Abstract
Adoptive immunotherapy is a potentially curative therapeutic approach for patients with advanced cancer. However, the in vitro expansion of antitumor T cells prior to infusion inevitably incurs differentiation towards effector T cells and impairs persistence following adoptive transfer. Epigenetic profiles regulate gene expression of key transcription factors over the course of immune cell differentiation, proliferation, and function. Using comprehensive screening of chemical probes with defined epigenetic targets, we found that JQ1, an inhibitor of bromodomain and extra-terminal motif (BET) proteins, maintained CD8+ T cells with functional properties of stem cell–like and central memory T cells. Mechanistically, the BET protein BRD4 directly regulated expression of the transcription factor BATF in CD8+ T cells, which was associated with differentiation of T cells into an effector memory phenotype. JQ1-treated T cells showed enhanced persistence and antitumor effects in murine T cell receptor and chimeric antigen receptor gene therapy models. Furthermore, we found that histone acetyltransferase p300 supported the recruitment of BRD4 to the BATF promoter region, and p300 inhibition similarly augmented antitumor effects of the adoptively transferred T cells. These results demonstrate that targeting the BRD4-p300 signaling cascade supports the generation of superior antitumor T cell grafts for adoptive immunotherapy.
Introduction
Adoptive immunotherapy is a promising therapeutic option for cancer patients. Antitumor T cell grafts can be expanded from tumor-infiltrating lymphocytes or tumor antigen–specific T cells in the peripheral blood (1, 2). Another strategy for generating T cell grafts is the genetic engineering of T cells with tumor-specific T cell receptors (TCRs) or chimeric antigen receptors (CARs) (3). Recent clinical trials have shown that adoptively transferred T cells generated with different approaches can induce clinically relevant responses for a variety of malignancies (4–11). However, although some of the patients can achieve complete eradication of the tumors, many of the patients with partial responses eventually relapse (4, 5, 7, 12, 13). The data from these clinical trials have suggested that persistence of the transferred T cells is highly correlated with treatment outcome (5, 14, 15). Ex vivo cultured T cells form surface marker patterns similar to those of memory T cells in vivo as follows: stem cell–like memory (TSCM), central memory (TCM), and effector memory (TEM) T cells. When adoptively transferred, T cells with TSCM and TCM phenotypes showed superior persistence and antitumor effects compared with T cells with the TEM phenotype in both mice and humans (15–19). However, the in vitro expansion of T cells is inevitably accompanied with their differentiation; TSCM and TCM cells differentiate toward TEM cells as they proliferate upon TCR and cytokine stimulation (20). Therefore, most of the T cell grafts currently used in adoptive T cell therapy trials comprise T cells with excessive differentiation.
Recent studies have highlighted that the differences in epigenetic architecture between each memory T cell subset are responsible for their distinct functions through the differential expression of multiple key transcription factors (21–26). Active or repressive epigenetic marks, including histone modifications and DNA methylation, are closely associated with transcriptional profiles at steady states and dynamic gene expression changes upon TCR stimulation. However, it remains largely unknown whether the exogenous manipulation of epigenetic states influences T cell differentiation status. In this study, we investigated the impact of epigenetic modification on memory T cell differentiation by using chemical probes with defined specificity for epigenetic enzymes and effector proteins. We found that JQ1, a specific inhibitor of bromodomain and extra-terminal motif (BET) proteins, supported the in vitro expansion of T cells with TSCM and TCM features. JQ1-treated T cells showed superior in vivo persistence and antitumor effects. These findings are applicable to adoptive immunotherapy for the generation of optimal T cell grafts.
Results
Screening of epigenetic targets that affect CD8+ T cell differentiation.
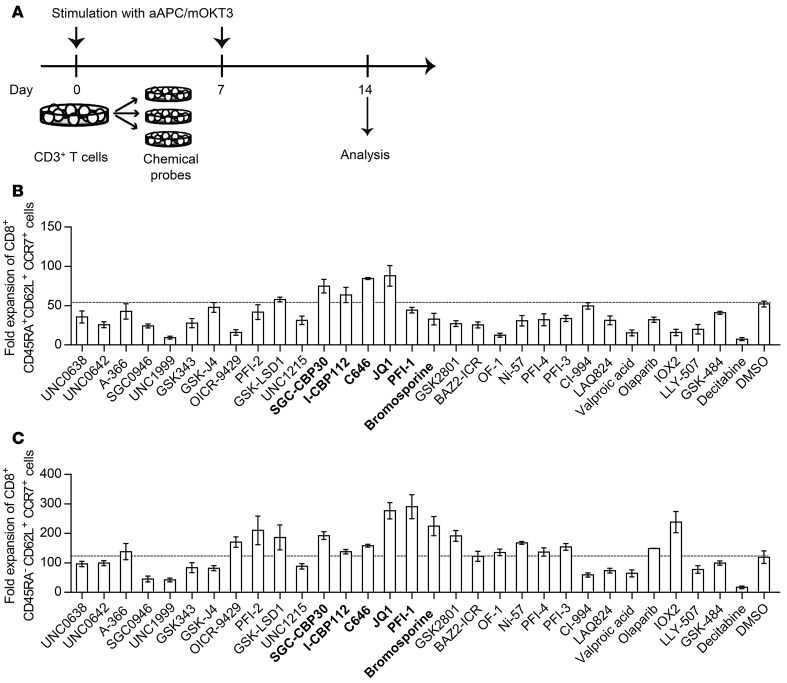
We previously developed artificial antigen-presenting cells (APCs) that express a membrane-bound form of the anti-CD3 monoclonal antibody in conjunction with the immunostimulatory molecules CD80 and CD83 (aAPC/mOKT3) (27). These cells robustly expanded polyclonal CD8+ T cells with memory T cell phenotypes in the presence of cocultured CD4+ T cells. Using this platform, we explored candidate epigenetic modulators that affect the differentiation status of CD8+ T cells without compromising their proliferation. Peripheral blood CD3+ T cells derived from a healthy donor were stimulated weekly with aAPC/mOKT3, and then individually treated with 31 chemical probes with defined epigenetic targets as listed in Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI86437DS1 (Figure 1A). Surface expression of CD45RA, CD62L, and CCR7 on CD8+ T cells was analyzed 14 days following the initial stimulation to identify the CD45RA+CD62L+CCR7+ or CD45RA–CD62L+CCR7+ T cell subsets, since these 2 populations exhibit superior in vivo persistence after adoptive transfer (18, 19, 28). We found that several inhibitors targeting p300, BET proteins, and histone deacetylases (HDACs) supported the maintenance of both CD45RA+CD62L+CCR7+ and CD45RA–CD62L+CCR7+ T cells (Supplemental Figures 1 and 2). In terms of the absolute cell number, treatment with JQ1, which targets BET proteins, resulted in the efficient expansion of both CD45RA+CD62L+CCR7+ and CD45RA–CD62L+CCR7+ T cells (Figure 1, B and C). Although the HDAC inhibitors CI-994, LAQ-824, and valproic acid markedly enhanced the frequency of CD45RA+/–CD62L+CCR7+ cells, they did not increase the absolute number of T cells with these phenotypes because of their considerable toxicity (Supplemental Figure 2). We also tested the effects of each chemical probe on CD3+CD45RO+ T cells, which represent antigen-primed memory T cells, and found that JQ1 treatment enhanced the frequency and fold expansion of CD45RA–CD62L+CCR7+ T cells most efficiently (Supplemental Figure 3). Based upon these observations, we focused on BET proteins as promising targets for modulating CD8+ T cell differentiation during ex vivo T cell culture.
Figure 1. Screening of epigenetic targets that support the maintenance of CD45RA+CD62L+CCR7+ and CD45RA–CD62L+CCR7+ T cell phenotypes.
(A) Peripheral blood CD3+ T cells were stimulated with artificial antigen-presenting cells that express a membrane-bound form of anti-CD3 mAb (clone OKT3), CD80, and CD83 (aAPC/mOKT3), and the cells were subsequently treated with epigenetic chemical probes. The surface expression of CD45RA, CD62L, and CCR7 on CD8+ T cells was analyzed 14 days following the initial stimulation. (B and C) The fold expansion of CD45RA+CD62L+CCR7+ cells (B) and CD45RA–CD62L+CCR7+ cells (C) within the CD8+ T cell population treated with each chemical probe 14 days following TCR stimulation is shown. Inhibitors of p300 and BET proteins are indicated in bold. The error bars represent the SD of 3 technical replicates. The dotted lines indicate the mean values in DMSO-treated control wells.
JQ1 maintains T cells with the functional and transcriptional properties of TSCM and TCM cells.
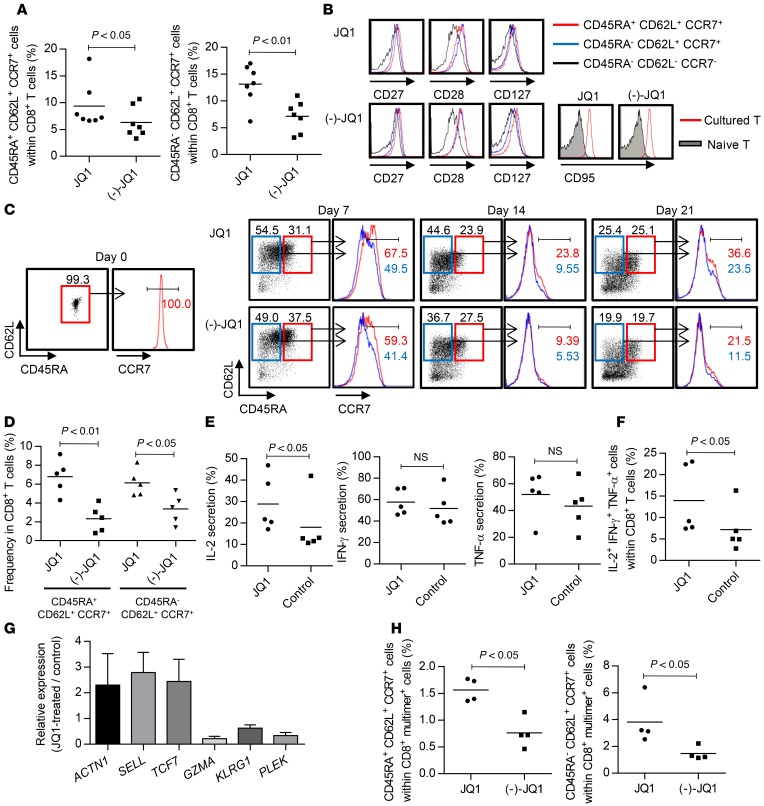
To further investigate the effect of BET protein inhibition on T cell differentiation, we used JQ1 as a representative probe that did not interfere with overall T cell viability or proliferation at 0.15 μM in our experimental system (Supplemental Figure 4, A and B). First, we confirmed that JQ1 treatment supported the maintenance of both CD45RA+CD62L+CCR7+ and CD45RA–CD62L+CCR7+ cells within the CD8+ T cell populations in multiple samples significantly better than (–)-JQ1, an inactive enantiomer of JQ1 (Figure 2A). Both subsets expressed CD27, CD28, and CD127 equally, and all of the cultured T cells were positive for CD95 (Figure 2B). Therefore, the CD45RA+CD62L+CCR7+ and CD45RA–CD62L+CCR7+ cells had phenotypes corresponding to TSCM and TCM cells, respectively, as described previously (28). To assess the effect of JQ1 on T cell differentiation in more detail, we cultured naive (CD45RA+CD62L+CCR7+) T cells in the presence or absence of JQ1. Although they progressively differentiated into T cells with a TEM phenotype upon stimulation, the TSCM and TCM marker phenotypes were significantly maintained with JQ1 treatment (Figure 2, C and D). Furthermore, JQ1-treated CD8+ T cells secreted more IL-2 compared with the control, while they preserved IFN-γ and TNF-α production, resulting in a higher frequency of T cells with polyfunctional cytokine secretion (Figure 2, E and F). These results reflect the attributes of TSCM and TCM cells (28, 29). Although CD4+ T cells maintained higher CD62L and CCR7 expression than CD8+ T cells, JQ1 treatment further enhanced this expression (Supplemental Figure 5, A and B). In addition, the JQ1-treated CD4+ T cells secreted more IL-2 upon TCR restimulation than the control CD4+ T cells (Supplemental Figure 5C). Furthermore, we assessed the expression profile of representative genes with differential expression among the TSCM, TCM, and TEM phenotypes (Supplemental Figure 6). JQ1-treated CD8+ T cells had gene expression signatures consistent with the TSCM and TCM phenotypes compared with the control T cells (Figure 2G). We obtained similar results when CD45RO+ T cells were treated with JQ1; JQ1-treated T cells significantly maintained CD8+ T cells with a TCM phenotype (Supplemental Figure 7A). Similarly, these T cells produced more IL-2 and TNF-α upon TCR engagement and had gene expression profiles consistent with those of the TCM phenotype compared with the control (Supplemental Figure 7, B–D). A similar effect of JQ1 was observed when T cells were stimulated with anti-CD3 antibody alone. In this case, the JQ1-T cells exhibited higher frequencies of TSCM and TCM populations and secreted more IL-2 than the (–)-JQ1–treated T cells (Supplemental Figure 8, A–D). These results suggest that JQ1 treatment can support the maintenance of TSCM and TCM phenotypes independently of costimulatory signals. Finally, we assessed the effect of JQ1 in the context of antigen-specific T cell stimulation using artificial APCs expressing HLA-A2 (aAPC/A2), which was previously developed to expand A2-restricted antigen-specific T cells (8, 30). Peripheral blood CD3+ T cells derived from HLA-A2–positive donors were stimulated weekly with aAPC/A2 pulsed with A2/MART127–35 peptide. Similar to the results observed for aAPC/mOKT3 stimulation, JQ1-treated T cells maintained TSCM and TCM phenotypes significantly better in the A2/MART1 multimer–positive CD8+ T cell population (Figure 2H and Supplemental Figure 9). These results indicate that JQ1 treatment enables the expansion of TSCM and TCM cells while inhibiting their differentiation into TEM cells in vitro.
Figure 2. JQI, a BET bromodomain inhibitor, maintains CD8+ T cells with features of stem cell–like and central memory T cells.
(A) Frequency of CD45RA+/–CD62L+CCR7+ cells within the CD8+ T cell population cultured for 14 days in the presence of JQ1 or (–)-JQ1 (n = 7, paired t test). (B) Expression of CD27, CD28, CD127, and CD95 in the CD8+ T cells cultured in the presence of JQ1 or (–)-JQ1. Representative FACS plots of the samples shown in A. (C and D) CD45RA+CD62L+CCR7+ T cells were stimulated weekly with aAPC/mOKT3 and cultured with or without JQ1. Representative FACS plots of CD45RA, CD62L, and CCR7 expression at the indicated time points (C) as well as the frequency of CD45RA+/–CD62L+CCR7+ cells within the CD8+ T cell population 21 days following initial stimulation (D) are shown (n = 5, paired t test). (E and F) The secretion of IL-2, IFN-γ, and TNF-α was evaluated by intracellular flow cytometry in CD8+CD45RA+CD62L+CCR7+ T cells cultured for 14 days with or without JQ1. The frequency of each cytokine-producing cell type (E) and those secreting all 3 cytokines (F) are shown (n = 5, paired t test). (G) Expression profiles of representative genes with differential expression between the TSCM/TCM and TEM phenotypes. Fold expression levels in JQ1-treated CD8+ T cells relative to those in the control T cells are shown (n = 4). Error bars indicate the SD. (H) CD3+ T cells isolated from HLA-A2+ donors were stimulated by artificial antigen-presenting cells that express HLA-A*02:01, CD80, and CD83 (aAPC/A2) and were pulsed with the MART127–35 peptide once per week. The frequency of CD45RA+/–CD62L+CCR7+ cells within the A2/MART1 multimer+ CD8+ T cell population 21 days after initial stimulation is shown (n = 4, paired t test). NS, not significant.
JQ1-treated T cells have superior in vivo persistence and antitumor effects in a CAR gene therapy model.
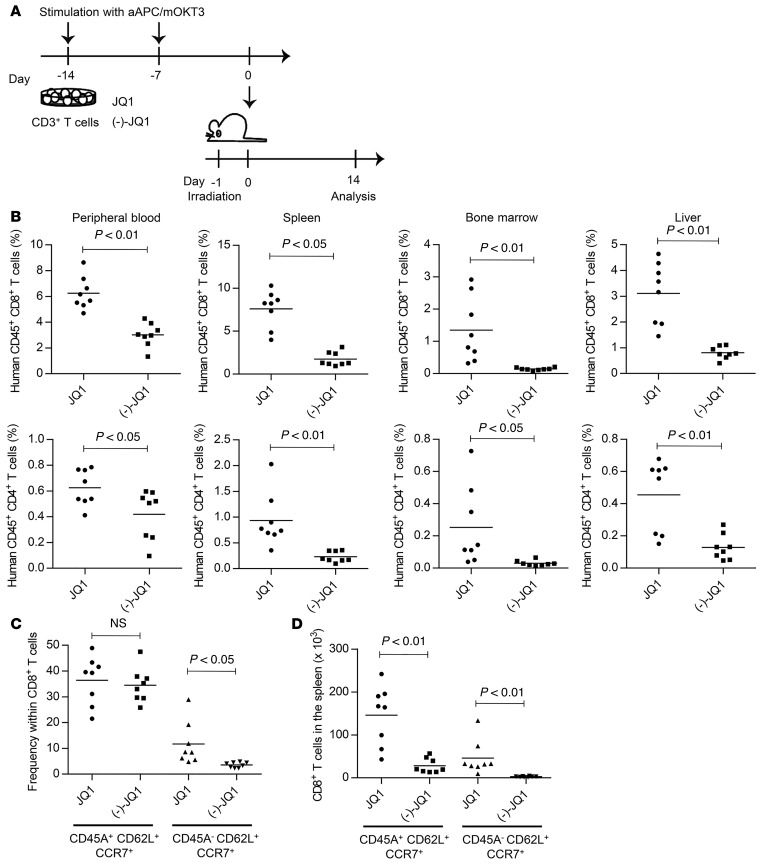
Based on the observation that JQ1 treatment led to the expansion of T cells with TSCM and TCM phenotypes, we assessed its effect on the in vivo persistence of cultured T cells. CD3+ T cells were cultured in the presence of JQ1 or (–)-JQ1 and infused into irradiated NSG mice (Figure 3A). JQ1-treated T cells showed significantly higher engraftment in the peripheral blood, spleen, liver, and bone marrow compared with the control for both CD4+ and CD8+ T cells (Figure 3B). A TSCM subset with comparable frequency was found within the JQ1- or (–)-JQ1–treated CD8+ T cell population engrafted in the spleen, whereas T cells with a TCM phenotype were slightly more frequent among JQ1-treated T cells (Figure 3C). In terms of the absolute number of persistent cells, significantly more TSCM and TCM cells were recovered from mice injected with JQ1-treated T cells (Figure 3D).
Figure 3. JQ1-treated T cells show enhanced in vivo persistence.
(A) CD3+ T cells were stimulated weekly with aAPC/mOKT3, cultured for 14 days in the presence of JQ1 or (–)-JQ1, and subsequently infused into irradiated NSG mice (5 million T cells per mouse). (B) Human CD4+ or CD8+ T cell chimerism in the indicated organs 14 days following adoptive transfer of T cells (n = 8, unpaired t test). (C and D) Frequency of CD45RA+/–CD62L+CCR7+ cells within the CD8+ T cell population (C) and total cell counts of each T cell subset within the spleen (D) (n = 8, unpaired t test). NS, not significant.
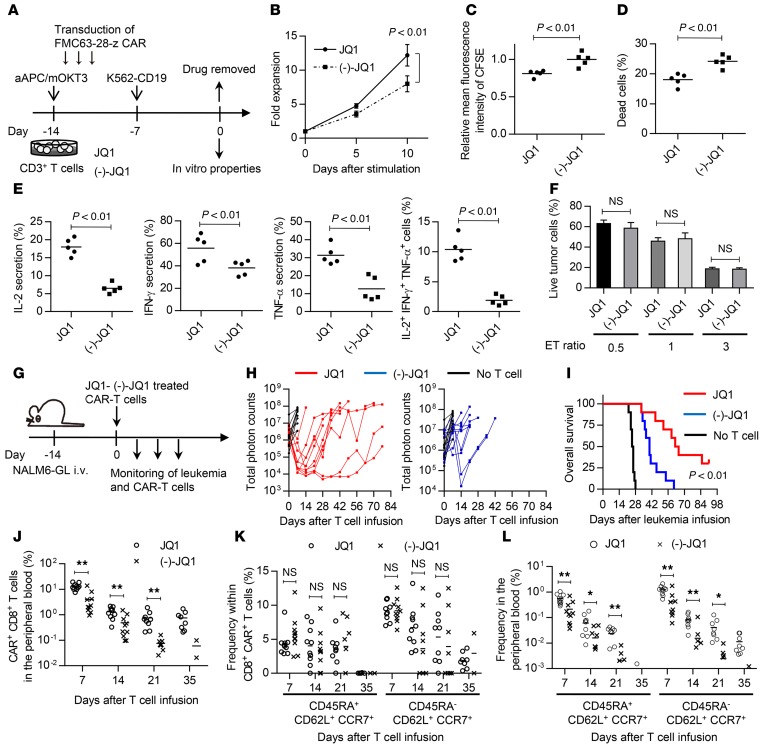
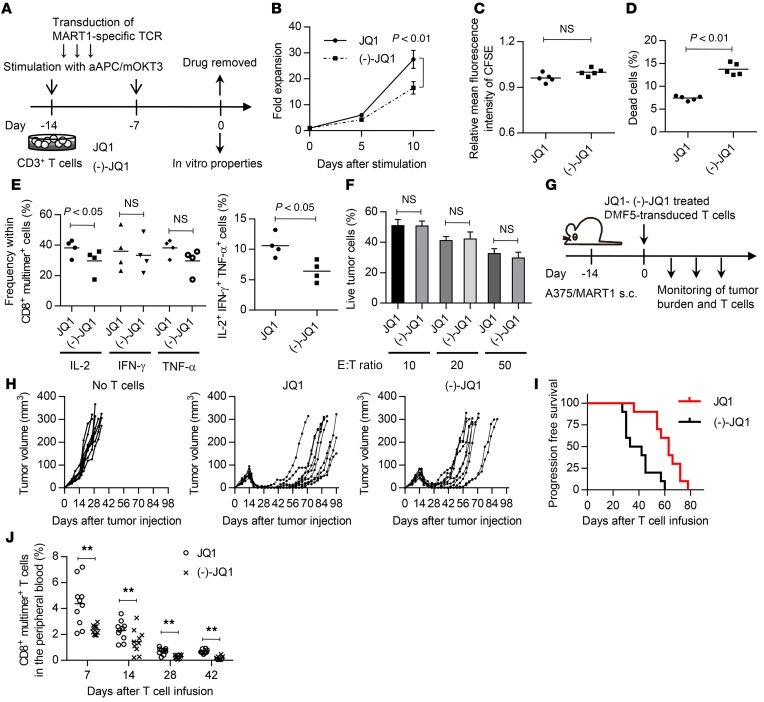
Next, we investigated the in vivo effect of JQ1 on antitumor T cells. Culturing CAR-T cells with IL-7 and IL-15 was recently found to preferentially expand the TSCM population, and those T cells have been shown to exhibit superior antitumor responses in vivo (19). We analyzed the in vitro and in vivo attributes of JQ1-treated CAR-T cells. CD3+ T cells were transduced with an anti-CD19 CAR construct encoding an FMC63-derived single-chain variable fragment (scFv) linked to CD28 and the CD3ζ chain (31). After a 2-week expansion in the presence of JQ1 or (–)-JQ1, the drugs were withdrawn, and the cells’ functional properties were compared (Figure 4A). When serially restimulated with the human erythroleukemic cell line K562 transduced with CD19 (K562-CD19), the JQ1-treated T cells showed superior proliferation compared with the control T cells (Figure 4B). They also exhibited more rapid cell division and reduced cell death upon stimulation (Figure 4, C and D). Moreover, JQ1-treated CAR-T cells showed improved cytokine production upon antigen encounter (Figure 4E). Interestingly, the IFN-γ, TNF-α, and IL-2 secretion levels were enhanced in JQ1-treated T cells. These phenotypes were maintained after repeated stimulation with K562-CD19 (Supplemental Figure 10). The generated CAR-T cells showed similar in vitro cytotoxic activity against the CD19+ acute lymphoblastic leukemia cell line NALM-6 transduced with EGFP fused with firefly luciferase (NALM6-GL; Figure 4F). These results are consistent with previous findings showing that the differentiation status of antitumor T cells does not necessarily affect their in vitro cytotoxic activity (16). In order to assess the in vivo antileukemic effects of JQ1-treated CAR-T cells, the CAR-T cells expanded in the presence of JQ1 or (–)-JQ1 were injected into NSG mice that had been transplanted with NALM6-GL (Figure 4G). All of the untreated mice died within 4 weeks after leukemia cell infusion. Serial imaging of the leukemia cells showed an obvious delay of leukemia progression in the mice transplanted with JQ1-treated CAR-T cells (Figure 4H and Supplemental Figure 11A). The mice transplanted with JQ1-treated CAR-T cells showed significantly longer overall survival compared with the control mice (Figure 4I). We also assessed the persistence of the transplanted CAR-T cells in the peripheral blood. The CAR-T cells progressively declined throughout the time course (Figure 4J and Supplemental Figure 11B). At autopsy, NALM6-GL maintained the expression of CD19 and constituted 30%–90% of the bone marrow cells and 20%–60% of the spleen cells (Supplemental Figure 12A). The residual T cells were mostly negative for CAR expression, and very few CAR+ T cells were recovered from the spleen and bone marrow (Supplemental Figure 12B and Supplemental Table 2). These results indicate that the cause of relapse was CAR-T cell disappearance rather than the loss of CD19 expression in leukemia cells. Nevertheless, the JQ1-treated T cells showed significantly better persistence than the control T cells at each time point. When evaluated in the peripheral blood, CAR-T cells with TSCM and TCM phenotypes gradually decreased and almost disappeared by day 35 after T cell transplantation, suggesting that they differentiated into TEM cells (Figure 4K). Since there was no significant difference in the frequency of these phenotypes within the CD8+CAR+ T cell population, the JQ1-treated T cells similarly differentiated into effector cells in vivo after withdrawal of the drug. Importantly, the overall frequency of these populations was higher in the JQ1-treated group at each time point (Figure 4L). These results demonstrate that JQ1 treatment enhances the in vivo persistence of CAR-T cells, which contributes to improved antitumor effects.
Figure 4. JQ1 treatment improves the antitumor activity of chimeric antigen receptor–transduced T cells in a mouse leukemia model.
(A) CD3+ T cells were retrovirally transduced with the anti-CD19 chimeric antigen receptor (CAR) consisting of anti-CD19 scFv, CD28, and CD3z, and the cells were cultured with JQ1 or (–)-JQ1. The drugs were removed on day 14, and the functional properties of the CAR T cells were analyzed. (B) CAR T cells generated as shown in A were serially stimulated with K562 transduced with CD19 (K562-CD19), and the fold expansion of the CD8+ CAR T cells was calculated at the indicated time points (n = 5, paired t test). Error bars indicate the SD. (C and D) JQ1-CAR T and (–)-JQ1-CAR T cells were labeled with CFSE and stimulated with K562-CD19. The relative mean fluorescence intensity of CFSE (C) and cell viability (D) were assessed 4 days following stimulation (n = 5, paired t test). (E) Cytokine secretion profiles of JQ1-CAR T and (–)-JQ1-CAR T cells upon stimulation with K562-CD19 (n = 5, paired t test). (F) CAR T cells were cocultured with the CD19-positive leukemia cell line NALM-6, which expresses EGFP-luciferase fusion protein (NALM6-GL), at the indicated ratio, and the residual NALM6-GL was analyzed by flow cytometry (n = 5, unpaired t test). (G) NSG mice were intravenously injected with NALM6-GL, then with CAR-transduced T cells 14 days after tumor injection. (H) Total photon counts measured from the in vivo bioluminescent imaging of luciferase activity at the indicated time points following infusion of T cells. (I) Kaplan–Meier curve for overall survival of the mice treated with CAR-transduced T cells (n = 10, log-rank test). Representative data from 2 experiments are shown. (J) Frequency of human CD45+CD8+ CAR-transduced T cells in the peripheral blood following T cell injection. (K and L) The persistence of CD45RA+/–CD62L+CCR7+ T cells was monitored in the peripheral blood. The frequencies within the CD8+ CAR+ T cell population (K) and overall peripheral blood cells (L) (n = 10, unpaired t test) are shown. *P < 0.05, **P < 0.01. NS, not significant.
JQ1-treated T cells have superior in vivo persistence and antitumor effects in a TCR gene therapy model.
We next investigated the influence of JQ1 treatment on the antitumor effects of TCR gene–transferred T cells. CD3+ T cells were transduced with HLA-A2/MART127–35-specific TCR genes (clone DMF5) and were expanded for 2 weeks in the presence or absence of JQ1 (Figure 5A). JQ1-treated DMF5-transduced T cells showed superior proliferation upon serial stimulation by aAPC/A2 pulsed with MART127–35 peptide (Figure 5B). Although no significant difference in the cellular division rate was found, the JQ1-treated T cells showed better viability than the (–)-JQ1–treated T cells upon stimulation (Figure 5, C and D). JQ1 treatment contributed to the maintenance of IL-2 secretion, and the frequency of cells producing multiple cytokines was significantly higher among the JQ1-treated T cells (Figure 5E). We also evaluated cytokine secretion in the context of repeated TCR stimulation and found that JQ1-treated T cells similarly maintained their IL-2 secretion capacity (Supplemental Figure 13). As seen in the CAR-transduced T cells, no significant difference existed in the cytotoxic activity against A375 melanoma cells transduced with full-length MART1 (A375-MART1) (Figure 5F) (32). To investigate the in vivo antitumor responses, the expanded T cells were injected into NSG mice that had been subcutaneously inoculated with A375-MART1 (Figure 5G). Although both JQ1- and (–)-JQ1–treated T cells were initially effective against tumors, they relapsed at later time points (Figure 5H). However, JQ1-treated T cells significantly delayed the progression of tumors compared with control T cells, indicating the impact of JQ1 treatment on the durability of antitumor effects (Figure 5I). Moreover, the frequency of CD8+ A2/MART1 multimer+ T cells in the peripheral blood was significantly higher in the JQ1-treated group (Figure 5J). These results demonstrate that JQ1 treatment promoted superior in vivo antitumor activity in TCR gene therapy, likely due to the improved persistence of the transferred T cells.
Figure 5. JQ1 treatment improves the antitumor activity of T cell receptor–transduced T cells.
(A) CD3+ T cells were retrovirally transduced with HLA-A2/MART127–35 TCR (clone DMF5) and cultured in the presence of JQ1 or (–)-JQ1. The drugs were removed on day 14, and the functional properties of the A2/MART1 T cells were analyzed. (B) DMF5-transduced T cells generated in the presence of JQ1 or (–)-JQ1 were serially stimulated with aAPC/A2 pulsed with MART127–35 peptide, and the fold expansion of the CD8+ A2/MART1 cells was calculated at the indicated time points (n = 4, paired t test). Error bars indicate the SD. (C and D) JQ1-DMF5 and (–)-JQ1-DMF5 T cells were labeled with CFSE and stimulated with aAPC/A2 pulsed with MART127–35 peptide. The relative mean fluorescence intensity of CFSE (C) and cell viability (D) were assessed 4 days following stimulation (n = 5, paired t test). (E) Cytokine secretion profiles of JQ1-DMF5 T cells and (–)-JQ1-DMF5 T cells upon stimulation with aAPC/A2 pulsed with the cognate peptide (n = 4, paired t test). (F) DMF5 T cells were cocultured with the melanoma cell line A375 transduced with full-length MART1 (A375/MART1) at the indicated ratio, and the residual cells were analyzed by flow cytometry (n = 5, unpaired t test). (G) The HLA-A2/MART1 T cells generated as shown in A were adoptively transferred into NSG mice subcutaneously injected with 0.5 million A375/MART1 cells. (H) Depiction of the tumor volume until 14 weeks after tumor cell injection (n = 10 each). (I) Kaplan–Meier curve for progression-free survival of the treated mice (n = 10). Representative data from 2 experiments are shown. (J) Frequency of human CD8+ A2/MART1 multimer+ T cells in the peripheral blood following T cell injection. **P < 0.01. NS, not significant.
JQ1 promotes the maintenance of TSCM and TCM phenotypes by suppressing BATF.
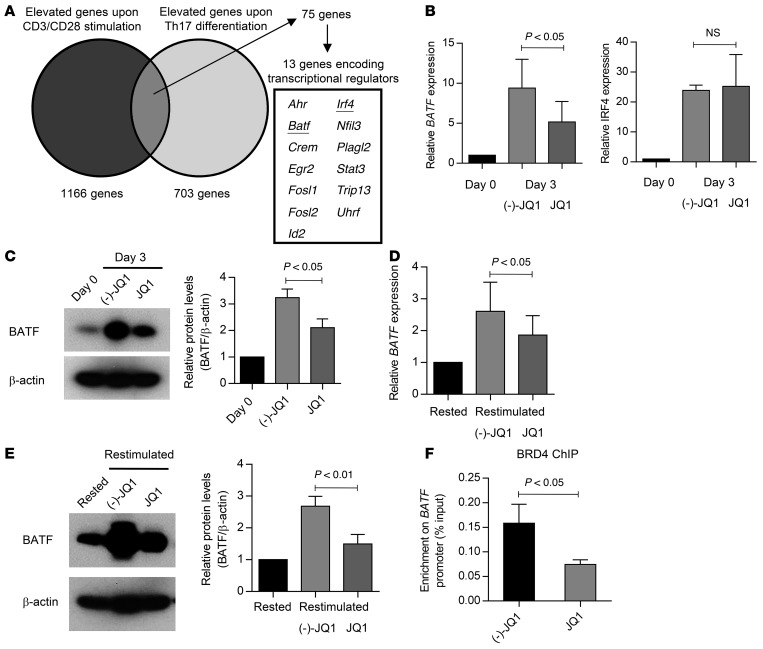
We next investigated the mechanism of how JQ1 treatment affects T cell differentiation. Because the effect of JQ1 was more prominent after repeated TCR stimulation (Figure 2C), we speculated that JQ1 treatment prevents T cell differentiation by inhibiting genes induced upon TCR stimulation. Mele and colleagues recently reported that JQ1 inhibits Th17 differentiation in both mouse and human CD4+ T cells by repressing multiple Th17-related genes (33). Based on these observations, we analyzed the previously published microarray data and extracted the transcription factors upregulated upon both TCR stimulation and Th17 differentiation (Figure 6A). Among these genes, we first focused on BATF and IRF4, which together play an important role in the effector CD8+ T cell differentiation (34). The expression of both genes was rapidly upregulated upon aAPC/mOKT3-mediated TCR stimulation (Figure 6B). Although JQ1 treatment significantly attenuated BATF expression, it did not affect IRF4 expression. The JQ1-mediated suppression of BATF expression was also confirmed at the protein level (Figure 6C). We observed that BATF expression was similarly upregulated upon restimulation of the cultured T cells with aAPC/mOKT3, which was suppressed by JQ1 treatment at both the mRNA and protein levels (Figure 6, D and E). To investigate whether BET proteins directly regulate BATF expression, we performed chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) with an anti-BRD4 antibody, one of the major bromodomain transcription factors targeted by JQ1. As shown in Figure 6F, JQ1-treated T cells exhibited significantly less relative enrichment of BRD4 in the promoter region of BATF than control T cells, suggesting that BRD4 binds to the BATF promoter region and contributes to its upregulation upon TCR stimulation.
Figure 6. BATF expression is downregulated by JQ1 treatment.
(A) Venn diagram showing the overlap between TCR-induced genes and genes upregulated upon Th17 differentiation. (B) Real-time quantitative PCR analysis of BATF and IRF4 expression in CD8+ T cells 3 days after stimulation with aAPC/mOKT3 in the presence of JQ1 or (–)-JQ1. Fold expression changes (normalized to UBC) compared with the CD8+ T cells before stimulation are shown (n = 4, paired t test). (C) Immunoblotting of BATF in the same samples as in B. Representative images and the quantified protein levels relative to β-actin are shown (n = 4, paired t test). Error bars depict the SD. (D) Real-time quantitative PCR analysis of BATF in CD8+ T cells rested overnight in cytokine-free media or restimulated with aAPC/mOKT3 in the presence of JQ1 or (–)-JQ1. Fold expression changes relative to rested samples are shown (n = 4, paired t test). (E) Immunoblotting of BATF in the same samples as in D. Representative images and quantified protein levels relative to β-actin are shown. (n = 4, paired t test). Error bars depict the SD. (F) CD8+ T cells treated with JQ1 or (–)-JQ1 were restimulated with aAPC/mOKT3, and the chromatin samples were immunoprecipitated with an anti-BRD4 antibody. The enrichment of BRD4 at the promoter region of BATF was measured with qPCR (n = 4, paired t test). Error bars indicate the SD. NS, not significant.
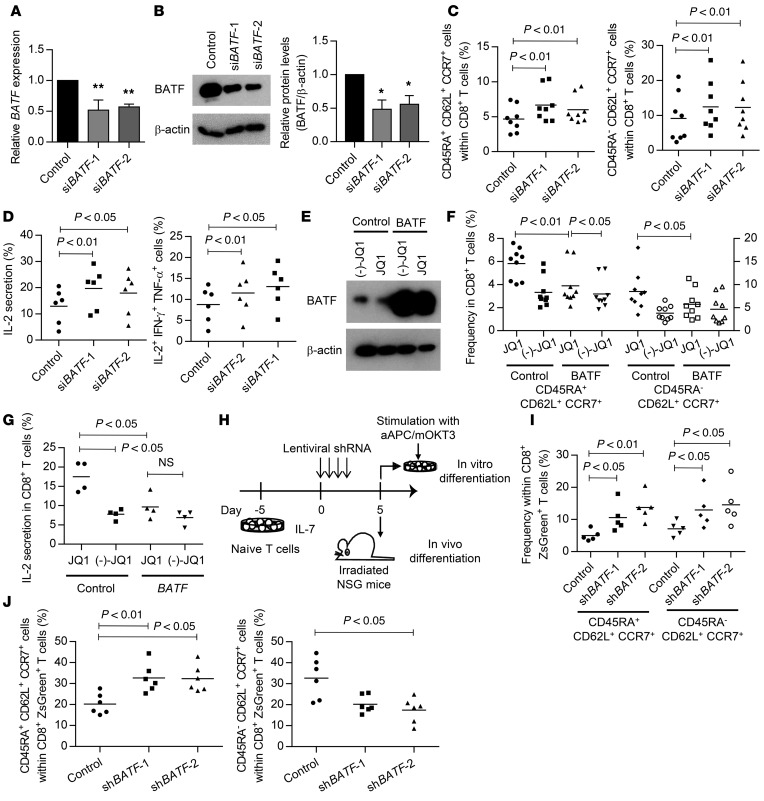
We next investigated whether the blockade of BATF expression contributed to the expansion of T cells with TSCM and TCM phenotypes. CD3+ T cells were stimulated with aAPC/mOKT3 and retrovirally transduced with an siRNA targeting BATF (Figure 7, A and B). The knockdown of BATF significantly enhanced the maintenance of the CD45RA+CD62L+CCR7+ and CD45RA–CD62L+CCR7+ populations following subsequent TCR stimulation (Figure 7C). These 2 populations similarly expressed CD27, CD28, and CD127 (Supplemental Figure 14). Moreover, the T cells with knockdown of BATF showed enhanced IL-2 secretion and cytokine polyfunctionality, as seen in the JQ1-treated T cells (Figure 7D). Conversely, when T cells were ectopically transduced with BATF cDNA, the frequency of T cells with TSCM and TCM phenotypes was partially but significantly reduced in the presence of JQ1 (Figure 7, E and F). The JQ1-treated BATF-overexpressing T cells exhibited decreased IL-2 secretion (Figure 7G). These results demonstrate that BATF upregulation is associated with the differentiation of CD8+ T cells into effector memory T cells.
Figure 7. Downregulation of BATF expression by JQ1 treatment contributes to the maintenance of the TSCM and TCM phenotypes.
(A and B) Validation of BATF knockdown with qPCR (A, n = 4) and immunoblotting (B, representative blots of 3 experiments and quantified protein levels). Error bars indicate the SD. *P < 0.05, **P < 0.01 in a 1-sample t test compared to control siRNA (Ctsh), which was assigned a value of 1. (C) CD3+ T cells were stimulated with aAPC/mOKT3 and transduced with control or siBATF and truncated nerve growth factor receptor (ΔNGFR). The frequency of CD45RA+/–CD62L+CCR7+ cells within the ΔNGFR+CD8+ T cell population 14 days after initial stimulation is shown (n = 8, paired ANOVA). (D) The IL-2, IFN-γ, and TNF-α secretion levels were assessed by intracellular flow cytometry in the ΔNGFR+CD8+ T cell population upon restimulation with aAPC/mOKT3. Frequencies of IL-2–secreting cells and cells producing all 3 cytokines are shown (n = 6, paired ANOVA). (E and F) CD3+ T cells were stimulated with aAPC/mOKT3 and transduced with the control ΔNGFR or BATF-IRES-ΔNGFR. The CD8+NGFR+ cells were isolated on day 7 and immunoblotted for BATF (E, representative blots of 4 experiments). Frequencies of CD45RA+CD62L+CCR7+ cells (left y axis) and CD45RA–CD62L+CCR7+ cells (right y axis) within the ΔNGFR+CD8+ T cell population were assessed 14 days following stimulation (F, n = 9, paired ANOVA). (G) IL-2 production of NGFR+CD8+ T cells upon restimulation with aAPC/mOKT3 was assessed by intracellular flow cytometry (n = 4, paired ANOVA). (H–J) Naive T cells were pretreated with IL-7, transduced with lentiviral shRNAs against BATF, and stimulated with aAPC/mOKT3 or transplanted into irradiated NSG mice. Frequency of CD45RA+/–CD62L+CCR7+ cells within the ZsGreen+CD8+ T cell population was evaluated 10 days after stimulation with aAPC/mOKT3 (I, n = 5, paired ANOVA) or 11 days following T cell transplantation into NSG mice (J, n = 6, paired ANOVA). NS, not significant.
Next, we studied the effect of BATF knockdown on memory T cell formation from naive T cells. We used a lentivirus shRNA-mediated knockdown system for transduction into naive T cells without TCR stimulation (Supplemental Figure 15, A and B). Isolated naive T cells were cultured in the presence of IL-7 and transduced with lentivirus vectors (Figure 7H). We confirmed that naive T cells were successfully transduced with the lentiviral plasmid without undergoing differentiation (Supplemental Figure 15C). Upon in vitro TCR stimulation with aAPC/mOKT3, they acquired a memory T cell phenotype (Supplemental Figure 16A). The BATF-suppressed CD8+ T cells showed significantly better retention of the TSCM and TCM phenotypes compared with the control T cells (Figure 7I). Next, we evaluated naive T cell differentiation in the in vivo setting. Naive T cells have been shown to differentiate into T cells with a memory phenotype through xenogeneic reactions within NSG mice (28). Therefore, we evaluated the impact of BATF knockdown on memory T cell formation by transplanting naive T cells lentivirally transduced to express BATF shRNA into irradiated NSG mice. To elicit the engraftment of T cells, 5 million cultured CD3+ T cells were coinjected into each mouse. The mice were sacrificed 11 days after transplantation, and the differentiation status of the engrafted T cells was assessed. Strikingly, T cells transduced with the shRNA against BATF possessed a higher frequency of TSCM within the CD8+ ZsGreen+ T cell population (Figure 7J and Supplemental Figure 16B). Conversely, the TCM phenotype was less frequent than in the control T cells. At later time points, the transplanted T cells further differentiated into TCM and TEM cells (Supplemental Figure 16C). These results indicate that BATF knockdown does not arrest but does delay differentiation into the TEM cell population both in vitro and in vivo.
We also explored the effects of JQ1 on the expression of other genes listed in Figure 6A and several representative transcription factors associated with effector differentiation in mice (Supplemental Figure 17A) (35–39). Among these genes, FOSL2, ID2, and PRDM1 exhibited significantly reduced expression after JQ1 treatment. Although we tested the effects of these genes individually by siRNA-mediated knockdown, they did not affect the frequency of T cells with a TSCM or TCM phenotype (Supplemental Figure 17, B and C). Additionally, we evaluated whether JQ1 treatment affects the activity of several essential pathways related to TCR and cytokine stimulation: ERK, MAPK, AKT, and S6K (Supplemental Figure 18, A and B). Upon JQ1 treatment, the phosphorylation of S6K, a target protein of the mammalian target of the rapamycin (mTOR) pathway, was significantly reduced. Interestingly, no significant difference was noted in the phosphorylation of AKT, an upstream enzyme regulating the mTOR pathway activity, suggesting that JQ1 regulated S6K phosphorylation downstream of the AKT pathway. Moreover, T cells transduced with lentiviral shRNAs against BATF also demonstrated a decreased level of pS6K compared with the control T cells (Supplemental Figure 18, C and D). These results are consistent with previous findings that the CD8+ T cells derived from BATF-deficient mice exhibited a decreased level of phosphorylated S6K upon effector differentiation and that the inhibition of the mTOR pathway through AKT inhibition or the mTOR inhibitor rapamycin contributed to the generation of T cells with TSCM and TCM phenotypes (34, 40, 41).
p300 recruits BRD4 to the BATF promoter region and induces BATF transcriptional activity.
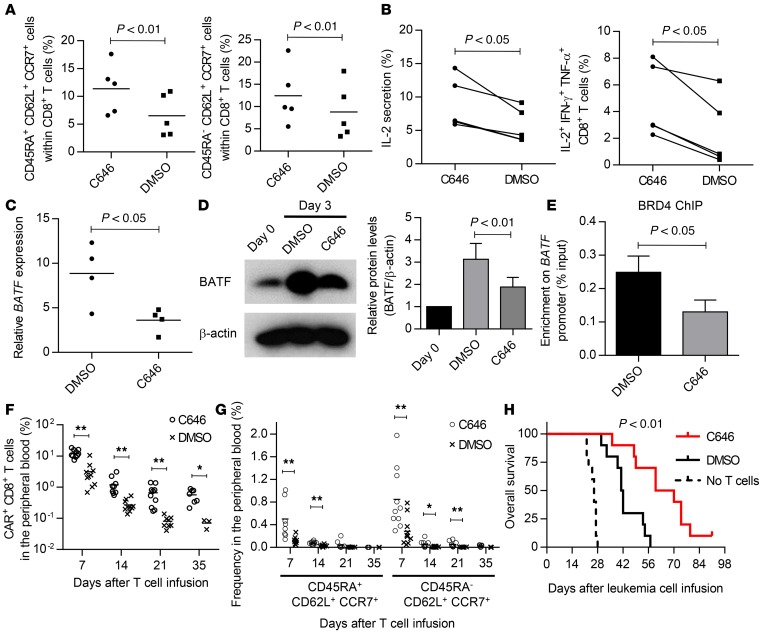
Finally, we investigated the mechanism of BET protein recruitment to the BATF promoter region. In addition to JQ1, we found that multiple p300 inhibitors maintained CD45RA+/–CD62L+CCR7+ cells in the initial screening experiment (Figure 1, B and C). We verified that the treatment of T cells with C646, a p300 inhibitor, significantly enhanced the maintenance of CD8+ T cells with TSCM and TCM phenotypes in multiple donor samples (Figure 8A). These T cells secreted more IL-2 and possessed polyfunctional cytokine secretion capacity (Figure 8B). Because p300 is one of the major histone acetyltransferases, we hypothesized that p300-mediated histone acetylation facilitates the binding of BRD4 to the BATF promoter. Similarly to JQ1, C646-treated CD8+ T cells exhibited significantly decreased expression of BATF upon TCR stimulation at both the mRNA and protein levels (Figure 8, C and D). Furthermore, the enrichment of BRD4 in the BATF promoter region was clearly reduced in the presence of C646 (Figure 8E), suggesting that p300 is associated with the BRD4 occupancy at the BATF promoter. We also assessed the impact of C646 treatment on in vivo antitumor responses using an anti-CD19 CAR treatment model. C646-treated anti-CD19 CAR-T cells showed superior persistence when transferred into mice harboring CD19+ NALM-6 cells (Figure 8F). C646 treatment also enhanced the overall persistence of T cells with TSCM and TCM phenotypes compared with the control at each time point (Figure 8G). No significant difference was found in the frequency of these subsets within the CD8+CAR+ T cell population (Supplemental Figure 19). Consistent with their better persistence, C646-treated CAR-T cells showed superior antileukemic effects when evaluated in terms of overall survival (Figure 8H). Collectively, these findings show that BET proteins contribute to the effector differentiation of cultured T cells in a p300-dependent manner, which impairs their overall survival potential during ex vivo T cell expansion. Pharmacological inhibition of either BET proteins or p300 significantly improves the maintenance of T cells with TSCM and TCM phenotypes and enhances the effect of adoptive immunotherapy.
Figure 8. p300 recruits BRD4 to the BATF promoter region.
(A) Frequency of CD45RA+/–CD62L+CCR7+ cells within the CD8+ T cell population cultured for 14 days in the presence of C646 or DMSO (n = 5, paired t test). (B) Frequency of IL-2–producing cells and cells secreting IL-2, IFN-γ, and TNF-α upon restimulation with aAPC/mOKT3 in C646- or DMSO-treated CD8+ T cells (n = 5, paired t test). (C) Expression of BATF in C646- or DMSO-treated CD8+ T cells 3 days following stimulation with aAPC/mOKT3 (n = 4, paired t test). Fold expression (normalized to UBC) compared with the CD8+ T cells before stimulation is shown. (D) Immunoblotting of BATF in the same samples as in C. Representative blots and quantified protein levels relative to β-actin (n = 4, paired t test). Error bars depict the SD. (E) CD8+ T cells treated with C646 or DMSO were restimulated with aAPC/mOKT3, collected 3 days following stimulation, and subjected to chromatin immunoprecipitation with an anti-BRD4 antibody. Enrichment at the BATF promoter region was measured by qPCR (n = 4, paired t test). Error bars indicate the SD. (F and G) CD3+ T cells were transduced with anti-CD19 CAR and expanded for 2 weeks in the presence of C646 or DMSO. NSG mice were intravenously injected with NALM6-GL and transplanted with CAR+ T cells 14 days after injection of the leukemic cells. Persistence of overall CAR+CD8+ T cells (F) and cells with TSCM and TCM phenotypes (G) in the peripheral blood at the indicated time points is shown. *P < 0.05, **P < 0.01. (H) Kaplan–Meier curves for overall survival of the NALM6-GL–injected mice are shown (n = 10, log-rank test). Representative data from 2 experiments.
Discussion
In this study, we demonstrated that the BET bromodomain inhibitor JQ1 supported the maintenance of T cells with TSCM and TCM phenotypes in vitro without affecting overall proliferation. In addition to surface marker expression patterns, JQ1-treated T cells showed gene expression and cytokine secretion profiles reflecting those specific T cell subsets. Through the comprehensive analysis of the downstream BET protein target genes, we observed that JQ1 inhibited the expression of BATF, which is regulated by BRD4 and p300 upon TCR stimulation. Importantly, JQ1-treated T cells demonstrated superior in vivo persistence and antitumor effects in multiple cancer immunotherapy models.
It is now emerging that the quality of T cells used for adoptive immunotherapy can be significantly improved by modulating particular signaling pathways such as PI3K/AKT, glycolytic pathways, and the Wnt/β-catenin pathway (40, 42, 43). In addition to previous findings, we provide evidence that the inhibition of BET proteins contributes to the generation of long-lived T cells. Importantly, JQ1 treatment was effective for both naive T cells and CD45RO+ memory T cells. Naive T cells and stem cell–like memory T cells are superior T cell sources when peripheral blood T cells are used for gene transfer. By contrast, most of the naturally occurring tumor-infiltrating lymphocytes are antigen primed and have differentiated into CD45RO+ memory T cells, in which maintenance of the TCM phenotype is the best strategy for generating optimal antitumor T cells (44–46). Our results suggest that JQ1 treatment should confer beneficial effects to a variety of T cell grafts.
The effects of JQ1 at least in part resulted from the transcriptional modulation of BATF. BATF is a transcription factor essential for the differentiation of CD8+ T cells into effector T cells by regulating the metabolic state of TCR-stimulated T cells (34, 47). In a murine virus infection model, Batf-deficient CD8+ T cells exhibit increased memory precursors and reduced effector functions. Although BATF is required for efficient effector T cell functions in vivo, we found that suppression of BATF expression is advantageous during ex vivo expansion of T cells. In addition, it has recently been shown that BATF expression induces exhaustive phenotypes and limits the IL-2 secretion of T cells, which is consistent with our data (48). Collectively, these data suggest that a transient blockade of the pathways or genes required for effector T cells generates better T cell grafts for adoptive immunotherapy. Pharmacological intervention is suitable for this purpose, which can transiently suppress effector T cell programs. The transferred T cells are then released from pharmacological inhibition and can acquire robust effector functions in vivo. In addition to BATF, there are several other transcription factors essential for effector T cell differentiation, including T-BET, Blimp-1, IRF4, and ID2 (34, 37–39). These factors have nonredundant roles in determining the fate of antigen-primed T cells, since genetic knockout of the individual genes results in significant changes in memory/effector establishment. Although we did not observe any significant influence of inhibiting several of these genes on phenotype markers, this lack of evidence does not necessarily exclude them from potential roles in memory T cell differentiation. Because these transcription factors form complex networks and often regulate each other, simultaneous interventions may be required to affect T cell differentiation. In addition, several other proteins may affect important steps in human memory T cell differentiation in the context of ex vivo T cell expansion. Further exploration of JQ1-induced gene expression alterations will be crucial to achieving a comprehensive understanding of the inhibition mechanism.
BET proteins might also play a role in effector T cell formation in vivo. Interestingly, recent studies using in vivo RNA interference screening have identified that knockdown of cyclin T1, a major component of the positive transcription elongation factor β (P-TEFβ), promotes the formation of memory precursors in vivo (49). Considering that BRD4 regulates P-TEFβ activity, the inhibition of BET proteins could also contribute to memory T cell formation, which should be evaluated in the in vivo setting. We also found that p300 helps to recruit BRD4 to the BATF promoter region. In fact, recent studies have demonstrated that BRD4 and p300 form a complex to transcriptionally activate several other genes such as MYC and FOS (50, 51). Our studies of pharmacological modulation of BET and p300 point to an acetylation-dependent signaling mechanism, since JQ1 directly inhibits BET protein binding to acetylated lysine residues in chromatin and C646 inhibits the catalytic acetyltransferase activity of p300. Interestingly, p300 also functions downstream of BATF in Th17 cells, as indicated by the change in the genome-wide occupancy of p300 in the absence of BATF (52). Further studies are warranted to investigate the bidirectional regulation between p300 and BATF.
In summary, we show that JQ1 treatment promoted the expansion of T cells with features of TSCM and TCM cells while inhibiting their differentiation into effector T cells via suppressing BATF. These findings are applicable to a broad range of adoptive immunotherapy for generating optimal T cell grafts.
Methods
In vitro culture of human T cells.
Healthy donor–derived peripheral blood mononuclear cells were isolated by Ficoll-Paque PLUS density gradient centrifugation (GE Healthcare). All donors were identified to be positive or negative for HLA-A*02:01 (A2) by high-resolution HLA DNA typing (American Red Cross). CD3+ or CD8+ T cells were purified through negative magnetic selection using a Pan T Cell Isolation Kit and CD8+ T Cell Isolation Kit (Miltenyi Biotec), respectively. CD45RO+ memory T cells were isolated from CD3+ T cells through negative selection with anti-CD45RA microbeads (Miltenyi Biotec). CD45RA+CD62L+CCR7+ naive T cells were separated by negative selection with anti-CD45RO microbeads and subsequent positive selection with anti-CD62L microbeads (Miltenyi Biotec). The coexpression of CCR7 in the isolated CD62L+ cells was confirmed by flow cytometry. Purified T cells were stimulated with aAPC/mOKT3 irradiated with 200 Gy at an effector to target (E:T) ratio of 3:1. On the following day, 100 IU/ml IL-2 and 10 ng/ml IL-15 (Peprotech) were added to the cultures. Culture media were replenished every 3 days. Unless otherwise noted, T cells were restimulated with aAPC/mOKT3 once per week. For antigen-specific stimulation of T cells, HLA-A*02:01 (A2)+ donor–derived T cells were weekly stimulated with aAPC/A2 pulsed with 10 μg/ml heteroclitic MART127–35 peptide at an E:T ratio of 10:1 as previously described (53). Synthetic peptides were obtained from ProImmune. For TCR stimulation without costimulatory signaling, CD3+ T cells were stimulated with anti-CD3 mAb (clone OKT3) coated on a plate at 5 μg/ml.
Cell lines.
Both aAPC/mOKT3 and aAPC/A2 are derived from the human erythroleukemic cell line K562 and were established as previously described (30). aAPC/A2 has successfully been used in the clinic (8). The CD19+ leukemia cell line NALM-6 was obtained from DSMZ. The melanoma cell line A375 was obtained from American Type Culture Collection (ATCC). All the cells were routinely checked for the presence of mycoplasma contamination using a PCR-based technology.
Retroviral transduction of T cells.
PG13 packaging cells stably transduced with each retrovirus plasmid were used for infection of T cells. Transduction was performed 2 days following stimulation with aAPC/mOKT3 for 3 consecutive days. The full-length BATF cDNA was cloned into the pMX-internal ribosome entry site (IRES)-truncated nerve growth factor receptor (ΔNGFR) (32, 54). HLA-A2/MART127–35 TCR (clone DMF5) genes were provided by Steven A. Rosenberg (NIH/NCI, Bethesda, MD) and subcloned into the pMX vector. The anti-CD19 CAR (FMC63 anti-CD19 scFv linked with CD28 and CD3z) was also subcloned into the pMX vector.
Lentiviral transduction of T cells.
For lentivirus production, 293T cells were transfected with the pLVX-shRNA2 plasmid (Clontech), psPAX2, and pMD2.G. The virus supernatants were collected and immediately used for transduction into purified naive T cells. To enhance the transduction efficiency, naive T cells were pretreated with 10 ng/ml IL-7 (Peprotech) for 5 days before transduction (55). The shRNA-transduced T cells were distinguished as ZsGreen+ cells.
Epigenetic chemical probes.
Epigenetic probes used in the experiments are listed in Table S1 and were used at the indicated concentrations. The (–)-JQ1 enantiomer was used at the same concentration as JQ1 (0.15 μM). Valproic acid and decitabine were purchased from Sigma-Aldrich. All the reagents were dissolved in DMSO except for UNC1215 and valproic acid, which were dissolved in water. DMSO was added at a final concentration of 0.01% (v/v), including the control.
Flow cytometry.
The following antibodies were used for flow cytometry: APC-Cy7-anti-CD4 (clone RPA-T4, BioLegend), PE-Cy7-anti-CD8 (clone SFCI21Thy2D3, Beckman Coulter), FITC-anti-CD45RA (clone MEM-56, Life Technologies), APC-Cy7-anti-CD45RA (clone HI100, BioLegend), Pacific Blue-anti-CCR7 (clone G043H7, BioLegend), APC-Cy7-anti-CD27 (clone O323, BioLegend), APC-anti-CD28 (clone CD28.2, BioLegend), PE-anti-CD127 (clone HIL-7R-M21, BD Biosciences), PerCP/Cy5.5-anti-CD95 (clone DX2, BD Biosciences), APC-anti-CD45 (clone HI30, BioLegend), PE-anti-CD271 (clone ME20.4, BioLegend), PE-Cy7-anti-CD271 (clone ME20.4, BioLegend), V450-anti-CD271 (clone C40-1457, BD Biosciences), FITC-anti-IL-2 (clone 5344.111, BD Biosciences), PE-anti-TNF-α (clone MAb11, BioLegend), PE-Cy7-anti-IFN-γ (clone 4S.B3, BioLegend), Alexa Fluor 488-anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (clone E10, Cell Signaling Technology), Alexa Fluor 647-anti-phospho-p38 MAPK (Thr180/Tyr182) (clone 28B10, Cell Signaling Technology), PE-anti-STAT5 (pY694) (clone 47/Stat5(pY694), BD Biosciences), anti-phospho-S6 ribosomal protein (Ser235/236) (catalog 2211; Cell Signaling Technology), anti-phospho-Akt (Thr308) (clone D25E6; Cell Signaling Technology), and anti-BATF antibody (clone D7C5; Cell Signaling Technology). R-PE-anti-rabbit IgG (H+L) (Jackson ImmunoResearch) was used as the secondary antibody following staining with anti–phospho-S6 ribosomal protein (Ser235/236), anti–phospho-Akt (Thr308), and anti-BATF antibody. The stained cells were analyzed with a FACS Canto II (BD Biosciences). The data analysis was performed with Flowjo software (Tree Star). For intracellular flow cytometric analysis of cytokine production, T cells were stimulated with aAPC/mOKT3 at an E:T ratio of 1:1, and incubated for 6 hours. Brefeldin A (BioLegend) was added to the cultures 2 hours following the stimulation. The cells were then fixed and permeabilized using a Cytofix/Cytoperm kit (BD Biosciences). For CFSE dilution assay, T cells were labeled with 5 μM CFSE (Life Technologies) before culture. Dead cells were discriminated with the LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (Life Technologies). The CAR-transduced T cells were stained with biotin-labeled protein L (Genscript), followed by streptavidin-PE (Life Technologies) or streptavidin-APC (Jackson ImmunoResearch).
Quantitative real-time PCR.
RNA was extracted with TRIzol (Life Technologies) and reverse transcribed using Superscript III (Life Technologies). Quantitative real-time PCR was performed on the CFX96 Real-Time PCR Detection System (Bio-Rad) using SYBR Select Master Mix (Life Technologies). Results were normalized to UBC, and relative expression levels were calculated using the 2-ΔΔCT method (56). The following primers were used for real-time PCR experiments (from 5′ to 3′): ACTN1 forward, GCCTCATCAGCTTGGGTTAT, and reverse, CATGATGCGGGCAAATTC; AHR forward, AGCCGGTGCAGAAAACAG, and reverse, CTATGCCGCTTGGAAGGAT; BATF forward, ACACAGAAGGCCGACACC, and reverse, CTTGATCTCCTTGCGTAGAGC; CREM forward, CAAATCCAAGACAAATGACCA, and reverse, GCCAGTCTGAGTCTGCACAT; EGR2 forward, TTGACCAGATGAACGGAGTG, and reverse, GGAGATCCAACGACCTCTTC; FOSL1 forward, AGGCCTTGTGAACAGGAGAC, and reverse, TTCTGCAGCTCCTCAATCTC; FOSL2 forward, GCCCAGTGTGCAAGATTAGC, and reverse, GGGCTCCTGTTTCACCACTA; GZMA forward, CCTGTGATTGGAATGAATATGGT, and reverse, AGGGCTTCCAGAATCTCCAT; ID2 forward, TGAACGACTGCTACTCCAAGC, and reverse, GGATTTCCATCTTGCTCACC; IRF4 forward, GCCAAGATTCCAGGTGACTC, and reverse, CTGGCTAGCAGAGGTTCTACG; KLRG1 forward, AACGGACAATCAGGAAATGAG, and reverse, CCTTGAGAAGTTTAGAGGTGATCC; NFIL3 forward, AACCCTCGATGGTGTCAAGT, and reverse, GCTCTCCTGCGTGTGTTCTA; PLAGL2 forward, CGGGAACACCTTTCTCAAAT, and reverse, GCTGAGGGCAGCTATATGGT; PLEK forward, ACCCTGCCTACCTGCACTAC, and reverse, ACAGCCTCTCAAGTGAATTGC; PRDM1 forward, ACGTGTGGGTACGACCTTG, and reverse, CTGCCAATCCCTGAAACCT; RUNX3 forward, TGGCATCGAACAGCATCTT, and reverse, GGGGAGGGAGGTGTGAAG; SELL forward, GGGGTGGACAATGCTCTG, and reverse, TAAGTCCAGCAGTCGGTTCC; STAT3 forward, AGGAGGCGTCACTTTCACTT, and reverse, GCTGCTGCTTTGTGTATGGT; STAT4 forward, CGTGGTCTTAACTCAGATCAACTC, and reverse, CATCACTGTAGCTAGATTGGACTGTAA; TBX21 forward, TGTGGTCCAAGTTTAATCAGCA, and reverse, TGACAGGAATGGGAACATCC; TCF7 forward, CGAGAAGAGCAGGCCAAGTA, and reverse, GCCTCTTCTTCTTCCCGTAGT; TRIP13 forward, ACGTCAACAGCAACCTCATC, and reverse, TGGGCTAACGCTTTACACAG; UBC forward, ATTTGGGTCGCGGTTCTTG, and reverse, TGCCTTGACATTCTCGATGGT; UHRF1 forward, AGAGCAAGAAGAAGGCGAAG, and reverse, CGATGGTACATTCCTTGGTG.
Mice.
In murine experiments, 6- to 10-week-old male NSG mice were used. In the experiments for evaluating in vivo persistence of the cultured T cells, mice were irradiated (1.5 Gy), and were injected with T cells on the following day. In the leukemia treatment model, the CD19+ NALM-6 leukemia cell line was transduced with pMX-EGFP-firefly luciferase for in vivo imaging studies, and 5 million of the cells were intravenously injected into NSG mice. CD3+ T cells were retrovirally transduced with anti-CD19 CAR and transplanted into the mice 14 days following NALM6-GL infusion. CAR-transduced T cells were stimulated with K562 transduced with CD19 seven days following the initial stimulation. Imaging of the tumor cell localization was performed with a Xenogen XPM-2 (Perkin Elmer). The mice were monitored for health and body weight and were euthanized by CO2 inhalation when they became moribund or when weight loss exceeded 20% of body weight before tumor injection. All the dead mice underwent autopsy and the cause of death was investigated. In the melanoma tumor model, 0.5 million A375 cells transduced with full-length MART1 were subcutaneously injected into NSG mice. CD3+ T cells were retrovirally transduced with anti–A2/MART1 TCR (clone DMF5), and intravenously infused into the mice 14 days after tumor inoculation. The volume of subcutaneous tumors was monitored at least twice weekly until they reached over 300 mm3. The mice were euthanized by CO2 inhalation when they became moribund, weight loss exceeded 20% of body weight before tumor injection, the tumor grew to 300 mm3, or an ulcer formed around the tumor. Progression-free survival was defined as the time to tumor regrowth exceeding the initial tumor volume. Mice were randomly assigned to treatment groups in each experiment. The incidence of xenogeneic graft-versus-host disease was monitored daily in each experiment, and the mice were sacrificed when they exhibited one of the following symptoms: more than 20% loss of initial body weight, pronounced lethargy, hunched posture, severe diarrhea, and severe dermatitis. In the experiments for evaluating naive T cell differentiation, 1 million purified naive CD8+ T cells transduced with shRNAs against BATF were transplanted into irradiated NSG mice. Five million cultured CD3+ T cells from the same donor were cotransplanted to support engraftment.
Microarray data analysis.
We analyzed previously published microarray gene expression data from the Gene Expression Omnibus (GEO) database (GSE23321, GSE11057, GSE13887, and GSE43955). The original CEL files were obtained from GEO and normalized using the JustRMA function from the Affy package provided by Bioconductor. The expression levels of individual genes were compared between 2 groups by using the RankProd analysis, and the differentially expressed genes as determined by false discovery rate less than 0.01 or less than 0.05 were extracted.
In vitro cytotoxicity assay.
The cytolytic activity of CAR-transduced and TCR-transduced T cells was analyzed as previously described with slight modification (57). Briefly, 1 × 105 CAR- or TCR-transduced T cells were cocultured with CFSE-labeled or EGFP+ target cells for 4 hours at the indicated ratio. The cells were then harvested and stained with the LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit. The absolute counts of viable target cells were determined by flow cytometry, and the frequency of surviving cells was calculated as a ratio to the cell counts incubated without T cells.
Immunoblotting.
For immunoblotting, cells were lysed in NP-40 buffer. Equal amounts of proteins were resolved by Tris-Glycine SDS-PAGE and transferred to Immobilon-P membranes (Millipore). The blots were probed with anti-BATF antibody (clone D7C5, Cell Signaling Technology) or anti–β-actin antibody (sc-47778, Santa Cruz Biotechnology) at 4°C overnight. The membranes were then washed and incubated with HRP-conjugated anti-rabbit IgG (sc-2004, Santa Cruz Biotechnology) or anti-mouse IgG (W402B, Promega) at room temperature for 1 hour. The signal was detected by Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare). Protein levels were quantified using ImageJ software (NIH).
siRNA interference.
For retrovirus-mediated knockdown experiments, the cluster sequence of the human primary microRNA (pri-miRNA) was cloned into the pMX-ΔNGFR retroviral plasmid and the original miRNA sequences were replaced by siRNA sequences as previously described (58). The target sequences for BATF, from 5′ to 3′ are as follows: ACTCATCTGATGATGTGAGAA (siBATF-1), TGATGATGTGAGAAGAGTTCA (siBATF-1), GCATTCCACCAACCTCATGTC (siBATF-1), TCATCTGATGATGTGAGAAGA (siBATF-2), CACAGAGGAACTGAAGTACTTC (siBATF-2), GGACTCATCTGATGATGTGAG (siBATF-2), GGATTATCCCGGGAACTTTGA (siFOSL2), CTGTCATCAAGCCCATCAGCA (siFOSL2), GCCTGCTATACAACATGAACG (siID2), GACCACCCTCAACACGGATAT (siID2), GGTTATTGGAGTGATGAGTAA (siPRDM1), AGAGTACAGCGTGAAAGAAAT (siPRDM1), TGTGAGCAGCATGAATGGCAT (siPRDM1), and AGAGATTTAGCAGCACCAGCA (siPRDM1). The following sequences were used as a control: TAAGGCTATGAAGAGATAC and TAGCGACTAAACACATCAA.
In the experiments using pLVX-shRNA against BATF, the following sequences were used as targets: GAGCGAAGACCTGGAGAAA (shBATF-1) and CACAGAGGAACTGAAGTAC (shBATF-2).
ChIP.
Cells were crosslinked in 1% formaldehyde for 10 minutes, and quenched with 125 mM glycine for 5 minutes. Fixed cells were lysed in NP-40 buffer with 0.1% SDS, and sonicated with the Sonic Dismembrator Model 100 (Fisher Scientific). Then, 4 μg of anti-BRD4 antibody (Abcam, ab75898) was preincubated with Protein-A Dynabeads (Life Technologies) in PBS with 0.5% BSA for 6 hours, washed 3 times, and incubated with the lysates overnight. After washing, samples were eluted in elution buffer (1% SDS, 100 mM NaHCO3). The eluates were then treated with RNase A (Fisher Scientific) for 3 hours and DNA was purified using a MinElute PCR Purification Kit (QIAGEN). The collected DNA samples were directly used for qPCR reactions. Data are shown as the ratio of the immunoprecipitated to input samples. The following primers were used for evaluating the enrichment in the BATF promoter region (920–983 bp upstream of the transcriptional start site of BATF): forward, CTGAGAACCCTACAAATGCG and reverse, TGACCGAGAGGGTAAGTGTA.
Statistics.
Statistical significance of differences between 2 groups was assessed with a 2-tailed paired or unpaired t test. Comparisons of more than 2 groups were performed by analysis of variance (ANOVA). Differences were considered statistically significant at a P value of less than 0.05. Error bars in the data represent SD of at least 3 biological replicates. Epigenetic probes were screened using T cells derived from 1 donor with 3 technical replicates. In mice tumor model experiments, the overall survival and progression-free survival of the mice treated with T cells were depicted by a Kaplan-Meier curve, and the survival difference between groups was compared with the log-rank test. All statistical analysis was performed using GraphPad Prism 6. No statistical method was used to predetermine sample size.
Study approval.
This study was performed in accordance with the Helsinki Declaration and approved by the Research Ethics Board of University Health Network, Toronto, Canada. Written informed consent was obtained from all healthy donors who provided peripheral blood samples. All animal experiments were approved by the Ontario Cancer Institute/Princess Margaret Cancer Centre Animal Care Committee at the University Health Network.
Author contributions
YK and NH designed the project. YK, MN, YY, TO, TG, MA, and KS performed the experiments. CHA provided critical reagents. YK, MOB, CHA, and NH analyzed the results and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the following grants and fellowships: NIH grant R01 CA148673 (to NH), Ontario Institute for Cancer Research Clinical Investigator Award IA-039 (to NH), Japan Society for the Promotion of Science Postdoctoral Fellowship for Overseas Researchers (to YK), the Princess Margaret Cancer Foundation (to MOB and NH), Guglietti Fellowship Award (to TO), and the Frederick Banting and Charles Best Canada Graduate Scholarship (to TG). The Structural Genomics Consortium (SGC) is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute, Innovative Medicines Initiative (EU/EFPIA) (ULTRA-DD grant 115766), Janssen, Merck & Co., Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and the Wellcome Trust.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(9):3479–3494. doi:10.1172/JCI86437.
Contributor Information
Yuki Kagoya, Email: ykagoya-tky@umin.ac.jp.
Munehide Nakatsugawa, Email: munakatsu@gmail.com.
Yuki Yamashita, Email: yyyuhki423@gmail.com.
Toshiki Ochi, Email: occhie_34@hotmail.com.
Tingxi Guo, Email: tguo@uhnres.utoronto.ca.
Mark Anczurowski, Email: mark.anczurowski@mail.utoronto.ca.
Kayoko Saso, Email: ksaso@uhnresearch.ca.
Naoto Hirano, Email: naoto.hirano@utoronto.ca.
References
- 1.Stagg J, Johnstone RW, Smyth MJ. From cancer immunosurveillance to cancer immunotherapy. Immunol Rev. 2007;220:82–101. doi: 10.1111/j.1600-065X.2007.00566.x. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10(9):909–915. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadelain M, Rivière I, Brentjens R. Targeting tumours with genetically enhanced T lymphocytes. Nat Rev Cancer. 2003;3(1):35–45. doi: 10.1038/nrc971. [DOI] [PubMed] [Google Scholar]
- 4.Dudley ME, et al. Adoptive cell therapy for patients with metastatic melanoma: evaluation of intensive myeloablative chemoradiation preparative regimens. J Clin Oncol. 2008;26(32):5233–5239. doi: 10.1200/JCO.2008.16.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg SA, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17(13):4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackensen A, Meidenbauer N, Vogl S, Laumer M, Berger J, Andreesen R. Phase I study of adoptive T-cell therapy using antigen-specific CD8+ T cells for the treatment of patients with metastatic melanoma. J Clin Oncol. 2006;24(31):5060–5069. doi: 10.1200/JCO.2006.07.1100. [DOI] [PubMed] [Google Scholar]
- 7.Morgan RA, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314(5796):126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler MO, et al. Establishment of antitumor memory in humans using in vitro-educated CD8+ T cells. Sci Transl Med. 2011;3(80): doi: 10.1126/scitranslmed.3002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davila ML, et al. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med. 2014;6(224): doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maude SL, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371(16):1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344(6184):641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins PF, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29(7):917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Besser MJ, et al. Adoptive transfer of tumor-infiltrating lymphocytes in patients with metastatic melanoma: intent-to-treat analysis and efficacy after failure to prior immunotherapies. Clin Cancer Res. 2013;19(17):4792–4800. doi: 10.1158/1078-0432.CCR-13-0380. [DOI] [PubMed] [Google Scholar]
- 14.Robbins PF, et al. Cutting edge: persistence of transferred lymphocyte clonotypes correlates with cancer regression in patients receiving cell transfer therapy. J Immunol. 2004;173(12):7125–7130. doi: 10.4049/jimmunol.173.12.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louis CU, et al. Antitumor activity and long-term fate of chimeric antigen receptor-positive T cells in patients with neuroblastoma. Blood. 2011;118(23):6050–6056. doi: 10.1182/blood-2011-05-354449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gattinoni L, et al. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J Clin Invest. 2005;115(6):1616–1626. doi: 10.1172/JCI24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinrichs CS, et al. IL-2 and IL-21 confer opposing differentiation programs to CD8+ T cells for adoptive immunotherapy. Blood. 2008;111(11):5326–5333. doi: 10.1182/blood-2007-09-113050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cieri N, et al. IL-7 and IL-15 instruct the generation of human memory stem T cells from naive precursors. Blood. 2013;121(4):573–584. doi: 10.1182/blood-2012-05-431718. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, et al. Closely related T-memory stem cells correlate with in vivo expansion of CAR.CD19-T cells and are preserved by IL-7 and IL-15. Blood. 2014;123(24):3750–3759. doi: 10.1182/blood-2014-01-552174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. 2012;12(4):269–281. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Araki Y, et al. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity. 2009;30(6):912–925. doi: 10.1016/j.immuni.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scharer CD, Barwick BG, Youngblood BA, Ahmed R, Boss JM. Global DNA methylation remodeling accompanies CD8 T cell effector function. J Immunol. 2013;191(6):3419–3429. doi: 10.4049/jimmunol.1301395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crompton JG, et al. Lineage relationship of CD8(+) T cell subsets is revealed by progressive changes in the epigenetic landscape. Cell Mol Immunol. 2016;13(4):502–513. doi: 10.1038/cmi.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russ BE, et al. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity. 2014;41(5):853–865. doi: 10.1016/j.immuni.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138(5):1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araki Y, Fann M, Wersto R, Weng NP. Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B) J Immunol. 2008;180(12):8102–8108. doi: 10.4049/jimmunol.180.12.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler MO, et al. Ex vivo expansion of human CD8+ T cells using autologous CD4+ T cell help. PLoS ONE. 2012;7(1): doi: 10.1371/journal.pone.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gattinoni L, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17(10):1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 30.Butler MO, Hirano N. Human cell-based artificial antigen-presenting cells for cancer immunotherapy. Immunol Rev. 2014;257(1):191–209. doi: 10.1111/imr.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholson IC, et al. Construction and characterisation of a functional CD19 specific single chain Fv fragment for immunotherapy of B lineage leukaemia and lymphoma. Mol Immunol. 1997;34(16–17):1157–1165. doi: 10.1016/S0161-5890(97)00144-2. [DOI] [PubMed] [Google Scholar]
- 32.Nakatsugawa M, et al. Specific roles of each TCR hemichain in generating functional chain-centric TCR. J Immunol. 2015;194(7):3487–3500. doi: 10.4049/jimmunol.1401717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mele DA, Salmeron A, Ghosh S, Huang HR, Bryant BM, Lora JM. BET bromodomain inhibition suppresses TH17-mediated pathology. J Exp Med. 2013;210(11):2181–2190. doi: 10.1084/jem.20130376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurachi M, et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol. 2014;15(4):373–383. doi: 10.1038/ni.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen KB, et al. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science. 2002;297(5589):2063–2066. doi: 10.1126/science.1074900. [DOI] [PubMed] [Google Scholar]
- 36.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12(11):749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27(2):281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutishauser RL, et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31(2):296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang CY, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12(12):1221–1229. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crompton JG, et al. Akt inhibition enhances expansion of potent tumor-specific lymphocytes with memory cell characteristics. Cancer Res. 2015;75(2):296–305. doi: 10.1158/0008-5472.CAN-14-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholz G, et al. Modulation of mTOR signalling triggers the formation of stem cell-like memory T cells. EBioMedicine. 2016;4:50–61. doi: 10.1016/j.ebiom.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sukumar M, et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest. 2013;123(10):4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gattinoni L, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med. 2009;15(7):808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klebanoff CA, et al. IL-15 enhances the in vivo antitumor activity of tumor-reactive CD8+ T cells. Proc Natl Acad Sci U S A. 2004;101(7):1969–1974. doi: 10.1073/pnas.0307298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berger C, Jensen MC, Lansdorp PM, Gough M, Elliott C, Riddell SR. Adoptive transfer of effector CD8+ T cells derived from central memory cells establishes persistent T cell memory in primates. J Clin Invest. 2008;118(1):294–305. doi: 10.1172/JCI32103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klebanoff CA, et al. Central memory self/tumor-reactive CD8+ T cells confer superior antitumor immunity compared with effector memory T cells. Proc Natl Acad Sci U S A. 2005;102(27):9571–9576. doi: 10.1073/pnas.0503726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Godec J, et al. Inducible RNAi in vivo reveals that the transcription factor BATF is required to initiate but not maintain CD8+ T-cell effector differentiation. Proc Natl Acad Sci U S A. 2015;112(2):512–517. doi: 10.1073/pnas.1413291112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quigley M, et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med. 2010;16(10):1147–1151. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen R, et al. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity. 2014;41(2):325–338. doi: 10.1016/j.immuni.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong MM, Byun JS, Sacta M, Jin Q, Baek S, Gardner K. Promoter-bound p300 complexes facilitate post-mitotic transmission of transcriptional memory. PLoS One. 2014;9(6): doi: 10.1371/journal.pone.0099989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell. 2015;58(6):1028–1039. doi: 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151(2):289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirano N, et al. Efficient presentation of naturally processed HLA class I peptides by artificial antigen-presenting cells for the generation of effective antitumor responses. Clin Cancer Res. 2006;12(10):2967–2975. doi: 10.1158/1078-0432.CCR-05-2791. [DOI] [PubMed] [Google Scholar]
- 54.Ochi T, et al. Optimization of T-cell reactivity by exploiting TCR chain centricity for the purpose of safe and effective antitumor TCR gene therapy. Cancer Immunol Res. 2015;3(9):1070–1081. doi: 10.1158/2326-6066.CIR-14-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verhoeyen E, Costa C, Cosset FL. Lentiviral vector gene transfer into human T cells. Methods Mol Biol. 2009;506:97–114. doi: 10.1007/978-1-59745-409-4_8. [DOI] [PubMed] [Google Scholar]
- 56.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 57.Jedema I, van der Werff NM, Barge RM, Willemze R, Falkenburg JH. New CFSE-based assay to determine susceptibility to lysis by cytotoxic T cells of leukemic precursor cells within a heterogeneous target cell population. Blood. 2004;103(7):2677–2682. doi: 10.1182/blood-2003-06-2070. [DOI] [PubMed] [Google Scholar]
- 58.Okamoto S, et al. Improved expression and reactivity of transduced tumor-specific TCRs in human lymphocytes by specific silencing of endogenous TCR. Cancer Res. 2009;69(23):9003–9011. doi: 10.1158/0008-5472.CAN-09-1450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.