik3c2b Mtm1 dKOs had levels similar to WT, indicating that ablation of Pik3c2b in Mtm1 KO mice restored PI3P levels to baseline. Notably, Pik3c2b knockout by itself did not significantly alter the total amount of PI3P in skeletal muscle; this is unlike what has been observed in other cell types (e.g., hepatocytes), where 60% reduction was found with Pik3c2b knockout (21).
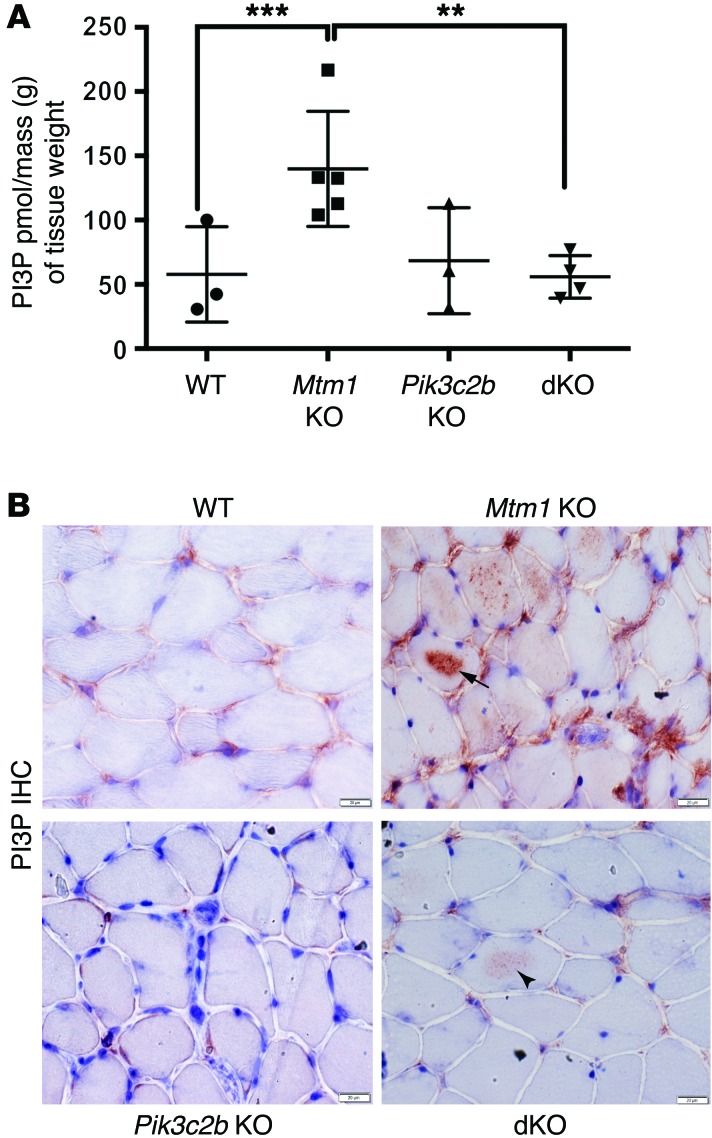
Figure 3. Restoration of PI3P levels in the Pik3c2b Mtm1 dKO mice.
(A) PI3P levels are normalized in the dKO animals, as determined using a PI3P ELISA kit [purified lipid (pmol)/mass (g) of muscle tissue]. PI3P levels in WT muscle = 58 ± 21 pmol/g (n = 3), Mtm1 KO = 140 ± 20 pmol/g (n = 5, ***P = 0.01 vs. WT, **P = 0.01 vs. dKO), Pik3c2b KO = 68 ± 24 pmol/g (n = 3), and dKO = 56 ± 8 pmol/g (n = 4, P = 0.87 vs. WT, 1-way ANOVA followed by Dunnett’s multiple comparison test). (B) PI3P subcellular localization by IHC on tibialis anterior muscle. Labeling in WT and Pik3c2b KO is observed faintly in the perinuclear and membrane compartment, while staining in Mtm1 KO is seen diffusely, with intense intracellular staining in the sarcolemma of several myofibers (arrow). dKO muscle shows staining similar to WT, though with areas of mildly increased labeling within the sarcolemma of selected myofibers (arrowhead). Scale bars: 20 μm.
We next examined PI3P subcellular distribution (Figure 3B). In WT muscle, anti-PI3P antibodies produced perinuclear labeling and patchy staining on the sarcolemmal membrane, a distribution consistent with endosomal localization. In Mtm1 KOs, PI3P staining was more intense and also found in a much broader distribution, both around the membrane and now within the sarcolemma. In keeping with our ELISA results, dKO staining was similar to WT, though there were a few areas of sarcolemmal accumulation (Figure 3B, arrowhead). These data reflect increased PI3P in Mtm1 KOs and restoration of PI3P to normal in dKOs, consistent with our hypothesis regarding an association between PI3P reduction and phenotypic improvement.
Pik3c2b haploinsufficiency significantly improves the Mtm1 KO phenotype.
We next examined whether there was a dosage effect to the rescue provided by Pik3c2b ablation by studying Pik3c2bfl/+ Mtm1 KO animals. As compared with Mtm1 KO alone, these animals showed a small but significant improvement in body weight and survival (Supplemental Figure 2, A and B). This improvement was mirrored by muscle histology (Supplemental Figure 2, C–G), where Pik3c2bf/+ Mtm1 KO muscle had increased fiber size, fewer central nuclei, and improved triad appearance as compared with Mtm1 KOs. There was also an intermediate but nonsignificant restoration of total PI3P levels (Supplemental Figure 2H). Taken together, these data show that heterozygous ablation of Pik3c2b promotes partial improvement of the Mtm1 KO phenotype.
Temporal knockout of Pik3c2b rescues the Mtm1 KO phenotype.
Because MTM patients manifest symptoms at birth, the most effective treatment(s) will improve disease after phenotype onset. To test whether this can be accomplished in MTM mice with Pik3c2b inhibition, we used a temporal KO strategy with a tamoxifen-inducible (TAM-inducible) Cre transgenic line. We generated Pik3c2b Mtm1 dKOs that expressed Tg(ER-Cre), and established that 1 week of TAM treatment was sufficient to promote Pik3c2b ablation and produce undetectable levels of PIK3C2B protein (Figure 4, A and B). To create conditional knockout of Pik3c2b and Mtm1 at an early symptomatic time point, we treated these animals with TAM starting at P21 (early-TAM-cKO mice). Early-TAM-cKO animals showed an initial reduction in body weight (due to TAM-related food aversion), but by 28 days regained their appetite, showing weights matching those of TAM-exposed WTs (Figure 4D). From this age onward, early-TAM-cKOs were indistinguishable from WT littermates, demonstrating normal motor function and muscle histology, and showing no reduction in survival (Figure 4, C, D, and H). The oldest early-TAM-cKOs currently are more than 160 days old; in contrast, the average age of mortality for TAM-treated Mtm1 KOs is 42 days (Figure 4C). By all methods of examination, therefore, early symptomatic targeting of Pik3c2b promotes full rescue of the Mtm1 KO phenotype.
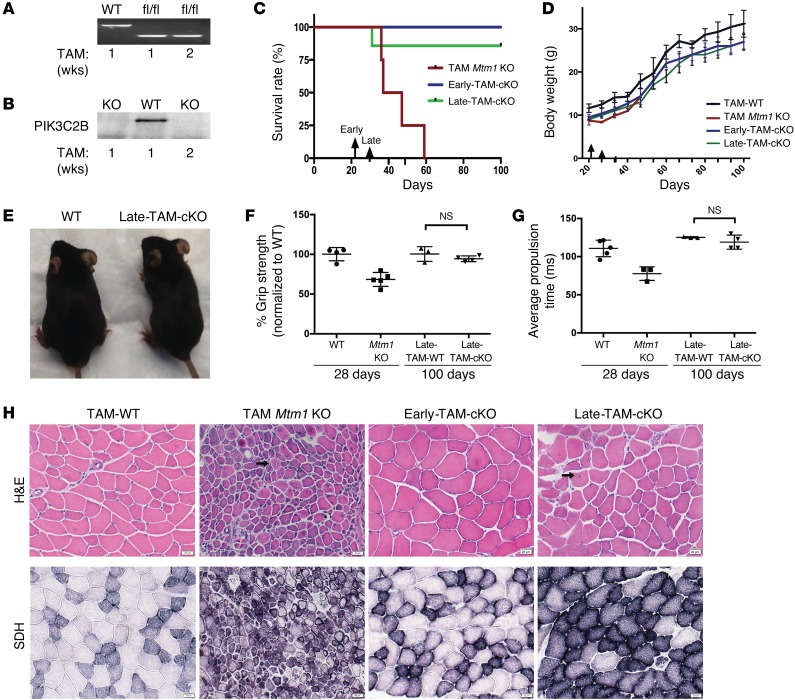
Figure 4. Temporal knockout of Pik3c2b at both early and late stages of disease rescues the Mtm1 disease phenotype.
(A) RT-PCR showing excision of floxed Pik3c2b. (B) Western blot reveals undetectable PIK3C2B protein in cKO muscle. (C) Early-TAM-cKO (induced by tamoxifen treatment starting at 21 days) or late-TAM-cKO (starting at 30 days) of Pik3c2b confers a survival benefit to Mtm1 KO mice. Mtm1 KO alone plus TAM median survival = 42 days, while both early-TAM-cKO (n = 3) and late-TAM-cKO (n = 7) animals live at least 100 days, with the oldest survivors more than 160 days old. (D) Body weights of early-TAM-cKO (n = 3), late-TAM-cKO (n = 7), and Mtm1 KO mice (n = 4) are initially similar to those of Mtm1 KOs, but from 51 days of age onward they have weights similar to WT. (E) Late-TAM-cKOs appear similar (thought slightly smaller) than WTs (age = 160 days). (F) Grip strength measured at 28 and 100 days of age. At 28 days of age, Mtm1 KOs have a significant decrease in mean grip strength (from Figure 1D). After induction of cKO at 30 days, however, grip strength of late-TAM-cKO (n = 4), as measured at 100 days of age, equals that of WT (94% ± 3.1% vs. WT 100% ± 7.5% [P = 0.38]). (G) Late-TAM-cKOs have normalized treadmill behavior. As demonstrated in Figure 1E, Mtm1 KOs have reduced average propulsion time at 28 days (77± 7.2 ms). However, conditional KO of Pik3c2b in Mtm1 KOs starting at 30 days results in normal propulsion at 100 days of age (119 ± 8 ms, n = 4, vs. 125 ± 0.7 ms for WT, n = 3, P = 0.27). (H) Early-TAM-cKOs have normal myofiber size and structure and an absence of central nuclei. Late-TAM-cKO muscle structure is also relatively restored, with slight reductions in myofiber size and small increases in central nuclei (arrows). Scale bars: 20 μm. Comparisons were done using ANOVA followed by Dunnett’s multicomparison test.
To test the ability of Pik3c2b KO to alter the MTM phenotype at late stages of the disease, we targeted Pik3c2b with TAM starting at P30 (late-TAM-cKOs) (Supplemental Video 4). Remarkably, late-TAM-cKO animals survive significantly longer than Mtm1 KOs alone (Figure 4C), with the oldest survivor now healthy and active at more than 160 days old (Supplemental Video 5). While these animals have reduced body weight as compared with WT (Figure 4, D and E), their motor function — which was reduced in Mtm1 KOs by the age of late-TAM exposure (Figure 1, D and E) — is restored to normal (Figure 4, F and G). Muscle structure is also relatively rescued (Figure 4H), with only slight alterations in myofiber size and percentage central nuclei (Supplemental Figure 3). Notably, for both early-TAM-cKO and late-TAM-cKO, despite the healthy appearance of the dKO mice, the study was terminated at 100 days in order to complete biochemical and histopathologic analyses. In all, these data show that Pik3c2b knockout can reverse the MTM phenotype at late stages of the disease.
Skeletal muscle–specific knockout of Pik3c3 results in a mild vacuolar myopathy.
In muscle, PI3P is synthesized by both PIK3C3 and PIK3C2B. We previously created muscle-specific Pik3c3 KO mice using Ckmm-Cre, but these animals die of complications related to severe cardiomyopathy (18). In order to test the effect of Pik3c3 reduction on Mtm1 KO animals, we re-derived the Pik3c3 animals with Cre driven by the Acta1 promoter to confine targeting to skeletal muscle (Supplemental Figure 4A). Skeletal muscle–specific Pik3c3 KOs (targeting confirmed by quantitative RT-PCR; Supplemental Figure 4B) are similar in appearance to WTs (Supplemental Figure 4C), and show no alteration in survival through the first 6 months of life. On muscle histology, Pik3c3 KOs exhibit mild dystrophic changes, with fiber size variability, internal nuclei, and vacuoles (Supplemental Figure 4E), changes consistent with a mild vacuolar myopathy. Notably, heterozygous Pik3c3fl/+ Acta1-Cre animals have no detectable clinical or histopathologic phenotype.
We have previously shown that muscle-specific knockout of Pik3c3 with Ckmm-Cre results in a significant decrease in total skeletal muscle PI3P levels (18). In our Acta1-Cre KOs, there was again a trend of reduced total PI3P, though with our current sample size it did not reach statistical significance (Supplemental Figure 4D). In addition, we examined PI3P distribution by immunostaining, and observed reduced or absent staining along the sarcolemmal membrane in Pik3c3 KO as compared with WT (Supplemental Figure 5, A and B). Notably, parallel analysis of PI3P in Pik3c2b KO muscle revealed intact membrane staining but reduced perinuclear nuclear expression (Supplemental Figure 5C). These data suggest that PI3P localization is differentially established and maintained by PIK3C3 and PIK3C2B.
Muscle-specific knockout of Pik3c3 exacerbates the Mtm1 KO phenotype.
We next crossed Pik3c3 KOs with Mtm1+/– females to derive animals targeted for both genes. Strikingly, Mtm1 KOs heterozygous for Pik3c3 showed evidence of more severe symptomatology (Supplemental Video 6), as reflected by smaller body weight and significantly decreased survival (Figure 5, A–C). Muscle structural analysis revealed changes similar to Mtm1 KOs, though with small, nonsignificant trends toward improvement in myofiber size and percentage of central nuclei (Figure 5, D–F). There was also an apparent reduction in PI3P, reflected both by immunostaining with anti-PI3P and through mass ELISA analysis (Figure 5, D and G). Notably, we have obtained only a single live-born Pik3c3 Mtm1 dKO, which displayed an extremely severe phenotype with early paralysis and death by age 3 weeks. Despite multiple breedings and sufficiently large litter sizes expected to produce dKOs, we have yet to obtain additional live-born dKOs, suggesting that these animals are likely embryonic lethal. Taken with the phenotype of the Pik3c3fl/+ Mtm1 KO animals, these data reveal that Pik3c3 ablation worsens the Mtm1 KO phenotype.
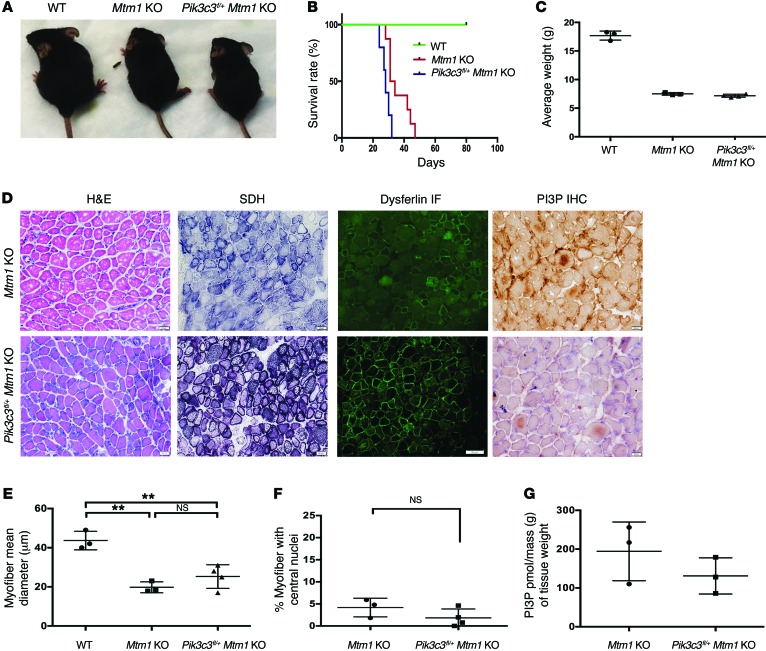
Figure 5. Skeletal muscle–specific knockout of Pik3c3 exacerbates the Mtm1 KO disease phenotype.
(A) Photomicrographs of littermates at 28 days of age. At this stage, Pik3c3fl/+ Mtm1 KOs exhibit more severe symptomatology, as reflected by smaller body weight, hunched posture, and splayed hindlimbs (indicative of limb weakness). (B) Kaplan-Meier survival curve reveals that Pik3c3fl/+ decreases survival of Mtm1 KO mice. Mtm1 KOs (n = 8) live to a median age of 32.5 days, while Pik3c3fl/+Mtm1 KOs (n = 5) have a median survival of 28 days (*P = 0.01). (C) Pik3c3fl/+Mtm1 KOs have reduced body weight as compared with WT, demonstrated by average body weight (in grams or g) at 27–30 days (WT = 17.7 ± 0.4 [n = 3], Mtm1 KO = 7.5 ± 0.1 [n = 3], Pik3c3fl/+Mtm1 KO = 7.2 ± 0.1 (n = 4), P < 0.0001). (D–F) Histological analysis by H&E, SDH, and dysferlin IF reveals that Pik3c3fl/+Mtm1 KO muscle structure is significantly abnormal (scale bars: 20 μm). Myofiber size is reduced, with the average myofiber diameter for Pik3c3fl/+Mtm1 KOs (25 ± 6.0 μm, n = 4) similar to that of Mtm1 KOs (20 ± 2.8 μm, n = 3) but significantly smaller as compared with WT (43 ± 4.8 μm, n = 3, **P < 0.01). (F) Central nucleation is similar between Pik3c3fl/+Mtm1 KO (1.8% ± 2.0% [n = 4]) and Mtm1 KO (4.2% ± 2.1% [n = 3, P = 0.2]). (G) PI3P levels are decreased in Pik3c3fl/+ Mtm1 KO animals, as determined qualitatively using IHC (D, black scale bars = 11 μm) and quantitatively using PI3P mass ELISA [purified lipid (pmol)/mass (g) of muscle tissue]. PI3P levels in Pik3c3f/+ Mtm1 KO muscle = 131 ± 38 pmol/g (n = 3), versus Mtm1 KO alone = 194 ± 62 pmol/g (n = 3, P = 0.3). All comparisons in figure were done using ANOVA followed by Dunnett’s multiple comparison test.
We previously reported that inhibition of autophagy significantly worsened the phenotype in an MTM zebrafish model (22), and have demonstrated that Pik3c3 knockout is associated with impaired autophagy (18). We thus were interested to examine autophagy in Pik3c3fl/+Mtm1 KO animals. As determined by LC3 Western blot analysis, neither Pik3c3fl/+ nor Mtm1 KO animals alone showed evidence of abnormal autophagy. On the other hand, there was evidence of significant alterations in LC3 levels in the muscle of the combined Pik3c3fl/+ Mtm1 KO animals (Supplemental Figure 6), suggesting autophagy is dysregulated in this setting.
mtm, a zebrafish model of MTM.
The ability of Pik3c2b knockout to suppress the Mtm1 KO phenotype suggests that inhibitors of PIK3C2B may be effective therapeutics for MTM. While there are no commercially available inhibitors specific to PIK3C2B, several pan-PI3K inhibitors display inhibitory activity against it (Supplemental Table 3 and ref. 23, 24). To test the potential efficacy of these inhibitors, we developed a new zebrafish model of MTM suitable for drug screening. This model was created using zinc finger nucleases that targeted exon 5 of the zebrafish mtm1 gene and produced an 8-bp deletion (Figure 6A). This deletion is predicted to cause a frameshift and early stop mutation. Zebrafish heterozygous for the mutation have no phenotype. Zebrafish homozygous for the mutation (mtmΔ8/Δ8 or mtm) manifest phenotypic changes starting at 3 days postfertilization (dpf). mtm zebrafish have progressive phenotypic alterations, including fin fold degeneration (Figure 6B and Supplemental Figure 7), pectoral fin abnormalities (Figure 6C), liver changes (Figure 6D), and early death (Figure 6E). Examination of muscle structure revealed triad abnormalities similar to those seen in human MTM (Figure 6F). Lastly, analysis of photomotor activation revealed significant reduction in motor behavior (Figure 6G).
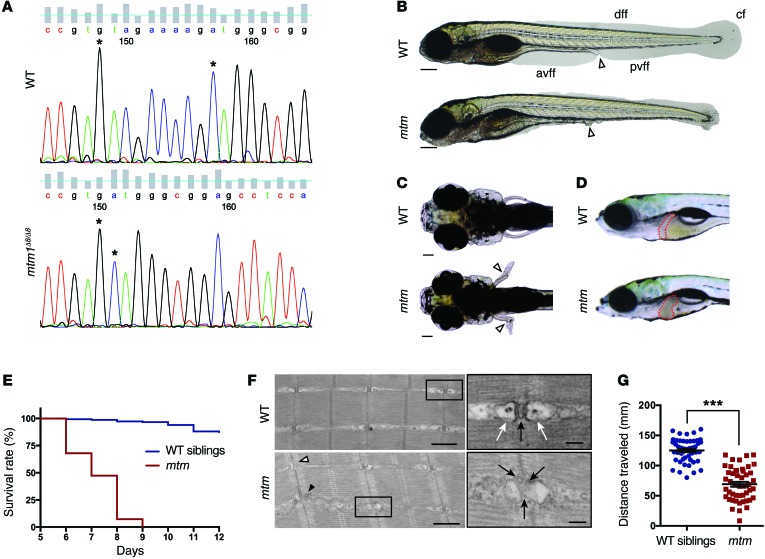
Figure 6. Phenotypic characterization of mtm, a novel zebrafish model of MTM.
(A) Sanger sequencing of an RT-PCR fragment from WT (top) and homozygous mtm1Δ8/Δ8 mutant (mtm; bottom) larvae showing an 8-bp deletion in exon 5 of mtm1. (B) Bright-field of WT siblings compared with mtm larvae at 4 dpf illustrating complete loss of the anterior and posterior ventral fin folds (white arrowhead indicates urogenital opening; scale bar: 200 μm). This represents a “severe” mutant phenotype. avff, anterior ventral fin fold; pvff, posterior ventral fin fold; dff, dorsal fin fold; cf, caudal fin. (C and D) By 7 dpf, mtm larvae have outwardly kinked pectoral fins (white arrowheads; scale bar: 100 μm) and abnormal liver appearance (liver outlined in red). (E) mtm larvae have median and maximum survival of 7 and 9 days, respectively, while most WT siblings survive into adulthood (n = 150 per group; P < 0.001, Mantel-Cox test). (F) At 5 dpf, electron micrographs show aberrant skeletal muscle ultrastructure in mtm larvae (scale bars: 500 nm; inset: 100 nm). WT larvae have normal triad structure where T-tubules (inset; black arrow) are apposed by terminal cisternae/SR (inset; white arrows), whereas mtm muscle has L-tubules (black arrowhead), triads lacking SR (white arrowhead), and fragmented T-tubules (inset; black arrows). (G) mtm larvae have significantly impaired motor function compared with WT. In a 30-second optovin-induced photoactivation period performed at 7 dpf, mutants only travel about half (55%) the distance covered by their siblings (***P < 0.001). n = 48 each; Student’s t test, 2-tailed. Error bars indicate SEM.
To confirm the specificity of the mtm phenotype, we performed RNA rescue using zebrafish mtm1 RNA. RNA expressing mtm embryos exhibited significant resolution of their phenotype (Supplemental Figure 8), confirming that the observed abnormalities are due to mtm1 mutation. In addition, we directly compared the changes seen in mtm mutants with those in our previously published morpholino model (9, 22). Morphants were indistinguishable in appearance from mtms (Supplemental Figure 9), corroborating both the specificity of the mutant phenotype and the accuracy of the morpholino model. Lastly, we examined the genetic interaction between pik3c2b and mtm1 by testing pik3c2b morpholinos in mtm mutants. We observed that pik3c2b knockdown, which reduced RNA levels only for the first 2–3 dpf because of the transient nature of morpholinos, partially suppressed the mtm1 mutant larval phenotype (Supplemental Figure 10), indicating cross-species conservation of PIK3C2B modification in the MTM disease process.
PI3K inhibitors ameliorate the mtm zebrafish phenotype.
To investigate the ability of PI3K inhibitors to ameliorate aspects of the mtm phenotype, we screened 5 inhibitors with varying activity against PIK3C2B. In the initial screen, we provided drugs from 1–4 dpf and then scored mutants for fin degeneration. We observed that 3 drugs, wortmannin, LY294002, and PI-103, significantly diminished the development of this phenotype (Figure 7A and Supplemental Figure 11). We next performed a prolonged treatment trial, and observed significant, dose-dependent improvements in mtm swim behavior and survival with both wortmannin and LY294002, and prolonged survival with PI-103 (Figure 7, B and C). In all, treatment with wortmannin, LY294002, or PI-103 significantly reduced the development and progression of the mtm phenotype. As these 3 chemicals show relatively specific activity toward PIK3C2B at the concentrations tested (Supplemental Table 1 and ref. 23), these results corroborate our Pik3c2b KO data and suggest that a selective PIK3C2B inhibitor would be a potential therapeutic agent for MTM treatment. Notably, inhibitors specifically targeted to either PIK3C3 (VPS34-IN1; ref. 25) or the class I PI3K (GDC-0941) did not appreciably improve any aspect of the mtm phenotype (Figure 7 and Supplemental Figure 11).
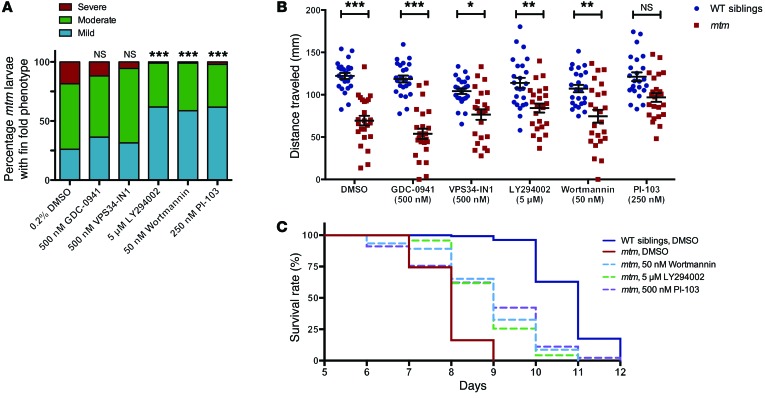
Figure 7. Treatment with pan-PI3K inhibitors improves the mtm zebrafish phenotype.
(A) Severity of fin fold phenotype in 4-dpf mtm mutants is significantly improved after treatment with 5 μM LY294002, 50 nM wortmannin, or 250 nM PI-103 (***P < 0.001), but not by inhibition of class III PI3K with 500 nM VPS34-IN1 (P = 0.4955) or class I PI3Ks with 500 nM GDC-0941 (P = 0.3494). From left to right: n = 92, 110, 108, 97, 92, 94; Kruskal-Wallis test, followed by Dunn’s post-test. (B) At 7 dpf, the mean distance traveled in a 30-second optovin-induced photoactivation period by mtm larvae treated with DMSO alone is 57% that of WT siblings (P < 0.001). Class I inhibition with GDC-0941 does not improve mtm motor function (45% WT, P < 0.001). In contrast, mtm motor function is improved relative to cognate-treated WT siblings by treatment with LY294002 (74% WT, P = 0.0059), wortmannin (70% WT, P = 0.0012), and, surprisingly, VPS34-IN1 (73% WT, P = 0.0155). Though these groups remained significantly different from WT, this improvement suggests an mtm-specific chemical effect for this phenotype (**P < 0.01, ***P < 0.001). Notably, mutants treated with PI-103 are statistically indistinguishable from PI-103–treated WT siblings (80% WT) and move significantly better than DMSO-treated mtm (*P < 0.0171). n = 24 each group; 2-way ANOVA, followed by Tukey’s post-test. (C) Kaplan-Meier survival curve showing that treatment with LY294002, wortmannin, and PI-103 improves median survival from 8 to 9 days and maximum survival from 9 to 11 to 12 days in comparison with DMSO-treated mtm larvae (P < 0.001 compared with mtm DMSO, Mantel-Cox or Gehan-Breslow-Wilcoxon tests). Inset legend top to bottom: n = 132, 43, 46, 47, 44.
Wortmannin treatment improves phenotypic features and survival of the Mtm1 KO mouse.
The ability of PI3K inhibitors with activity against PIK3C2B to ameliorate the phenotype of mtm1 KO zebrafish suggests that such chemicals may also improve the MTM mouse model. Wortmannin has been used extensively in mice, with well-established dose parameters and safety profile, and thus we tested this PI3K inhibitor in our Mtm1 KO mice. We treated both WT and Mtm1 KO mice daily using oral gavage starting at P21. Treated KO animals appeared healthier and more active than untreated littermates, and were obviously distinguishable by qualitative observation (Supplemental Video 7) While body weight was not improved by wortmannin (Supplemental Figure 12A), analysis of grip strength and treadmill performance (32 days of age, n = 2) of treated Mtm1 KOs showed restoration of these parameters to WT levels (Supplemental Figure 12, B and C). Most importantly, wortmannin provided a significant improvement in survival, with treated Mtm1 KOs living an average of 10 days longer than untreated KOs (n = 6, P < 0.001; Figure 8A), and with the longest survivor living until 50 days of age. Muscle histopathology was also improved, with fewer central nuclei and larger fiber size as compared with age-matched untreated KOs (Figure 8B). In addition, PI3P expression, as determined by immunostaining, was reduced in wortmannin-treated MTM animals, with a clear qualitative reduction in PI3P particularly in the central compartment of the myofiber. Taken together, these data demonstrate a significant improvement with wortmannin, and suggest that PI3K inhibition is a viable potential therapeutic approach for MTM.
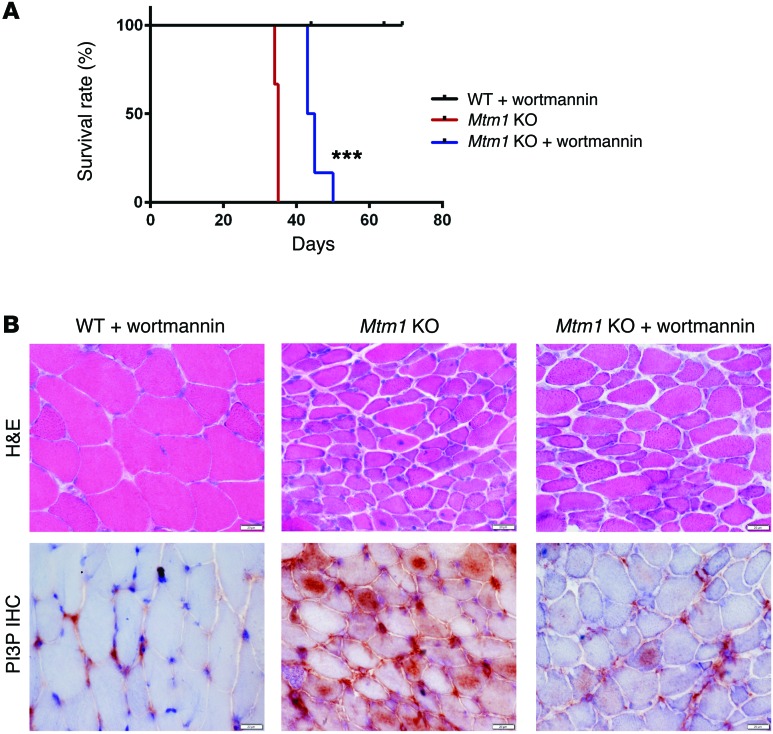
Figure 8. Wortmannin treatment prolongs survival and improves the phenotype of Mtm1 KO mice.
(A) Kaplan-Meier survival curve. As compared with a placebo DMSO control treatment (n = 4), wortmannin treatment increases median survival of Mtm1 KO mice from 34.5 to 44 days, with maximum survival changed from 35 to 50 days (n = 6, ***P < 0.001). (B) Histopathological assessment of TA muscle tissue of 32-day-old animals reveals that muscle fibers from Mtm1 KO mice treated with wortmannin have modest improvements in myofiber size and central nucleation (top panel, n =2 per treatment group, scale bars: 20 μm). Immunostaining of PI3P on cross section of TA muscle (60×) reveals reduced labeling in wortmannin-treated Mtm1 KO as compared with the intense staining seen in placebo-exposed Mtm1 KO muscle (bottom panel, n =2 per treatment group, scale bars: 20 μm).
Discussion
In this study, we demonstrate the remarkable ability of PIK3C2B inhibition to not only prevent but also reverse the phenotypic consequences of Mtm1 mutation in mice and zebrafish. To our knowledge, Pik3c2b is the first genetic modifier identified for Mtm1 mutation, and our study is the first to demonstrate in any preclinical model of PIP metabolic disease that genetic ablation of a reciprocal or balancing PIP enzyme can completely rescue a disease phenotype. Because of this, our study has several important implications.
First, we identify a new therapeutic strategy for MTM, a devastating childhood disease with no existing treatments. Given that loss of Pik3c2b by itself in mice has no discernible phenotype (19), PIK3C2B inhibition is an extremely attractive target that may work either as a monotherapy or as a complement to existing efforts under investigation such as MTM1 gene therapy and MTM1 enzyme replacement therapy (26, 27). It is particularly noteworthy that Pik3c2b targeting rescues at even late stages of the MTM mouse disease process, suggesting it may be applicable as a treatment for all MTM patients regardless of age and clinical severity. Notably, to our knowledge, Pik3c2b is the first gene modifier uncovered for Mtm1 mutation and MTM, and our study highlights the continued importance of identifying and targeting such modifiers as a means for understanding disease pathomechanisms and discovering novel pathways for therapeutic intervention.
At present, there are no inhibitors exclusively targeting PIK3C2B that have been tested in animal models or in clinical trials, and the existing PI3K inhibitors that target PIK3C2B typically have activity against PIK3C3 as well. This likely explains why we observe a significant but not complete amelioration of disease phenotype in our zebrafish model with LY294002, wortmannin, and PI-103, and a modest but significant prolongation in survival in our mouse model with wortmannin, as these drugs inhibit not only PIK3C2B but also PIK3C3 and the class I PI3K. It is also consistent with our observation that the drugs that target class I or III kinases (but not class II) have no impact on the mtm zebrafish phenotype. Therefore, a key future aim is to develop an inhibitor(s) that exclusively targets PIK3C2B with minimal or no action against other PI3Ks and that has a suitable safety profile for clinical translation. Such inhibitors may have benefit beyond treatment for MTM, as recent studies have implicated PIK3C2B activation in tumor progression (28), hepatitis C replication (29), and diabetes (21).
Second, we believe our study provides a roadmap for future therapy development for the broad (and growing) class of disorders of PIP metabolism. This group of conditions includes not only several rare, monogenetic diseases such as hemimegalencephaly and Yunis-Varon syndrome, but also more common “sporadic” diseases such as Alzheimer disease and several human cancers. We recognize that 2 critical questions raised by our data are (a) whether our observations are restricted to MTM1 and PIK3C2B, or instead are more generally applicable to other PIP kinase/phosphatase pairings with reciprocal activity, and (b) whether the modifying effect of Pik3c2b ablation is due exclusively to reducing PI3P or instead is related to a change(s) in another pathway shared between PIK3C2B and MTM1.
Previous studies have touched on the potentially complex nature of PIP kinase/phosphatase interactions. Vaccari et al. defined a complicated interplay between FIG4, MTMR2, and the regulation of PI(3,5)P2 (30, 31). In mice and in cell culture, heterozygous reduction of Fig4 and knockdown of the kinase PIK FYVE both improve the neuropathy associated with knockout of the PIP phosphatase Mtmr2 (a model for MTMR2-related Charcot-Marie-Tooth disease), supporting the concept that PIP “rebalancing” may have utility across the broad spectrum of PIP diseases. However, in the converse setting, Mtmr2 knockout exacerbates the neuronal degeneration observed in Fig4 mutants, suggesting that these homeostatic interactions may be tissue and system dependent. Notably, unlike Mtmr2 KO mice, we previously reported that Fig4 haploinsufficiency fails to improve the Mtm1 KO phenotype (32). In another informative study, Di Paolo et al. found that targeting synaptojanin 1 in Pip5k1c KO mice restored PI(4,5)P2 levels but did not improve the neuronal phenotype (33), suggesting that simply rebalancing PIP levels alone may not be sufficient for symptomatic rescue, and that a specific and precise pairing between kinase and phosphatase is required. This is supported by our data, where Pik3c2b targeting improves the Mtm1 phenotype while Pik3c3 mutation does the opposite.
Our findings thus add to the growing body of work suggesting complex relationships between MTM1 and the kinases that synthesize PI3P. The fact that PIK3C2B inhibition suppresses the loss of MTM1 but that inhibiting PIK3C3 exacerbates the phenotype is noteworthy. This may imply that PIK3C2B and MTM1 act on a pool of PI3P isolated or distinct from that generated by PIK3C3, a concept substantiated by work in vitro (34), in Drosophila (16), and in Caenorhabditis elegans (17), and by our PI3P immunolocalization data from the Pik3c3 and Pik3c2b KOs. If separately regulated pools exist, the exact roles and reasons for these distinct microdomains of PI3P in skeletal muscle are not understood. A recent study from Haucke and colleagues sheds some light on this question. They demonstrate that PI3P and PI4P act sequentially in the procession of endosomal sorting and exocytosis, show that MTM1 and the PI4 kinase PI4K2α regulate this process, and identify abnormalities in exocytosis in cells from MTM patients (35).
Future studies combining PI3P markers with in vivo models such as our zebrafish and mouse mutants should help to continue unraveling these interactions. Notably, a potential explanation for why Pik3c3 mutation worsens the Mtm1 phenotype relates to the fact that PIK3C3 is required for autophagy in muscle (18, 36). We have previously demonstrated in zebrafish that loss of mtm1 does not significantly affect autophagy, but that secondary impairment in autophagic flux in mtm1 morphants promotes a dramatic worsening of the phenotype (22), and also shown that knockout in muscle of Pik3c3 significantly impairs autophagy (18). Thus a similar phenomenon may be at play in our Pik3c3 Mtm1 dKO mice. This hypothesis is supported by our data, which revealed alterations in the autophagy marker LC3 in Pik3c3+/– Mtm1 KO muscle, as well as by several lines of evidence, including the demonstration by Devereaux et al. that Pik3c3 knockdown causes a secondary but massive destabilization of autophagic complexes (34) and by Fetalvero et al. that Mtm1 KO mice have improved muscle mass when treated with an mTOR inhibitor (AZD8055) that increases autophagic flux (37).
In conclusion, we demonstrate that PIK3C2B inhibition results in both prevention and rescue of the MTM phenotype in 2 vertebrate models. This has important therapeutic and pathomechanistic implications for MTM, as well as for the broader field of PIP metabolic diseases, where novel treatment strategies are desperately needed.
Methods
Experimental design.
The objective of this study was to test the hypothesis that kinase inhibition can rebalance PI3P levels and restore normal muscle structure and function in Mtm1 KO mice and zebrafish. We did this through genetic studies in mice using conditional knockout lines of Pik3c2b and Pik3c3, where gene targeting of these kinases was introduced in Mtm1 KO mice either spatially (i.e., muscle-specific KO) or temporally (i.e., in postsymptomatic animals using TAM-inducible Cre). We confirmed genetic modification and tested chemical inhibition using a newly developed zebrafish mtm1 mutant and a panel of known PI3K inhibitors.
Generation of mutant mouse strains.
Mtm1 KO mice were obtained via collaboration from Ana Buj-Bello. Floxed Pik3c2b and Cre mice [B6.FVB(129S4)-Tg(Ckmm-Cre) 5Khn/J, B6.Cg-Tg(ACTA1-cre)79Jme/J, and Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J] were obtained from Jackson Laboratory. Floxed Pik3c3 mice were a gift from Fan Wang (Duke University, Trinity, NC), as described previously (38), and Tg(ER-Cre) mice were a gift from James Rutka (SickKids, Toronto, Canada). Mice were crossed to make either Pik3c2bfl/+ Ckmm-Cre or Pik3c3fl/+ Acta-Cre heterozygous mice, and in-crossed to produce double floxed alleles and thus muscle-specific conditional KO animals. To generate the double-KO male mice (dKOs), Mtm1+/– females were crossed onto the Pik3c2b KO or Pik3c3 KO background. To generate temporal knockout of Pik3c2b in the Mtm1 background, mice with the floxed Pik3c2b allele and Tg(ER-Cre) were fed a TAM diet starting at 21 or 30 days for 1 or 2 weeks. TAM-citrate mixture at 40 mg/kg body weight was incorporated into the standard rodent diet premixed with approximately 5% sucrose as a palatability enhancer (Teklad Diets; Harlan Laboratories) for 1 or 2 weeks. Body weights and food consumption were monitored 3 times weekly. Mouse genotypes were established using previously published methodology (19, 20, 38). Supplemental Table 1 shows all mouse lines used for this study.
Wortmannin trial in Mtm1 KO mice.
We initiated a wortmannin versus DMSO treatment trial starting at 21 days of age. Animals were dosed once daily except on weekends, and given drug or DMSO until the end point (defined as lack of movement, significant weight loss, or hindlimb paralysis) was reached. Wortmannin (Cayman Chemicals) was stored at –20°C in a 10-mg/ml solution in DMSO. Before administration, mice were weighed and a fresh aliquot of wortmannin was diluted 1:1,000 in PBS. Oral gavage was used to administer doses using a calibrated All-Plastic Norm-Ject Syringe (Fisher Scientific) containing 150 μg drug/kg body weight in the morning hours. Control animals were given a corresponding solution of DMSO diluted 1:1,000 in PBS at the same time. A new syringe was used for each animal, and the animal was held to ensure solution was swallowed. Notably, wortmannin versus DMSO administration was performed in a blinded fashion by a trained animal technician based on animal identification number and without knowledge of the genotype.
Quantitative RT-PCR.
Total mRNA was isolated from quadriceps muscle homogenates using Qiagen RNeasy kits and reverse-transcribed with iScript (BioRad Laboratories). Quantitative PCR was performed using SYBR Green reagent on ABI equipment (Invitrogen). Primers for Pik3c2b were as follows: 5′-ACCTGCAACTGTTCCTCCAC-3′ (forward) and 5′-ACAGGGCTTCAGCACAAAGT-3′ (reverse). Primers for Pik3c3 transcript analysis were published previously (18).
Western blot analysis.
Protein was isolated in RIPA buffer from quadriceps muscle of KO and age-matched littermates. Primary antibodies for PIK3C2B (catalog 611342, BD Biosciences), β-actin (catalog 4967, Cell Signaling Technology), and LC3 (catalog NB100-2331, Novus Biologicals Canada) were applied overnight at 4°C at 1:1,000 dilution in 3% BSA, 5% nonfat milk, 0.1% Tween-20. Indirect analysis of protein expression was performed using Bio-Rad Image Lab Software.
Histology and staining analysis.
Flash-frozen muscle tissue from tibialis anterior, 8-mm cross sections, was stained with Mayer’s H&E or succinate dehydrogenase (SDH). Micrographs were captured with an Infinity1 camera (Lumenera Corp.) through an Olympus BX43 light microscope. Quantification of the number of centrally nucleated fibers and myofiber size was performed manually from H&E photographs taken at ×40 magnification. For indirect immunofluorescence, frozen sections were blocked for 1 hour, then incubated overnight at 4°C with Dysferlin antibody (catalog ab124684, Abcam), diluted 1:100 in blocking solution. Secondary antibodies (Alexa Fluor 488; Invitrogen) were applied for 1 hour at room temperature at 1:1,000 dilution. PI3P staining was performed using routine IHC on frozen sections using Acetone fixation at –20°C for 20 minutes. Sections were blocked in 1% TBS solution, 10% goat serum, and incubated in the primary PI3P antibody 1:50 (catalog Z-P003, Echelon Biosciences) for 1 hour at 37°C. Biotinylated secondary antibody 1:100 (ABC kit; Vector Laboratories) was applied for 30 minutes at 37°C, and incubated in the Avidin-Biotin detection system for 30 minutes at 37°C. Signal was detected using DAB, counterstained in hematoxylin, and mounted with Permount (Thermo Fisher Scientific).
PI3P ELISA.
Lipid analysis was performed by indirect analysis of whole quadriceps muscle lysates using Echelon’s PI3P Mass ELISA kit (K-3300) (Echelon Biosciences), as previously described (10).
Transmission electron microscopy.
Mouse muscles for transmission electron microscopy were fixed in 2% glutaraldehyde containing 0.1 M sodium cacodylate buffer overnight at 4°C, then taken to the Advanced Bioimaging Center (Mount Sinai, Toronto, Canada). For zebrafish, 5-dpf larvae were anesthetized in tricaine and fixed in Karnovsky’s fixative overnight at 4°C. After rinsing in buffer, tissue was postfixed in 1% osmium tetroxide in buffer, dehydrated, and embedded in Quetol-Spurr resin. Sections 90 nm thick were cut on an RMC MT6000 ultramicrotome, stained with uranyl acetate and lead citrate, and viewed with an FEI Tecnai 20 microscope.
Phenotypic studies.
Motor functional analysis was performed in the Clinical Phenogenomics Core facility at the Toronto Centre for Phenogenomics per standard protocol using the Bioseb-Bio-GS3 for grip strength and the Clever Sys Inc-ExerGait for gait analysis.
Generation of mtm1 mutants.
An mtm1 mutant line was generated using zinc finger nucleases (ZFN; Sigma-Aldrich) targeting exon 5 of mtm1. The sequences for the ZFN binding site and cut site (PZFN1 left half site/cut site/PZFN2 right half site) were 5′-CTTAGGTGTCATAAGCCGT/gtagaa/AAGATGGGCGGAGCC-3′ (Sigma-Aldrich). One-cell-stage, AB WT embryos were injected with 4.6 nl ZFN solution with a Nanoject (Drummond Scientific). Potential founders (F0) were outcrossed to AB WT fish. Genomic DNA was isolated from single F1 embryos at 4 dpf, and mtm1 sequence spanning the ZFN target site was amplified with the following primers: F: 5′-TGGCCTAATGATTTTTGCACTGTTC-3′; and R: 5′-TGAAGAAGCAGCTGCATTAAAAGTTCT-3′.
Amplicons were then sequenced with primer F: 5′-TTACAGTATGTGCAGGATTGCTGGT-3′ to identify the presence of any mutations. Carriers were identified and outcrossed ultimately to the F4 generation. In-cross progeny from the F4 generation were characterized in this study.
Genotyping zebrafish for mtm1.
The following primer pair was used to specifically amplify the WT allele: F: 5′-TTACAGTATGTGCAGGATTGCTGGT-3′; and R: 5′-GACTGGAGGCTCCGCCCATCTTT-3′. The 8-bp deletion allele was amplified using the following primer pair: F: 5′-CCTTAGGTGTCATAAGCCGTGAT-3′; and R: 5′-AGAACTTTTAATGCAGCTGCTTCTTCA-3′.
Zebrafish phenotypic analysis.
F5 larvae from heterozygous carriers were used for analysis of fin fold, pectoral fin, and liver phenotypes. Mutants were segregated at 3 dpf by morphology under a dissection microscope. To document mtm phenotypes, larvae were anesthetized with approximately 0.6 mM tricaine methanesulfonate and mounted in 3% methylcellulose on glass slides, and bright-field images were taken with a microscope (Olympus BX43). Fin fold morphology was scored at 4 dpf (unless otherwise noted) on a 3-point ordinal scale that includes mild, moderate, and severe categories (Supplemental Figure 6). For statistical analysis, all “mild” mutants were counted as 1, “moderate” as 2, and “severe” as 3 to generate an average rank score. Appropriate nonparametric analyses were used to compare the distribution of ranked scores between groups and statistical significance determined using GraphPad.
mRNA rescue of mtm mutants.
Full-length zebrafish mtm1 ORF fused to EGFP and cloned into a pCS2 plasmid vector (9) was linearized and in vitro transcribed using the mMessage mMachine SP6 kit (Ambion). Embryos were collected from pairwise matings of F4 mtm1+/Δ8 for injections at the 1-cell stage. mRNA was prepared at approximately 66.7 ng/μl in sterile water with phenol red. A total of 200 pg mRNA was delivered directly into the cell with a Picopump (World Precision Instruments) and needle calibrated to a 3-nl drop size. At 1 dpf, all larvae were dechorionated. Injected embryos were sorted into GFP+ and GFP– groups. At 4 dpf, larvae were anesthetized with tricaine and severity of fin degeneration scored on the 3-point ordinal scale (see above). Bright-field images were taken of each individual larva, and genotypes were confirmed by PCR.
Morpholino knockdown of mtm1 and pik3c2b.
Morpholinos (MOs) were designed to the exon 3 splice acceptor site of mtm1 (Ex3-SA; 5′-CAATGTTCCTGCGTGTGTTGACAGG-3′) and exon 24 splice donor site of pik3c2b (Ex24-SD; 5′-AGCTCTTTCAGTGCTCCTACCGTTT-3′). An MO designed to a random sequence (5′-CCTCTTACCTCAGTTACAATTTATA-3′) with no homology by Basic Local Alignment Search Tool (BLAST) analysis in the zebrafish genome was used as a control (GeneTools LLC). AB zebrafish were used for injections. For mtm1 knockdown, 1.5 nl of 0.6 mM (~7.5 ng) control MO or mtm1 Ex3-SA MO was delivered into the yolk of 1-cell stage embryos with a Picopump (World Precision Instruments) and needle calibrated to a 0.5-nl drop size (22). For pik3c2b MO experiments, 3 nl of 0.16 mM (~4 ng) either control or pik3c2b Ex24-SD MO was used. At 4 dpf, larvae were anesthetized and fin morphology scored on an ordinal scale from WT-like to severe as described above.
Chemical treatments.
The following chemicals and concentrations were used: 5–10 μM LY294002 (Calbiochem), 50–100 nM wortmannin (Calbiochem), 100–500 nM VPS34-IN1 (Calbiochem), 100–500 nM GDC-0941 (APExBIO), and 100–500 nM PI-103 (APExBIO). Chemical stocks were prepared in DMSO and added 1:500 (0.2% of final volume) to egg water with no methylene blue added to prepare working concentrations. Groups of approximately 50 embryos (1 dpf) were distributed into sterile 10-cm tissue culture dishes (Corning) containing 40 ml of egg water plus chemical. Every day, 50% of the egg water was discarded and replaced with freshly prepared egg water plus chemical. Culture dishes were wrapped in aluminum foil and incubated at 28.5°C.
Photochemical activation of motor behavior.
To assess motor function of the mtm mutant, an assay was developed using optovin analogs identified by Kokel et al. (39). Optovin is a reversible TRPA1 ligand that elicits motor excitation following exposure to light. In our study, optovin analog 6b8 (ID 5707191; ChemBridge) was used to promote motor activity. Optovin 6b8 stock was prepared to 10 mM in DMSO and 10 μM working concentrations by addition of stock 1:1,000 in egg water, and 150 μl egg water plus optovin was aliquoted to 96-well plates. Next, single 7-dpf larvae were added to wells using a glass pipette. Larvae were deposited into wells by surface tension or by free swimming, both of which negligibly changed the total volume in the well. To ensure data were reproducible, plates were covered with foil to protect them from light and larvae incubated at 28.5°C for 5 minutes before motor behavior analysis. The time between addition of the first and last larvae to their wells was between 5 and 10 minutes. After 5 minutes of incubation, motor behavior of larvae was monitored for 30 seconds using the ZebraBox platform (ViewPoint). Parameters were set to capture 10 seconds of baseline behavior in the dark followed by 10 seconds of exposure to white light to elicit ambulatory movement and finally 10 seconds of recovery behavior in the dark. The average total distance traveled during the 30-second cycle was used to compare groups (i.e., mtm mutants vs. siblings and mtm + DMSO vehicle control vs. mtm + chemicals). These data were treated as continuous and analyzed using statistics software (GraphPad Prism).
Statistics.
Postcapture analysis, including tests of statistical significance, was performed using Microsoft Excel 2008 (Microsoft) and GraphPad Prism version 5 for Mac OSX (GraphPad Software). The difference between groups was assessed by ordinary 1-way ANOVA followed by Dunnett’s multiple comparisons test or 2-tailed Student’s t test (for 2-sample comparisons). For survival curves, the log-rank (Mantel-Cox) test was performed. Differences were considered to be statistically significant at P less than 0.05 (*), P < 0.01 (**), or P < 0.001 (***). All data unless otherwise specified are presented as mean ± SEM.
Study approval.
All procedures involving mice were performed in compliance with the Animals for Research Act of Ontario and the Guidelines of the Canadian Council on Animal Care using an institutionally approved protocol reviewed by the Centre for Phenogenomics (Toronto, Canada) Animal Care Committee. All zebrafish experiments were performed in accordance with the policies and guidelines of the Canadian Council on Animal Care and the University of Toronto using standard care practices and an institutionally reviewed and approved animal use protocol.
Author contributions
NS performed the majority of the mouse experiments, while JRV performed most zebrafish studies. HG, AR, XL, and NME all contributed technical assistance to the murine studies, and AED and HG assisted with zebrafish work. CD and GD performed PIP measurements and assisted in interpretation of PIP data. SVB helped with force generation studies. ABB contributed the Mtm1 KO mice and critical comments on the manuscript. ELF was instrumental to the design and execution of the experiments. JJD designed and conceived the study, interpreted all data, and formulated the manuscript.
Supplementary Material
Acknowledgments
We thank the University of Michigan and the Toronto Centre for Phenogenomics mouse facilities for assistance with mouse husbandry. We thank Fan Wang for the gift of the floxed Pik3c3 mice and James Rutka for the Tg(ER-Cre) mice. We acknowledge Carey Backus for assistance with mouse breeding, Andrea Pang for help with mouse genotyping, Nika Maani, Lindsay Smith, and Ramil Noche for assistance with zebrafish experiments, and Sarah Smith for critical reading of the manuscript. This study was supported through multiple funding sources, including the University of Michigan Department of Pediatrics (to JJD), the Taubman Center at the University of Michigan (to JJD and ELF), the Muscular Dystrophy Association (to JJD), an NIH K08 career development award to JJD, the Myotubular Trust (to JJD), and the Canadian Institutes for Health Research (to JJD).
Footnotes
Conflict of interest: A. Buj-Bello is an inventor of a patent for gene therapy for myotubular myopathy and a scientific advisor for Audentes Therapeutics.
Reference information:J Clin Invest. 2016;126(9):3613–3625. doi:10.1172/JCI86841.
Contributor Information
Nesrin Sabha, Email: nesrin.sabha@sickkids.ca.
Hernan Gonorazky, Email: hernan.gonorazky@sickkids.ca.
Aaron Reifler, Email: reiflera@med.umich.edu.
Ann E. Davidson, Email: ann.davidson@sickkids.ca.
Xingli Li, Email: lixingli@med.umich.edu.
Gilbert Di Paolo, Email: gd2175@cumc.columbia.edu.
Eva L. Feldman, Email: efeldman@umich.edu.
References
- 1.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443(7112):651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 2.Krauss M, Haucke V. Phosphoinositide-metabolizing enzymes at the interface between membrane traffic and cell signalling. EMBO Rep. 2007;8(3):241–246. doi: 10.1038/sj.embor.7400919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivière JB, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44(8):934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow CY, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84(1):85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azzedine H, et al. Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early-onset glaucoma. Am J Hum Genet. 2003;72(5):1141–1153. doi: 10.1086/375034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yap TA, Bjerke L, Clarke PA, Workman P. Drugging PI3K in cancer: refining targets and therapeutic strategies. Curr Opin Pharmacol. 2015;23:98–107. doi: 10.1016/j.coph.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laporte J, et al. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet. 1996;13(2):175–182. doi: 10.1038/ng0696-175. [DOI] [PubMed] [Google Scholar]
- 8.McEntagart M, et al. Genotype-phenotype correlations in X-linked myotubular myopathy. Neuromuscul Disord. 2002;12(10):939–946. doi: 10.1016/S0960-8966(02)00153-0. [DOI] [PubMed] [Google Scholar]
- 9.Dowling JJ, et al. Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 2009;5(2): doi: 10.1371/journal.pgen.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierson CR, et al. Modeling the human MTM1 p.R69C mutation in murine Mtm1 results in exon 4 skipping and a less severe myotubular myopathy phenotype. Hum Mol Genet. 2012;21(4):811–825. doi: 10.1093/hmg/ddr512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amoasii L, et al. Myotubularin and PtdIns3P remodel the sarcoplasmic reticulum in muscle in vivo. J Cell Sci. 2013;126(pt 8):1806–1819. doi: 10.1242/jcs.118505. [DOI] [PubMed] [Google Scholar]
- 12.Hnia K, Vaccari I, Bolino A, Laporte J. Myotubularin phosphoinositide phosphatases: cellular functions and disease pathophysiology. Trends Mol Med. 2012;18(6):317–327. doi: 10.1016/j.molmed.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Al-Qusairi L, et al. T-tubule disorganization and defective excitation-contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc Natl Acad Sci U S A. 2009;106(44):18763–18768. doi: 10.1073/pnas.0900705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Qusairi L, Laporte J. T-tubule biogenesis and triad formation in skeletal muscle and implication in human diseases. Skelet Muscle. 2011;1(1): doi: 10.1186/2044-5040-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razidlo GL, Katafiasz D, Taylor GS. Myotubularin regulates Akt-dependent survival signaling via phosphatidylinositol 3-phosphate. J Biol Chem. 2011;286(22):20005–20019. doi: 10.1074/jbc.M110.197749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Velichkova M, et al. Drosophila Mtm and class II PI3K coregulate a PI(3)P pool with cortical and endolysosomal functions. J Cell Biol. 2010;190(3):407–425. doi: 10.1083/jcb.200911020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu N, Shen Q, Mahoney TR, Neukomm LJ, Wang Y, Zhou Z. Two PI 3-kinases and one PI 3-phosphatase together establish the cyclic waves of phagosomal PtdIns(3)P critical for the degradation of apoptotic cells. PLoS Biol. 2012;10(1): doi: 10.1371/journal.pbio.1001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reifler A, et al. Conditional knockout of pik3c3 causes a murine muscular dystrophy. Am J Pathol. 2014;184(6):1819–1830. doi: 10.1016/j.ajpath.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harada K, Truong AB, Cai T, Khavari PA. The class II phosphoinositide 3-kinase C2beta is not essential for epidermal differentiation. Mol Cell Biol. 2005;25(24):11122–11130. doi: 10.1128/MCB.25.24.11122-11130.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buj-Bello A, et al. The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc Natl Acad Sci U S A. 2002;99(23):15060–15065. doi: 10.1073/pnas.212498399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alliouachene S, et al. Inactivation of the class II PI3K-C2β potentiates insulin signaling and sensitivity. Cell Rep. 2015;13(9):1881–1894. doi: 10.1016/j.celrep.2015.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dowling JJ, Low SE, Busta AS, Feldman EL. Zebrafish MTMR14 is required for excitation-contraction coupling, developmental motor function and the regulation of autophagy. Hum Mol Genet. 2010;19(13):2668–2681. doi: 10.1093/hmg/ddq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight ZA, et al. A pharmacological map of the PI3-K family defines a role for p110α in insulin signaling. Cell. 2006;125(4):733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis MI, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29(11):1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 25.Bago R, et al. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem J. 2014;463(3):413–427. doi: 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Childers MK, et al. Gene therapy prolongs survival and restores function in murine and canine models of myotubular myopathy. Sci Transl Med. 2014;6(220): doi: 10.1126/scitranslmed.3007523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Childers MK, Beggs AH, Buj-Bello A. Gene replacement rescues severe muscle pathology and prolongs survival in myotubularin-deficient mice and dogs. Ann Transl Med. 2015;3(17): doi: 10.3978/j.issn.2305-5839.2015.10.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russo A, Okur MN, Bosland M, O’Bryan JP. Phosphatidylinositol 3-kinase, class 2 beta (PI3KC2β) isoform contributes to neuroblastoma tumorigenesis. Cancer Lett. 2015;359(2):262–268. doi: 10.1016/j.canlet.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maehama T, Fukasawa M, Date T, Wakita T, Hanada K. A class II phosphoinositide 3-kinase plays an indispensable role in hepatitis C virus replication. Biochem Biophys Res Commun. 2013;440(1):150–156. doi: 10.1016/j.bbrc.2013.09.048. [DOI] [PubMed] [Google Scholar]
- 30.Vaccari I, et al. Genetic interaction between MTMR2 and FIG4 phospholipid phosphatases involved in Charcot-Marie-Tooth neuropathies. PLoS Genet. 2011;7(10): doi: 10.1371/journal.pgen.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaccari I, et al. Loss of Fig4 in both Schwann cells and motor neurons contributes to CMT4J neuropathy. Hum Mol Genet. 2015;24(2):383–396. doi: 10.1093/hmg/ddu451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reifler A, et al. Murine Fig4 is dispensable for muscle development but required for muscle function. Skelet Muscle. 2013;3(1): doi: 10.1186/2044-5040-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Paolo G, et al. Impaired PtdIns(4,5)P2 synthesis in nerve terminals produces defects in synaptic vesicle trafficking. Nature. 2004;431(7007):415–422. doi: 10.1038/nature02896. [DOI] [PubMed] [Google Scholar]
- 34.Devereaux K, et al. Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PLoS One. 2013;8(10): doi: 10.1371/journal.pone.0076405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ketel K, et al. A phosphoinositide conversion mechanism for exit from endosomes. Nature. 2016;529(7586):408–412. doi: 10.1038/nature16516. [DOI] [PubMed] [Google Scholar]
- 36.Jaber N, Dou Z, Lin RZ, Zhang J, Zong WX. Mammalian PIK3C3/VPS34: the key to autophagic processing in liver and heart. Autophagy. 2012;8(4):707–708. doi: 10.4161/auto.19627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fetalvero KM, et al. Defective autophagy and mTORC1 signaling in myotubularin null mice. Mol Cell Biol. 2013;33(1):98–110. doi: 10.1128/MCB.01075-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Budolfson K, Wang F. Pik3c3 deletion in pyramidal neurons results in loss of synapses, extensive gliosis and progressive neurodegeneration. Neuroscience. 2011;172:427–442. doi: 10.1016/j.neuroscience.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kokel D, et al. Photochemical activation of TRPA1 channels in neurons and animals. Nat Chem Biol. 2013;9(4):257–263. doi: 10.1038/nchembio.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.