Abstract
Diabetes strongly impacts protein metabolism, particularly in skeletal muscle. Insulin and IGF-1 enhance muscle protein synthesis through their receptors, but the relative roles of each in muscle proteostasis have not been fully elucidated. Using mice with muscle-specific deletion of the insulin receptor (M-IR–/– mice), the IGF-1 receptor (M-IGF1R–/– mice), or both (MIGIRKO mice), we assessed the relative contributions of IR and IGF1R signaling to muscle proteostasis. In differentiated muscle, IR expression predominated over IGF1R expression, and correspondingly, M-IR–/– mice displayed a moderate reduction in muscle mass whereas M-IGF1R–/– mice did not. However, these receptors serve complementary roles, such that double-knockout MIGIRKO mice displayed a marked reduction in muscle mass that was linked to increases in proteasomal and autophagy-lysosomal degradation, accompanied by a high-protein-turnover state. Combined muscle-specific deletion of FoxO1, FoxO3, and FoxO4 in MIGIRKO mice reversed increased autophagy and completely rescued muscle mass without changing proteasomal activity. These data indicate that signaling via IR is more important than IGF1R in controlling proteostasis in differentiated muscle. Nonetheless, the overlap of IR and IGF1R signaling is critical to the regulation of muscle protein turnover, and this regulation depends on suppression of FoxO-regulated, autophagy-mediated protein degradation.
Introduction
Muscle wasting and the associated decrease in muscle strength cause significant morbidity and predict risk of mortality in elderly individuals (1). A diverse range of insults including aging, immobility, cancer, and uncontrolled diabetes can cause muscle wasting. Type 2 diabetes and insulin resistance in older individuals has been shown to accelerate the decline in muscle strength (2, 3), adding muscle dysfunction to the long list of diabetic complications that contribute to morbidity and mortality.
Insulin is a major regulator of muscle metabolism, enhancing glucose uptake in the postprandial state. Insulin has also been shown to control muscle protein synthesis and degradation (4, 5), and these effects on proteostasis are mimicked by insulin-like growth factor-1 (IGF-1) (6). Indeed, excess IGF-1 causes muscle hypertrophy, and IGF-1 expression in muscle increases in response to resistance exercise, lending to its reputation as a critical factor for muscle growth (7, 8). Insulin and IGF-1 signal via highly homologous tyrosine kinase receptors, both of which are expressed in muscle. Studies using human myoblasts and myotubes suggest that muscle expresses higher levels of IGF-1 receptor (IGF1R) relative to insulin receptor (IR) (9), but in vivo studies suggest that both may be involved in maintenance of muscle mass. For example, genetic inactivation of IGF1R in muscle has been reported to cause a mild decrease in muscle fiber size, and deletion of IR in muscle has been suggested to promote age-related muscle atrophy (10, 11). We recently showed that combined loss of IR and IGF1R in muscle induces a marked decrease in muscle mass (12), indicating that signaling via IR and/or IGF1R is necessary for normal muscle growth.
The precise mechanisms underlying the decrease in muscle size and downstream targets of insulin and IGF-1 action involved in muscle proteostasis have not been identified. After stimulation with their respective ligand, IR and IGF1R activate common downstream molecular pathways, including both the IRS/PI3K/Akt and MAPK pathways (13). Activation of Akt mimics many of the physiologic effects of insulin and IGF-1 in muscle, including enhanced glucose uptake by increasing translocation of the glucose transporter GLUT4 and increased growth via mTOR activation (14). However, the extent to which these pathways are involved in muscle growth is unclear, since muscle-specific knockout of both IR and IGF1R has profound effects on muscle mass, despite the fact that whole-body glucose tolerance is maintained (12).
FoxO transcription factors are important transcriptional targets of insulin and IGF-1. In liver, FoxOs mediate fasting-induced gluconeogenesis, and this is reversed by nuclear exclusion following insulin stimulation (15). This negative regulation of FoxOs by insulin/IGF-1 is a highly evolutionarily conserved pathway as evidenced by studies in Caenorhabditis elegans that show that deletion of the FoxO homolog DAF-16 reverses longevity and many of the metabolic changes that occur in worms in which activity of the IR/IGF1R homolog DAF-2 is reduced (16). In muscle, FoxOs have been implicated in the control of both proteasomal and autophagy-lysosomal degradation, the 2 critical protein degradation pathways in muscle, by controlling of the expression of many E3-ubiquitin ligases and autophagy genes (17, 18). These studies imply that FoxO transcription factors can control muscle atrophy, but how these interact with upstream signals from IR and IGF1R to regulate muscle protein homeostasis remains to be fully elucidated.
The aim of the current study was to determine the relative roles of IR and IGF1R signaling in muscle protein turnover and decipher the downstream mediators of insulin/IGF-1 in this process. We show that muscle-specific deletion of Igf1r alone has little effect on muscle mass, whereas deletion of IR reduces mass by 20% and combined deletion of IR and Igf1r results in a marked decrease in muscle mass due to an induction of both proteasomal- and autophagy-lysosomal–mediated protein degradation. This is completely dependent on FoxO-mediated signals, as combined muscle-specific deletion of FoxO1/3/4 isoforms in the context of IR/Igf1r deletion rescues these mice. Thus, IR and IGF1R play overlapping roles in control of muscle proteostasis, and this regulation is due to inhibition of FoxO-mediated protein degradation. These results demonstrate how IR/IGF1R signaling controls muscle protein turnover and elucidate how diabetes and insulin resistance can contribute to muscle wasting and its associated morbidity.
Results
IGF1R is expressed in undifferentiated myoblasts, but IR expression is higher in differentiated muscle.
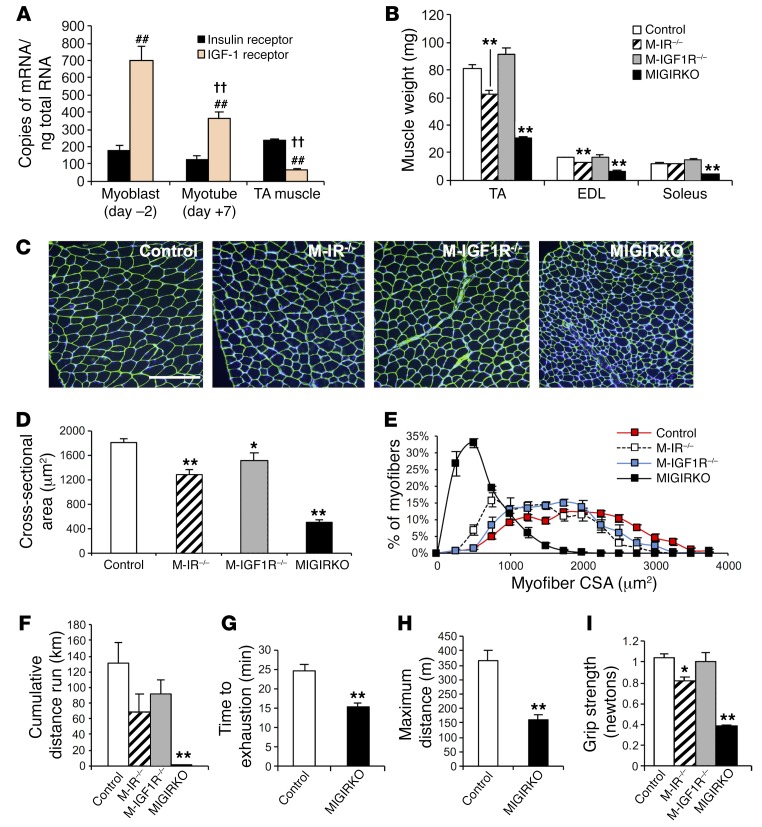
Human myoblasts and myotubes express both IR and IGF1R. Previous reports suggest that myoblasts express 6-fold higher levels of IGF1R compared with IR (9), yet muscle-specific deletion of Igf1r or IR alone in mouse models has little effect on muscle size (10, 11). To evaluate the absolute expression of IR and IGF1R in differentiated muscle and compare it with that in undifferentiated myoblasts, we measured mRNA copy number for IR and Igf1r in mouse myoblasts, myotubes, and tibialis anterior (TA) muscle by quantitative PCR (qPCR) using cDNA standards for each of these receptors. These results showed that primary mouse myoblasts in culture (differentiation day –2) contain about 700 copies of Igf1r mRNA per nanogram of total RNA compared with only 180 copies of IR mRNA (Figure 1A), recapitulating previous results (9). In myotubes (differentiation day +7), there was a 48% decrease in Igf1r mRNA copy number with little change in IR mRNA, and Igf1r mRNA was further decreased in mature TA muscle, such that the ratio of IR/Igf1r mRNA increased from 0.25 in early myoblasts to 3.6 in mature muscle. This was consistent across all muscle groups tested (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI86522DS1). Western blot confirmed that the protein expression of IGF1R decreased with myoblast differentiation while protein levels of IR increased (Supplemental Figure 1B). Thus, during muscle differentiation, there is a switch such that in myoblasts IGF1R predominates over IR, whereas in differentiated muscle IR predominates over IGF1R.
Figure 1. IR predominates over IGF1R in differentiated muscle, and combined deletion of IR and Igf1r in muscle dramatically decreases muscle fiber size and muscle function.
(A) Copy number of insulin receptor (IR) and IGF-1 receptor (Igf1r) mRNA was measured by qPCR in undifferentiated (day –2) primary mouse myoblasts, differentiated myotubes (day +7) (n = 3 experiments), and tibialis anterior (TA) muscle from 8-week-old male mice (n = 7). Copy number was quantitated using a standard curve obtained by serial dilution of IR and IGF1R cDNA plasmids (##P < 0.01 vs. insulin receptor in same group; ††P < 0.01 vs. IGF-1 receptor in each other group). (B) Muscle weights from 8-week-old knockout and control mice (n = 21 controls and 5–10 knockouts). (C) Laminin staining of TA muscle cross sections (scale bar: 200 μm). (D and E) Muscle fiber cross-sectional area (D) and myofiber size distribution (E) of TA from C (n = 5–11). (F) Distance run in kilometers (km) by females during a 30-day voluntary wheel cage running experiment (n = 16 controls and 4–8 knockouts). (G and H) Time to exhaustion (G) and maximum distance run (H) during an acute treadmill test of MIGIRKO and control mice (n = 6). (I) Forelimb grip strength in control, M-IR–/–, M-IGF1R–/–, and MIGIRKO mice (n = 16 controls and 4–8 knockouts). (*P < 0.05, **P < 0.01 vs. control, t test for 2 groups, ANOVA for 3 or more.)
Loss of IR and IGF1R in muscle in vivo leads to marked decreases in muscle mass, impaired muscle function, and a switch to a more oxidative fiber type.
To determine the relative contribution of IR and IGF1R signaling to muscle mass, protein turnover, and function, we generated mice with muscle-specific deletion of IR (IRfl/fl Acta1-Cre, hereafter referred to as M-IR–/–), Igf1r (Igf1rfl/fl Acta1-Cre, hereafter referred to as M-IGF1R–/–), or both (MIGIRKO) (Supplemental Figure 1C). Similar to our previous findings, MIGIRKO mice showed a significant 60% decrease in muscle mass, and a 40% decrease in muscle size normalized to body weight (Figure 1B and Supplemental Figure 1, D and E; and ref. 12). By comparison, M-IR–/– mice showed a modest 20% decrease in muscle mass, which occurred primarily in glycolytic muscles, such as TA and extensor digitorum longus (EDL), and M-IGF1R–/– mice showed no change in muscle mass. Analysis of cross-sectional area and size distribution of myofibers in TA muscle showed that these changes in muscle mass were due to decreases in muscle fiber size, which were especially decreased in MIGIRKO muscle (Figure 1, C–E).
Functionally, this decrease in muscle mass resulted in a complete loss of voluntary wheel cage running activity in MIGIRKO mice. Both M-IR–/– and M-IGF1R–/– mice also tended to run less than controls, but in neither case did this reach statistical significance (Figure 1F). In a forced-running acute treadmill test, MIGIRKO mice also performed poorly with a 38% decrease in running time and a 56% reduction in maximum distance run compared with controls (Figure 1, G and H). Muscle grip strength was also decreased by 60% in MIGIRKO mice and by 20% in M-IR–/– mice compared with controls (Figure 1I). This decrease in exercise capacity and muscle strength was associated with marked atrophy of diaphragm muscle (Supplemental Figure 1F), leading to the impaired survival previously reported (12).
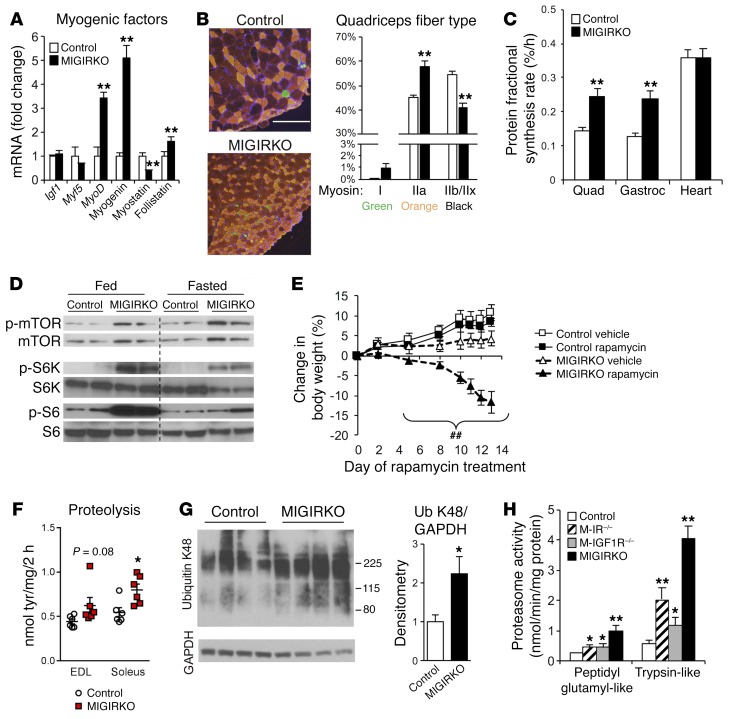
MIGIRKO muscle showed no change in mRNA expression of Igf1r or Myf5, but a 3- to 5-fold increase in levels of the myogenic factors MyoD and myogenin. The hypertrophic factor follistatin was increased 50%, and expression of the muscle growth inhibitor myostatin was decreased 60% (Figure 2A), which did not account for the small muscle size. Interestingly, quadriceps muscle from MIGIRKO showed increased type IIa oxidative fiber density and decreases in glycolytic fibers myosin IIb and IIx, indicating a switch toward an oxidative phenotype of the muscle (Figure 2B). Likewise, soleus muscle from MIGIRKO mice contained more myosin I fibers and no myosin IIb or IIx (Supplemental Figure 1G). This oxidative switch in muscle fiber type is supported by our previous histologic staining showing that MIGIKRO muscle contains more succinate dehydrogenase–positive oxidative fibers than controls (12).
Figure 2. MIGIRKO mice display evidence of oxidative muscle and a high-protein-turnover state with increased protein synthesis and degradation.
(A) qPCR of myogenic factors in quadriceps from MIGIRKO and control mice (n = 8). (B) Fiber type density in quadriceps (n = 6, scale bar: 200 μm). (C) Protein fractional synthesis rates were measured using 13C-labeled phenylalanine in quadriceps (Quad), gastrocnemius (Gastroc), and cardiac (Heart) muscle (n = 11). (D) Western blots for phosphorylation of mammalian target of rapamycin (mTOR), S6 kinase (S6K), and S6 protein in quadriceps from MIGIRKO and control mice either fed or fasted overnight. (E) Percent change in body weight during 14-day rapamycin treatment in MIGIRKO and control mice (n = 4–5). (F) Proteolysis measured by tyrosine release in ex vivo muscle from control and MIGIRKO female mice (n = 6). (G) Western blot and densitometry of K48-linked polyubiquitin (Ub) proteins in gastrocnemius from MIGIRKO and control mice treated with MG132 for 3 days. (H) Proteasome activity in gastrocnemius lysates from control, M-IR–/–, M-IGF1R–/–, and MIGIRKO mice measured by breakdown of fluorescently labeled peptidyl glutamyl-like (LLE) or trypsin-like (LSTR) substrates (n = 5–8). (*P < 0.05, **P < 0.01 vs. control, t test with 2 groups or ANOVA with 4 groups; ##P < 0.01 vs. MIGIRKO vehicle, ANOVA.)
MIGIRKO muscle displays a high-protein-turnover state with increased protein synthesis and degradation.
To assess protein fractional synthesis rates (FSRs) of muscle protein in vivo, control and MIGIRKO mice were given a flooding dose of 13C-labeled phenylalanine, and its incorporation into muscle protein was assessed by mass spectrometry as previously described (19). Despite the smaller fiber size, FSR of muscle protein was increased by 70%–80% in quadriceps and gastrocnemius muscles from MIGIRKO, whereas there was no change in FSR in heart or liver (Figure 2C and Supplemental Figure 2A). Phosphorylation of mTORC1, a critical regulator of protein synthesis in virtually all cells (20), and its downstream targets S6 kinase and S6 was markedly increased in MIGIRKO muscle compared with controls under fed conditions, and even slightly increased after an overnight fast (Figure 2D), consistent with a high rate of protein synthesis. Treatment of MIGIRKO mice with the mTOR inhibitor rapamycin for 14 days resulted in a significant loss of body weight in MIGIRKO mice but not controls (Figure 2E), further demonstrating that MIGIRKO mice rely on mTOR signaling and a high protein synthetic rate to maintain even a reduced muscle mass. Also consistent with the great dependence on protein synthesis in the knockout mice, rapamycin treatment decreased S6 phosphorylation and tended to decrease muscle mass more in MIGIRKO (27%–46%) than controls (6%–26%) (Supplemental Figure 2, B and C).
MIGIRKO mice display increases in proteolysis and ubiquitin-proteasomal activity in the fed state.
Protein synthesis is always competing against protein degradation in maintenance of muscle mass. In MIGIRKO mice, reduced muscle mass despite increased muscle protein synthesis indicates a greater increase in protein degradation than synthesis. To determine whether protein degradation was increased in MIGIRKO muscle, we determined rates of proteolysis in soleus and EDL muscle isolated from fed animals. Tyrosine release from soleus was significantly increased in MIGIRKO mice, and tyrosine release tended to increase in EDL muscle (Figure 2F). Lysine-48–linked polyubiquitin chains, which are targeted for proteasomal degradation (21), were increased in muscle from MIGIRKO mice after 3 days of treatment with MG132, a proteasome inhibitor (Figure 2G), and mimicked total polyubiquitin chains (Supplemental Figure 3A). Proteasomal activity using 2 different substrates was increased approximately 2-fold in both M-IR–/– and M-IGF1R–/–, and was further increased up to 5-fold in MIGIRKO muscle compared with controls (Figure 2H). This increase in activity was associated with increased protein levels of subunits of the 19S regulatory complex and the 20S catalytic core of proteasomes under both fed and fasted conditions (Supplemental Figure 3B). Increased protein degradation can release amino acids, which can feed back to activate mTOR and increase protein synthesis (22). Total essential amino acid levels determined by metabolomics analysis were increased in MIGIRKO muscle compared with controls (Supplemental Figure 2D). Branch chain amino acids and aromatic amino acids were unchanged, but Asp, Glu, and Cys were increased in MIGIRKO, and Pro was decreased (Supplemental Figure 2, E–G). Taken together, these data indicate that protein degradation pathways are increased to a greater degree than synthesis when IR and Igf1r is deleted in muscle, leading to amino acid generation and a high-protein-turnover state.
Autophagy is increased in MIGIRKO muscle.
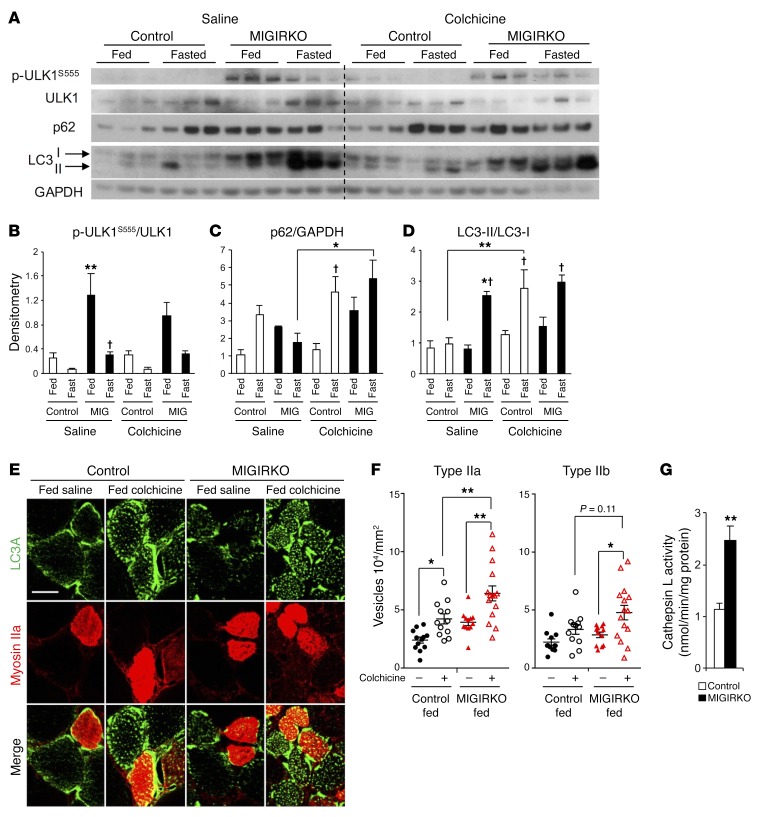
Another important pathway for protein degradation and turnover in muscle is autophagy. Autophagy can be induced by starvation (macroautophagy), but is also part of regular organelle maintenance or clearance of aggregates (mitophagy, aggrephagy) (23). Autophagy is controlled by many cellular signaling events. During starvation, autophagy is induced in muscle by FoxO transcription factors and AMPK signaling, whereas when nutrients are abundant, autophagy is suppressed by mTOR signaling. Critical markers of autophagy are phosphorylation of ULK1S555 (AMPK site), accumulation of p62 proteins, and the cleaved-lipidated LC3-II proteins (24). To determine autophagy flux in MIGIRKO mice, we measured levels of these mediators and markers of autophagy in muscle of randomly fed and overnight-fasted mice, with or without treatment with colchicine, a known inhibitor of autophagosome-lysosome fusion in muscle (25). Phosphorylation of ULK1S555 was markedly increased in fed MIGIRKO mice compared with fed control mice, and decreased in both after fasting (Figure 3, A and B), consistent with IR/IGF1R-dependent and -independent pathways of control. Colchicine treatment did not affect p-ULK1S555 levels. Consistent with increased autophagocytic flux, colchicine treatment of MIGIRKO mice fasted overnight showed significantly higher levels of p62 in comparison with saline-treated mice (Figure 3, A and C). Upon initiation of autophagy, LC3 isoforms are cleaved and lipidated from the precursor LC3-I to the active LC3-II. Thus, autophagocytic flux can be determined by measurement of the induction of LC3-II or the ratio of LC3-II to LC3-I. In both the fed and fasted states, total protein levels of LC3-II and LC3-II normalized to GAPDH were markedly increased in MIGIRKO muscle compared with controls (Figure 3A and Supplemental Figure 3D), and this was not due to a specific isoform, as both LC3A and LC3B were upregulated in MIGIRKO (Supplemental Figure 3C). Upon fasting, MIGIRKO mice showed a 3-fold increase in LC3-II whether it was normalized to LC3-I or GAPDH, indicating a block in autophagy in the fasted state, likely due to massive activation of the autophagy-lysosome system that exceeds clearance capacity (Figure 3, A and D, and Supplemental Figure 3D). Colchicine treatment tended to increase the LC3-II/LC3-I ratio in randomly fed MIGIRKO mice, and significantly increased the LC3-II/LC3-I ratio in muscle from fasted control animals to the same level as in the MIGIRKOs.
Figure 3. Activation of autophagy is increased in MIGIRKO muscle, especially in the fed state.
(A) Western blots for autophagy intermediates in quadriceps from fed or 16-hour-fasted MIGIRKO and control mice treated with either saline or 0.4 mg/kg colchicine for 2 days (the blots are from parallel samples run on separate gels). (B–D) Densitometric analysis of p-ULK1S555/ULK1 (B), p62 normalized to GAPDH (C), or ratio of LC3-II/LC3-I (D) from A (n = 3–6 per group). Note samples 4 and 16 are switched in the LC3 Western blot, but correctly identified for densitometry. (E) LC3A (green) and myosin IIa (red) immunostaining of quadriceps from fed MIGIRKO and control mice (scale bar: 20 μm). (F) Quantification of LC3A-positive vesicles per cross-sectional area in type IIa or IIb fibers from quadriceps in E (n = 3 fibers each from 4–5 mice per group). (G) Cathepsin L activity in gastrocnemius lysates (n = 6–8). (*P < 0.05, **P < 0.01 vs. control under same conditions or as specified; †P < 0.05, fed vs. fasted, t test for 2 groups, ANOVA for more.)
As noted previously, myosin fiber typing indicated a shift from glycolytic to oxidative muscle in MIGIRKO mice. To determine whether the increased protein levels of LC3 were distributed equally among different fiber types, we costained quadriceps muscles from control and MIGIRKO mice using antibodies against LC3A and myosin IIa, a marker of oxidative fibers. LC3A staining of controls showed individual fibers that stained positive, while others showed no staining or minimal staining (Figure 3E). Costaining with myosin IIa revealed that all oxidative type IIa fibers also stained positive for LC3A. In addition, some but not all myosin IIa–negative fibers were also LC3A positive. Overall, quantification showed that 70% of all fibers (per high-power field) in MIGIRKO muscle were LC3A positive compared with only 56% LC3A-positive fibers in controls (Supplemental Figure 3E). Electron microscopy of MIGIRKO muscle also revealed numerous autophagosomes in MIGIRKO with very few in controls (Supplemental Figure 3F).
We hypothesized that autophagy flux through lysosomal degradation is increased in MIGIRKO mice in the fed state, but upon fasting, induction of autophagy exceeds the capacity to clear autophagosomes. To assess autophagy flux in the fed state, we measured LC3A-positive vesicles in cross sections of quadriceps muscle from randomly fed mice treated with saline or colchicine (Figure 3E). Accumulation of LC3A vesicles in oxidative type IIa fibers was increased with colchicine treatment in controls and increased further in MIGIRKO muscle (Figure 3F). In glycolytic type IIb fibers, where the induction of autophagy is reported to be more robust with fasting (26), MIGIRKO mice again showed an increased vesicle accumulation with colchicine that tended to increase higher than that in the control colchicine group. Activity of the lysosomal protease cathepsin L was also increased nearly 2-fold in MIGIRKO muscles (Figure 3G). Taken together, these data indicate that deletion of IR and Igf1r in muscle induces marked alterations in autophagy flux such that in the fed state, MIGIRKO mice display increased rates of muscle autophagy flux, but upon fasting, autophagy induction increases and outstrips clearance, leading to accumulation of autophagy intermediates in MIGIRKO mice.
Loss of IR induces mild muscle atrophy and weakness as mice age.
To ascertain the relative roles of IR versus IGF1R in modulation of autophagy in muscle with aging, we measured autophagy markers and muscle size and function in aged (52-week-old) M-IR–/– and M-IGFR–/– mice. At this age, muscle from M-IR–/– mice showed dysregulated autophagy with increased LC3-II (Supplemental Figure 4A). Similarly to younger mice, 1-year-old M-IR–/– mice also showed significant decreases in muscle size and grip strength, but no changes on an acute treadmill test (Supplemental Figure 4, B–D). By contrast, M-IGFR–/– mice showed normal grip strength and performance on a treadmill test, and we observed no increase in autophagy or decrease in muscle mass even with aging.
MIGIRKO mice show nuclear accumulation of FoxO isoforms and increased expression of FoxO target genes.
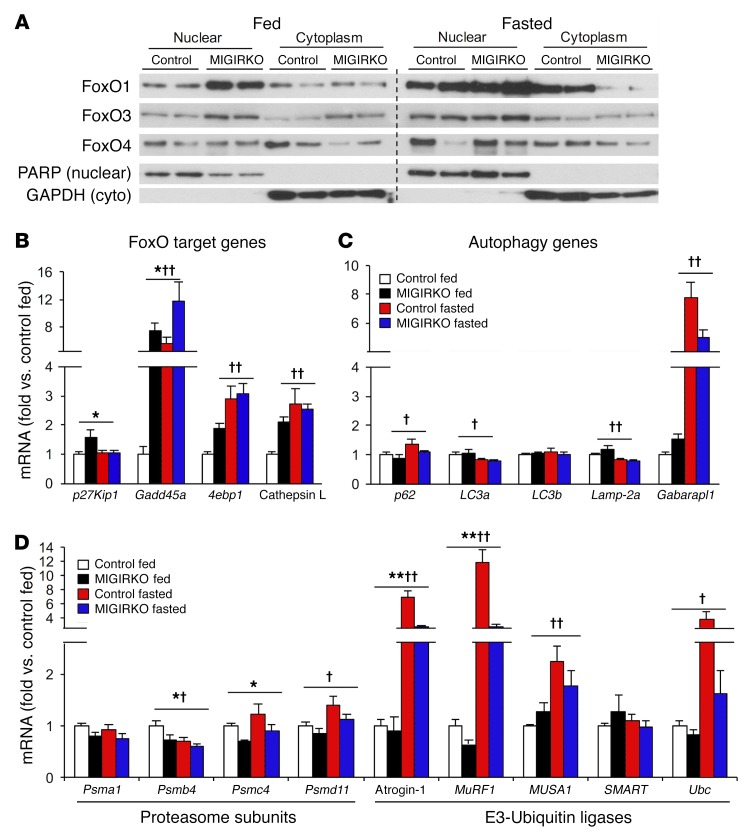
FoxO transcription factors have been shown to control muscle atrophy in response to starvation and denervation (27). Following insulin or IGF-1 stimulation, FoxOs are phosphorylated by Akt and excluded from the nucleus, thus inhibiting their transcriptional activity. Conversely, loss of IR and IGF1R could lead to FoxO accumulation in the nucleus and increased transcription of FoxO target genes. Subcellular fractionation of muscle from fed MIGIRKO mice revealed that FoxO1 and FoxO3 were increased in nuclear fractions compared with controls (Figure 4A). Upon fasting, which lowers circulating insulin levels, there was an increase in all FoxO isoforms in nuclear fractions from MIGIRKO mice, rising to similar levels to those seen in fasted controls.
Figure 4. FoxO transcription factors display nuclear accumulation in fed MIGIRKO mice with increases in transcription of FoxO target genes.
(A) Western blot of FoxO1, FoxO3, and FoxO4 in nuclear and cytoplasmic fractions of muscle from fed or overnight-fasted MIGIRKO and control mice (n = 2 per group). PARP and GAPDH are used as nuclear and cytoplasmic markers, respectively. (B and C) qPCR of FoxO target genes (B) and autophagy intermediates (C) in TA from MIGIRKO and control mice either randomly fed or fasted overnight. (D) qPCR of proteasome subunits and E3-ubiquitin ligases in TA from MIGIRKO and control mice either randomly fed or fasted overnight (n = 4–8 per group). (*P < 0.05, **P < 0.01 control vs. MIGIRKO; †P < 0.05, ††P < 0.01 fed vs. fasted, 2-way ANOVA.)
In agreement with the increase in FoxO nuclear localization, expression of FoxO target genes (p27Kip1, Gadd45a, 4E binding protein [4ebp1, encoding 4E-BP1], and cathepsin L) (28) was increased in muscle from fed MIGIRKO mice (Figure 4B). Likewise, fasting increased Gadd45a, 4ebp1, and cathepsin L mRNAs in controls to the same level as in MIGIRKO, again matching the pattern of FoxO isoform nuclear localization under these conditions. Despite the marked increases in LC3 proteins in MIGIRKO muscle, mRNA levels of LC3a, LC3b, and Lamp-2a were not changed in fed or fasted mice, and Gabarapl increased upon fasting in both control and MIGIRKO (Figure 4C). In contrast, proteasomal subunit mRNA levels increased marginally or not at all with fasting (Figure 4D), and mRNA levels of the E3-ubiquitin ligases atrogin-1, MuRF1, and MUSA1 were not different between control and MIGIRKO mice in the fed state. Upon fasting, atrogin-1, MuRF1, and MUSA1 increased in both control and MIGIRKO mice, although in MIGIRKO, levels were significantly lower than in control. Thus, muscle from MIGIRKO mice displays nuclear accumulation of FoxO isoforms and increased gene expression of many, but not all, FoxO target genes, and these increases are seen primarily in the fed state.
Short-term inducible knockout of IR and IGF1R in muscle recapitulates muscle atrophy and autophagy of MIGIRKO mice.
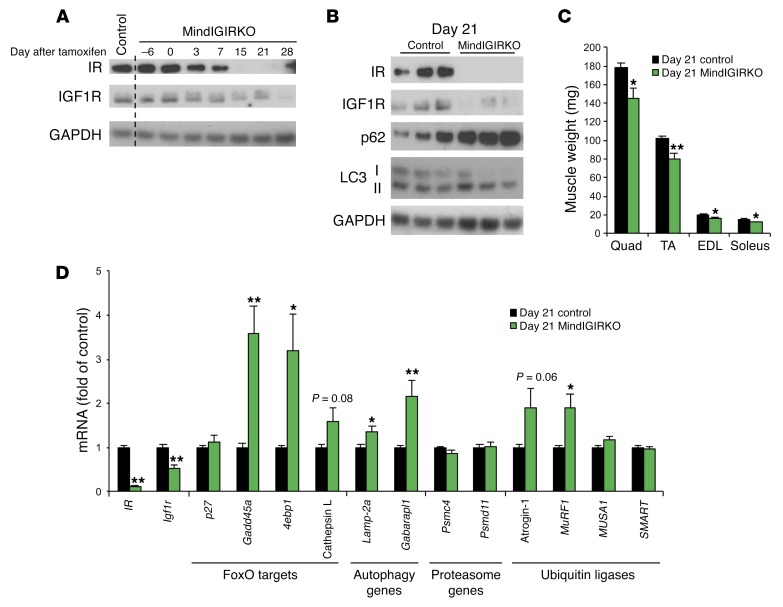
In MIGIRKO mice, gene inactivation occurs during mid-embryogenesis (day 9.5) (29). To evaluate the effect of short-term loss of IR and IGF1R signaling on autophagy and muscle size in adult mice, we created an inducible knockout of IR and IGF1R using mice harboring an HSA-CreERT2 transgene crossed to the IRfl/fl Igf1rfl/fl background. As early as 3 days after the tamoxifen treatment protocol, 3-month-old MindIGIRKO (muscle inducible IGF1R IR knockout) mice showed an approximately 50% reduction of IR and IGF1R protein compared with controls, and by 15 to 21 days, both IR and IGF1R fell to almost undetectable levels (Figure 5A). Also by day 21, levels of the autophagy proteins p62 and LC3-II were increased (Figure 5B), and MindIGIRKO mice displayed a 20% decrease in muscle mass (Figure 5C). qPCR at this time also showed increased mRNA levels of Gadd45a, 4E binding protein, Lamp-2a, Gabarapl, and MuRF1 (Figure 5D). These data indicate that loss of IR and IGF1R in muscle in adult animals results in rapid increases in expression of a majority of FoxO target genes, increased autophagy and muscle atrophy.
Figure 5. Short-term inducible deletion of IR and IGF1R in muscle induces atrophy, autophagy, and FoxO target gene expression.
(A) Western blot analysis of IR and IGF1R in TA muscle from control and MindIGIRKO (muscle inducible IGF1R IR knockout) mice sacrificed at various times before or after a 6-day treatment of tamoxifen. Day 0 is the last day of tamoxifen injection. (B) Western blots for autophagy intermediates in TA muscle from control or MindIGIRKO mice 21 days after the last injection of tamoxifen. (C) Dissected muscle weights at day 21 in control or MindIGIRKO mice. (D) qPCR for mRNA levels of IR, Igf1r, FoxO target genes, autophagy genes, proteasome subunit genes, and ubiquitin ligase genes in TA muscle at day 21 in control or MindIGIRKO mice. (n= 5-6). (*P < 0.05, **P < 0.01 vs. day 21 control, Student’s t test.)
Deletion of FoxO1/3/4 isoforms in muscle reverses autophagy in MIGIRKO mice, but does not change proteasomal activity.
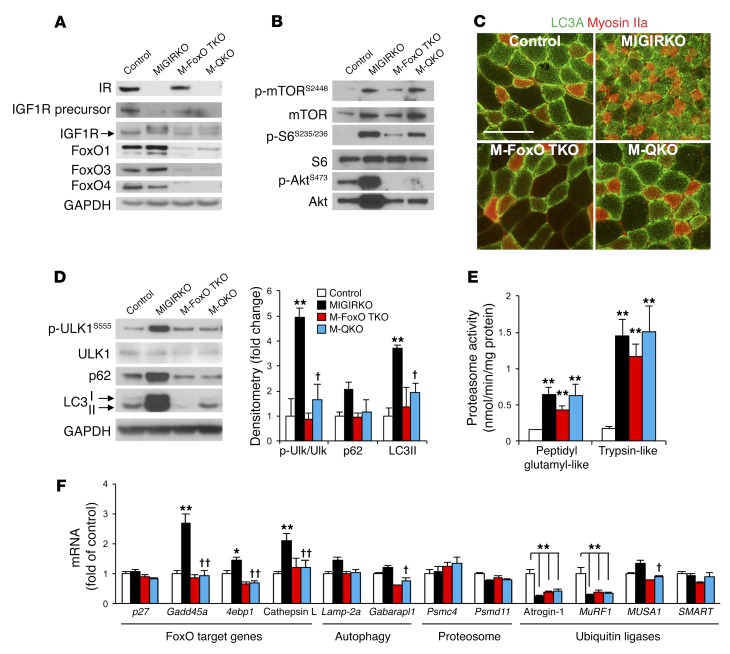
To determine the role of FoxO transcription factors in muscle atrophy and increased autophagy in MIGIRKO mice, we crossed MIGIRKO mice with mice in which FoxO1, FoxO3, and FoxO4 genes were floxed to delete all the major isoforms of FoxO expressed in muscle. Mice in which 5 separate genes — IR, Igf1r, FoxO1, FoxO3, and FoxO4 — were specifically deleted in muscle (muscle quintuple-knockout mice, hereafter referred to as M-QKO mice; Figure 6A and Supplemental Figure 5A) were born in normal Mendelian ratios, and appeared normal both on external inspection and following dissection compared with littermate controls and with muscle FoxO1/3/4 triple-knockout mice (M-FoxO TKO) (Supplemental Figure 5B). Phosphorylation of mTOR and S6 tended to increase in M-QKO and M-FoxO TKO mice compared with controls, but p-S6 was attenuated in comparison with MIGIRKO mice (Figure 6B and Supplemental Figure 5C). Phosphorylation of Akt was blunted in M-QKO and M-FoxO TKO mice compared with controls, indicating an increased reliance on mTOR signaling when FoxOs are deleted in muscle. The proportion of LC3A immunofluorescent-positive myocytes returned to normal in the M-QKO muscle (Figure 6C and Supplemental Figure 5D). Western blot analysis of muscle lysates revealed that the increases in phosphorylation of ULK1S555 and increased levels of p62 and LC3-II protein observed in MIGIRKO mice were normalized to the level of controls in M-QKO mice (Figure 6D), indicating a return to normal of the increased autophagy. Total polyubiquitin proteins tended to increase in MIGIRKO muscle even when not treated with MG132, but polyubiquitin proteins in M-FoxO TKO and M-QKO muscle were indistinguishable from controls (Supplemental Figure 6A). On the other hand, proteasomal activity using 2 different substrates, which was increased in MIGIRKO mice, remained elevated in M-FoxO TKO and M-QKO muscle (Figure 6E). This was in part reflected in muscle amino acid levels that were elevated in M-QKO muscle compared with controls, although not to the degree of MIGIRKO muscle (Supplemental Figure 6B). Amino acid levels in M-FoxO TKO muscle were increased, although this did not reach statistical significance. Several amino acids that were elevated in MIGIRKO returned to control levels in M-QKO, including Trp, Asn, Ser, and Thr (Supplemental Figure 6, C–E). However, other amino acids remained elevated or only partially rescued, including Ile, Leu, Arg, Lys, Cys, and Gly, but Glu was increased in all knockout models. Not surprisingly, FoxO target genes that were increased in MIGIRKO muscle (Gadd45a, 4E binding protein, cathepsin L) returned to the level of control mice when FoxOs were deleted in M-QKO, as did Gabarapl and MUSA1 (Figure 6F). Interestingly, after just 3 hours of fasting, atrogin-1 and MuRF1 were significantly decreased in MIGIRKO muscle and remained low in M-FoxO TKO and M-QKO muscle. Taken together, these data show that IR and IGF1R control muscle protein turnover primarily by acting to suppress FoxO-mediated autophagy, whereas the increase in proteasomal activity is not regulated by FoxO proteins.
Figure 6. Deletion of FoxO1/3/4 in MIGIRKO mice reverses autophagy and transcription of FoxO target and autophagy genes.
(A) Western blot of IR, IGF1R, and FoxO isoforms in TA muscle from control, MIGIRKO, muscle FoxO1/3/4 triple-knockout (M-FoxO TKO), and muscle quintuple-knockout (M-QKO) mice fasted for 3 hours. (B) Western blot of mTOR, S6, and Akt signaling in quadriceps (n = 5–6; for quantification see Supplemental Figure 5C). (C) LC3A (green) and myosin IIa (red) immunostaining of deep portion of TA muscle from control and knockout mice (scale bar: 100 μm). (D) Western blot and densitometric analysis of autophagy intermediates in control, MIGIRKO, M-FoxO TKO, and M-QKO mice (n = 3). (E) Proteasome activity in muscle lysates from control, MIGIRKO, M-FoxO TKO, and M-QKO mice (n = 6). (F) qPCR for mRNA of FoxO target genes, autophagy genes, proteasome subunit genes, and ubiquitin ligase genes in TA muscle from control, MIGIRKO, M-FoxO TKO, and M-QKO mice fasted for 3 hours (n = 6). (*P < 0.05, **P < 0.01 vs. control; †P < 0.05, ††P < 0.01 vs. MIGIRKO, ANOVA.) Blots are from parallel samples run on separate gels.
Deletion of FoxO1/3/4 isoforms in muscle rescues atrophy and muscle function in MIGIRKO mice.
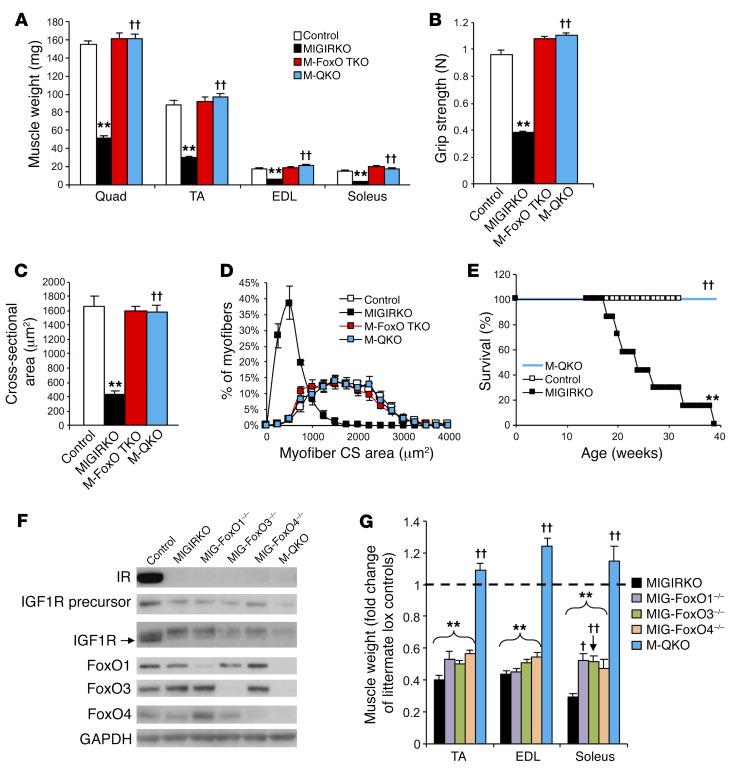
In addition to the reversal of the increased autophagy, M-QKO mice demonstrated a complete rescue of the muscle atrophy observed in the MIGIRKO mice. Thus, muscle mass and body weight in M-QKO were equal to those in controls and M-FoxO TKO (Figure 7A and Supplemental Figure 5B). Grip strength, which was reduced by 60% in MIGIRKO mice, was completely restored in M-QKO (Figure 7B). Myofiber cross-sectional area and myofiber size distribution measured by immunofluorescent staining in M-QKO mice were likewise rescued to the level of control mice (Figure 7, C and D). The restoration of muscle size and function also rescued the impaired survival of MIGIRKO mice (Figure 7E).
Figure 7. Deletion of FoxO1/3/4 in MIGIRKO mice reverses muscle atrophy and muscle dysfunction, but deletion of a single FoxO isoform does not.
(A) Muscle weights from control, MIGIRKO, M-FoxO TKO, and M-QKO mice (n = 6–9). (B) Grip strength in control, MIGIRKO, M-FoxO TKO, and M-QKO mice (n = 12 pooled controls and 5–9 KO mice; MIGIRKO data are the same as in Figure 1I). (C and D) Myofiber cross-sectional (CS) area (C) and myofiber distribution (D) of laminin-stained TA sections from control, MIGIRKO, M-FoxO TKO, and M-QKO mice (n = 4). (E) Survival curves for control, MIGIRKO, and M-QKO female mice (n = 6–7). (F) Western blot of IR, IGF1R, and FoxO isoforms in TA from control, MIGIRKO, MIGIRKO plus FoxO1 knockout (MIG-FoxO1–/–), MIGIRKO plus FoxO3 knockout (MIG-FoxO3–/–), MIGIRKO plus FoxO4 knockout (MIG-FoxO4–/–), and M-QKO mice. (G) Dissected muscle weights normalized to littermate lox controls from MIGIRKO, MIG-FoxO1–/–, MIG-FoxO3–/–, MIG-FoxO4–/–, and M-QKO mice (n = 4–10 per group). (**P < 0.01 vs. control; †P < 0.05, ††P < 0.01 vs. MIGIRKO, ANOVA.) Blots are from parallel samples run on separate gels.
The effects of FoxO deletion to rescue the muscle phenotype required all 3 FoxO proteins. Deletion of only FoxO1 in MIGIKRO (MIG-FoxO1–/–) did not significantly change muscle atrophy in TA or EDL muscle (Figure 7, F and G). Likewise, deletion of only FoxO3 in MIG-FoxO3–/–, or only FoxO4 in MIG-FoxO4–/–, did not rescue muscle atrophy of MIGIRKO mice, nor did it reverse the increases in autophagy markers in TA muscle (Supplemental Figure 7A). MIG-FoxO1–/– and MIG-FoxO3–/– mice did show mildly increased soleus size compared with MIGIRKO, such that the 70% reduction in MIGIRKO soleus mass was decreased to only a 40% reduction in MIG-FoxO1–/– and MIG-FoxO3–/– mice. This may be due to the differential expression of FoxO4, which was lower in soleus and diaphragm compared with other muscles, whereas FoxO1 and FoxO3 are equally expressed in all muscle types (Supplemental Figure 7B). Combined inactivation of both FoxO1 and FoxO3 in MIGIRKO mice was able to increase soleus mass compared with that in MIGIRKOs, but did not rescue TA and EDL atrophy (Supplemental Figure 7, C–E). Thus, muscle atrophy that occurs in response to IR and Igf1r deletion is rescued completely with inactivation of FoxO1/3/4, but inactivation of any 1 of the 3 FoxO isoforms is insufficient to reverse the loss of muscle mass in MIGIRKO mice.
Discussion
Insulin and IGF-1 are known to modulate a number of intracellular pathways affecting protein metabolism, including mTOR and FoxO, but the relative roles of each hormone in muscle hypertrophy and atrophy and the most critical downstream targets that mediate these effects on protein turnover are not known. Our study demonstrates that in skeletal muscle insulin receptors are more important than IGF-1 receptors in maintaining mass, such that deletion of IR results in a 20% loss of muscle mass, while loss of Igf1r has no effect on muscle mass. However, IR and IGF1R have overlapping signaling and function, such that in either single knockout, enough signal is generated to maintain most muscle mass. This overlapping role of IR and IGF1R is evident by the fact that deletion of both receptors in MIGIRKO mice induces a marked 60% reduction of muscle mass, accompanied by loss of muscle strength and endurance, leading to death by 6 months of age, due to atrophy of diaphragm muscle. This loss of muscle mass is characterized by a high rate of protein turnover in the fed state with both increased protein synthesis and degradation. This atrophy can be observed as early as 3 weeks after the deletion of IR and Igf1r in adult MindIGIRKO animals, where it results in an approximately 20% loss of muscle mass. The induction of autophagy and muscle atrophy in MIGIRKO mice is rescued when FoxO1/3/4 are deleted in muscle. Thus, insulin and IGF-1 act through overlapping pathways to regulate muscle protein turnover in the fed state, especially autophagy, and this regulation of autophagy depends completely on the ability of the hormones to suppress FoxO transcription factors.
IGF-1 is thought to be the primary mediator of muscle growth, since overexpression of IGF-1 is sufficient to induce muscle hypertrophy and muscle-specific expression of a dominant-negative IGF1R impairs muscle growth early in development (8, 30). On the other hand, insulin also plays an important role in muscle protein synthesis and degradation (31), and uncontrolled diabetes can lead to accelerated loss of muscle mass and strength (3). Our data suggest that the loss of muscle mass in uncontrolled type 1 diabetes is due specifically to the loss of the ligand, insulin, and may be less influenced by other metabolic changes such as hyperglycemia. Skeletal muscle expresses both IR and IGF1R, and we now show that IGF1R is dominant in early differentiation states, but IR is dominant in mature muscle. Using in vivo genetics, we demonstrate that IR contributes more to muscle mass, such that deletion of IR leads to a greater decrease in muscle mass than deletion of Igf1r.
IR and IGF1R can form hybrid receptors. However, deletion of Igf1r, which would eliminate these hybrids, shows no effect on muscle mass. Thus, hybrid receptors likely play either no role or a minor role in muscle proteostasis. Although these in vivo genetic studies may not be exact mirrors of diabetes or obesity-related insulin resistance, they demonstrate the important roles of insulin and IGF-1 signaling in muscle homeostasis. Since insulin can activate IGF1R and IGF-1 can activate IR, and since signaling downstream of these receptors is similar, our data demonstrate that in the absence of IGF1R, IR can help maintain muscle mass and vice versa. However, combined deletion of IR and Igf1r allows for unrestrained autophagy in the fed state, causing marked loss of muscle mass and reduced survival. How this relates to muscle loss with aging or to sex-specific differences in longevity related to IGF-1 signaling remains to be determined.
Our study demonstrates that FoxO transcription factors are critical mediators of muscle atrophy in response to reduced insulin and IGF-1 signaling. In the insulin and IGF-1 signaling cascades, FoxO proteins are phosphorylated by Akt, which excludes them from the nucleus. Since muscle expresses FoxO1, FoxO3, and FoxO4, it requires knockout of all 3 FoxOs to completely rescue the loss of muscle mass that occurs with deletion of IR and Igf1r in muscle. This overlapping role of the 3 FoxO proteins is supported by recent studies suggesting that deletion of all 3 FoxOs is needed to prevent starvation- or denervation-induced muscle atrophy (27, 32). Both insulin and IGF-1 concentrations decline with fasting in rodents, but insulin is the primary hormone that declines with fasting in humans, indicating that a reduction in insulin signaling is likely the reason for starvation-induced muscle atrophy and thus explaining why, in rodents, deletion of FoxOs prevents this muscle loss. However, the attenuation of denervation-induced atrophy by FoxO deletion is likely more complex, since denervation does not acutely change systemic insulin or IGF-1 levels, although changes in local IGF-1 production or IGF-1/insulin resistance in muscle could still be contributing factors.
The major transcriptional targets of FoxOs include the E3-ubiquitin ligases (so-called “atrogenes”) and other autophagy intermediates that contribute to the control of protein turnover by IR and IGF1R. Although we do not see a significant increase in E3-ligase mRNA levels in fed MIGIRKOs and even observe a decrease in E3-ligase mRNA levels in muscle from fasted MIGIRKOs relative to controls, this is not unexpected, since models of chronic muscle atrophy (>21 days denervation, immobilization, or long-term spinal cord injury) in both humans and mice display no change or even a decrease in the key atrophy markers atrogin-1 and MuRF-1 (33–36). However, FoxOs are the critical targets of IR/IGF1R, as deletion of these proteins rescued muscle atrophy and decreased E3-ligase expression. Indeed, we see a significant decrease in atrogin-1 and MuRF1 mRNA levels in M-FoxO TKO and M-QKO compared with controls, as well as a decrease in MUSA1 in comparison with MIGIRKO mice, which helps to explain decreased ubiquitin-proteasome degradation in muscle from these mice. Nonetheless, changes in protein degradation pathways upon deletion of IR and Igf1r are not completely explained by transcriptional regulation. For example, mRNA levels of LC3 isoforms and p62 were unchanged in MIGIRKO, but the protein levels were markedly increased. We have previously shown a similar discrepancy in mRNA and protein levels of IRS, Akt, and GLUT transporter isoforms in MIGIRKO muscle (12). Interestingly, proteasome subunit mRNA levels were unchanged in MIGIRKO, and proteasomal activity was increased even when FoxOs were deleted and muscle size was rescued in M-FoxO TKO and M-QKO mice. Exactly how this occurs is unknown, but FoxOs are posttranslationally modified in a number of different ways, including phosphorylation, acetylation, and ubiquitination (37). This can influence their transcriptional activity and modify protein interactions outside the nucleus. One study implicates acetylation of cytoplasmic FoxO1 in the control of autophagy (38). Additionally, the autophagy inducer TP53INP2 is regulated in diabetes, but whether its expression is regulated by FoxOs is unknown (39). Future work should focus on the effect of posttranslational modifications of FoxOs as well as protein-protein interactions with FoxOs during muscle wasting in hopes of better understanding the control of protein turnover by insulin, IGF-1, and FoxOs.
Although both IR and IGF1R signaling can activate mTORC1 via Akt (7, 8), mTORC1 is paradoxically activated when the 2 receptors are deleted. This leads to increased protein synthetic rate, which helps, but is insufficient to offset the effects of the increased protein degradation. Indeed, inhibition of mTORC1 with rapamycin induces rapid deterioration of muscle mass in MIGIRKO mice, indicating that these mice are dependent on mTOR signaling for the small amount of muscle growth that they can achieve. The activation of mTOR in MIGIRKO mice is likely the result of enhanced amino acid–induced activation, since release of amino acids by proteasomal degradation can feed back to initiate mTORC1 signaling (22). Consistent with this hypothesis, amino acid levels in MIGIRKO muscle were elevated. In addition, we have previously shown that Akt protein and phospho-/activated Akt are paradoxically increased in MIGIRKO muscle (12), and this could activate mTOR by suppression of the TSC1/TSC2 complex, ultimately leading to increased protein synthesis that does not completely balance increased degradation, resulting in muscle atrophy and a high-protein-turnover state. The fact that the increase in proteolysis and amino acid release occurs in the fed state in MIGIRKO mice is important, since caloric restriction also activates proteolysis, but does not activate mTOR, indicating that interaction between IR/IGF1R and mTOR signaling, as well as dietary status, can influence protein turnover in muscle.
In summary, muscle wasting is an important complication of diabetes, especially long-standing or poorly controlled diabetes. We now demonstrate that this is likely due to decreased signaling via IR, but that both receptors play overlapping roles in the maintenance of muscle mass. Furthermore, we show that the IR/IGF1R signaling cascades maintain muscle mass via suppression of FoxO1/3/4-mediated autophagy and protein degradation. These data indicate that insulin and IGF-1 are critical hormonal regulators of muscle mass and proteostasis. Understanding how these hormones influence FoxO biology will help to identify new targets with therapeutic potential to benefit patients with muscle wasting, diabetes, and metabolic disease.
Methods
Animal care and use
Male mice were used for studies unless indicated. M-IR–/–, M-IGF1R–/–, and MIGIRKO mice were generated using Acta1-Cre (stock 006149; Jackson Laboratory), as previously described (12). Since no differences were observed among the IRfl/fl, Igf1rfl/fl, IRfl/+ Igf1rfl/fl, and IRfl/fl Igf1rfl/fl mice, the results in these controls were pooled. MindIGIRKO (muscle inducible IGF1R IR knockout) mice were generated using muscle-specific, tamoxifen-inducible HSA-CreERT2 (40) mice crossed to IRfl/fl Igf1rfl/fl. We thank Pierre Chambon (Institute for Genetics and Cellular and Molecular Biology, Strasbourg, France) for generating the HSA-CreERT2 mice and allowing us to collaborate with E. Dale Abel to obtain these mice. Muscle-specific FoxO1/3/4 triple-knockout (M-FoxO TKO) mice were generated by crossing of Acta1-Cre and FoxO1/3/4 triple-floxed mice, provided by Domenico Accili. M-QKO (muscle quintuple-knockout) mice, in which 5 separate genes were deleted in muscle — IR, Igf1r, FoxO1, FoxO3, and FoxO4 — were generated by crossing of MIGIRKO with FoxO1/3/4 triple-floxed mice.
Diets and treatments
Animals were maintained on a standard chow diet (Lab Diet 9F, 5020). Fed mice were allowed ad libitum access to food and sacrificed at 9:00 am. For fasting studies, mice were transferred to a new cage without food for 16 hours before sacrifice. For rapamycin treatments, mice were injected i.p. with rapamycin (2 mg/kg/d) or vehicle (0.2% sodium carboxymethylcellulose, 0.25% polysorbate-80 in water) daily for up to 2 weeks as previously described (41). For MG132 treatments, mice were injected i.p. with MG132 (2.5 mg/kg/d in 20% DMSO/saline; NC9038428; Cayman Chemical) or 20% DMSO/saline daily for 3 days. For colchicine treatments, mice were injected i.p. with colchicine (0.4 mg/kg/d; C9754; Sigma-Aldrich) or saline daily for 2 days as previously described (25). Inducible recombination in 12-week-old MindIGIRKO mice was achieved with 5 i.p. injections over 6 days of 100 mg/kg tamoxifen (T5648; Sigma-Aldrich) dissolved in peanut oil.
Primary mouse myoblast isolation
Primary mouse myoblasts were isolated as previously described (42). Briefly, skeletal muscle from forelimbs and hind limbs of 2- to 3-week-old IRfl/fl Igf1rfl/fl mice was minced and digested in 2.5 mM CaCl2 with 2.4 U/ml dispase (Grade II; Roche) and 1% collagenase B (Roche) for 20 minutes, with gentle pipetting twice. Homogenates were centrifuged at 1,000 g and the pellets resuspended in Ham’s/F-10 medium supplemented with 20% FBS, 2.5 ng/ml bFGF (human recombinant; Sigma-Aldrich), 4× penicillin/streptomycin, 1× Normocin (Life Technologies) and plated on plastic for 2 hours to deplete fibroblasts. The supernatant was then transferred and maintained on collagen-coated dishes (Biocoat; Corning). For differentiation experiments, proliferating myoblasts (differentiation day –2) were harvested, or allowed to reach confluence, then switched to differentiation medium (DMEM [Gibco] plus 2% horse serum) for 7 days (differentiation day +7). Myotubes were easily identifiable by microscopy, and spontaneous contractions were observed by day +7.
Physiologic measurements
For voluntary wheel cage running, animals were housed individually in wheel cages (Nalgene) with available access to physical activity at all times. Wheel cage revolutions were monitored every day, and the accumulated running distance was calculated at the end of 30 days. Acute treadmill tests and grip strength tests were performed by the Joslin Diabetes Research Center. Mice were acclimated to the treadmill for 20 minutes on 2 consecutive days without starting of the treadmill. On the third day, the treadmill was started at 5 m/min and increased by 5 m/min every 5 minutes until exhaustion. Grip strength was measured using Chatillon DFE II series (2lbF, 10N; Ametek). Amino acid levels were measured in mixed quadriceps and gastrocnemius muscle at the Mayo Clinic Metabolomics Resource Core using time-of-flight mass spectrometry.
Protein fractional synthesis rate
Protein fractional synthesis rate (FSR) of muscle, heart, and liver was measured after an i.p. injection of 15 mg/kg 13[C6]-labeled phenylalanine 20 minutes before sacrifice as previously described (19). Tissues were snap-frozen, the total proteins were extracted, and incorporation of 13[C6]Phe was determined by tandem mass spectrometry. Tissue fluid enrichment was used as the precursor pool for FSR calculations.
Proteolysis assay
Proteolysis was measured as previously described (43), with the following modifications. Soleus and EDL muscles were isolated from 8-week-old female mice and preincubated in 1 ml of KRB buffer (in mM: 117 NaCl, 4.7 KCl, 2.5 CaCl2, 1.2 KH2PO4, 1.2 MgSO4, 24.6 NaHCO3, and 5 glucose) for 30 minutes with constant bubbling of 95% O2/5% CO2 before transferring to fresh 1 ml of KRB containing 0.5 mM cycloheximide to inhibit protein synthesis for 2 hours. Incubation buffer was collected, and tyrosine concentration was determined in 0.5 ml of incubation buffer as previously described (44) and was normalized to muscle weight.
Proteasome and cathepsin activity assays
Proteasome activity was determined from muscle homogenates using substrates for peptidylglutamyl-like and trypsin-like activity of the 26S proteasome as previously described (45, 46) with the following modifications. Frozen powdered muscle was homogenized in 50 mM Tris-HCl, 5 mM MgCl2, 250 mM sucrose, 2 mM ATP, 1 mM DTT, 0.5 mM EDTA, pH 7.5, then centrifuged (600 g for 20 minutes) at 4°C to pellet contractile proteins and membranes. The supernatant was removed and centrifuged (16,000 g for 10 minutes at 4°C) to yield the final supernatant for proteasome activity. The pellet was washed twice in 150 mM KCl, 20 mM sodium phosphate, pH 6.0, and these lysosomal-enriched fractions were saved for cathepsin L activity. Fifteen micrograms of protein from the proteasome supernatant was mixed with Z-Leu-Leu-Glu-7-amido-4-methylcoumarin (LLE) (C0483; Sigma-Aldrich) or with Boc-Leu-Ser-Thr-Arg-7-amido-4-methylcoumarin (LSTR) (B4636; Sigma-Aldrich; 100 μM final concentration) in assay buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 40 mM KCl, 2 mM ATP, 1 mM DTT, 0.5 mg/ml BSA) in a 96-well black plate, and fluorescence was monitored (360 nm excitation, 460 nm emission) every 3 minutes for 1.5 hours at 37°C. Enzyme activity (Vmax) was determined as the change in fluorescence during the linear phase of the reaction and compared against a standard curve of 7-amido-4-methylcoumarin (A9891; Sigma-Aldrich). Lysosomal-enriched fractions were measured for cathepsin L activity as previously described (46) using Z-Phe-Arg-7-amido-4-methylcoumarin as a substrate.
Histology
Frozen cross sections of TA or quadriceps muscle were immunofluorescently stained for laminin, costained for myosin IIa and myosin I, or costained for LC3A and myosin IIa using the protocol previously described (47). See Supplemental Table 2 for antibodies used.
Quantification of muscle histology
Cross-sectional area.
Images of laminin-stained fibers were taken in the same superficial area of TA muscles, then changed to grayscale using Adobe Photoshop 6.0. ImageJ64 software (NIH) was used to threshold images, and the Analyze Particles tool was used to find all cross sections between 500 and 106 square pixels with circularity of 0.5 to 1.0. Pixel area was converted to square micrometers by quantification of the length of the 200-μm scale bar in pixels (conversion for these images was 1 pixel2 = 0.04410194 μm2).
Myosin fiber type density.
Images of myosin I– and myosin IIa–costained cross sections were taken in soleus or in the deep portion of quadriceps muscles with identifiable landmarks (interior fascia and intact exterior surface) indicating intact muscle without sectioning of artifacts. Four hundred fifty to 800 fibers were manually counted and normalized to total number of fibers in the field of interest.
LC3A vesicles.
Confocal images of LC3A- and myosin IIa–costained quadriceps muscle cross sections were taken in the same deep portion of the muscle. ImageJ64 software was used to manually draw the circumference of either myosin IIa–positive or myosin IIa–negative (type IIb) fibers, and pixel cross-sectional area was measured. Pixel area was converted to square micrometers by quantification of the length of the 20-μm scale bar in pixels (conversion for these images was 1 pixel2 = 0.19753086 μm2). Fibers were then thresholded in ImageJ64; the Analyze Particles tool was used to find all vesicles between 4 and 120 square pixels, and this number was normalized to cross-sectional area. Up to 3 type IIa and 3 type IIb fibers were quantified per mouse and used for statistical analysis.
Muscle fractionation
Mixed hind-limb muscle was fractionated as previously described (12), with the following modifications. Homogenates were centrifuged at 1,000 g (pellet), then at 16,000 g to remove mitochondria and plasma membranes yielding the cytosolic fraction as the supernatant. The pellet was then resuspended in 2 ml of homogenization buffer, filtered using a 40-μm nylon mesh cell filter (Fisher) to remove unbroken cells/debris. The filtrate was again centrifuged at 1,000 g, yielding a nuclear fraction that was resuspended in RIPA buffer (Millipore) for Western analysis.
qPCR and plasmids
Mouse IGF1R and IR cDNA plasmids were ordered from Origene (MC224356, MC224342) and cloned into a pBabe-Hygromycin vector, and Qiagen Maxiprep was performed to isolate plasmid DNA content that was measured using a nanodrop spectrophotometer to yield nanograms of DNA, which was converted into copies of plasmid based on molecular weight. Serial dilutions of each plasmid were used to generate standard curves for calculation of IR or Igf1r mRNA copy number normalized to total RNA. Total RNA was extracted from muscle using Qiazol reagent (Qiagen), then reverse transcribed into cDNA (Applied Biosystems) according to the manufacturer’s protocols. qPCR was carried out using SYBR Green (Bio-Rad) with primers as detailed in Supplemental Table 1, and normalized to Tata binding protein (TBP).
Western analysis
Powdered muscle tissue was homogenized in RIPA buffer with protease and phosphatase 2 and 3 inhibitors (Sigma-Aldrich). Lysates were subjected to SDS-PAGE and blotted using antibodies listed in Supplemental Table 2. Full uncut gels are available in the online Supplemental Materials.
Statistics
All data are presented as mean ± SEM. Student’s 2-tailed t test was performed for comparison of 2 groups, and ANOVA was performed for comparison of 3 or more groups to determine significance. For fasted studies, 2-way ANOVA was performed. A P value less than 0.05 was considered significant.
Study approval
Animal studies were performed according to protocols approved by the Institutional Animal Care and Use Committee.
Author contributions
BTO designed the study, researched data, and wrote the manuscript. KYL, SS, and JF researched data, helped write the manuscript, and helped design experiments. KK, MTK, MFH, and KIS researched data and helped design experiments. MR, ROP, and LJG helped design experiments. WC and AK generated plasmids. EDA and DA provided critical reagents. KSN provided reagents, helped design experiments, and helped write the manuscript. CRK designed the study and helped write the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01 DK031036 (to CRK), R01 AR42238 (to LJG), and R01 DK41973 and U24 DK100469 (to KSN). BTO was funded by a K08 training award from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the NIH (K08 DK100543), Mayo Clinic Metabolomics Resource Core grant U24 DK100469 from the NIDDK, which originates from the NIH Director’s Common Fund, and Mayo Clinic Clinical and Translational Science Awards grant UL1 TR000135 from the National Center for Advancing Translational Sciences of the NIH. KIS was funded by a K01 award from the NIDDK of the NIH (K01 DK105109). AK was supported by a German Research Foundation (DFG) fellowship Kl2399-1/1 grant. The Joslin Diabetes Research Center core facility was used for part of this work (P30 DK36836).
Footnotes
KYL’s present address is: Department of Biomedical Sciences, Ohio University, Athens, Ohio, USA.
JF’s present address is: Section of Molecular Physiology, August Krogh Centre, Department of Nutrition, Exercise and Sports, University of Copenhagen, Copenhagen, Denmark.
KIS’s present address is: Dorothy M. Davis Heart and Lung Research Institute, Department of Physiology and Cell Biology, The Ohio State University Wexner Medical Center, Columbus, Ohio, USA.
AK’s present address is: German Institute of Human Nutrition (DIfE), Nuthetal, Germany; and National Center for Diabetes Research (DZD), Neuherberg, Germany.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(9):3433–3446. doi:10.1172/JCI86522.
Contributor Information
Katherine Klaus, Email: klaus.katherine@mayo.edu.
Samir Softic, Email: samir.softic@childrens.harvard.edu.
Joachim Fentz, Email: jfentz@nexs.ku.dk.
Matthew M. Robinson, Email: robinson.matthew@mayo.edu.
Weikang Cai, Email: weikang.cai@joslin.harvard.edu.
Andre Kleinridders, Email: andre.kleinridders@dife.de.
E. Dale Abel, Email: dale-abel@uiowa.edu.
Domenico Accili, Email: da230@cumc.columbia.edu.
K. Sreekumaran Nair, Email: nair@mayo.edu.
C. Ronald Kahn, Email: c.ronald.kahn@joslin.harvard.edu.
References
- 1.Newman AB, et al. Strength, but not muscle mass, is associated with mortality in the health, aging and body composition study cohort. J Gerontol A Biol Sci Med Sci. 2006;61(1):72–77. doi: 10.1093/gerona/61.1.72. [DOI] [PubMed] [Google Scholar]
- 2.Park SW, et al. Decreased muscle strength and quality in older adults with type 2 diabetes: the health, aging, and body composition study. Diabetes. 2006;55(6):1813–1818. doi: 10.2337/db05-1183. [DOI] [PubMed] [Google Scholar]
- 3.Park SW, et al. Accelerated loss of skeletal muscle strength in older adults with type 2 diabetes: the health, aging, and body composition study. Diabetes Care. 2007;30(6):1507–1512. doi: 10.2337/dc06-2537. [DOI] [PubMed] [Google Scholar]
- 4.Fulks RM, Li JB, Goldberg AL. Effects of insulin, glucose, and amino acids on protein turnover in rat diaphragm. J Biol Chem. 1975;250(1):290–298. [PubMed] [Google Scholar]
- 5.Meek SE, Persson M, Ford GC, Nair KS. Differential regulation of amino acid exchange and protein dynamics across splanchnic and skeletal muscle beds by insulin in healthy human subjects. Diabetes. 1998;47(12):1824–1835. doi: 10.2337/diabetes.47.12.1824. [DOI] [PubMed] [Google Scholar]
- 6.Goldspink G. Changes in muscle mass and phenotype and the expression of autocrine and systemic growth factors by muscle in response to stretch and overload. J Anat. 1999;194(pt 3):323–334. doi: 10.1046/j.1469-7580.1999.19430323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rommel C, et al. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3(11):1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 8.Schiaffino S, Mammucari C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skelet Muscle. 2011;1(1): doi: 10.1186/2044-5040-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palsgaard J, Brown AE, Jensen M, Borup R, Walker M, De Meyts P. Insulin-like growth factor I (IGF-I) is a more potent regulator of gene expression than insulin in primary human myoblasts and myotubes. Growth Horm IGF Res. 2009;19(2):168–178. doi: 10.1016/j.ghir.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Mavalli MD, et al. Distinct growth hormone receptor signaling modes regulate skeletal muscle development and insulin sensitivity in mice. J Clin Invest. 2010;120(11):4007–4020. doi: 10.1172/JCI42447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Neill ED, et al. Absence of insulin signalling in skeletal muscle is associated with reduced muscle mass and function: evidence for decreased protein synthesis and not increased degradation. Age (Dordr) 2010;32(2):209–222. doi: 10.1007/s11357-009-9125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Neill BT, Lauritzen HP, Hirshman MF, Smyth G, Goodyear LJ, Kahn CR. Differential role of insulin/IGF-1 receptor signaling in muscle growth and glucose homeostasis. Cell Rep. 2015;11(8):1220–1235. doi: 10.1016/j.celrep.2015.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7(2):85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 14.Izumiya Y, et al. Fast/Glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab. 2008;7(2):159–172. doi: 10.1016/j.cmet.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007;6(3):208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389(6654):994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 17.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–412. doi: 10.1016/S0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanza IR, et al. Chronic caloric restriction preserves mitochondrial function in senescence without increasing mitochondrial biogenesis. Cell Metab. 2012;16(6):777–788. doi: 10.1016/j.cmet.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newton K, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134(4):668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 22.Quy PN, Kuma A, Pierre P, Mizushima N. Proteasome-dependent activation of mammalian target of rapamycin complex 1 (mTORC1) is essential for autophagy suppression and muscle remodeling following denervation. J Biol Chem. 2013;288(2):1125–1134. doi: 10.1074/jbc.M112.399949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(19):1845–1846. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- 24.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ju JS, Varadhachary AS, Miller SE, Weihl CC. Quantitation of “autophagic flux” in mature skeletal muscle. Autophagy. 2010;6(7):929–935. doi: 10.4161/auto.6.7.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milan G, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat Commun. 2015;6: doi: 10.1038/ncomms7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Vos KE, Coffer PJ. The extending network of FOXO transcriptional target genes. Antioxid Redox Signal. 2011;14(4):579–592. doi: 10.1089/ars.2010.3419. [DOI] [PubMed] [Google Scholar]
- 29.Miniou P, Tiziano D, Frugier T, Roblot N, Le Meur M, Melki J. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res. 1999;27(19): doi: 10.1093/nar/27.19.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernández AM, Dupont J, Farrar RP, Lee S, Stannard B, Le Roith D. Muscle-specific inactivation of the IGF-I receptor induces compensatory hyperplasia in skeletal muscle. J Clin Invest. 2002;109(3):347–355. doi: 10.1172/JCI13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nair KS, Ford GC, Ekberg K, Fernqvist-Forbes E, Wahren J. Protein dynamics in whole body and in splanchnic and leg tissues in type I diabetic patients. J Clin Invest. 1995;95(6):2926–2937. doi: 10.1172/JCI118000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang H, et al. mTORC1 promotes denervation-induced muscle atrophy through a mechanism involving the activation of FoxO and E3 ubiquitin ligases. Sci Signal. 2014;7(314): doi: 10.1126/scisignal.2004809. [DOI] [PubMed] [Google Scholar]
- 33.Bertaggia E, Coletto L, Sandri M. Posttranslational modifications control FoxO3 activity during denervation. Am J Physiol Cell Physiol. 2012;302(3):C587–C596. doi: 10.1152/ajpcell.00142.2011. [DOI] [PubMed] [Google Scholar]
- 34.Léger B, et al. Atrogin-1, MuRF1, and FoXO, as well as phosphorylated GSK-3beta and 4E-BP1 are reduced in skeletal muscle of chronic spinal cord-injured patients. Muscle Nerve. 2009;40(1):69–78. doi: 10.1002/mus.21293. [DOI] [PubMed] [Google Scholar]
- 35.Suetta C, et al. Aging affects the transcriptional regulation of human skeletal muscle disuse atrophy. PLoS One. 2012;7(12): doi: 10.1371/journal.pone.0051238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanson AM, Harrison BC, Young MH, Stodieck LS, Ferguson VL. Longitudinal characterization of functional, morphologic, and biochemical adaptations in mouse skeletal muscle with hindlimb suspension. Muscle Nerve. 2013;48(3):393–402. doi: 10.1002/mus.23753. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Wang Y, Zhu WG. Applications of post-translational modifications of FoxO family proteins in biological functions. J Mol Cell Biol. 2011;3(5):276–282. doi: 10.1093/jmcb/mjr013. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12(7):665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 39.Sala D, et al. Autophagy-regulating TP53INP2 mediates muscle wasting and is repressed in diabetes. J Clin Invest. 2014;124(5):1914–1927. doi: 10.1172/JCI72327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuler M, Ali F, Metzger E, Chambon P, Metzger D. Temporally controlled targeted somatic mutagenesis in skeletal muscles of the mouse. Genesis. 2005;41(4):165–170. doi: 10.1002/gene.20107. [DOI] [PubMed] [Google Scholar]
- 41.Wende AR, et al. Enhanced cardiac Akt/protein kinase B signaling contributes to pathological cardiac hypertrophy in part by impairing mitochondrial function via transcriptional repression of mitochondrion-targeted nuclear genes. Mol Cell Biol. 2015;35(5):831–846. doi: 10.1128/MCB.01109-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol. 1994;125(6):1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price SR, et al. Muscle wasting in insulinopenic rats results from activation of the ATP-dependent, ubiquitin-proteasome proteolytic pathway by a mechanism including gene transcription. J Clin Invest. 1996;98(8):1703–1708. doi: 10.1172/JCI118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waalkes TP, Udenfriend S. A fluorometric method for the estimation of tyrosine in plasma and tissues. J Lab Clin Med. 1957;50(5):733–736. [PubMed] [Google Scholar]
- 45.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Meth Enzymol. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 46.Klaude M, Mori M, Tjäder I, Gustafsson T, Wernerman J, Rooyackers O. Protein metabolism and gene expression in skeletal muscle of critically ill patients with sepsis. Clin Sci. 2012;122(3):133–142. doi: 10.1042/CS20110233. [DOI] [PubMed] [Google Scholar]
- 47.Lee KY, et al. Tbx15 controls skeletal muscle fibre-type determination and muscle metabolism. Nat Commun. 2015;6: doi: 10.1038/ncomms9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.