Abstract
Huntington’s disease (HD) is a progressive, adult-onset neurodegenerative disease caused by a polyglutamine (polyQ) expansion in the N-terminal region of the protein huntingtin (HTT). There are no cures or disease-modifying therapies for HD. HTT has a highly conserved Akt phosphorylation site at serine 421, and prior work in HD models found that phosphorylation at S421 (S421-P) diminishes the toxicity of mutant HTT (mHTT) fragments in neuronal cultures. However, whether S421-P affects the toxicity of mHTT in vivo remains unknown. In this work, we used murine models to investigate the role of S421-P in HTT-induced neurodegeneration. Specifically, we mutated the human mHTT gene within a BAC to express either an aspartic acid or an alanine at position 421, mimicking tonic phosphorylation (mHTT-S421D mice) or preventing phosphorylation (mHTT-S421A mice), respectively. Mimicking HTT phosphorylation strongly ameliorated mHTT-induced behavioral dysfunction and striatal neurodegeneration, whereas neuronal dysfunction persisted when S421 phosphorylation was blocked. We found that S421 phosphorylation mitigates neurodegeneration by increasing proteasome-dependent turnover of mHTT and reducing the presence of a toxic mHTT conformer. These data indicate that S421 is a potent modifier of mHTT toxicity and offer in vivo validation for S421 as a therapeutic target in HD.
Introduction
Huntington’s disease (HD) is a devastating neurodegenerative disorder with no cure or disease-altering treatment. HD patients present clinically with any combination of abnormal movements, dementia, and affective changes and eventually succumb to premature death (1, 2). Pathological inspection of brains of HD patients reveals profound atrophy and cell death of medium spiny neurons (MSNs) of the striatum, with cortical loss as the disease progresses. Surviving neurons often have abnormal nuclear and/or cytoplasmic protein aggregates, a pathological hallmark of the disease (3).
HD is inherited in an autosomal dominant fashion, and genetic linkage studies identified huntingtin (HTT) as the gene whose mutation causes HD (4). HTT codes for a widely expressed 350-kDa protein with a polyglutamine (polyQ) repeat of variable length near its N-terminus. Although its predominant function is unclear, HTT has been implicated in multiple cellular processes, including intracellular signaling, metabolism, gene expression, intracellular transport, and ciliogenesis (5–7). Expansion of its polyQ stretch beyond 36 repeats causes the protein to misfold and confers a toxic gain of function (8). However, loss of function of the WT allele might also contribute to HD (9). How this polyQ-expanded, mutant form of HTT (mHTT) causes selective and progressive dysfunction and neurodegeneration is unclear.
HD is 1 of at least 9 genetic diseases caused by an abnormal polyQ stretch, highlighting the toxicity of polyQ-mediated protein misfolding in disease pathogenesis (10). However, host protein context has a vital role in pathogenesis. Posttranslational modifications (PTMs) influence the toxicity of several disease-associated polyQ proteins, including HTT (11, 12). For instance, several PTMs localized to the N-terminal 17 amino acids of HTT modulate its toxicity (13–18). These residues, immediately preceding the polyQ stretch, are well positioned to alter the expansion’s propensity to cause protein misfolding. However, other HTT PTMs farther away from the polyQ repeat region by primary sequence also modify mHTT toxicity (19–29).
Serine 421 of HTT is found in an RXRXXS motif and forms a consensus Akt phosphorylation site that is highly conserved in vertebrates (19). S421 also can be phosphorylated by the serum- and glucocorticoid-induced kinase (SGK) (30). Studies of HTT phosphorylated at S421 (S421-P) in cell culture suggest it modulates the regulatory role for HTT in vesicular trafficking (31–33) and is associated with a decrease in buildup of nuclear mHTT fragments (34) and NMDA-mediated excitotoxicity (35). PolyQ expansion decreases the proportion of HTT phosphorylated at S421 (36, 37), and intriguingly, phosphorylation in specific brain tissues correlates inversely with susceptibility to degeneration in HD.
In a primary striatal neuron model of HD based on the overexpression of an HTT-N480-68Q fragment (38), manipulations that increase Akt activity increase S421-P and reduce toxicity of HTT-N480-68Q. Mimicking tonic phosphorylation with a serine–to–aspartic acid mutation (HTT-N480-68Q-S421D) is itself sufficient to abrogate toxicity, whereas toxicity persists if phosphorylation is blocked with a serine-to-alanine mutation (HTT-N480-68Q-S421A) (19). The same phosphomimetic mutation or either genetic or pharmacological inhibition of calcineurin, a putative S421 phosphatase, reduces the toxicity of lentivirus-mediated expression of HTT-N480-68Q in rat striatum (37).
Although these findings suggest a key connection between S421-P and HTT toxicity, critics have pointed out that there is no evidence that an N-terminal 480–amino acid fragment is normally generated during HD. In addition, this fragment is missing nearly 85% of the HTT protein, including influential C-terminal domains, such as the nuclear export domain (39), highly conserved protein interaction domains known as HEAT repeats (6), and a series of conserved proteolytic cleavage sites, including 1 for caspase-6 (12). Given the extensive in vivo data indicating that nuclear localization, protein interactions, and proteolytic cleavage are critical events in HTT pathogenesis, it is unclear whether and how manipulating HTT-S421 in vivo affects pathogenesis. The fact that all the work on the role of S421-P with the N-terminal 480–amino acid fragment was done using overexpression with strong nonendogenous viral promoters added to doubts about the physiological significance of S421 to HD and whether it is a therapeutic target. HTT has important and varied roles in embryonic development (40), but it is unknown whether S421 affects these functions. Similarly, HD differentially affects tissues in the CNS and periphery, and whether S421 has a role determining cell specificity and network dysfunction is unknown.
Therefore, to determine whether and to what extent S421 regulates HTT toxicity and to test S421 as a therapeutic target for HD, we generated transgenic mice that express human HTT regulated by its endogenous promoter with S421 mutated to mimic or prevent phosphorylation. Specifically, we altered the human HTT gene within the BAC used to create the BACHD mouse model of HD (41) to express HTT-S421D-97Q or HTT-S421A-97Q. We show that mimicking phosphorylation strongly ameliorates mHTT-induced behavioral dysfunction and neurodegeneration, but neuronal dysfunction persists when S421 phosphorylation is blocked. Our data also suggest that S421 phosphorylation mitigates neurodegeneration by increasing proteasome-dependent turnover of mHTT and reducing levels of a toxic mHTT conformer. These data indicate that S421 is a potent modifier of mHTT toxicity and offer in vivo validation for S421 as a therapeutic target in HD.
Results
Generating and characterizing BAC transgenic mice expressing mHTT with an S421 mutation.
To study the role of HTT-S421-P in a mammalian model system, we used the BACHD mouse model of HD. BACHD mice express human mHTT with a pathogenic 97Q repeat stretch under the control of its endogenous promoter. As they age, these mice show robust behavioral and histopathological phenotypes (15, 41).
With the BAC used to generate the BACHD model, we performed 2 parallel versions of Red/ET triple recombination, followed by FLP recombination to remove the selectable marker (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI80339DS1). The mutated BACs express aspartic acid or alanine at position 421 (Figure 1A). We verified recombineering by PCR amplification of the local region and sequencing. We sequenced the 5′ and 3′ ends of the HTT gene to confirm that the 97 mixed CAA-CAG codons encoding the expanded polyQ repeat were unaltered without other unintended mutations. The structural integrity of the BACs was confirmed by pulsed-field gel electrophoresis. Finally, purified and linearized BAC DNA (Supplemental Figure 1C) was microinjected into FvB/N pronuclei to generate transgenic founders.
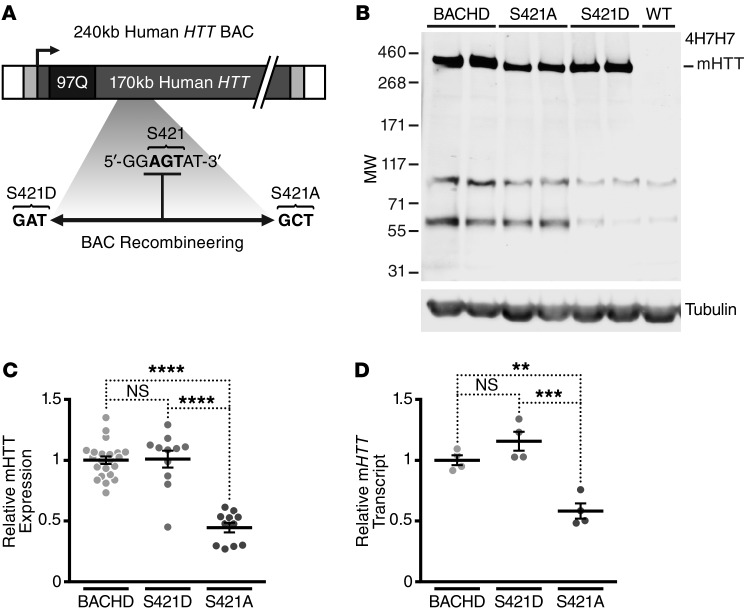
Figure 1. Generation and characterization of mHTT transgenic mice with phosphomimetic (S421D) and phosphoresistant (S421A) mutations at S421.
(A) Schematic of the recombineered BAC cassette, in which S421D or S421A mutations within the original BACHD construct were generated. (B) Representative Western blot demonstrating expression levels of mHTT from cortical lysates of 2-month-old BACHD, mHTT-S421D, and mHTT-S421A mice. The blot was probed with anti-expanded polyQ mAb 4H7H7 and anti–γ-tubulin as a loading control. (C) Quantification of expression levels of mHTT from cortical lysates of 2-month-old BACHD, mHTT-S421D, and mHTT-S421A mice. Values are based on the mean of 3 independent 4H7H7 blots, each with lysates from at least 3 mice per line compared across different blots by normalization to BACHD samples. Each value was first normalized for input using the anti–γ-tubulin control. F = 36.71, S421A vs. BACHD/S421D, P < 0.0001. (D) Quantification of the levels of cortical mHTT transcript in BACHD, mHTT-S421D, and mHTT-S421A mice by qRT-PCR. The results are from 4 independent samples per transgenic line, each run in quadruplicate. Values are normalized to BACHD mice. F = 51.8, P < 0.0001; S421A vs. BACHD/S421D, P < 0.0001. **P < 0.01; ***P < 0.001; ****P < 0.0001; all by 1-way ANOVA.
Seven founders with the BACHD-S421D transgene (mHTT-S421D mice) and 2 with the BACHD-S421A transgene (mHTT-S421A mice) were obtained. One mHTT-S421D line and 1 mHTT-S421A line expressed sufficient mHTT to make valid comparisons with HD transgenic lines with similar levels of HTT. To control for genetic background, these 2 lines were expanded by breeding with FvB/NJ WT mice, the same inbred strain used to characterize the original BACHD mice. Again, we confirmed the proper incorporation of the entire gene and S421 mutation without changes in the polyQ repeat length.
To characterize the mice, we analyzed mHTT protein levels in the F1 progeny of the founder lines. Levels of mHTT protein are generally directly proportional to their toxicity (41–43). Thus, we looked for lines that expressed mHTT at levels similar to those of BACHD mice. We used Western blots to evaluate steady-state levels of soluble mHTT levels in cortical lysates from 2-month-old mice. Because all lines have the same polyQ repeat length, we used the mouse mAb 4H7H7, which selectively recognizes pathogenic polyQ stretches (15). One line of hemizygous mHTT-S421D mice expressed mHTT protein at levels similar to those of the original BACHD model (Figure 1, B and C). We confirmed this finding with 1C2, another mouse mAb that recognizes abnormal polyQ stretches (Supplemental Figure 2). We analyzed levels of cortical mHTT transcript by quantitative real-time PCR (qRT-PCR). mHTT-S421D mice had levels of mHTT mRNA approximately equivalent to those of BACHD mice (Figure 1D).
We performed a similar analysis for the mHTT-S421A mutant mice. One hemizygous line expressed approximately 45% of the mHTT protein of BACHD mice (Figure 1, B and C). Levels of transcript were also consistent with this determination (Figure 1D). This level of mHTT expression is similar to that of BACHD-L mice, a previously reported line that expresses mHTT at lower levels than the primary BACHD model but still shows pathology (Supplemental Figure 2 and ref. 41). Thus, although the lower levels of mHTT in mHTT-S421A mice preclude a direct comparison with BACHD mice, the line has sufficient expression to assess phenotype and control for nonspecific effects of the mutation.
We next determined whether S421 alters steady-state levels of soluble mHTT protein during aging in a way that could account for any altered phenotype. We used Western blots to compare mHTT levels in the cortex of BACHD, mHTT-S421D, and mHTT-S421A mice at 1 and 12 months of age. Like the BACHD line, mHTT-S421D or mHTT-S421A mice showed no clear differences in protein levels with age (Supplemental Figure 3A). Overexposing the blots showed no non-obvious changes in the production of soluble mHTT polyQ fragments associated with the S421 mutation at either age (Supplemental Figure 3B).
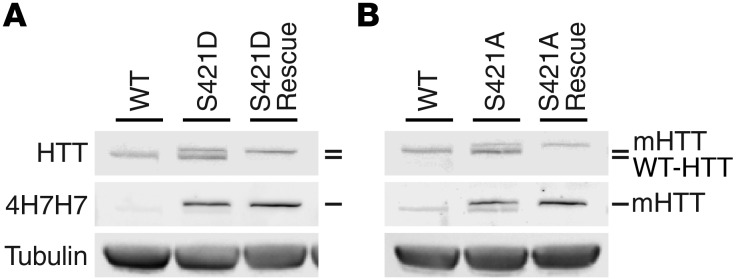
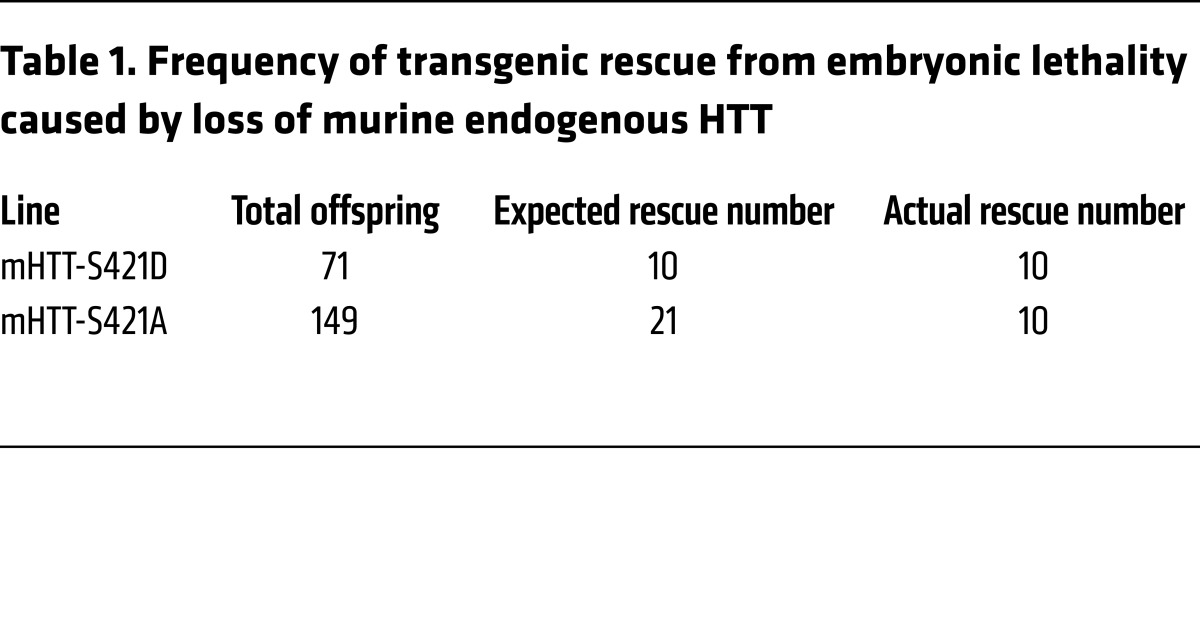
Finally, we determined whether mutation of S421 affects HTT’s essential functions. Complete loss of HTT is embryonic lethal (44), but mHTT is sufficient for murine development and neurogenesis (41, 45, 46) when expressed by its endogenous promoter and regulatory regions. To test whether S421 mutation permits HTT to mediate its critical role in embryonic development, we crossed mHTT-S421D and mHTT-S421A transgenic mice with mice heterozygous for endogenous murine Htt (technically, Hdh) for 2 successive rounds. We genotyped the progeny at weaning age and found that the mHTT-S421D and mHTT-S421A transgenes rescued embryonic lethality. We confirmed these results by immunoblotting for transgenic and endogenous HTT in cortical lysates. Mice rescued by transgenic mHTT-S421D (Figure 2A) or mHTT-S421A (Figure 2B) expressed mHTT but not endogenous murine HTT. This breeding scheme should produce transgene-positive mice on an Htt-null background at an expected Mendelian ratio of 1 of every 7 animals. Consistent with an important role for phosphorylation at S421 in HTT function, mHTT-S421A–rescued mice were generated at less than the expected ratio (Table 1), suggesting a decrease in fitness during the embryonic or neonatal period. We obtained 10 rescue mice from each of the mHTT-S421D and mHTT-S421A lines, and the mice showed no obvious abnormalities through at least 6 months of age.
Figure 2. Essential functions of HTT in development can be rescued by expression of transgenic mHTT with phosphomimetic or phosphoresistant mutations at S421.
Western blots of cortical lysates probed with anti-HTT (mAb 2166), anti-expanded polyQ (4H7H7), and anti–γ-tubulin confirm that mice rescued by phosphomimetic (A) or phosphoresistant (B) mHTT transgene express only mHTT and lack endogenous murine HTT. Some residual signal after membrane stripping can be appreciated in the 4H7H7 blots.
Table 1. Frequency of transgenic rescue from embryonic lethality caused by loss of murine endogenous HTT.
Phosphomimetic mutation at S421 strongly ameliorates motor and psychiatric-like deficits caused by the expression of mHTT.
The neuronal dysfunction elicited by mHTT in BACHD mice causes early and severe motor phenotypes, with psychiatric-like deficits later (15, 41, 47). To determine whether phosphorylation at S421 affects mHTT-induced neuronal dysfunction in vivo, we determined the behavioral phenotypes of mHTT-S421D mice.
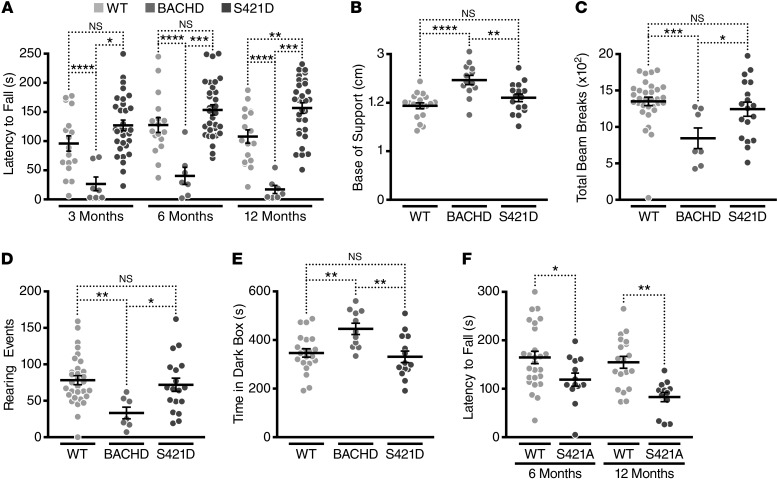
We assessed mouse motor function on the accelerating rotarod. We used a 2-way mixed-factorial design to analyze repeated performances at 3, 6, and 12 months of age in a cohort of BACHD, mHTT-S421D, and WT mice (Figure 3A). Analysis by 2-way repeated-measures ANOVA revealed significant effects of genotype (F(2,52) = 26.15, P < 0.0001) and age (F(2,104) = 4.58, P = 0.0124) on the behavioral phenotype. Next, we used Bonferroni post hoc tests to compare each genotype with each of the others at all ages tested (Supplemental Table 1). BACHD mice had motor dysfunction at all ages tested, falling off of the rotarod more quickly than WT littermate controls. mHTT-S421D mice were indistinguishable from the WT littermate controls at 3 and 6 months of age and showed only a modest deficit at 12 months of age. At all 3 ages tested, mHTT-S421D mice performed better than the BACHD mice. Thus, mutating S421 to mimic tonic phosphorylation mitigated mHTT-induced deficits in accelerating-rotarod performance compared with that of BACHD mice expressing similar levels of mHTT.
Figure 3. mHTT-S421D mice, but not mHTT-S421A mice, have less severe motor and psychiatric-like behavioral deficits than BACHD mice.
(A) Analysis of motor performance on the accelerating rotarod of a cohort of BACHD (n = 7), mHTT-S421D (n = 17), and WT (n = 31) mice at 3, 6, and 12 months of age. (B) Analysis of hind-limb gaits of BACHD, mHTT-S421D, and WT mice at 12 months of age with the CatWalk XT (Noldus). (C and D) Comparison of total activity (C) and rearing activity (D) in BACHD, mHTT-S421D, and WT mice in the open field for 10 minutes at 12 months of age. (E) Comparison of anxiety-like behavior in the light-dark box over 10 minutes in BACHD, mHTT-S421D, and WT mice at 12 months of age. (F) Analysis of motor performance on the accelerating rotarod of a cohort of mHTT-S421A (n = 13) and WT (n = 19) mice at 6 and 12 months of age. In A and F, 2-way repeated-measures ANOVA statistical analyses were used, with 1-way ANOVA used for B–E. Bonferroni post hoc tests were used for all pairwise comparison. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We wondered whether changes in body weight might explain our rotarod results. Weight can influence rotarod performance generally (48), and overexpression of HTT and mHTT in BAC and yeast artificial chromosome (YAC) transgenic mice causes an obesity-with-age phenotype (41, 49). Like the BACHD mice, mHTT-S421D mice were heavier than WT littermate controls at all 3 ages tested (Supplemental Figure 4). In fact, mHTT-S421D mice gained weight similarly to BACHD mice at 3 and 6 months of age but did not display any deficits at these ages. We conclude that the amelioration of rotarod deficits cannot be explained by differences in body weight.
Since each behavioral assay has its own inherent limitations, we sought to assay motor function at 12 months of age with several complementary tests. Gait abnormalities occur in HD mouse models (50, 51), and automated gait analysis with the CatWalk (Noldus) might be a more sensitive test than the rotarod (52). Accordingly, we analyzed mean hind-limb gait (distance between hind paws) in our cohort of BACHD, mHTT-S421D, and WT mice at 12 months of age (Figure 3B), and there were considerable differences among genotypes (1-way ANOVA: F(2,45) = 12.27, P < 0.0001). Consistent with postural instability, BACHD mice maintained a wider base of support than WT and mHTT-S421D mice. No differences were detected between WT and mHTT-S421D mice.
Next, we used an automated open-field assay to assess spontaneous locomotor activity, exploratory drive, and anxiety-like behavior. For total movement (Figure 3C), 1-way ANOVA revealed differences among genotypes (F(2,53) = 9.031, P = 0.0004). BACHD mice were much less active than mHTT-S421D or WT animals. In contrast, there were no differences between WT and mHTT-S421D animals. As a measure of exploratory drive and anxiety-like behavior, we also compared the number of rearing events over the same period (Figure 3D). Once again, the genotypes differed (1-way ANOVA: F(2,53) = 4.910, P = 0.0111). BACHD mice reared fewer times than WT or mHTT-S421D mice, but there were no differences between WT and mHTT-S421D mice. Finally, as a test of anxiety-like behavior, we quantified a ratio of the times the mice spent in the center and the periphery of the box. Mice prefer the more protected periphery to the more exposed center, and BACHD mice spend less time in the center (53), consistent with greater anxiety. In our cohort, BACHD mice spent less time in the center than WT and mHTT-S421D mice (Supplemental Figure 5A), but it did not reach statistical significance.
BACHD mice also exhibit a higher level of anxiety-like behavior than WT controls in the light-dark box and elevated plus maze assays (15, 47). We compared the same cohort of mice in the light-dark box to determine whether the mHTT-S421D mutation modifies this phenotype and found differences among genotypes (Figure 3E) (1-way ANOVA: F(2,43) = 7.146, P = 0.0021). Consistent with increased levels of anxiety, BACHD mice spent more time inside the dark box than WT and mHTT-S421D mice. mHTT-S421D mice showed a complete rescue of this phenotype. Surprisingly, we did not detect a rescue of phenotype with the elevated plus maze (Supplemental Figure 5B).
Depression is a particularly debilitating component of HD, and depressive-like phenotypes have been reported in BACHD mice (54, 55). In a second cohort of mice, we tested for anhedonia with the sucrose preference test and for behavioral despair in the Porsolt forced swim test at 3–4 months of age. These assays reveal depressive-like phenotypes in BACHD mice as early as 2 months of age (54). However, no major phenotypic differences were detected in our cohort (Supplemental Figure 5, C–E).
BACHD mice may also show impaired sensorimotor gating at later ages (55, 56). We tested for this phenotype in our second cohort in the prepulse inhibition of startle assay, but we again found no meaningful differences (Supplemental Figure 5F).
Finally, to control for nonspecific effects associated with mutation at S421, we performed behavioral assays with the mHTT-S421A animals.
These mice express mHTT at levels similar to those of the reported BACHD-L line, and since BACHD-L mice show impaired motor coordination on the accelerating rotarod at 6 months of age (41), mHTT-S421A mice would be expected to do so as well. As expected, no deficits were detected in these mice at 3 months of age (data not shown). Effects of genotype and age were found by comparison of data at 6 and 12 months of age (Figure 3F) by 2-way repeated-measures ANOVA (genotype: F(1,30) = 11.50, P = 0.0020; age: F(1,30) = 6.69, P = 0.0148). mHTT-S421A mice fell from the rotarod more quickly than WT control mice at both ages.
These behavioral studies show that phosphomimetic mutation at S421 results in a partial or complete amelioration of motor and psychiatric-like deficits induced by mHTT. This effect cannot be explained as simply resulting from any mutation at this site unrelated to phosphomimicry, since mice with the mHTT-S421A mutation to prevent phosphorylation display motor deficits consistent with their lower transgene expression levels.
Phosphomimetic mutation at S421 prevents striatal neurodegeneration caused by mHTT in vivo.
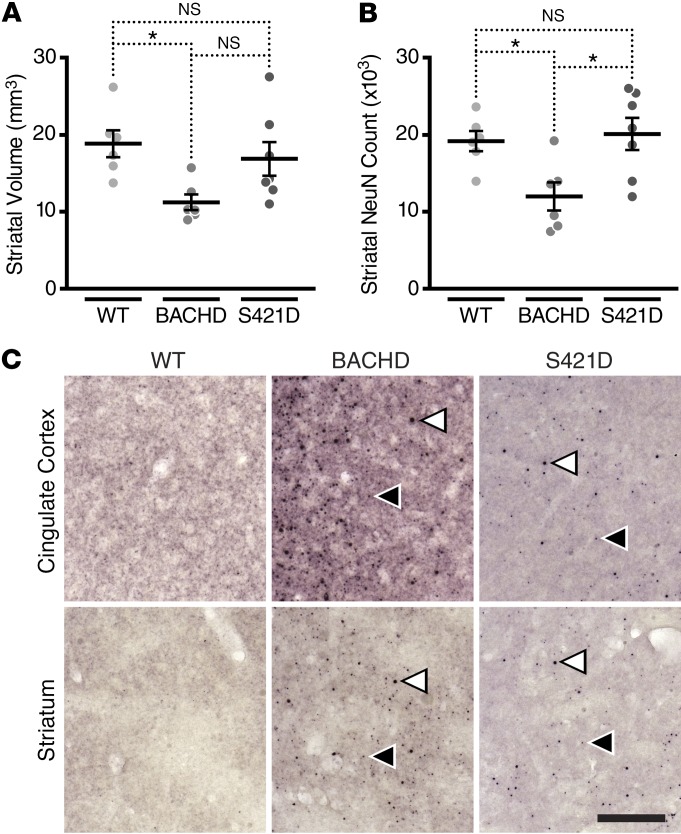
We next sought to determine whether the phosphomimetic mutation influences neuropathology in BACHD brains at 12 months of age. BACHD mice have atrophy of the forebrain without accompanying changes in the cerebellum (41), similar to the pattern of degeneration in human HD. We used unbiased stereology to measure striatal volume in brains from an independent cohort of BACHD, mHTT-S421D, and WT mice (Figure 4A and Supplemental Figure 6A). BACHD mice had smaller striatal volumes than WT mice, but mHTT-S421D mice were similar to the WT controls.
Figure 4. Phosphomimetic mutation at S421 rescues the neurodegeneration caused by expression of mHTT at 12 months of age without accompanying loss of inclusion body formation.
(A) Measurement of striatal volume by unbiased stereology in BACHD (n = 6), mHTT-S421D (n = 7), and WT (n = 6) mice. (B) Measurement of striatal NeuN counts with correction for volume changes in the same cohort. (C) Immunohistochemistry with polyclonal sheep antibody S830 reveals prominent inclusion bodies in BACHD and mHTT-S421D striatum and cingulate cortex but not in WT littermate controls. Example smaller (black) and larger (white) inclusion bodies are indicated by arrowheads. The scale bar represents 220 μm. In A and B, 1-way ANOVA statistical analysis was used with Bonferroni post hoc tests for all pairwise comparisons. *P < 0.05.
We also counted striatal NeuN-positive cells to determine whether expression of mHTT caused any detectable loss, taking care to correct our calculations for changes in volume already measured (Figure 4B). BACHD mice had fewer striatal NeuN-positive cells than WT mice and mHTT-S421D mice, but mHTT-S421D mice were similar to WT controls.
We repeated these studies in the cortex (Supplemental Figure 6, B and C). While the volumes and NeuN-positive-cell counts of mHTT-S421D mice were indistinguishable from those of WT controls, there was a trend toward lower volumes and NeuN counts in BACHD mice. However, the differences did not achieve statistical significance with this sample size, consistent with cortical degeneration being less profound than striatal degeneration in HD.
These data indicate that expression of mHTT causes atrophy and NeuN-positive-cell loss in the striatum, while phosphomimetic mutation at S421 abolishes this neurodegeneration in vivo.
Formation of mHTT-induced inclusion bodies in vivo occurs with phosphomimetic mutation at S421.
The formation of inclusion bodies (IBs) by mHTT is a hallmark of HD pathology, and these findings have been recapitulated in mouse models. Aged BACHD mice develop IBs that are similar to those in the human disease and can be visualized histologically with certain HTT antibodies.
Staining with the polyclonal antibody S830 (55, 57) revealed prominent IBs in the cortex and striatum of BACHD but not WT mice (Figure 4C). mHTT-S421D mice also had a significant burden of IBs, although the levels were less than those observed in the BACHD animals. This finding is consistent with previous results from primary neurons overexpressing HTT-N480-68Q (19). Thus, despite the substantial amelioration of mHTT-induced behavioral dysfunction and neurodegeneration in mHTT-S421D mice, widespread formation of IBs still occurs, further supporting the notion that IBs are not the cause of HD (38, 58, 59).
Phosphomimetic mutation at S421 decreases steady-state levels of soluble mHTT in the striatum.
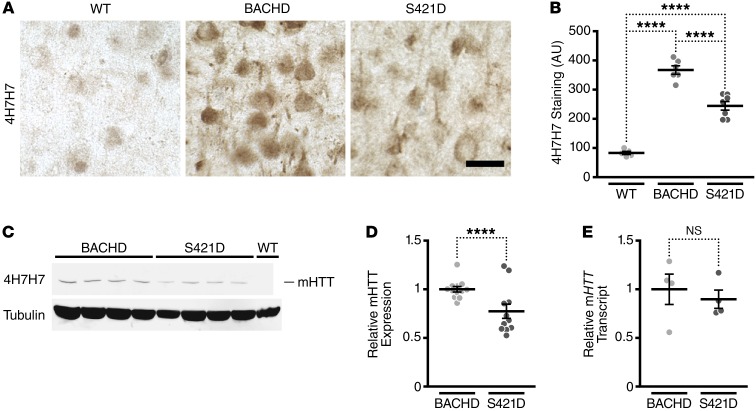
While analyzing striatal histopathology, we observed that slices from mHTT-S421D striata were less immunoreactive with mAb 4H7H7 (Figure 5, A and B). In the native state, mAb 4H7H7 recognizes in situ a compact structure of the polyQ expansion in HTT. Small-angle x-ray scattering data suggest that HTT adopts multiple conformations and that the compact structure recognized by mAb 4H7H7 is preferentially adopted when the length of the polyQ expansion is in the range that causes HD (60). The compact structure recognized by mAb 4H7H7 in situ predicts at a single-cell level whether and when neurons undergo degeneration induced by mHTT, suggesting that malfolded HTT conformers contribute to toxicity (61).
Figure 5. Phosphomimetic mutation at S421 decreases steady-state levels of mHTT in the striatum.
(A and B) Immunohistochemistry of striatal sections of BACHD (n = 6), mHTT-S421D (n = 7), and WT (n = 5) mice with mAb 4H7H7 (A) and subsequent quantification of signal intensity (B). Scale bar: 20 μm. (C) Representative Western blot of BACHD and mHTT-S421D striatal lysates with 4H7H7 and anti–γ-tubulin as a loading control. (D) Quantification of expression levels of mHTT in striatal lysates of BACHD and mHTT-S421D mice. Values are based on the mean of 3 independent 4H7H7 blots of 4 mice per transgenic line, compared across different blots by normalization to BACHD samples. Each value was first normalized for input using the anti–γ-tubulin controls. (E) Quantification of the levels of striatal mHTT transcript in BACHD and mHTT-S421D mice by qRT-PCR. The results are from 4 independent samples per transgenic line, each run in quadruplicate. Values are normalized to BACHD. Means were tested with an unpaired t test in D and E. ****P < 0.0001.
The reduction in mAb 4H7H7 staining was surprising, since we showed earlier that BACHD and S421D express similar levels of mHTT protein and mRNA in the cortex (Figure 1). The amount of epitope recognized by mAb 4H7H7 in situ can be estimated from the level of HTT and the length of the polyQ expansion it contains (61). To determine whether the reduction in mAb 4H7H7 staining could be accounted for by reduced expression of HTT in the striatum, we compared steady-state levels of soluble mHTT in striatal lysates. We found that mHTT-S421D mice have a modest but reproducible and significant reduction in soluble mHTT protein, compared with BACHD mice (Figure 5, B and C). To determine whether these lower protein levels arise from reduced transcription, we examined striatal mHTT mRNA levels. We also verified mHTT mRNA levels in the cerebellum and brainstem to ensure that our findings were not an artifact of the transgene insertion site, which could present as anomalous expression patterns. As in our previous cortical analyses (Figure 1D), we found that levels of mHTT mRNA in the striatum, cerebellum, and brainstem were indistinguishable statistically in BACHD and mHTT-S421D mice (Figure 5E and Supplemental Figure 7), indicating that the observed changes in mHTT protein cannot be explained by changes in the transcription or stability of the transgene mRNA. Thus, the modest reduction in striatal mHTT protein levels and disproportionately greater reduction in 4H7H7 epitope suggests that S421-P independently affects mHTT conformation and mHTT protein metabolism.
Phosphomimetic mutation at S421 increases the turnover of an N-terminal fragment of HTT in striatal cells.
Having found that the S421D mutation decreased steady-state striatal levels of mHTT in vivo, we returned to the validated in vitro system to exploit the rigor and level of control it provides to further investigate the underlying mechanisms by which S421-P affects HTT levels. Ectopic expression also avoids any possible confounds in the murine system from BAC integration into the host chromatin or from developmental expression of mHTT. Further, it would confirm that any changes in mHTT steady-state levels could be attributed specifically to mutation of S421.
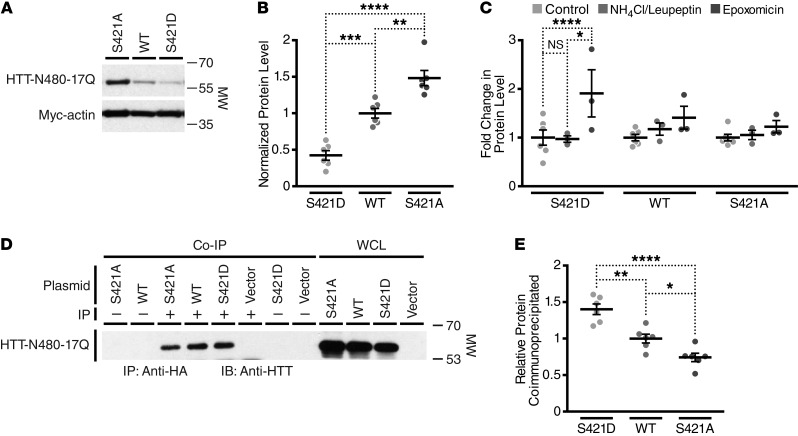
Accordingly, we expressed a myc-actin control and HTT-N480-17Q-S421, -S421D, or -S421A in striatal cell line St14A, as these cells display several properties of striatal MSNs, including expression of the striatal marker DARPP-32 and responsiveness to dopamine receptor agonists (62). We transiently cotransfected 1 of the 3 HTT-S421 variants along with a myc-actin control and lysed the cells 48 hours later. In agreement with our in vivo results, phosphomimetic mutation at S421 decreased steady-state levels of HTT (Figure 6, A and B). Furthermore, cells transfected with the S421A variant that cannot be phosphorylated showed an increase in steady-state HTT levels over nonmutated HTT-S421 (Figure 6, A and B). These results suggest that phosphorylation of HTT at S421 leads to an increase in its turnover.
Figure 6. Phosphomimetic mutation at S421 increases the turnover of an N-terminal fragment of HTT in striatal cells via the proteasome.
(A) Representative Western blot of St14A cell lysates generated 48 hours after transfection with HTT-N480-17Q (probed with mAb 5492) and myc-actin. (B) Quantification of the effect of S421 mutation on HTT-N480 levels in St14A cell lysate. (C) Quantification of the fold change in levels of HTT-N480 with and without S421 mutation after treatment with the specific proteasome inhibitor epoxomicin or lysosome inhibitors ammonium chloride and leupeptin. Values in B and C are based on the mean of 3 independent experiments after normalization for transfection efficiency and loading with the myc-actin control. (D) Representative Western blot after coimmunoprecipitation of HTT-N480-17Q with anti-HA rabbit polyclonal or IgG control from St14A cell lysates after transfection of HTT-N480-17Q, myc-actin, and HA-ubiquitin and treatment with epoxomicin and PR619 (an inhibitor of deubiquitinases) to enhance accumulation of ubiquitin. One-twentieth whole-cell lysates (WCL) were run as controls on the same gel. (E) Quantification of the effect of S421 mutation on HTT-N480 coimmunoprecipitation by HA-ubiquitin in St14A cell lysate. Values are expressed as a normalized ratio of immunoprecipitated to whole-cell control levels per construct. Final data represent 2 independent experiments each run in triplicate. Statistical analysis in each case was by 1-way ANOVA with Bonferroni post hoc tests for all pairwise comparisons. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We next sought to dissect whether the increase in protein turnover associated with phosphomimetic HTT-N480-17Q-S421D might be attributed to enhanced degradation by the proteasome or the lysosome. We treated St14A cells with the specific proteasome inhibitor epoxomicin for 4 hours or with the lysosome inhibitors ammonium chloride and leupeptin for 16 hours and compared HTT-N480-17Q accumulation in terms of fold change over control treatment. Proteasomal inhibition led to trends for a greater buildup of HTT-N480-17Q than inhibition of the lysosome for each of the 3 constructs tested. Levels of the HTT-N480-17Q-S421D construct approximately doubled with proteasome inhibition but were unaffected by lysosome inhibition (Figure 6C). The phosphomimetic polypeptide’s combination of decreased steady-state levels and significant accumulation upon epoxomicin treatment implies that S421D mutation leads to a significant increase in flux of HTT-N480-17Q through the proteasome.
Next, we cotransfected St14A cells with the HTT-N480 constructs, myc-actin, and HA-ubiquitin and proceeded to coimmunoprecipitate HTT-N480 with ubiquitin. To increase our sensitivity for detecting ubiquitinated HTT, we treated the cells with epoxomicin to inhibit the proteasome and with PR619, an inhibitor of deubiquitinases (63). Phosphomimetic mutation at S421 increased the relative amount of HTT-N480 that coimmunoprecipitated with ubiquitin, whereas less nonmutated HTT-N480 coimmunoprecipitated and even less of the phosphoresistant HTT-N480 coimmunoprecipitated (Figure 6, D and E). Importantly, no HTT was detected in the vector control and IgG control conditions, indicating that no nonspecific binding of HTT to the beads occurred. This result is consistent with our model in which HTT-S421-P leads to an increase in HTT ubiquitination with a subsequent increase in its turnover via the proteasome, resulting in the observed lower steady-state levels of mHTT-S421D in mouse striatum and HTT-N480-S421D in St14A cells.
Discussion
In this study, we show that phosphomimetic mutation at S421 strongly ameliorates the toxicity of mHTT in vivo. A single–amino acid change made S421 behave as if it were tonically phosphorylated and was sufficient to prevent onset of a variety of behavioral deficits and the selective striatal neurodegeneration induced by mHTT in the BACHD mice. It did not interfere with the ability of the mHTT transgene to rescue the embryonic lethality of HTT knockout, indicating that it permits HTT to subserve its essential functions during development. mHTT-S421D mice and BACHD mice had similar levels of HTT mRNA and soluble protein in the cortex, but mHTT-S421D mice had decreased steady-state levels of protein and 4H7H7 epitope in the striatum, offering a clue about a mechanism by which S421D mitigates HD-related phenotypes in vivo. Biochemical analyses in vitro confirmed that the S421D mutation decreased the steady-state levels of soluble HTT in a striatal cell line via increased HTT flux through the proteasome. These results validate S421-P as a therapeutic target in HD and support strategies to increase mHTT turnover in the striatum as a promising treatment.
S421 is phosphorylated by Akt, a serine/threonine-specific protein kinase that is centrally involved in a variety of cellular signaling cascades, including those that regulate protein turnover (64). Intriguingly, phosphorylation at Akt consensus sites might have pathophysiological relevance not only to mHTT, but to other polyQ disease-associated proteins as well. For example, abnormal polyQ expansions in ataxin-1 and the androgen receptor cause spinocerebellar ataxia type 1 and spinal and bulbar muscular atrophy, respectively, and both proteins contain Akt consensus sites. In each case, the presence or absence of phosphorylation influences markedly the toxicity of the host protein (65–68). The effect seems to be related to alterations in protein turnover (65, 69). Every known polyQ disease-associated protein has at least 1 predicted Akt consensus phosphorylation site (Supplemental Table 2 and ref. 70), raising the possibility of an evolutionarily conserved posttranslational mechanism to regulate disease-associated polyQ proteins.
Previous studies shed light on HTT’s regulation by protein-turnover pathways (71–73), and we showed that the S421D mutation decreases striatal steady-state mHTT levels. How might S421 mediate this effect? Reports of phosphorylation-dependent ubiquitination in other proteins (74) offer 1 plausible direct mechanism whereby S421-P could increase HTT flux through the proteasome. Alternatively or in parallel, phosphorylation might increase the ability of HTT to bind ubiquitinated proteins via indirect means, as with p62 (75). S421 is also situated in a region of HTT rich in PTMs (12), so 1 or more of these PTMs might contribute to regulation of HTT catabolism. HTT acetylation at K444 may target HTT to autophagosomes for degradation (26), whereas our work suggests that S421-P increases HTT flux through the proteasome.
One enticing hypothesis is that regulating these 2 relatively nearby PTMs determines whether HTT is degraded by the proteasome or autophagy (76). Since mHTT may cause impaired loading of autophagosomes (77), its targeting to the proteasome might be imperative for preventing neuronal dysfunction and death. Serines 431 and 432 of HTT may also be phosphorylation sites whose mutation influences mHTT accumulation, and K444 may be ubiquitinated as well as acetylated (29). These findings support the hypothesis that PTMs in this region of HTT are crucial regulators of HTT clearance. Finally, the general age-dependent decline in proteasome function (78) and inability to process the same levels of flux could explain why the mHTT-S421D mice finally developed rotarod deficits at 12 months of age.
Our study also determined that preventing phosphorylation at S421 did not increase mHTT toxicity. The behavioral deficits and pathology induced by comparable levels of mHTT persisted in transgenic mice in which phosphorylation at this site was blocked with the corresponding (S421A) mutation. Disease-associated expansion of the polyQ repeat reduces HTT phosphorylation at S421 (36, 37). If baseline levels of S421-P of mHTT are sufficiently low, blocking it altogether with the S421A mutation may have little additional effect. By the same logic, permanent addition of a negative charge to mimic tonic phosphorylation would be a substantial perturbation. That is, the decrease of S421-P caused by polyQ expansion could be considered a toxic loss of function that is reversed by phosphomimetic S421D mutation. In agreement, the mHTT-S421D mice demonstrated remarkable amelioration of the HD phenotype, and the striatum showed a complete rescue of neurodegeneration. Previous in vitro work studying N-terminal fragments of mHTT reported either similar or increased toxicity with introduction of the S421A mutation, depending on the model system used (19, 34, 37). This partial discrepancy with our in vivo findings underscores the importance of complementing in vitro discoveries with in vivo validation.
Why were the ameliorating effects of mHTT-S421D most apparent in the striatum? Circuit-specific factors, such as the predominance of inhibitory GABAergic MSNs (79), likely play a role. Additionally, MSNs depend on other brain regions for the delivery of an important extracellular growth factor, brain-derived neurotrophic factor (BDNF) (80), whose downstream effects include activation of Akt. These and other factors likely explain why, under normal conditions, the striatum shows less phosphorylation at S421 than other brain regions less affected in HD (36). Yet mHTT-S421D might decrease steady-state levels of mHTT in brain regions beyond the striatum. At baseline, the cortex clears soluble mHTT–exon 1 more effectively than the striatum (81), so any added effect of the phosphomimetic mutation would be more difficult to detect. Nevertheless, comparison of the mHTT-S421D line with BACHD mice revealed a distinct trend toward increased cortical levels of mHTT mRNA with no such trend noted at the protein level, consistent with the possibility that S421D increases mHTT turnover in the cortex as well.
In this context, it is intriguing to consider the effect of the S421D mutation on the amount of mHTT in a conformation recognized by 4H7H7. We found that striatal immunohistochemistry staining with mAb 4H7H7 of mHTT-S421D proteins fixed in their native conformations was down by 30% compared with the BACHD controls, but total levels of HTT-S421D detected by denaturing Western blots were down by only 15%. While there are considerable limitations to the quantifications, this discrepancy could be revealing. Recently, disease-associated polyQ stretches were shown to adopt a compact hairpin structure in situ (60), whose presence quantitatively and significantly predicts neurodegeneration; this structure is recognized by mAb 4H7H7 (61). Hence, the aforementioned discrepancy could mean that S421 regulates HTT conformation directly or that the fraction of HTT that is lowered in the striatum has particularly adopted the compact hairpin structure recognized by mAb 4H7H7. Since this conformation predicts toxicity, neurons might be accelerating the clearance of this subset of HTT molecules to enhance their survival.
Our work does not exclude the possibility that other mechanisms are important in orchestrating the phosphomimetic mutation’s ability to diminish mHTT toxicity. For example, S421-P may restore an mHTT-induced vesicular trafficking defect (32). However, such deficits have not been reported in BACHD mice and thus cannot be appropriately assessed in our model system.
In conclusion, this study demonstrates the significance of S421 to the toxicity of mHTT in vivo. Our findings indicate that increasing levels of S421-P in HD patients could be a viable therapeutic target for this devastating disease. Akt’s oncogenic potential (64) would need to be considered carefully in any approach that involved direct activation of Akt. A related kinase, SGK, also is capable of phosphorylating HTT at S421 (30), perhaps representing another pharmacological target. Alternatively, future work focused on the effects of S421D mutation on mHTT’s local structure and binding partners might lead to the identification of additional targets with more attractive profiles for a small-molecule drug.
Methods
Supplemental Methods are available online with this article; doi:10.1172/JCI80339DS1.
BAC recombineering
S421A/D point mutations were inserted into the original HTT BAC (41) by 1 Red/ET triple recombination step (Gene Bridges GmbH). Correct removal of the FRT selection marker via FLP recombination with preservation of the point mutation was verified by PCR and sequencing of regions around the mutation, exon 1, and native stop codon.
Generation and breeding of transgenic mice
Final BACs were prepared, confirmed to be free of degraded DNA by pulsed-field gel electrophoresis, and microinjected as described previously (15, 82) into FvB/N pronuclei by the Gladstone Transgenic Core. All mice were maintained on the FvB/NJ background. Hdh+/– mice were a gift from Scott Zeitlin (University of Virginia School of Medicine, Charlottesville, Virgina, USA; ref. 44).
Genotyping and sequencing
Routine genotyping of HTT mice and sequencing of the 5′-UTR, exon 1, and CAG-CAA repeat region were as described previously (15). For the region around S421, PCR used the following primers with an annealing temperature of 57°C: 5′-CGAGCTTCTGCAAACCCTGAC-3′ and 5′-TTGGCAAGGAAGATGGAATGCAG-3′. The PCR product was purified and then sequenced by Elim Biopharmaceuticals.
Preparation of brain lysate and Western blotting
Mouse brains were dissected on ice, snap-frozen on dry ice, and stored at –80°C until use. Brains were homogenized with a pellet pestle (Kontes) as described previously (41). Protein concentrations of the soluble supernatants were determined using the BCA Protein Assay (Thermo Fisher Scientific).
For Western blots, samples (40 μg) were prepared and run on NuPAGE (Invitrogen) 3%–8% Tris-acetate or 4%–12% Bis-Tris precast gels according to the manufacturer’s instructions. Immunoblots were probed with 4H7H7 (1:5,000; ref. 15), 1C2 (1:3,000; MAB1574; Millipore), anti-HTT 2166 (1:3,000; mAb2166; Millipore), or anti–γ-tubulin (1:20,000; T6557; Sigma-Aldrich). Images were captured with the Odyssey CLx (LI-COR Biosci).
Preparation of RNA extracts and qRT-PCR
Previously undisturbed mice in their home cage were anesthetized with isoflurane and decapitated. Brains were dissected on ice, snap-frozen with liquid nitrogen, and stored at –80°C until further processing. RNA was isolated and purified with the miRNeasy Mini kit (Qiagen) according to the manufacturer’s instructions, with frozen tissue being disrupted and homogenized with the TissueLyser II (Qiagen).
RNA (300 ng) was converted to cDNA using the TaqMan Reverse Transcription kit (Invitrogen) and a 1:1 mix of random hexamers and oligo-dT (16). qRT-PCR with SYBR Green (Applied Biosystems) was performed on 1:40 dilutions of the samples using a 7900HT Fast Real-Time PCR system (Applied Biosystems). We generated primers that anneal specifically to human HTT mRNA using QuantPrime (83) (forward, 5′-ATCCCGGTCATCAGCGACTATC-3′; reverse, 5′-GCTTGTAATGTTCACGCAGTGG-3′). Samples were normalized using primers for mouse β-actin (QuantiTect primers; Qiagen). The standard curve method was used to analyze the data.
Behavioral testing
All testing was performed during the light phase of the light cycle, and animals were given at least 45 minutes to acclimate to the testing room. Approximately equal numbers of males and females were used per each genotype. Data were collected and scored blindly to genotype. All apparatuses were cleaned with 70% ethanol between each run.
Accelerating rotarod.
Mice were trained on a rotarod (Med Associates) at 16 rpm for 3 trials. The subsequent 2 days, mice were tested twice a day, 3 times each session, as the rotarod accelerated from 4 to 40 rpm over 5 minutes. Only mice with data collected at each time point were analyzed.
Open field.
Total and rearing activities were assessed in an automated clear plastic chamber (41 × 41 × 30 cm) Flex-Field/Open-Field Photobeam Activity System (San Diego Instruments). Total beam breaks and rears (the number of times the mouse stood erect on its hind legs) were measured automatically by the instrument over 10 minutes.
Gait assessment.
Mice were placed on the CatWalk XT (Noldus) and removed after 3 runs that were judged compliant by the software (version 9.1) with its default settings. Paw identification was performed automatically by the software with human review of each call to confirm accuracy.
Light-dark box.
Anxiety-like behavior was assessed in the open-field chamber with an added dark box insert (catalog 7001-0364; San Diego Instruments) over 10 minutes under standard lighting conditions.
Elevated plus maze.
Mice were given 10 minutes to explore the maze (Hamilton-Kinder) with arms elevated 63 cm above the ground under dim lighting conditions. Total distance traveled and time spent in the open and closed arms were calculated based on infrared photobeam breaks.
Sucrose preference test.
Mice were single housed at least 24 hours before testing and habituated to drinking water from 2 bottles placed in the top of their cage. Preweighed bottles, 1 containing normal drinking water and 1 containing 2% sucrose solution, were placed into the cage the morning of the first day of testing. Bottles were weighed twice daily as close to the beginning and end of the 12-hour light cycle as possible for 3 days.
Porsolt forced swim test.
A clear plastic cylinder 45 cm high and 20 cm in diameter was filled two-thirds full with water at 20°C to 21°C. Mice were individually placed in the cylinder and allowed to swim for 6 minutes while being video-recorded. Mice were scored for immobility during the last 4 minutes of the test.
Prepulse inhibition of startle test.
Testing was performed in a small isolated chamber (9.5 × 11 × 6 in.), free from external movement and noise (Kinder Scientific). Mice were given 5 minutes to acclimate to the restraining chamber and 70 dB background noise before stimulus testing began. Mice were exposed to a series of acoustic pulses for 20 minutes in which some pulses are preceded by a weaker acoustic signal (prepulse [pp]) or no stimulus at all at random intervals for 80 trials (24 trials at 40 ms, 120 dB; 14 trials at 4 dBpp [dB intensity of the prepulse above the 68dB white noise background], 40 ms, 120 dB; 14 trials at 12 dBpp, 40 ms, 120dB; 14 trials at 20dBpp, 40 ms, 120 dB; 14 trials with no stimulation). Average and maximum amplitudes of pulses (and those with preceding prepulses) were measured automatically by the device.
Immunohistochemistry and stereology
Mice anesthetized with avertin were perfused with saline using an automated pump. The right hemibrain was immersion-fixed in 4% paraformaldehyde in PBS at 4°C for 48 hours. The volume of the neocortex was estimated in sagittal brain sections sectioned with the vibratome at 50 μm, as described previously (84, 85). For neuron counts, the total number of NeuN-positive cells was averaged and expressed as relative numbers per area after accounting for any changes in volume.
Immunohistochemical staining of mHTT IBs
Multiple perfusion-fixed brains were embedded in gelatin blocks, postfixed, and freeze-cut into coronal 35-μm sections in 24 sequential series (NeuroScience Associates). These were stored at –20°C in cryoprotectant as individual series of sections and stained for mHTT-derived aggregates with an affinity-purified preparation of the sheep anti-HTT antibody S830 (30 ng/ml; refs. 55, 57; gift from G. Bates, King’s College London, London, United Kingdom) under uniform modified free-floating conditions (86). Binding was visualized using ultrasensitive HRP-based Ni2+/DAB immunohistochemistry. Images were collected on a Spot Slider camera on a Leica DMR microscope. Each field is 600 × 600 μm.
Analysis of HTT steady-state levels and clearance
HTT-N480-17Q constructs (19) were cotransfected with Lipofectamine 2000 (Invitrogen) into St14A cells (ref. 62; gift from E. Cattaneo, University of Milan, Milan, Italy) along with myc-actin (ref. 87; gift from H. Rommelaere, Ghent University, Ghent, Belgium) to control for transfection efficiency. When indicated, cells were treated for 4 hours with DMSO or 100 nM epoxomicin (Sigma-Aldrich) in DMSO or for 16 hours with water or 20 nM ammonium chloride/100 μM leupeptin (Sigma-Aldrich) in water. The short incubation time for epoxomicin treatment was used to eliminate the possibility of nonspecific effects of epoxomicin on the lysosome. Cell culture, lysis, and Western analysis were as described previously (16) with slight modifications. NuPAGE Novex 4%–12% Bis-Tris precast gels with MOPS running buffer (Invitrogen) were used with 20 μg of sonicated whole-cell lysate. Quantitative densitometric analysis was performed on digitalized images of immunoblots using ImageJ (NIH). Water and DMSO control condition measurements were found to have no significant differences and were combined.
Huntingtin coimmunoprecipitation
St14A cells were transiently transfected as described above with addition of HA-ubiquitin (ref. 88; gift from B. Carter, Vanderbilt University, Nashville, Tennessee, USA). Immunoprecipitation was performed as described previously (73) with the following modifications and brief summary: Cells were treated for 4 hours with epoxomicin (100 nM) and 1 hour with PR619 (100 μM; Sigma-Aldrich) before lysis. Twenty micrograms of the lysate was set aside for whole-cell controls. Four hundred micrograms of the lysate was diluted into 500 μl of IP buffer, combined with 1 μl of rabbit polyclonal anti-HA H6908 (Sigma-Aldrich) and 30 μl of Dynabeads (Thermo Fisher Scientific), and rotated at 4°C overnight. After washing and elution, the eluted lysate was run on gels as described above and immunoblotted with rabbit polyclonal anti-HTT VB3130 (Viva Bioscience). Quantification was performed as above and expressed as a ratio of immunoprecipitated to whole-cell levels per construct.
Statistics
All quantified data are presented as mean ± SEM. For all statistical analysis, Prism 5 software (GraphPad) was used with a significance level set at 0.05. For group comparisons, 1- or 2-way ANOVAs were performed as indicated with Bonferroni post hoc analysis for pairwise comparisons unless otherwise stated. When appropriate, means of 2 groups were compared with an unpaired 2-way t test.
Study approval
All mice were bred and maintained under standard conditions consistent with NIH guidelines and approved by the UCSF Institutional Animal Care and Use Committee.
Author contributions
IHK and SF wrote the manuscript. IHK and AST were responsible for BAC mutation and purification, with advice and supervision from XG and XWY. IHK performed most of the behavioral experiments, with some performed by the Gladstone core facility. IHK analyzed all behavioral experiments with advice from the Gladstone core facility. IHK and HZ performed and analyzed brain dissections, qRT-PCR, and Western blots. EM performed stereology and immunohistochemistry with 4H7H7. AO performed and interpreted inclusion body staining with S830. JSS and AL conceptualized and performed all experiments in St14A cells and conceived of the hypothesis of an indirect role of HTT-S421-P in HTT proteasomal turnover. SH and FS provided the HTT-N480 constructs. ACD, HZ, and IHK performed plasmid mutation and cloning. KFW contributed to the development of mechanistic hypotheses. SF supervised the entire project.
Supplementary Material
Acknowledgments
We thank members of the Finkbeiner laboratory for helpful discussions. We thank Hui (Annie) Wang, André Zandona, Matthew Campioni, and Ana Osório Oliveira for assistance with mouse husbandry and logistics. We thank Michael Gill, Ravi Ponnusamy, Andrew Zwilling, and Nino Devidze as well as Iris Lo, Jeffrey Simms, Carlee Toddes, Allyson Davis, and Ryan Craft of the Gladstone Institutes Behavioral Core for training and assistance with the implementation and analysis of behavioral assays. We thank Gary Howard for editorial assistance and Kelley Nelson for administrative support. We thank Gregory Cox for helpful advice. We thank Gillian Bates for the S830 antisera, Elena Cattaneo for the St14A cells, and Scott Zeitlin for the Hdh knockout mice. Work in the laboratory of S. Finkbeiner was supported by National Institute of Neurological Disorders and Stroke (NINDS) grants 3R01 NS039074 and R01 NS083390, the Taube/Koret Center for Neurodegenerative Disease, and the Hellman Family Foundation Alzheimer’s Disease Research Program. I.H. Kratter was also supported by NINDS award 5F31 NS077543. I.H. Kratter, H. Zahed, and A.C. Daub were supported by the UCSF Medical Scientist Training Program. The animal care facility was partly supported by an NIH Extramural Research Facilities Improvement Program Project (C06 RR018928). X.W. Yang and X. Gu are supported by the NIH/NINDS grants R01 NS049501 and R01 NS074312, the Hereditary Disease Foundation, and CHDI Foundation Inc. J.S. Steffan and A. Lau are supported by NINDS grant NS072453 and CHDI Foundation Inc. E. Masliah is supported by NIH grants AG18440, AG010435, and NS057096.
Footnotes
IHK’s present address is: Western Psychiatric Institute and Clinic, University of Pittsburgh Medical Center, Pittsburgh, Pennsylvania, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(9):3585–3597. doi:10.1172/JCI80339.
Contributor Information
Ian H. Kratter, Email: Ian.Kratter@ucsf.edu.
Hengameh Zahed, Email: hzahed@gmail.com.
Alice Lau, Email: allau@uci.edu.
Andrey S. Tsvetkov, Email: Andrey.S.Tsvetkov@uth.tmc.edu.
Aaron C. Daub, Email: acdaub@gmail.com.
Xiaofeng Gu, Email: xgu@mednet.ucla.edu.
Frédéric Saudou, Email: frederic.saudou@inserm.fr.
Sandrine Humbert, Email: sandrine.humbert@inserm.fr.
X. William Yang, Email: xwyang@mednet.ucla.edu.
Eliezer Masliah, Email: emasliah@ucsd.edu.
Steven Finkbeiner, Email: sfinkbeiner@gladstone.ucsf.edu.
References
- 1.Walker FO. Huntington’s disease. Lancet. 2007;369(9557):218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 2.Finkbeiner S. Huntington’s disease. Cold Spring Harb Perspect Biol. 2011;3(6): doi: 10.1101/cshperspect.a007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57(5):369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 4.A novel gene containing a trinucleotide repeat that is expanded unstable on Huntington’s disease chromosomes The Huntington’s Disease Collaborative Research Group. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 5.Borrell-Pagès M, Zala D, Humbert S, Saudou F. Huntington’s disease: from huntingtin function and dysfunction to therapeutic strategies. Cell Mol Life Sci. 2006;63(22):2642–2660. doi: 10.1007/s00018-006-6242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imarisio S, et al. Huntington’s disease: from pathology and genetics to potential therapies. Biochem J. 2008;412(2):191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 7.Keryer G, et al. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J Clin Invest. 2011;121(11):4372–4382. doi: 10.1172/JCI57552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gil JM, Rego AC. Mechanisms of neurodegeneration in Huntington’s disease. Eur J Neurosci. 2008;27(11):2803–2820. doi: 10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- 9.Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington’s disease. Nat Rev Neurosci. 2005;6(12):919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 10.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 11.Pennuto M, Palazzolo I, Poletti A. Post-translational modifications of expanded polyglutamine proteins: impact on neurotoxicity. Hum Mol Genet. 2009;18(R1):R40–R47. doi: 10.1093/hmg/ddn412. [DOI] [PubMed] [Google Scholar]
- 12.Ehrnhoefer DE, Sutton L, Hayden MR. Small changes, big impact: posttranslational modifications and function of huntingtin in Huntington disease. Neuroscientist. 2011;17(5):475–492. doi: 10.1177/1073858410390378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steffan JS, et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304(5667):100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 14.Aiken CT, et al. Phosphorylation of threonine 3: implications for Huntingtin aggregation and neurotoxicity. J Biol Chem. 2009;284(43):29427–29436. doi: 10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu X, et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64(6):828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson LM, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187(7):1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atwal RS, et al. Kinase inhibitors modulate huntingtin cell localization and toxicity. Nat Chem Biol. 2011;7(7):453–460. doi: 10.1038/nchembio.582. [DOI] [PubMed] [Google Scholar]
- 18.Di Pardo A, et al. Ganglioside GM1 induces phosphorylation of mutant huntingtin and restores normal motor behavior in Huntington disease mice. Proc Natl Acad Sci U S A. 2012;109(9):3528–3533. doi: 10.1073/pnas.1114502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humbert S, et al. The IGF-1/Akt pathway is neuroprotective in Huntington’s disease and involves Huntingtin phosphorylation by Akt. Dev Cell. 2002;2(6):831–837. doi: 10.1016/S1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- 20.Wellington CL, et al. Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington’s disease. J Neurosci. 2002;22(18):7862–7872. doi: 10.1523/JNEUROSCI.22-18-07862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo S, Vacher C, Davies JE, Rubinsztein DC. Cdk5 phosphorylation of huntingtin reduces its cleavage by caspases: implications for mutant huntingtin toxicity. J Cell Biol. 2005;169(4):647–656. doi: 10.1083/jcb.200412071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham RK, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125(6):1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 23.Schilling B, et al. Huntingtin phosphorylation sites mapped by mass spectrometry. Modulation of cleavage and toxicity. J Biol Chem. 2006;281(33):23686–23697. doi: 10.1074/jbc.M513507200. [DOI] [PubMed] [Google Scholar]
- 24.Yanai A, et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat Neurosci. 2006;9(6):824–831. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anne SL, Saudou F, Humbert S. Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. J Neurosci. 2007;27(27):7318–7328. doi: 10.1523/JNEUROSCI.1831-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong H, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137(1):60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324(5932):1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller JP, et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington’s disease. Neuron. 2010;67(2):199–212. doi: 10.1016/j.neuron.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong G, Callegari E, Gloeckner CJ, Ueffing M, Wang H. Mass spectrometric identification of novel posttranslational modification sites in Huntingtin. Proteomics. 2012;12(12):2060–2064. doi: 10.1002/pmic.201100380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rangone H, et al. The serum- and glucocorticoid-induced kinase SGK inhibits mutant huntingtin-induced toxicity by phosphorylating serine 421 of huntingtin. Eur J Neurosci. 2004;19(2):273–279. doi: 10.1111/j.0953-816X.2003.03131.x. [DOI] [PubMed] [Google Scholar]
- 31.Zala D, Colin E, Rangone H, Liot G, Humbert S, Saudou F. Phosphorylation of mutant huntingtin at S421 restores anterograde and retrograde transport in neurons. Hum Mol Genet. 2008;17(24):3837–3846. doi: 10.1093/hmg/ddn281. [DOI] [PubMed] [Google Scholar]
- 32.Pineda JR, Pardo R, Zala D, Yu H, Humbert S, Saudou F. Genetic and pharmacological inhibition of calcineurin corrects the BDNF transport defect in Huntington’s disease. Mol Brain. 2009;2: doi: 10.1186/1756-6606-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colin E, et al. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27(15):2124–2134. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warby SC, Doty CN, Graham RK, Shively J, Singaraja RR, Hayden MR. Phosphorylation of huntingtin reduces the accumulation of its nuclear fragments. Mol Cell Neurosci. 2009;40(2):121–127. doi: 10.1016/j.mcn.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Metzler M, et al. Phosphorylation of huntingtin at Ser421 in YAC128 neurons is associated with protection of YAC128 neurons from NMDA-mediated excitotoxicity and is modulated by PP1 and PP2A. J Neurosci. 2010;30(43):14318–14329. doi: 10.1523/JNEUROSCI.1589-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warby SC, et al. Huntingtin phosphorylation on serine 421 is significantly reduced in the striatum and by polyglutamine expansion in vivo. Hum Mol Genet. 2005;14(11):1569–1577. doi: 10.1093/hmg/ddi165. [DOI] [PubMed] [Google Scholar]
- 37.Pardo R, et al. Inhibition of calcineurin by FK506 protects against polyglutamine-huntingtin toxicity through an increase of huntingtin phosphorylation at S421. J Neurosci. 2006;26(5):1635–1645. doi: 10.1523/JNEUROSCI.3706-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95(1):55–66. doi: 10.1016/S0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 39.Xia J, Lee DH, Taylor J, Vandelft M, Truant R. Huntingtin contains a highly conserved nuclear export signal. Hum Mol Genet. 2003;12(12):1393–1403. doi: 10.1093/hmg/ddg156. [DOI] [PubMed] [Google Scholar]
- 40.Reiner A, Dragatsis I, Zeitlin S, Goldowitz D. Wild-type huntingtin plays a role in brain development and neuronal survival. Mol Neurobiol. 2003;28(3):259–276. doi: 10.1385/MN:28:3:259. [DOI] [PubMed] [Google Scholar]
- 41.Gray M, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28(24):6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham RK, et al. Levels of mutant huntingtin influence the phenotypic severity of Huntington disease in YAC128 mouse models. Neurobiol Dis. 2006;21(2):444–455. doi: 10.1016/j.nbd.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Miller J, Arrasate M, Shaby BA, Mitra S, Masliah E, Finkbeiner S. Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into huntington’s disease molecular pathogenesis. J Neurosci. 2010;30(31):10541–10550. doi: 10.1523/JNEUROSCI.0146-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat Genet. 1995;11(2):155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 45.White JK, et al. Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nat Genet. 1997;17(4):404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- 46.Hodgson JG, et al. A YAC mouse model for Huntington’s disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23(1):181–192. doi: 10.1016/S0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- 47.Menalled L, et al. Systematic behavioral evaluation of Huntington’s disease transgenic and knock-in mouse models. Neurobiol Dis. 2009;35(3):319–336. doi: 10.1016/j.nbd.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McFadyen MP, Kusek G, Bolivar VJ, Flaherty L. Differences among eight inbred strains of mice in motor ability and motor learning on a rotorod. Genes Brain Behav. 2003;2(4):214–219. doi: 10.1034/j.1601-183X.2003.00028.x. [DOI] [PubMed] [Google Scholar]
- 49.Van Raamsdonk JM, et al. Body weight is modulated by levels of full-length huntingtin. Hum Mol Genet. 2006;15(9):1513–1523. doi: 10.1093/hmg/ddl072. [DOI] [PubMed] [Google Scholar]
- 50.Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J Comp Neurol. 2003;465(1):11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 51.Heng MY, et al. Early autophagic response in a novel knock-in model of Huntington disease. Hum Mol Genet. 2010;19(19):3702–3720. doi: 10.1093/hmg/ddq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vandeputte C, et al. Automated quantitative gait analysis in animal models of movement disorders. BMC Neurosci. 2010;11: doi: 10.1186/1471-2202-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aharony I, et al. A Huntingtin-based peptide inhibitor of caspase-6 provides protection from mutant Huntingtin-induced motor and behavioral deficits. Hum Mol Genet. 2015;24(9):2604–2614. doi: 10.1093/hmg/ddv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hult Lundh S, Nilsson N, Soylu R, Kirik D, Petersén Å. Hypothalamic expression of mutant huntingtin contributes to the development of depressive-like behavior in the BAC transgenic mouse model of Huntington’s disease. Hum Mol Genet. 2013;22(17):3485–3497. doi: 10.1093/hmg/ddt203. [DOI] [PubMed] [Google Scholar]
- 55.Pouladi MA, et al. Marked differences in neurochemistry and aggregates despite similar behavioural and neuropathological features of Huntington disease in the full-length BACHD and YAC128 mice. Hum Mol Genet. 2012;21(10):2219–2232. doi: 10.1093/hmg/dds037. [DOI] [PubMed] [Google Scholar]
- 56.Menalled L, et al. Systematic behavioral evaluation of Huntington’s disease transgenic and knock-in mouse models. Neurobiol Dis. 2009;35(3):319–336. doi: 10.1016/j.nbd.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sathasivam K, et al. Centrosome disorganization in fibroblast cultures derived from R6/2 Huntington’s disease (HD) transgenic mice and HD patients. Hum Mol Genet. 2001;10(21):2425–2435. doi: 10.1093/hmg/10.21.2425. [DOI] [PubMed] [Google Scholar]
- 58.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 59.Slow EJ, et al. Absence of behavioral abnormalities and neurodegeneration in vivo despite widespread neuronal huntingtin inclusions. Proc Natl Acad Sci U S A. 2005;102(32):11402–11407. doi: 10.1073/pnas.0503634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peters-Libeu C, et al. Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure. J Mol Biol. 2012;421(4–5):587–600. doi: 10.1016/j.jmb.2012.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller J, et al. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat Chem Biol. 2011;7(12):925–934. doi: 10.1038/nchembio.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ehrlich ME, et al. ST14A cells have properties of a medium-size spiny neuron. Exp Neurol. 2001;167(2):215–226. doi: 10.1006/exnr.2000.7551. [DOI] [PubMed] [Google Scholar]
- 63.Udeshi ND, et al. Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol Cell Proteomics. 2012;11(5):148–159. doi: 10.1074/mcp.M111.016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol. 2012;4(9): doi: 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen HK, et al. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113(4):457–468. doi: 10.1016/S0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- 66.Emamian ES, et al. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38(3):375–387. doi: 10.1016/S0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- 67.Lin HK, Yeh S, Kang HY, Chang C. Akt suppresses androgen-induced apoptosis by phosphorylating and inhibiting androgen receptor. Proc Natl Acad Sci U S A. 2001;98(13):7200–7205. doi: 10.1073/pnas.121173298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palazzolo I, et al. Akt blocks ligand binding and protects against expanded polyglutamine androgen receptor toxicity. Hum Mol Genet. 2007;16(13):1593–1603. doi: 10.1093/hmg/ddm109. [DOI] [PubMed] [Google Scholar]
- 69.Palazzolo I, et al. Overexpression of IGF-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron. 2009;63(3):316–328. doi: 10.1016/j.neuron.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31(13):3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steffan JS. Does Huntingtin play a role in selective macroautophagy? Cell Cycle. 2010;9(17):3401–3413. doi: 10.4161/cc.9.17.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng S, Clabough EB, Sarkar S, Futter M, Rubinsztein DC, Zeitlin SO. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 2010;6(2): doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ochaba J, et al. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc Natl Acad Sci U S A. 2014;111(47):16889–16894. doi: 10.1073/pnas.1420103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28(5):730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 75.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44(2):279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 76.Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol. 2010;2(12): doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez-Vicente M, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13(5):567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keller JN, Gee J, Ding Q. The proteasome in brain aging. Ageing Res Rev. 2002;1(2):279–293. doi: 10.1016/S1568-1637(01)00006-X. [DOI] [PubMed] [Google Scholar]
- 79.Lobo MK. Molecular profiling of striatonigral and striatopallidal medium spiny neurons past, present, and future. Int Rev Neurobiol. 2009;89:1–35. doi: 10.1016/S0074-7742(09)89001-6. [DOI] [PubMed] [Google Scholar]
- 80.Altar CA, et al. Anterograde transport of brain-derived neurotrophic factor and its role in the brain. Nature. 1997;389(6653):856–860. doi: 10.1038/39885. [DOI] [PubMed] [Google Scholar]
- 81.Tsvetkov AS, et al. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol. 2013;9(9):586–592. doi: 10.1038/nchembio.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gong S, Yang XW. Modification of bacterial artificial chromosomes (BACs) and preparation of intact BAC DNA for generation of transgenic mice. Curr Protoc Neurosci. 2005;Chapter 5: doi: 10.1002/0471142301.ns0521s31. [DOI] [PubMed] [Google Scholar]
- 83.Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B. QuantPrime--a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics. 2008;9: doi: 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zwilling D, et al. Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell. 2011;145(6):863–874. doi: 10.1016/j.cell.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ubhi K, et al. Neurodegeneration in a transgenic mouse model of multiple system atrophy is associated with altered expression of oligodendroglial-derived neurotrophic factors. J Neurosci. 2010;30(18):6236–6246. doi: 10.1523/JNEUROSCI.0567-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Osmand AP, Berthelier V, Wetzel R. Imaging polyglutamine deposits in brain tissue. Meth Enzymol. 2006;412:106–122. doi: 10.1016/S0076-6879(06)12008-X. [DOI] [PubMed] [Google Scholar]
- 87.Rommelaere H, Waterschoot D, Neirynck K, Vandekerckhove J, Ampe C. A method for rapidly screening functionality of actin mutants and tagged actins. Biol Proced Online. 2004;6:235–249. doi: 10.1251/bpo94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Geetha T, Kenchappa RS, Wooten MW, Carter BD. TRAF6-mediated ubiquitination regulates nuclear translocation of NRIF, the p75 receptor interactor. EMBO J. 2005;24(22):3859–3868. doi: 10.1038/sj.emboj.7600845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.