Abstract
Leptospirosis is a bacterial zoonosis of major concern on tropical islands. Human populations on western Indian Ocean islands are strongly affected by the disease although each archipelago shows contrasting epidemiology. For instance, Mayotte, part of the Comoros Archipelago, differs from the other neighbouring islands by a high diversity of Leptospira species infecting humans that includes Leptospira mayottensis, a species thought to be unique to this island. Using bacterial culture, molecular detection and typing, the present study explored the wild and domestic local mammalian fauna for renal carriage of leptospires and addressed the genetic relationships of the infecting strains with local isolates obtained from acute human cases and with Leptospira strains hosted by mammal species endemic to nearby Madagascar. Tenrec (Tenrec ecaudatus, Family Tenrecidae), a terrestrial mammal introduced from Madagascar, is identified as a reservoir of L. mayottensis. All isolated L. mayottensis sequence types form a monophyletic clade that includes Leptospira strains infecting humans and tenrecs on Mayotte, as well as two other Malagasy endemic tenrecid species of the genus Microgale. The lower diversity of L. mayottensis in tenrecs from Mayotte, compared to that occurring in Madagascar, suggests that L. mayottensis has indeed a Malagasy origin. This study also showed that introduced rats (Rattus rattus) and dogs are probably the main reservoirs of Leptospira borgpetersenii and Leptospira kirschneri, both bacteria being prevalent in local clinical cases. Data emphasize the epidemiological link between the two neighbouring islands and the role of introduced small mammals in shaping the local epidemiology of leptospirosis.
Author Summary
Islands are exceptionally prone to species introduction, including pathogens with detrimental public health consequences for which the invasive alien species could act as reservoirs. Our study shows how the local non-native mammal fauna of Mayotte Island is associated with the introduction and epidemiology of human leptospirosis, a zoonosis strongly affecting tropical islands. Data presented herein identify Tenrec ecaudatus (Family Tenrecidae), an omnivorous species introduced from neighbouring Madagascar, as a reservoir of the recently named Leptospira mayottensis. Further, we suggest a Malagasy origin of L. mayottensis, which occurs naturally within the Tenrecidae radiation endemic to Madagascar. Finally, we provide evidence that dogs and rats are reservoirs for other Leptospira lineages of medical importance on Mayotte. Altogether, our study highlights the impact of species introduction on human health and further suggests that the biogeography of microorganisms in insular ecosystems, including pathogenic endemic lineages, should be considered from evolutionary and medical perspectives.
Introduction
Leptospirosis is a bacterial zoonosis caused by pathogenic spirochetes of the genus Leptospira. Human infection is common in tropical countries where warm and humid conditions favour the survival of Leptospira spp. in water and soil. Furthermore, human populations in these countries often live in rural areas or in impoverished urban zones with inadequate sanitation and can be at higher risk of exposure to the infection via the contaminated environment. A wide variety of mammal species can be infected by Leptospira spp. but not all act as reservoirs. Animal reservoirs support the chronic colonization of their renal tubules by a biofilm of leptospires and thus, release over prolonged periods the bacteria into the environment via their urine. Rodents and particularly rats, especially Rattus spp., are considered as the principal reservoir of these pathogenic bacteria and the main source of human infection, a role likely favoured by the worldwide distribution and commensal behaviour of some invasive species in this genus.
Two methods are currently used to identify leptospires: a phenotypic classification based on the microscopic agglutination test (MAT) that recognizes over 25 serogroups and 300 serovars [1] and a more recently introduced genetic classification based on bacterial DNA sequences that allows the identification of 20 Leptospira spp., nine of which are pathogenic [2]. Genotyping through Multiple Locus Sequence Typing (MLST) based on a selected set of housekeeping genes [3] has become a method of choice to identify Leptospira at the species and infra-species levels. Indeed, molecular typing allows deciphering the molecular epidemiology of the disease, sharing data among laboratories investigating distant geographic regions and analyzing the evolution of host-parasite relationships using robust molecular characters [4].
The incidence of human leptospirosis has been reported to be highest on tropical islands and this observation holds true for the southwestern Indian Ocean (SWIO) region [5]. Poorly documented on Mauritius and Madagascar, the incidence of the disease in the Seychelles ranks first worldwide among surveyed countries [5], while on Reunion Island, a French overseas department, the rate of human leptospirosis has been reported to be nearly 20 times higher than in continental France [6].
Mayotte, the most southern island of the Comoros Archipelago is a French overseas department of about 375 km², located in the northern entrance of the Mozambique Channel about 300 km off the northwestern coast of Madagascar. Human leptospirosis is highly prevalent on the island with an annual incidence in 2013 estimated at 35 per 100,000 [6]. Most interestingly, Leptospira strains isolated from patients, hereafter referred to clinical isolates, showed a much larger species diversity on Mayotte than on Reunion Island: 16 different sequence types (STs) were identified by MLST on Mayotte [7], while only three on Reunion Island [8]. Further, pathogenic leptospires causing acute human infections on Mayotte were identified as Leptospira interrogans, Leptospira borgpetersenii, Leptospira kirschneri and members of a previously undefined phylogroup that has recently been named as Leptospira mayottensis [7,9]. By contrast, only L. interrogans and L. borgpetersenii have been reported on Reunion Island [8]. The high diversity of leptospires on Mayotte, unique so far within the SWIO region, likely reflects local eco-epidemiologic specificities of animal reservoirs. With the exception of bats, no native small mammals occur on Mayotte [7]. Although one study has previously investigated animal reservoirs on Mayotte [7], it targeted Rattus rattus, the only rat species known on the island, for kidney carriage of pathogenic leptospires, whereas other species of the local wild and domestic fauna were screened by MAT, a serological test that provides evidence of previous infections but does not assess the actual carrier state of the investigated animals.
Through the investigation of the local wild and domestic fauna, the present study aimed at identifying animal reservoirs of leptospires on Mayotte, with a particular focus on L. mayottensis. This leptospiral species has been recently detected in wild small mammals endemic to neighbouring Madagascar [4] and, hence, it can be hypothesized that the geographic proximity and various socio-economic exchanges between these two islands could have facilitated the introduction of this pathogenic bacteria to Mayotte.
Methods
Ethics statement
All animal procedures carried out in this study were performed in accordance with the European Union legislation for the protection of animals used for scientific purposes (Directive 2010/63/EU). The ethical terms of the research protocol were approved by the CYROI Institutional Animal Care and Use Committee (Comité d’Ethique du CYROI n° 114, IACUC certified by the French Ministry of Higher Education and Research) under accreditation 03387 (LeptOI) and 03584 (BatMan).
Animal sampling
Mammals were trapped on Mayotte during three field sessions carried out from July 2012 to December 2014. Rattus rattus and Tenrec ecaudatus were captured using live traps baited with grilled coconut at eight and five localities, respectively. Sampling sites were selected in order to maximize the species diversity and geographic distribution of mammals in our sample. Trapping localities included a variety of different ecological settings across the island (see S1 Fig). In a local laboratory, R. rattus were sacrificed by cervical dislocation and T. ecaudatus by percussive blow to the head. For each individual, tissue was quickly collected from kidney, lung and spleen, pooled together and immediately stored in dry vials at -80°C. Insectivorous bats, Chaerephon sp. (Family Molossidae), were captured at dusk or at night using mist nets placed either in the vicinity of synanthropic roost sites or across flyways, while frugivorous bats, Pteropus seychellensis comorensis (Family Pteropodidae), were captured at night using mist nets set around fruiting trees. Captured bats were individually placed in clean cloth bags and brought back to a local laboratory. In most cases, animals spontaneously urinated after their removal from holding bags allowing the collection of urine. Bats were subsequently released at dusk at the initial capture site. Dogs (Canis lupus familiaris) were only manipulated and sampled by a local veterinary doctor; kidneys were obtained from animals that were euthanized for medical purposes, when possible urine collected from these same animals and immediately kept in dry vials at -80°C. In addition, urine samples were obtained from non-euthanized dogs through aseptic urethral catheterization. Kidneys from zebus (Bos indica) were purchased at the slaughterhouse of Mayotte (Kaweni) or obtained during traditional butchering. All samples were sent in liquid nitrogen to Reunion Island for analyses.
Leptospira spp. isolation
In addition to the sampling and storage at -80°C of organs and urine mentioned above, a few urine droplets and/or a small piece of freshly sampled kidney (crushed under sterile conditions) were used individually to inoculate three distinct culture media: (i) Ellinghausen-McCullough-Johnson-Harris (EMJH) liquid medium (Difco, Detroit, MI, USA) supplemented with Albumin Fatty Acid Supplement (AFAS; Royal Tropical Institute, Amsterdam, Netherlands) [10] (ii) EMJH liquid medium supplemented with AFAS, rabbit serum and fetal calf serum (1% each); and (iii) semisolid Fletcher medium (Difco, Detroit, MI, USA) supplemented with rabbit serum (8%). All media were supplemented with 5-fluorouracil (5-FU) at a final concentration of 200 μg.mL-1. Cultures were incubated at 28°C, visually checked for the presence of leptospires using a dark field microscope once a week for four months and positive cultures were further sub-cultured in fresh EMJH liquid medium deprived of 5-FU. DNA was extracted from 1 mL of each positive culture using the EZ1 Biorobot with Qiagen EZ1 DNA Tissue kits (Qiagen, Les Ulis, France).
Detection of pathogenic leptospires
Approximately 1 mm³ of pooled organs (kidney, lung and spleen) from each R. rattus and T. ecaudatus specimen and only kidney from dogs and zebus were dissected on sterile ice and further processed as previously described [11]. Thirty microliters of urine from bats and dogs were combined with 120 μL of Dulbecco’s modified medium (GIBCO, Grand Island, NY, USA) and 50 μL of ATL buffer (Qiagen, Les Ulis, France). Subsequently, total nucleic acids were extracted from urine or homogenized tissues by using an EZ1 extraction robot and the EZ1 Virus Mini Kit version 2.0. A reverse transcription step was performed on total nucleic acids with GoScript Reverse Transcriptase (Promega, Madison, WI, USA) to obtain cDNA. Detection of a portion of the 16S rRNA gene of pathogenic Leptospira spp. was then carried out on 5 μL of cDNA using a probe-specific real-time Polymerase Chain Reaction system (RT-PCR) [12].
Leptospira spp. genotyping
Leptospira spp. in samples testing positive by RT-PCR and/or culture were genotyped using a previously described MLST scheme encompassing six genes: secY, adk, rrs2, icdA, lipL32 and lipL41 [13], and recently optimized to improve the amplification of SWIO lineages [4]. RT-PCR is more sensitive than any MLST PCR (likely because of the very short length of RT-PCR amplicon), and some samples that were positive by the former detection system were MLST negative. In an attempt to further characterize Leptospira spp. from these particular samples, we used an alternative primer set (LA-LB) targeting a shorter piece of rrs2 [14]. The amplification of each marker was realized with GoTaq Hot Start Green Master Mix 2X (Promega, Madison, WI, USA) and further sequenced on both strands by direct Sanger sequencing (Genoscreen, Lille, France) using the same amplification primer sets. All sequences were deposited on GenBank under the following accession numbers: KT338823-KT338942, KT725237-KT725243 and KX427207-KX427227.
Phylogenetic analyses and genetic diversity
Accessible sequences from clinical Leptospira isolates from Mayotte were included in the study [15]. Since icdA sequences were not provided for these clinical isolates, we used five out of the six markers of the MLST scheme in our analyses. In addition, we added bacterial sequences obtained from endemic terrestrial small mammals from Madagascar, Microgale cowani, Microgale dobsoni, Microgale majori, Microgale longicaudata and Microgale principula (Family Tenrecidae, Subfamily Oryzorictinae). The complete MLST of these Leptospira strains were sequenced in a previous study [4] and from cultures realized during field missions carried out in the Central Highlands of Madagascar, Réserve Spéciale d’Ambohitantely, in March and October 2014 [16].
Phylogenetic analyses were performed on each gene separately and subsequently on concatenated sequences using the best model of sequence evolution determined by jModelTest v.0.1.1[17] for each dataset. Bayesian analyses were performed with MrBayes 3.1.2 [18] and consisted of two independent runs of four incrementally heated Metropolis Coupled Markov Chain Monte Carlo (MCMCMC) starting from a random tree. MCMCMC was run for 10,000,000 generations with trees and associated model parameters sampled every 100 generations. The convergence level of each phylogeny was validated by an average standard deviation of split frequencies inferior to 0.05. The initial 10% of trees from each run were discarded as burn-in and the consensus phylogeny along with posterior probabilities were obtained from the remaining trees. Bayesian trees were visualized and rooted to midpoint with FigTree v.1.3.1 (Andrew Rambaut, Institute of Evolutionary Biology, University of Edinburgh, 2006 to 2009; http://tree.bio.ed.ac.uk/).
Genetic diversity was compared between the identified clades in the multilocus phylogeny by estimating the nucleotide diversity (π) within each clade from the concatenated sequences using DNASP v.5.10.01[19].
Results
Altogether, 486 samples of tissue and/or urine collected on Mayotte from 289 Rattus rattus, 37 Tenrec ecaudatus, 53 Canis lupus familiaris, 18 Bos indica, 69 Chaerephon sp. and 20 Pteropus seychellensis comorensis were screened for the presence of Leptospira spp. by RT-PCR and/or culture. The results are summarized on Table 1.
Table 1. Animals from the wild and domestic fauna of Mayotte tested for carriage of Leptospira spp. by real-time PCR, culture, and genotyping.
| Host species | Number of animals | Number of animals | Number of Leptospira | Number of typed | Leptospira spp. | |||
|---|---|---|---|---|---|---|---|---|
| tested by RT-PCR | tested by culture | spp. positive animals | Leptospira strains | |||||
| (OP, K, U)* | (K, U)* | (%) | Lb | Lm | Li | Lk | ||
| Rattus rattus | 289 | 81 | 46 (15.9) | 21 | 13 | 0 | 3 | 5 |
| (OP = 289) | (K = 81) | (38PCR+/Culture- | ||||||
| 8PCR+/Culture+) | ||||||||
| Tenrec | 37 | 36 | 10 (27.0) | 8 | 0 | 8 | 0 | 0 |
| ecaudatus | (OP = 37) | (K = 36, U = 17, 17†) | (2PCR+/Culture- | |||||
| 8PCR+/Culture+) | ||||||||
| Bos indica | 18 | 0 | 1 PCR+ (5.6) | 0 | 0 | 0 | 0 | 0 |
| (K = 18) | ||||||||
| Canis lupus | 53 | 0 | 7 PCR+ (13.2) | 6 | 3 | 0 | 0 | 3 |
| familiaris | (K = 38, U = 45, 30†) | |||||||
| Chaerephon | 69 | 69 | 0 | 0 | 0 | 0 | 0 | 0 |
| sp. | (U = 69) | (U = 69) | ||||||
| Pteropus | 20 | 20 | 2 PCR+ (10.0) | 1 | 0 | 0 | 0 | 1 |
| seychellensis | (U = 20) | (U = 20) | ||||||
| comorensis | ||||||||
Lb: L. borgpetersenii, Lm: L. mayottensis, Li: L. interrogans, Lk: L. kirschneri.
* OP: organ pool, K: kidney, U: urine.
† Number of animals with paired sampling (OP/K+U)
Leptospira spp. detection by RT-PCR and culture
Pathogenic Leptospira spp. were detected by RT-PCR in 27.0% of T. ecaudatus, 15.9% of R. rattus, 13.2% of dogs, 10.0% of P. seychellensis comorensis and 5.6% of zebus. All urine samples from Chaerephon sp. tested negative by RT-PCR. Bacterial cultures were attempted using 117 freshly sampled kidney and 94 urine samples. Overall, leptospires culture was successful for samples testing positive through RT-PCR only. Among the 81 R. rattus tested by culture inoculated with kidney, 14 were positive by RT-PCR, of which eight allowed positive cultures (57.1%). Similarly, eight of the 10 RT-PCR-positive samples (80.0%) from T. ecaudatus allowed isolation of leptospires by culture. Two of them (MDI295 and MDI321) yielded positive cultures from both urine and kidney samples. Lastly, no isolate grew in urine cultures from RT-PCR positive P. seychellensis comorensis samples. No culture was attempted when sampling dogs or zebus.
Leptospira species diversity in wild and domestic mammals
We were able to PCR amplify all six loci of the MLST scheme using DNA extracted from all 16 Leptospira cultures, obtained from eight R. rattus and eight T. ecaudatus samples. Interestingly, for T. ecaudatus, we failed to amplify the lipL41 gene with the conventional primers from Ahmed et al.[13], and used alternative primers designed by Dietrich et al. [4]. As expected, PCR targeting MLST loci were more arduous when using DNA extracted from tissue and urine of PCR-positive samples for which culture had failed. For instance, no successful PCR targeting MLST loci was recorded for the two RT-PCR positive T. ecaudatus failing to yield positive culture. Using the MLST scheme, only rrs2 locus was amplified from four PCR-positive R. rattus, while two loci (secY and rrs2) were amplified from two other R. rattus. Similarly, only rrs2 was amplified from six out of the seven RT-PCR-positive dog samples and from one out of the two RT-PCR-positive P. seychellensis comorensis samples. Lastly, no sequence could be obtained from the single zebu sample testing positive by RT-PCR. In addition, for seven PCR-positive R. rattus samples failing to produce any data using the MLST scheme, we successfully amplified and sequenced the rrs2 gene using an alternative primer set (LA-LB). Altogether, we could identify leptospiral diversity at the species level for 21 R. rattus, eight T. ecaudatus, six dogs and one frugivorous bat from Mayotte (see Table 1). Rattus rattus-borne Leptospira spp. appeared genetically diverse as L. borgpetersenii, L. kirschneri and L. interrogans were identified for thirteen, 5 and 3 samples, respectively. The sequencing of rrs2 locus in dog samples identified the infecting Leptospira spp. as L. borgpetersenii (n = 3) and L. kirschneri (n = 3). We also identified one of the two PCR-positive frugivorous bats as infected by L. kirschneri. Most importantly, all Leptospira strains isolated and/or genotyped from T. ecaudatus samples were identified as L. mayottensis.
Phylogenetic analyses of occurring Leptospira spp
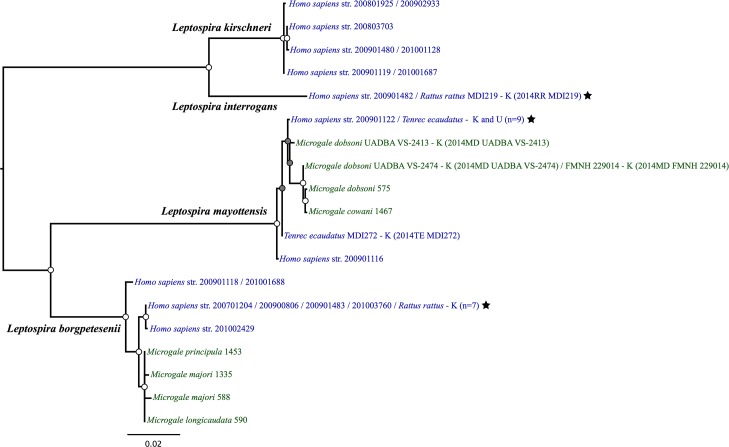
We constructed a concatenated phylogeny (2215 bp) by using bacterial sequences from the 16 animal Leptospira isolates from Mayotte of the present study, 17 previously described clinical isolates from Mayotte [20] and nine Leptospira strains infecting Malagasy endemic Tenrecidae (six previously published [4] and three obtained from three distinct animals sampled in Réserve Spéciale d’Ambohitantely). The phylogeny presented on Fig 1 shows that all bacterial sequences obtained from T. ecaudatus clustered into the monophyletic L. mayottensis clade that includes two clinical L. mayottensis isolates (str. 200901116 and str. 200901122). This clade also contains Leptospira sequence types (STs) obtained from Microgale cowani and M. dobsoni, these latter species being tenrecids endemic to Madagascar. Out of the eight sequences obtained from T. ecaudatus in Mayotte, seven were identical to each others and similar to clinical L. mayottensis str. 200901122 (100% of pairwise identity based on 2215 bp) and the last one (MDI272) was closely related to the clinical L. mayottensis str. 200901116 (99.70% of pairwise identity based on 2215 bp). Of note, the couple of kidney/urine cultures obtained from two T. ecaudatus (MDI295 and MDI321) yielded identical sequence. Leptospira mayottensis infecting Malagasy Microgale spp. displayed a 27-fold higher nucleotide diversity (π = 0.00298) compared to that infecting T. ecaudatus from Mayotte in four different sampling sites (π = 0.00011, S1 Fig). The phylogeny confirms R. rattus as carriers of L. interrogans and L. borgpetersenii. For L. interrogans, the same ST is shared by one R. rattus (MDI219) and the L. interrogans clinical isolate str. 200901482 (100% of pairwise identity based on 2215 bp). For L. borgpetersenii, one clade of this bacterial species included seven sequences obtained from R. rattus that were almost identical (99.99% pairwise identity, 2215 bp) to five previously described clinical L. borgpetersenii isolates. Noteworthy, sequences embedded in this clade were clearly distinct from L. borgpetersenii sequences obtained from Malagasy tenrecids. The nucleotide diversity of L. borgpetersenii from Malagasy tenrecids (π = 0.00181) was higher than that found in R. rattus from Mayotte (π = 0, one single ST).
Fig 1. Bayesian phylogenetic tree of pathogenic Leptospira species from Mayotte (blue) and Madagascar (green) based on concatenated sequences of five genes (secY, adk, lipL32, lipL41 and rrs2, total size: 2215 bp).
The analysis was realized under the GTR+G substitution model. At the nodes, grey and white circles indicate posterior probabilities superior to 0.70 and 0.90, respectively. Strain numbers of cultures produced herein are indicated in parentheses, “K” and “U” designating the sequences obtained from kidney or urine, respectively. Stars indicate lineages common to humans and animals (Rattus rattus or Tenrec ecaudatus). For Homo sapiens str. 200901122, the same sequence type was found in nine cultures from T. ecaudatus (2014TE MDI222, 2014TE MDI224, 2014TE MDI294U, 2014TE MDI295, 2014TE MDI295U, 2014TE MDI306, 2014TE MD308, 2014TE MDI321 and 2014TE MDI321U). For H. sapiens str. 200701204, 200900806, 200901483 and 201003760, the same sequence type found was in seven cultures from R. rattus (2014RR MDI247, 2014RR MDI250, 2014RR MDI251, 2014RR MDI259, 2014RR MDI260, 2014RR MDI284 and 2014RR MDI291). Specimen system: MDI = CRVOI specimen catalogue during field trips to Mayotte; FMNH = Field Museum of Natural History, Chicago; UADBA = Université d’Antananarivo, Département de Biologie Animale, Madagascar; for the other bacterial sequences from H. sapiens and Microgale spp. see Bourhy et al. 2012 [15] and Dietrich et al. 2014 [4]. Museum numbers for Microgale spp.: 575 = UADBA 30869; 588 = UADBA 30289; 590 = UADBA 30291; 1335 = UADBA 32122; 1453 = UADBA 32125; 1467 = UADBA 32101.
Although full MLST was not successful for bats, dogs and for some of the R. rattus, we still used rrs2 sequences alone in order to disentangle the role of these animals in human leptospirosis on Mayotte. For instance, the single leptospiral rrs2 (obtained using MLST scheme primers) sequence obtained from a frugivorous bat (P. seychellensis comorensis) was genetically related to clinical L. kirschneri isolates (98.70% pairwise identity, based on 452 bp) although nucleotide divergence at this locus together with PCR failure on all other loci suggested that L. kirschneri detected in frugivorous bats was actually divergent from clinical samples (see S2 Fig). Sequences obtained from dogs using the same primers revealed perfect identity with L. borgpetersenii (n = 3) and L. kirschneri (n = 2) clinical isolates from Mayotte. Lastly, as these rrs2 primers failed to produce amplification on some rat samples, we used an alternative primer set (LA-LB) that allowed amplification of a shorter rrs2 sequence, revealing five identical L. kirschneri sequences that, importantly, diverged from clinical isolates (3 mismatches out of 245 bp), thus in favour of a predominant role of dogs in L. kirschneri transmission to humans.
Discussion
Recent reports of human leptospirosis in the SWIO have stressed specificities singularizing Mayotte from the other islands of the region. Leptospira spp. infecting humans on Mayotte are diverse and belong to four bacterial species of which one, Leptospira mayottensis, was recently elevated to the rank of new species [9]. This situation clearly contrasts from that occurring on Reunion Island where Leptospira interrogans and Leptospira borgpetersenii are the only two species reported in human cases [8,21].
Our screening of Mayotte fauna allowed identifying 36 Leptospira strains as L. interrogans, L. borgpetersenii, L. mayottensis and L. kirschneri. We revealed either perfect or nearly perfect identity between (i) the single L. interrogans ST obtained from patients and Rattus rattus (100% identity), (ii) the prevailing L. borgpetersenii ST obtained from patients and R. rattus (100% identity), and (iii) both L. mayottensis STs obtained from patients and Tenrec ecaudatus (100% identity and 99.7% identity). The sequencing of rrs2 locus suggests that L. kirschneri reported in clinical cases originates from dogs although this phylogeny, based on the single rrs2 locus, known to display low polymorphism [22], is clearly weaker than the phylogeny based on full MLST. Finally, although a previous study reported a seroprevalence of 10% in Pteropus seychellensis comorensis in Mayotte [7], here all urine samples from bats tested negative for Leptospira sp. Although we cannot exclude that we missed positive samples due to our limited number of samples and the dynamic nature of leptospires excretion in bats over time [23], the single L. kirschneri rrs2 sequence obtained from a P. seychellensis comorensis specimen was clearly distinct from the lineages involved in human disease, suggesting that local bat species do not appear to play a significant epidemiological role in the transmission of leptospirosis. Considering bats being the only indigenous mammal species occurring on Mayotte, and dogs, R. rattus and T. ecaudatus the more widely represented mammal species in the environment [7], these results highlight the role of introduced wild and domestic small mammal species in the epidemiology of leptospirosis on Mayotte, and are specifically discussed hereafter.
Firstly and most importantly, our results suggest that T. ecaudatus is a main reservoir of L. mayottensis as this species was detected only in this host (infection rate: 27.0%) and not in the 289 R. rattus investigated herein. Although a previous study reported the detection of L. mayottensis in two out of 20 positive R. rattus [7], our results on a much larger sampling suggests that this species is not a significant reservoir of L. mayottensis. Moreover, all Leptospira strains infecting T. ecaudatus on Mayotte were strictly typed as L. mayottensis, although a larger sample size would be needed to confirm that L. mayottensis is the only Leptospira species infecting T. ecaudatus.
Secondly, the genetic analyses suggest that L. mayottensis likely originates from neighbouring Madagascar. Indeed, all L. mayottensis sequences cluster into a monophyletic well-supported clade including sequences obtained from three terrestrial wild small mammal species, namely T. ecaudatus, Microgale dobsoni and Microgale cowani, all belonging to the highly diversified Tenrecidae family representing an adaptive radiation of at least 32 endemic species, which colonized Madagascar some 25–30 million years ago [24]. This introduction scenario is further supported by the narrow genetic diversity of L. mayottensis in T. ecaudatus from Mayotte, based on positive samples from four different geographical sites. However, additional sampling of T. ecaudatus in Madagascar is necessary to confirm this hypothesis.
Thirdly, R. rattus is a major reservoir of pathogenic leptospires on Mayotte as also reported worldwide. Unexpectedly, the screening of a large number of rat samples (n = 289) from different localities (n = 8) shows that R. rattus on Mayotte is only marginally infected by L. interrogans. This feature contrasts with many investigations reported worldwide, including on the neighbouring islands of Madagascar and Reunion Island where rats are only infected with L. interrogans [8,25]. This peculiarity of Mayotte supports a massive contamination of the environment on this island by a diversity of Leptospira species, notably L. borgpetersenii detected in 62.0% of analysed R. rattus. Noteworthy, all L. borgpetersenii and L. interrogans obtained from R. rattus, showed perfect identity with human isolates based on the multilocus analysis. Although we identified L. kirschneri in five R. rattus, sequence analyses (rrs2) are rather supportive of a predominant role of dogs as reservoirs of L. kirschneri lineages of local medical importance. However, the absence of culture did not allow full genotyping of dog-hosted leptospires and their implication in human transmission needs further investigations.
Fourthly, L. borgpetersenii STs isolated from acute human cases on Mayotte group with two clearly distinct clades. The first lineage can be traced to R. rattus and is interestingly also closely related to a clade previously described as associated with endemic Malagasy Microgale species [4], indicating a possible host shift from endemic (Tenrecidae) to introduced (rats) mammals. The second L. borgpetersenii lineage, represented by two human isolates (str. 200901118 and 201001688), appears related to L. borgpetersenii lineages reported worldwide, e.g. Denmark, China, and Slovakia [4], but not in any wild and domestic animals sampled herein. This cosmopolite lineage may have been introduced through a reservoir, which is yet to be identified.
Finally, the remarkably high success rate to grow in culture L. mayottensis from T. ecaudatus (8/10, 80.0%) and L. borgpetersenii from R. rattus (7/13, 53.8%) indicates some specific biological peculiarities of these strains on Mayotte. Indeed, using the same protocol, we could grow in culture on Madagascar only three strains of L. mayottensis (obtained from kidney extracts from M. dobsoni, included on Fig 1), while other Leptospira species were found in terrestrial Malagasy wild animals [4,25]. Hence, this property to readily grow on liquid culture medium may reflect some adaptive advantage to survive and/or multiply in the environment. Interestingly, L. mayottensis was reported to grow over a wide range of temperature including 11°C and 37°C in contrast to other pathogenic strains [9].
Islands are considered as exceptionally prone to the successful introduction and subsequent invasive nature of exotic animals and plants [26] and have been studied in detail because of adaptive features of species in insular contexts. Our data document that a pathogen most certainly originally endemic to Madagascar [4] has been introduced to the small island of Mayotte where it expanded to become a significant emerging human pathogen. From an evolutionary perspective, our study highlights that beside macro-organism diversity, the associated micro-biodiversity, including endemic microbial lineages, deserves to be included in studies of invasive biology in the context of island biogeography. For instance, although we cannot exclude that other animal species may excrete L. mayottensis on Mayotte Island, the absence of L. mayottensis in other animal species together with the monophyly of L. mayottensis clade, composed of lineages strictly infeoded to tenrecids, support that this new Leptospira species has actually coevolved during the Malagasy radiation of tenrecs, as recently proposed for bat-borne Leptospira lineages in Madagascar [22].
Supporting Information
Numbers correspond to the 18 sampling sites where Rattus rattus and Tenrec ecaudatus (white) and bats (orange) were trapped. Map was created with QGIS 2.8.1 (QGIS Development Team, 2016, QGIS Geographic Information System, Open Source Geospatial Foundation Project). Photography of Mayotte: BD Topo IGN, 2008.
(TIFF)
The figures A and B display Bayesian phylogenetic trees of pathogenic Leptospira from Mayotte (blue) and Madagascar (green) based on 452 bp (57 taxa, HKY+I+G) (A) and 245 bp (64 taxa, K80+I) (B) of the rrs2 gene. At the nodes, the black numbers indicate posterior probabilities. The sequences highlighted in grey (A) and red (B) represent PCR positive samples for which only the rrs2 gene was obtained. Strain numbers of cultures produced herein are indicated in parentheses, “K” and “U” designating sequences obtained from kidney or urine, respectively. Specimen system: MDI and MAY = CRVOI specimen catalogue during field trips to Mayotte; all Canis lupus familiaris were sampled during field trips to Mayotte; FMNH = Field Museum of Natural History, Chicago; UADBA = Université d’Antananarivo, Département de Biologie Animale, Madagascar; for the other bacterial sequences from Homo sapiens and Microgale spp. see Bourhy et al. 2012 [15]and Dietrich et al. 2014 [4]. Museum numbers for Microgale spp.: 575 = UADBA 30869; 588 = UADBA 30289; 590 = UADBA 30291; 1335 = UADBA 32122; 1453 = UADBA 32125; 1467 = UADBA 32101.
(TIFF)
Acknowledgments
Permits were issued by Direction de l’Environnement, de l’Aménagement et du Logement (DEAL) of Mayotte. We thank Office National des Forêts, Conseil Général of Mayotte and Association Naturalistes Environnement et Patrimoine de Mayotte for allowing animal capture on conservation sites. For Madagascar samples, permits were provided by Malagasy national authorities: Ministère de l’Environnement et des Forêts, Madagascar National Parks and the Département de Biologie Animale of the Université d’Antananarivo (N°238/14/MEEF/SG/DGF/DCB.SAP/SCB). We are also grateful to Catherine Dioniso in helping to locate some bat roost sites. COOPADEM provided the field group laboratory space on Mayotte and we would like to thank Marion Pannequin and Laure Dommergues particularly. We also thank Voahangy Soarimalala for helping in preparation of samples at Réserve Spéciale Ambohitantely. We deeply think Dr Bénédicte Contamin and Fondation Mérieux in Madagascar for providing logistical supports. Computations were performed on the “Université de La Réunion” supercomputer facility. We are thankful to both anonymous reviewers who thoroughly examined the original manuscript and contributed to substantial improvements.
Data Availability
All Leptospira sequences were deposited on GenBank under the following accession numbers: KT338823-KT338942, KT725237-KT725243 and KX427207-KX427227.
Funding Statement
This study was supported by funds from ERDF-POCT Réunion, LeptOI (#32913), ParamyxOI (#33857); Centre National de la Recherche Scientifique - Institut Ecologie et Environnement (ECOSAN BatMan) and Agence Régionale de la Santé Ocean Indien. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14: 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerqueira GM, Picardeau M. A century of Leptospira strain typing. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2009;9: 760–768. 10.1016/j.meegid.2009.06.009 [DOI] [PubMed] [Google Scholar]
- 3.Ahmed A, Thaipadungpanit J, Boonsilp S, Wuthiekanun V, Nalam K, Spratt BG, et al. Comparison of two multilocus sequence based genotyping schemes for Leptospira species. PLoS Negl Trop Dis. 2011;5: e1374 10.1371/journal.pntd.0001374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dietrich M, Wilkinson DA, Soarimalala V, Goodman SM, Dellagi K, Tortosa P. Diversification of an emerging pathogen in a biodiversity hotspot: Leptospira in endemic small mammals of Madagascar. Mol Ecol. 2014;23: 2783–2796. 10.1111/mec.12777 [DOI] [PubMed] [Google Scholar]
- 5.Pappas G, Papadimitriou P, Siozopoulou V, Christou L, Akritidis N. The globalization of leptospirosis: worldwide incidence trends. Int J Infect Dis. 2008;12: 351–357. 10.1016/j.ijid.2007.09.011 [DOI] [PubMed] [Google Scholar]
- 6.Picardeau M, Bourhy P. Annual activity report. National Reference Center for Leptospirosis. Institut Pasteur [Internet]. 2014. Available: https://www.pasteur.fr/fr/sante/centres-nationaux-reference/les-cnr/leptospirose/rapports-d-activite
- 7.Desvars A, Naze F, Vourc’h G, Cardinale E, Picardeau M, Michault A, et al. Similarities in Leptospira serogroup and species distribution in animals and humans in the Indian ocean island of Mayotte. Am J Trop Med Hyg. 2012;87: 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guernier V, Lagadec E, Cordonin C, Minter GL, Gomard Y, Pagès F, et al. Human leptospirosis on Reunion Island, Indian Ocean: are rodents the (only) ones to blame? PLOS Negl Trop Dis. 2016;10: e0004733 10.1371/journal.pntd.0004733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourhy P, Collet L, Brisse S, Picardeau M. Leptospira mayottensis sp. nov., a pathogenic Leptospira species isolated from humans. Int J Syst Evol Microbiol. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellinghausen HC, Mccullough WG. Nutrition of Leptospira Pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and a medium of bovine albumine and polysorbate 80. Am J Vet Res. 1965;26: 45–51. [PubMed] [Google Scholar]
- 11.Wilkinson DA, Temmam S, Lebarbenchon C, Lagadec E, Chotte J, Guillebaud J, et al. Identification of novel paramyxoviruses in insectivorous bats of the Southwest Indian Ocean. Virus Res. 2012;170: 159–163. 10.1016/j.virusres.2012.08.022 [DOI] [PubMed] [Google Scholar]
- 12.Smythe LD, Smith IL, Smith GA, Dohnt MF, Symonds ML, Barnett LJ, et al. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect Dis. 2002;2: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed N, Devi SM, Valverde M de los A, Vijayachari P, Machang’u RS, Ellis WA, et al. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob. 2006;5: 28 10.1186/1476-0711-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lourdault K, Aviat F, Picardeau M. Use of quantitative real-time PCR for studying the dissemination of Leptospira interrogans in the guinea pig infection model of leptospirosis. J Med Microbiol. 2009;58: 648–655. [DOI] [PubMed] [Google Scholar]
- 15.Bourhy P, Collet L, Lernout T, Zinini F, Hartskeerl RA, van der Linden H, et al. Human Leptospira isolates circulating in Mayotte (Indian Ocean) have unique serological and molecular features. J Clin Microbiol. 2012; 10.1128/JCM.05931-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman SM, Andriniaina HRR, Soarimalala V, Beaucournu J-C. The fleas of endemic and introduced small mammals in central highland forests of Madagascar: faunistics, species diversity, and absence of host specificity. J Med Entomol. 2015;52: 1135–1143. 10.1093/jme/tjv113 [DOI] [PubMed] [Google Scholar]
- 17.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25: 1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- 18.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- 19.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25: 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 20.Bourhy P, Collet L, Clément S, Huerre M, Ave P, Giry C, et al. Isolation and characterization of new Leptospira genotypes from patients in Mayotte (Indian Ocean). PLoS Negl Trop Dis. 2010;4: e724 10.1371/journal.pntd.0000724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naze F, Desvars A, Picardeau M, Bourhy P, Michault A. Use of a new high resolution melting method for genotyping pathogenic Leptospira spp. PloS One. 2015;10: e0127430 10.1371/journal.pone.0127430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomard Y, Dietrich M, Wieseke N, Ramasindrazana B, Lagadec E, Goodman SM, et al. Malagasy bats shelter a considerable genetic diversity of pathogenic Leptospira suggesting notable host-specificity patterns. FEMS Microbiol Ecol. 2016; 10.1093/femsec/fiw037 [DOI] [PubMed] [Google Scholar]
- 23.Dietrich M, Wilkinson DA, Benlali A, Lagadec E, Ramasindrazana B, Dellagi K, et al. Leptospira and paramyxovirus infection dynamics in a bat maternity enlightens pathogen maintenance in wildlife. Environ Microbiol. 2015; 10.1111/1462-2920.12766 [DOI] [PubMed] [Google Scholar]
- 24.Olson LE. Tenrecs. Curr Biol. 2013;23: R5–R8. 10.1016/j.cub.2012.11.015 [DOI] [PubMed] [Google Scholar]
- 25.Rahelinirina S, Léon A, Harstskeerl RA, Sertour N, Ahmed A, Raharimanana C, et al. First isolation and direct evidence for the existence of large small-mammal reservoirs of Leptospira sp. in Madagascar. PloS One. 2010;5: e14111 10.1371/journal.pone.0014111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elton CS. The ecology of invasions by animals and plants University Of Chicago Press; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Numbers correspond to the 18 sampling sites where Rattus rattus and Tenrec ecaudatus (white) and bats (orange) were trapped. Map was created with QGIS 2.8.1 (QGIS Development Team, 2016, QGIS Geographic Information System, Open Source Geospatial Foundation Project). Photography of Mayotte: BD Topo IGN, 2008.
(TIFF)
The figures A and B display Bayesian phylogenetic trees of pathogenic Leptospira from Mayotte (blue) and Madagascar (green) based on 452 bp (57 taxa, HKY+I+G) (A) and 245 bp (64 taxa, K80+I) (B) of the rrs2 gene. At the nodes, the black numbers indicate posterior probabilities. The sequences highlighted in grey (A) and red (B) represent PCR positive samples for which only the rrs2 gene was obtained. Strain numbers of cultures produced herein are indicated in parentheses, “K” and “U” designating sequences obtained from kidney or urine, respectively. Specimen system: MDI and MAY = CRVOI specimen catalogue during field trips to Mayotte; all Canis lupus familiaris were sampled during field trips to Mayotte; FMNH = Field Museum of Natural History, Chicago; UADBA = Université d’Antananarivo, Département de Biologie Animale, Madagascar; for the other bacterial sequences from Homo sapiens and Microgale spp. see Bourhy et al. 2012 [15]and Dietrich et al. 2014 [4]. Museum numbers for Microgale spp.: 575 = UADBA 30869; 588 = UADBA 30289; 590 = UADBA 30291; 1335 = UADBA 32122; 1453 = UADBA 32125; 1467 = UADBA 32101.
(TIFF)
Data Availability Statement
All Leptospira sequences were deposited on GenBank under the following accession numbers: KT338823-KT338942, KT725237-KT725243 and KX427207-KX427227.