Abstract
In spite of improvements in surgical technology, the resectability and curability of intrahepatic cholangiocarcinoma (ICC) are still low. Our previous study showed that the strong Sal-like protein 4 (Sall4)-positive cases had shorter overall survival compared to Sall4-negative cases, indicating an oncogenic role of Sall4 in ICC. In this study, we aimed to explore the precise mechanism of Sall4 on ICC cell invasion and metastasis. We evaluated the expression of Sall4, PTEN, and Bmi-1 in 28 cases of adjacent tissues and 175 cases of ICC tissues by using immunohistochemical staining. We found that the expression of Sall4 and Bmi-1 was significantly increased in ICC tissues compared with the adjacent tissues, while PTEN expression was reduced in ICC tissues compared with the adjacent tissues, and there was a reverse relationship between Sall4 and PTEN in ICC, whereas there was a positive correlation in Sall4 and Bmi-1 expression in ICC. In addition, overall survival analysis showed that ICC patients with low PTEN exhibited a worse prognosis than ICC patients with high PTEN, and lower Bmi-1 expression showed a better prognosis than ICC patients with high Bmi-1. By a battery of experiments in vitro, we demonstrated that Sall4 promotes ICC cell proliferation, and progression of ICC might be through PTEN/PI3K/Akt and Bmi-1/Wnt/β-catenin signaling and enhancing epithelial–mesenchymal transition process. Thus, Sall4 may be a potential target for the treatment of ICC metastasis.
Keywords: intrahepatic cholangiocarcinoma, Sal-like protein 4, PTEN, Bmi-1, metastasis, EMT
Introduction
Intrahepatic cholangiocarcinoma (ICC) has a poor prognosis and a high incidence in the biliary tree.1 In spite of the improvement in surgical technology, the resectability and curability of ICC are still low,2 and due to the high invasiveness of ICC, survival time after resection is short.3 Thus, it is important to develop new and specific biomarkers to aid in the diagnosis of early-stage ICC, and subsequently help guide the stratification of patients.
Sal-like protein 4 (Sall4) is a stem cell marker, and plays a role in maintaining self-renewal.4 Upregulation of Sall4 is found in hepatocellular carcinoma, breast cancer, and endometrial cancer, predicting a poor prognosis.5–7 Our previous study showed that Sall4-positive ICC cases had significantly more frequent lymph nodal metastasis, vascular invasion, and nerve invasion. Furthermore, the strong Sall4-positive cases had shorter overall survival compared to Sall4-negative cases, indicating an oncogenic role of Sall4 in ICC.8
Lu et al showed that Sall4, as a transcription factor, repressed phosphatase and tensin homolog (PTEN) expression by binding to the promoter regions of PTEN, a tumor-suppressor gene that was associated with myeloid leukemia and cystic kidneys.9 Overexpression of Bmi-1, a oncogenic gene, was positively correlated with Sall4 in acute myeloid leukemia.10 And transcription from the Bmi-1 promoter was strikingly activated by Sall4 in a dose-dependent manner. Reducing Sall4 expression by siRNA also resulted in significant downregulation of Bmi-1, while constitutively overexpressing human Sall4 upregulated Bmi-1 expression in transgenic mice.11 Thus, Sall4 is able to simultaneously bind to the promoter of PTEN and Bmi-1 in hematological tumor cells. However, the precise mechanism of Sall4 on ICC cell invasion and metastasis remains unknown.
In this study, we evaluated the expression of Sall4, PTEN, and Bmi-1 in ICC tissues by using immunohistochemical (IHC) staining and analyzed their correlation, and the relationship between Sall4, PTEN, and Bmi-1 expression and overall survival of patients in ICC.
Materials and methods
Tissues
This study was approved by the Institutional Review Board of the Third Xiangya Hospital, Central South University, Changsha, People’s Republic of China. All subjects or their caregivers gave written informed consent. All of the total 175 ICC tissue sections and 28 adjacent tissues of cancer were the same as we used in the previous study.8 The clinicopathologic characteristics of the ICC samples, including tumor size, location, histological grade, clinical stage, hepatitis B virus infection, lymph node metastasis, vascular invasion, and nerve invasion were described as our previous study.8 All these samples were formalin fixed and paraffin embedded.
IHC staining assay
The expression of Sall4, PTEN, and Bmi-1 was evaluated by using IHC staining. The IHC staining was performed as we previously described. Briefly, the sections were deparaffinized and hydrated, and then were subjected to heat-induced antigen retrieval with citrate buffer. Then the sections were incubated with primary antibodies (anti-Sall4, 1:100; anti-PTEN, 1:200; anti-Bmi-1, 1:200, Sigma-Aldrich, St Louis, MO, USA) overnight at 4°C. After that, the sections were incubated with secondary antibody for 60 minutes at 37°C. The signaling was visualized by using substrate diaminobenzidine and counterstained with hematoxylin.
For evaluating the expression of Sall4, PTEN, and Bmi-1 in ICC tissues, extent and intensity were assessed at each spot. All tissue sections were analyzed and scored independently by three experienced pathologists (FH, GD, WN). PTEN and Bmi-1 staining intensity was scored as 0 (negative, −), 1 (positive, weak, +), 2 (positive, moderate, ++), and 3 (positive, strong, +++). The extent of staining was scored as 0–1.0 (0%–100%). The final staining score (0–3) was calculated as the multiplication of the intensity and extent scores. The expression of PTEN and Bmi-1 was scored as high expression (≥1) or low expression (<1). To compare the expression of Sall4, PTEN, and Bmi-1 between normal and tumor tissues, the score of Sall4, PTEN, and Bmi-1 expression was normalized to their average score in normal tissues.
Cell culture
The human ICC cell line ICC-9810, was purchased from the Institute of Cell Biology at the Chinese Academy of Sciences (Shanghai, People’s Republic of China). ICC-9810 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM), to which was added 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), at 37°C in a humidified incubator containing 5% CO2.
Transfection
In order to knock down Sall4 or PTEN, or to overexpress Bmi-1, we transfected the siRNA or expressed vector into ICC-9810 cells by using lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instruction. Sall4 siRNA, PTEN siRNA, and nonspecific siRNA were purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA); Bmi-1 expressed vector was purchased from Qiagen NV (Venlo, the Netherlands). The transfected cells were used for further analysis.
Real-time quantitative polymerase chain reaction analysis
Trizol reagent (Invitrogen, Waltham, MA, USA) was used to extract total ribonucleic acid from the indicated cells according to the manufacturer’s instructions. One-step real-time (RT)-PCR master mixes (Thermo Fisher Scientific) was used for real-time PCR to detect the mRNA levels of PTEN, PI3K, AKT, Bmi-1, Wnt3a, β-catenin, and GSK3β. The primers were used as the following: PTEN, sense, GGACCTCTTGTGAGTCCACG, antisense, TGGTTAC CGGAGCCCATAGA; PI3K, sense, CAAACCGGAAAAC CATGCCA, antisense, TGACACAGCGGATTGAACGA; AKT, sense, GGCAAACCAGGCAGTTCAAG, antisense, CAGGAATCTTGAGCAGGGGG; Bmi-1, sense, AGACAA GATGCAGCGACTGT, antisense, CATGTGTGGCG GATCCTGAA; Wnt3a, sense, CAAGATGGCGTGCA CAAACA, antisense, AGGAGTTCTTGCTCCACACG; β-catenin, sense, GCTCCACACCGTGACCATAA, antisense, TTTCATCTGAGGGTGCGCTT; GSK3β, sense, GACTAAGGTCTTCCGACCCC, antisense, TTAGCATCTGACGCTGCTGT; β-actin, sense, AGGGGCCGGACTCGTCATACT, antisense, GGCGGCACCACCATGTACCCT. The target genes expression was normalized by β-actin. All the quantitative polymerase chain reaction data were processed using 2−ΔΔCT method.
Western blot
Radioimmunoprecipitation assay lysis buffer was used to extract protein from indicated cells. Bradford protein assay kit (Beyotime, Shanghai, People’s Republic of China) was used to measure the protein concentration. Total 60 μg of protein was separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and blotted onto 0.22 μm nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk for 2 hours and incubated with primary antibodies (anti-Sall4, anti-PTEN, anti-phosphorylated PTEN, and anti-Bmi-1 [Sigma-Aldrich]; anti-PI3K, anti-pPI3K, anti-AKT, anti-pAKT, anti-Wnt3a, anti-β-catenin, anti-GSK3β, and anti-GSK3β [Cell Signaling Technology, Danvers, MA, USA]; anti-E-cadherin, anti-N-cadherin, anti-fibronectin, anti-vimentin [Abcam, Cambridge, UK]; and anti-GAPDH [Proteintech, Wuhan, People’s Republic of China]) overnight at 4°C. The membranes were washed with tris-buffered saline containing 0.1% Tween 20, and then incubated with appropriate horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit, 1:2,000; goat anti-mouse, 1:5,000; Boster, Wuhan, People’s Republic of China) for 1 hour at 37°C. Enhanced chemiluminescence reagent was used to detect the signal on the membrane. The data were analyzed and normalized to the expression of the internal control (glyceraldehyde 3-phosphate dehydrogenase).
Luciferase reporter gene assays
The PTEN promoter (−2,000 to 20+) or Bmi-1 promoter (−1,800 to 30+) region was constructed into the pGL3-basic vector (Promega Corporation, Fitchburg, WI, USA) generating pGL3-basic-PTEN or pGL3-basic-Bmi-1 performed by GeneCopoeia (Guangzhou, People’s Republic of China). All vector sequences were confirmed by DNA sequencing. Cells were transfected with the pGL3-basic-PTEN or pGL3-basic-Bmi-1 reporter plasmid together with different amounts of Sall4 expressed vector (0.5 or 1 μg). The cells transfected with pGL3-basic were used as negative control. After 48 hours of transfection, the cells were collected and lysed. Ten millilitres of the supernatants was used to detect luciferase activities by using luciferase reporter gene assay kit (BioVision, Milpitas, CA, USA) on a microplate luminometer.
Scratch assay
Cells in each group were collected and resuspended in completed DMEM medium. Each well of a 6-well plate was seeded with 2×105 cells and continually cultured to reach 100% confluence. And the cells were scratched with the head of a 20 μL tip, and washed with serum-free medium. These cells were cultured in 3% fetal bovine serum DMEM medium. Then these cells from each group were photographed for analysis.
Transwell assay
The indicated cells were starved for 24 hours, and then resuspended in serum-free medium and added to the upper chamber that had been coated with 1 mm matrigel (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). The lower chamber was filled with completed DMEM medium. After 48 hours’ culture, cells attached to the bottom were fixed and stained with crystal violet for 30 minutes and dried in air, and then the stained cells were photographed. After that, the crystal violet stain on cells was dissolved by 10% acetic acid. The optical density at 570 nm of crystal violet was detected by an enzyme immunoassay analyzer.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6 software (Graphpad Software, Inc., La Jolla, CA, USA) and the data are presented as the mean ± standard deviation. An unpaired two-tailed Student’s t-test or one-way analysis of variance with Bonferroni t post-test was used to analyze the data depending on conditions. The OS estimates over time were calculated using the Kaplan–Meier method with log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
The correlation of Sall4, PTEN, and Bmi-1 in ICC
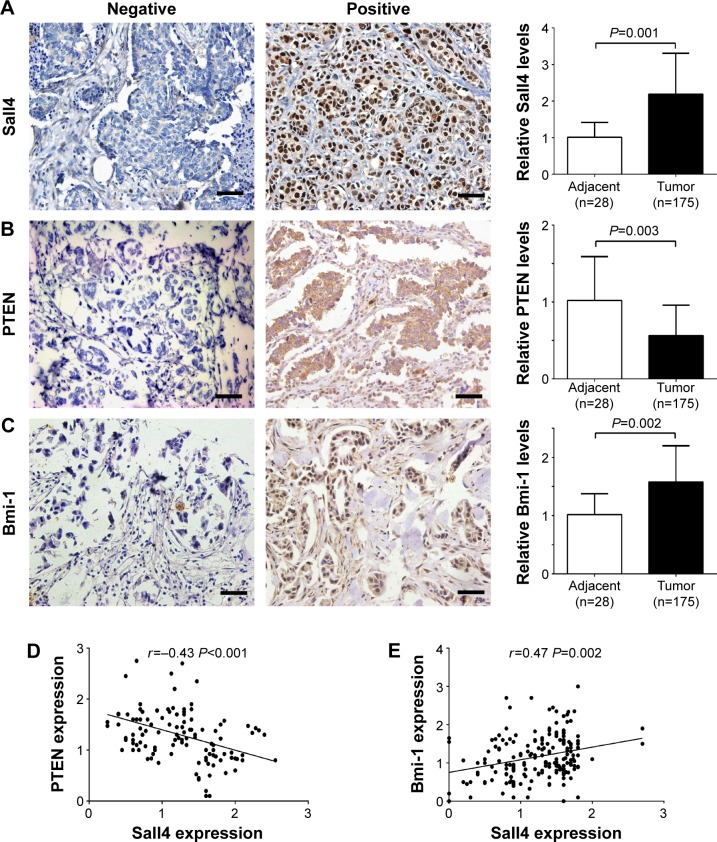
To investigate the role of Sall4, PTEN, and Bmi-1 in ICC, we collected 28 cases of adjacent tissues and 175 cases of ICC tissues and all of the tissues were included in tissue microarrays. The results of immunostaining showed that the expression of Sall4 and Bmi-1 was significantly increased in ICC tissues compared with the adjacent tissues, while PTEN expression was reduced in ICC tissues compared with the adjacent tissues (Figure 1A–C). Furthermore, we analyzed their relationship by Pearson analysis. We found that there was a reverse relationship between Sall4 and PTEN in ICC, whereas there was a positive correlation in Sall4 expression and Bmi-1 expression in ICC (Figure 1D and E).
Figure 1.
The expression of Sall4, PTEN, and Bmi-1 in intrahepatic cholangiocarcinoma and adjacent tissues.
Notes: (A) Representative images of Sall4 expression with negative and positive staining (left), and quantification of relative expression in tumor and adjacent tissues. Bar =100 μm; magnification 100×. (B) Representative images of PTEN expression with negative and positive staining (left), and quantification of relative expression in tumor and adjacent tissues. Scale bar =100 μm; magnification 100×. (C) Representative images of Bmi-1 expression with negative and positive staining (left), and quantification of relative expression in tumor and adjacent tissues. Scale bar =100 μm; magnification 100×. (D) Pearson correlation analysis for the correlation between Sall4 and PTEN in intrahepatic cholangiocarcinoma. (E) Pearson correlation analysis for the correlation between Sall4 and Bmi-1 in intrahepatic cholangiocarcinoma. Data are expressed by mean ± SD.
Abbreviation: SD, standard deviation.
Association between the expression of Sall4, PTEN, and Bmi-1 proteins and overall survival in patients with ICC
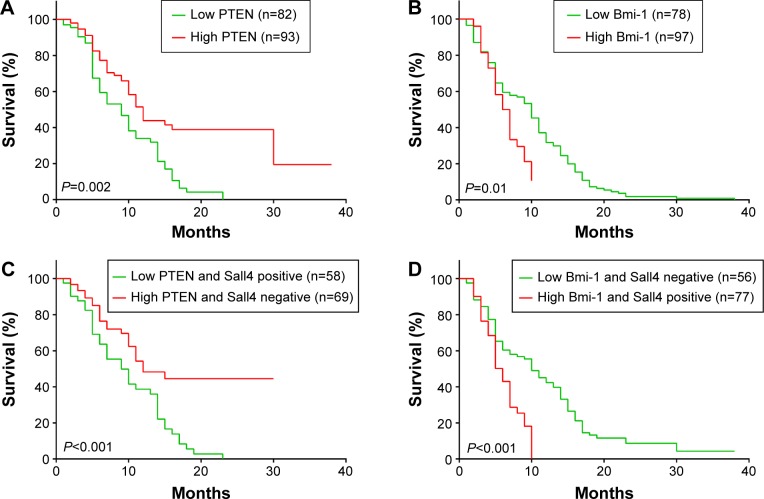
In the previous study, we have shown that high Sall4 expression predicted a poor survival in patients with ICC. We recently further analyzed the association between PTEN and Bmi-1 expression and clinical outcome in patients with ICC. All 175 ICC patients were included in the survival curves. The survival rate of patients with high PTEN expression was significantly increased compared with the survival of patients with low PTEN expression (P=0.002, Figure 2A), while high Bmi-1 expression predicted a poor outcome in ICC patients (P=0.01, Figure 2B). Furthermore, we investigated the association between PTEN/Bmi-1 and clinical outcome in Sall4 positive or negative ICC patients. The survival rate of Sall4 negative ICC patients with high PTEN expression was significantly increased compared to the survival of Sall4 positive patients with low PTEN expression (P<0.001, Figure 2C). On the other hand, high Bmi-1 expression predicted a poor outcome in Sall4-positive ICC patients (P<0.001, Figure 2D). These results indicate a close relationship between Sall4 and PTEN, and Bmi-1 in prognostic prediction of ICC patients.
Figure 2.
Kaplan–Meier overall survival curves of intrahepatic cholangiocarcinoma (ICC) patients.
Notes: (A) Kaplan–Meier analysis for PTEN expression in ICC patients. (B) Kaplan–Meier analysis for Bmi-1 expression in ICC patients. (C) Kaplan–Meier analysis for PTEN and Sall4 expression in ICC patients. (D) Kaplan–Meier analysis for Bmi-1 and Sall4 expression in ICC patients.
Abbreviation: ICC, intrahepatic cholangiocarcinoma.
Sall4 regulates promoter activity of PTEN and Bmi-1 in ICC cells
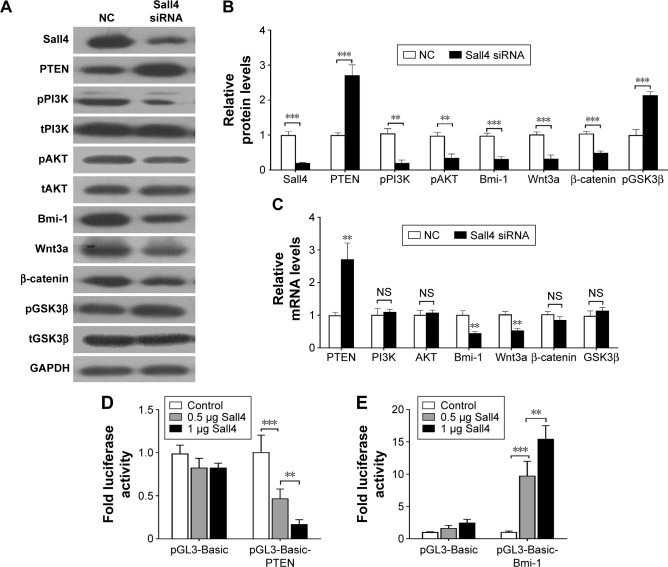
The correlation of Sall4, PTEN, and Bmi-1 prompted us to investigate whether Sall4 could regulate the expression of PTEN and Bmi-1. We knocked down Sall4 expression by transfecting Sall4 siRNA in ICC-9810 cells. Western blot analysis showed that knockdown of Sall4 increased PTEN expression and induced inactivation of PI3K/AKT signaling. In contrast, downregulation of Sall4 resulted in decrease of Bmi-1 expression and inactivated Wnt3a/β-catenin signaling, and also induced phosphorylation of GSK3β (Figure 3A and B). In addition, we found that knockdown of Sall4 also increased the mRNA levels of PTEN, but reduced the mRNA levels of Bmi-1 (Figure 3C). Furthermore, to investigate whether Sall4 controlled the expression of PTEN and Bmi-1 via regulating the promoter activity, we constructed the luciferase reporter gene containing PTEN promoter or Bmi-1 promoter, and then co-transfected them with Sall4 expressed vector. We found that the promoter activity of PTEN was significantly decreased by Sall4 in a dose-dependent manner, and the promoter activity of Bmi-1 was dramatically induced by Sall4 with increasing dose (Figure 3D and E). These results indicated that Sall4 regulates the expression of PTEN and Bmi-1 by activating or inactivating their promoter activity, and subsequently regulates PI3K/AKT and Wnt3a/β-catenin signaling.
Figure 3.
Knockdown of Sall4 regulates PTEN/PI3K/AKT and Bmi-1/Wnt3a/β-catenin signaling.
Notes: (A) Western blot analysis for Sall4, PTEN/PI3K/AKT, Bmi-1/Wnt3a/β-catenin and GSK3β expression after Sall4 knockdown in ICC-9810 cells. (B) Quantification of the bands of Western blot in (A). (C) Quantitative polymerase chain reaction analysis for PTEN, PI3K, AKT, Bmi-1, Wnt3a, β-catenin, and GSK3β mRNA levels after Sall4 knockdown in ICC-9810 cells. (D) Luciferase reporter gene assay using the reporter vector containing the promoter of PTEN gene. ICC-9810 cells were co-transfected with pGL3-basic-PTEN together with different amounts of a Sall4 expression vector. Sall4 reduces luciferase expression in a concentration-dependent manner in ICC-9810 cells. (E) ICC-9810 cells were co-transfected with pGL3-basic-Bmi-1 together with different amounts of a Sall4 expression vector. Sall4 induces luciferase expression in a concentration-dependent manner in ICC-9810 cells. Data are mean ± SD from three independent experiments. **P<0.01, ***P<0.001.
Abbreviations: mRNA, messenger RNA; NC, negative control; NS, not significant; SD, standard deviation.
Knockdown of PTEN or overexpression of Bmi-1 reverses Sall4-mediated effects on cell migration and invasion in ICC-9810 cells
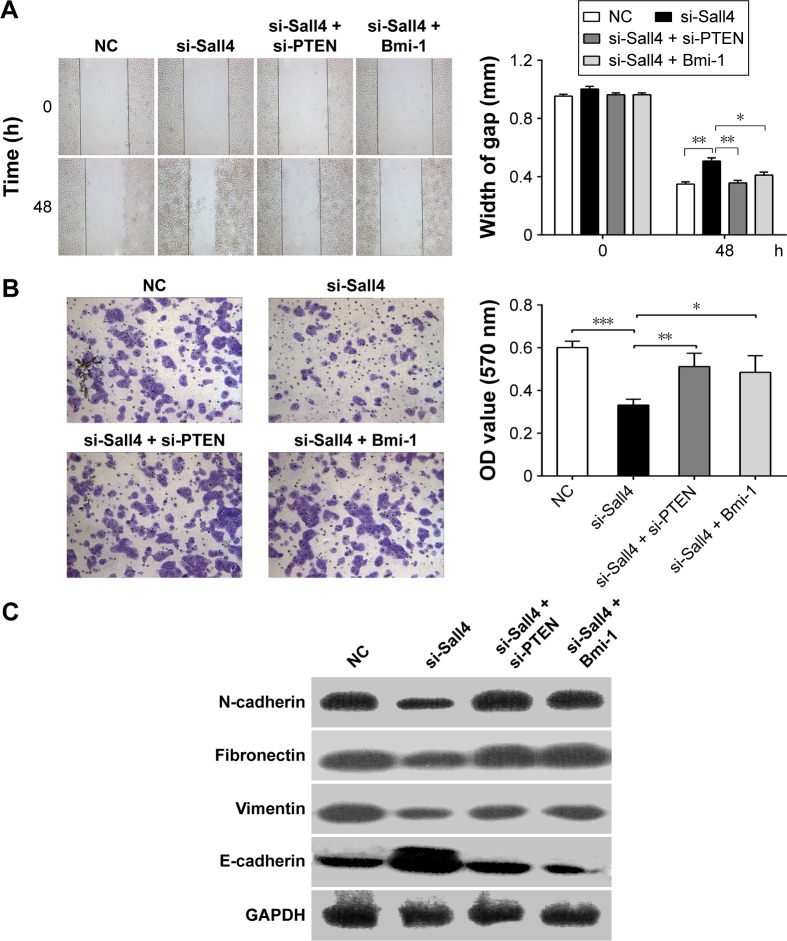
We have demonstrated that siRNA-induced Sall4 down-regulation showed an inhibitory effect on ICC metastasis. Recently, we further investigated the underlying mechanism of Sall4 on ICC metastasis. As shown in Figure 4A and B, the cell migration and invasion were all suppressed in ICC-9810 cells transfected with Sall4 siRNA, when compared with the negative control. However, knockdown of PTEN or overexpression of Bmi-1 reversed the effects of Sall4 siRNA transfection in ICC-9810 cells. These findings suggest that siRNA-induced Sall4 downregulation can inhibit malignant phenotypes of ICC cells via PTEN and Bmi-1 signaling. We further determined the expression of some key genes involved in epithelial–mesenchymal transition (EMT). As shown in Figure 4C, the expression of E-cadherin was notably upregulated, but the levels of N-cadherin, fibronectin, and vimentin were significantly reduced after knockdown of Sall4 in ICC-9810 cells. However, the effects of Sall4 knockdown were rescued by PTEN siRNA or Bmi-1-expressed vector transfection. These findings suggest that the inhibitory effects of Sall4 downregulation on ICC metastasis are involved in EMT through PTEN and Bmi-1.
Figure 4.
Knockdown of PTEN or overexpression of Bmi-1 reverses Sall4-mediated effects on cell migration and invasion in ICC-9810 cells.
Notes: (A) Wound healing assay was conducted to determine the cell migratory capacity of ICC-9810 cells co-transfected with Sall4 siRNA and PTEN siRNA or Bmi-1 expressed vector, non-specific siRNA was used as NC. 100× magnification. (B) Transwell assay was conducted to determine the cell invasive capacity of ICC-9810 cells co-transfected with Sall4 siRNA and PTEN siRNA or Bmi-1 expressed vector, nonspecific siRNA was used as NC. 100× magnification. (C) Western blot analysis for E-cadherin, fibronectin, vimentin, and N-cadherin expression after indicated treatment in ICC-9810 cells. Data are mean ± SD from three independent experiments. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: ICC, intrahepatic cholangiocarcinoma; mRNA, messenger RNA; NC, negative control; OD, optical density; SD, standard deviation; siRNA, small interfering RNA.
Discussion
ICC, a fatal primary liver cancer, arises from the peripheral intrahepatic bile duct epithelium.12 Although ICC incidences only account for 5%–10% of all primary liver cancers in the Western and Asian countries, it has been increasing steadily and substantially over the past decades and has a poor prognosis and high mortality rate because of lack of appropriate markers for early detection.13 Thus, to achieve earlier and more accurate diagnosis for better therapeutic outcome, knowledge of the mechanisms that trigger the tumorigenesis of ICC is essential.
The oncofetal protein Sall4 has been shown to play an important role in hepatocarcinogenesis and acts as a highly sensitive and specific marker for primary and metastatic gonadal and extragonadal yolk sac tumor.14,15 In our previous study, we found that Sall4 had a higher expression in ICC cases than in adjacent cancer tissues, which is associated with a worse prognosis; and Sall4 expression in ICC was significantly correlated with vascular and nerve invasion, and lymph node metastasis.8 These findings indicate that Sall4 is an indicator of aggressiveness and poor prognosis in ICC.
We analyzed the correlation between PTEN and Sall4 in ICC tissues by immunohistochemistry and found that PTEN expression was lower in ICC cases than in adjacent cancer tissues. Moreover, PTEN expression was negatively correlated with Sall4 expression. Importantly, ICC patients with low PTEN exhibited a worse prognosis than ICC patients with high PTEN. PTEN is a tumor suppressor that acts by inactivating PI3K/AKT signaling pathways. Loss of functional PTEN was found in many solid tumors, which led to increased activity of AKT and promoted both cell survival and proliferation through phosphorylation and inactivation of multiple downstream mediators.16 PI3K and AKT have a central role in the regulation of diverse biological functions, including cell metabolism, protein synthesis, cell survival, and inhibition of apoptosis.17 The upregulation of phosphorylated PI3K and AKT has been involved in tumorigenesis and metastasis in many different types of tumors.18 We previously observed that Sall4 siRNA transfection inhibited cell proliferation in ICC cancer cells.8 Here, we found that knockdown of Sall4 increased PTEN expression, and subsequently resulted in inactivation of PI3K and AKT. Moreover, Sall4 reduced the activity of PTEN promoter in a dose-dependent manner. Thus, combined with our recent results, it is reasonable that knockdown of Sall4 inhibits ICC tumor growth via upregulation of PTEN and inactivation of PI3K/AKT signaling.
In contrast to PTEN, we found that Bmi-1 expression was higher in ICC cases than in adjacent cancer tissues, which was positively correlated with Sall4 expression, which was also found in placental tissue and umbilical cord blood. Patients with lower Bmi-1 expression showed a better prognosis than ICC patients with high Bmi-1 expression. Bmi-1 gene was initially isolated as an oncogene in a retroviral-induced B- and T-cell leukemia, and it plays an essential role in leukemia stem cells.19,20 Bmi-1 deletion can inhibit tumor stem cell self-renewal and prevent leukemic recurrence.21 In addition, overexpression of Bmi-1 was found in many cancers and was used as an important marker for predicting the progression to acute myeloid leukemia,22 consistent with our recent findings in ICC. Upregulation of Sall4 is able to increase the levels of histone H3–K4 and H3–K79 methylation in the Bmi-1 promoter in leukemic cells.11 We also found that knockdown of Sall4 directly increased Bmi-1 expression by activating the promoter activity of Bmi-1 in ICC cells, indicating that Bmi-1 was a direct Sall4 target gene in several types of cancers. In addition, Bmi-1 can decrease GSK3β, the component of Axin/GSK3β/APC complex, to prevent the degradation of β-catenin, leading to activation of Wnt/β-catenin signaling in breast cancer.23,24 Interestingly, β-catenin is inactivated as a substrate of AKT.24 Thus, there is a crosstalk between Bmi-1/Wnt/β-catenin and PTEN/PI3K/AKT signaling, which are simultaneously regulated by Sall4. Sall4 promotes ICC cell proliferation, and progression of ICC might be via increasing Bmi-1 expression and Wnt/β-catenin signaling.
ICC is a highly malignant neoplasm, which is associated with EMT characterized by loss of epithelial phenotype and acquisition of a mesenchymal phenotype.25 During the process of EMT, loss of E-cadherin expression (the epithelial marker), gain of vimentin, fibronectin, and N-cadherin (the mesenchymal markers), and abnormal nuclear location of β-catenin are observed.26 β-catenin not only plays an important role in Wnt signaling pathways but also contributes to E-cadherin-mediated cell–cell adhesion and directly induces EMT.27 It was reported that reduced E-cadherin and increased vimentin and N-cadherin expression were correlated with lymphatic metastasis and poorer overall survival and disease-free survival of ICC, and β-catenin with vimentin or fibronectin was found to show an association with lymph node metastasis in ICC.28,29 In this study, we found that knockdown of Sall4 was able to increase E-cadherin expression and reduce the expression of vimentin, fibronectin, and N-cadherin, indicating the inhibition of EMT process. However, knockdown of PTEN or overexpression of Bmi-1 reversed downregulation of Sall4-mediated EMT inhibition, suggesting that Sall4 might enhance EMT process via PTEN and Bmi-1 signaling.
In conclusion, we demonstrate that Sall4, PTEN, and Bmi-1 expression is closely associated with tumor progression and prognosis in ICC and knockdown of Sall4 inhibits malignant phenotypes of ICC through regulating PTEN/PI3K/Akt and Wnt/β-catenin signaling and repressing the EMT process. Thus, Sall4 may be a potential target for the treatment of ICC metastasis.
Acknowledgments
This work was supported by the Natural Science Foundation of Hunan Province, People’s Republic of China (Grant number 2015JJ4057) and the Fundamental Research Funds for Central Universities of Central South University (Grant number 2016zzts160).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zhang H, Yang T, Wu M, Shen F. Intrahepatic cholangiocarcinoma: epidemiology, risk factors, diagnosis and surgical management. Cancer Lett. 2015 Sep 25; doi: 10.1016/j.canlet.2015.09.008. Epub. [DOI] [PubMed] [Google Scholar]
- 2.Yang L, Shan J, Shan L, Saxena A, Bester L, Morris DL. Trans-arterial embolisation therapies for unresectable intrahepatic cholangiocarcinoma: a systematic review. J Gastrointest Oncol. 2015;6(5):570–588. doi: 10.3978/j.issn.2078-6891.2015.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cillo U, Spolverato G, Vitale A, et al. Liver resection for advanced intrahepatic cholangiocarcinoma: a cost-utility analysis. World J Surg. 2015;39(10):2500–2509. doi: 10.1007/s00268-015-3150-1. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Yuan X, Zhu W, Qian H, Xu W. SALL4: an emerging cancer biomarker and target. Cancer Lett. 2015;357(1):55–62. doi: 10.1016/j.canlet.2014.11.037. [DOI] [PubMed] [Google Scholar]
- 5.Yong KJ, Chai L, Tenen DG. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 2013;369(12):1171–1172. doi: 10.1056/NEJMc1308785. [DOI] [PubMed] [Google Scholar]
- 6.Yue X, Xiao L, Yang Y, et al. High cytoplasmic expression of SALL4 predicts a malignant phenotype and poor prognosis of breast invasive ductal carcinoma. Neoplasma. 2015;62(6):980–988. doi: 10.4149/neo_2015_119. [DOI] [PubMed] [Google Scholar]
- 7.Li A, Jiao Y, Yong KJ, et al. SALL4 is a new target in endometrial cancer. Oncogene. 2015;34(1):63–72. doi: 10.1038/onc.2013.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng G, Zhu L, Huang F, et al. SALL4 is a novel therapeutic target in intrahepatic cholangiocarcinoma. Oncotarget. 2015;6(29):27416–27426. doi: 10.18632/oncotarget.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Jeong HW, Kong N, et al. Stem cell factor SALL4 represses the transcriptions of PTEN and SALL1 through an epigenetic repressor complex. PLoS One. 2009;4(5):e5577. doi: 10.1371/journal.pone.0005577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen Q, Liu S, Hu J, et al. The differential expression pattern of the BMI-1, SALL4 and ABCA3 genes in myeloid leukemia. Cancer Cell Int. 2012;12(1):42. doi: 10.1186/1475-2867-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Chai L, Liu F, et al. Bmi-1 is a target gene for SALL4 in hematopoietic and leukemic cells. Proc Natl Acad Sci U S A. 2007;104(25):10494–10499. doi: 10.1073/pnas.0704001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lafaro KJ, Cosgrove D, Geschwind JF, Kamel I, Herman JM, Pawlik TM. Multidisciplinary care of patients with intrahepatic cholangiocarcinoma: updates in management. Gastroenterol Res Pract. 2015;2015:860861. doi: 10.1155/2015/860861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Adra DP, Gill RS, Axford SJ, Shi X, Kneteman N, Liau SS. Treatment of unresectable intrahepatic cholangiocarcinoma with yttrium-90 radioembolization: a systematic review and pooled analysis. Eur J Surg Oncol. 2015;41(1):120–127. doi: 10.1016/j.ejso.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yong KJ, Gao C, Lim JS, et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 2013;368(24):2266–2276. doi: 10.1056/NEJMoa1300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Liu A, Peng Y, et al. Diagnostic utility of SALL4 in extragonadal yolk sac tumors: an immunohistochemical study of 59 cases with comparison to placental-like alkaline phosphatase, alpha-fetoprotein, and glypican-3. Am J Surg Pathol. 2009;33(10):1529–1539. doi: 10.1097/PAS.0b013e3181ad25d5. [DOI] [PubMed] [Google Scholar]
- 16.Yothaisong S, Dokduang H, Techasen A, et al. Increased activation of PI3K/AKT signaling pathway is associated with cholangiocarcinoma metastasis and PI3K/mTOR inhibition presents a possible therapeutic strategy. Tumour Biol. 2013;34(6):3637–3648. doi: 10.1007/s13277-013-0945-2. [DOI] [PubMed] [Google Scholar]
- 17.Lee D, Do IG, Choi K, et al. The expression of phospho-AKT1 and phospho-MTOR is associated with a favorable prognosis independent of PTEN expression in intrahepatic cholangiocarcinomas. Mod Pathol. 2012;25(1):131–139. doi: 10.1038/modpathol.2011.133. [DOI] [PubMed] [Google Scholar]
- 18.Chen MH, Chiang KC, Cheng CT, et al. Antitumor activity of the combination of an HSP90 inhibitor and a PI3K/mTOR dual inhibitor against cholangiocarcinoma. Oncotarget. 2014;5(9):2372–2389. doi: 10.18632/oncotarget.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy LS, Lobelle-Rich PA, Overbaugh J. flvi-2, a target of retroviral insertional mutagenesis in feline thymic lymphosarcomas, encodes bmi-1. Oncogene. 1993;8(7):1833–1838. [PubMed] [Google Scholar]
- 20.Yong AS, Stephens N, Weber G, et al. Improved outcome following allogeneic stem cell transplantation in chronic myeloid leukemia is associated with higher expression of BMI-1 and immune responses to BMI-1 protein. Leukemia. 2011;25(4):629–637. doi: 10.1038/leu.2010.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishida Y, Maeda A, Chachad D, et al. Preclinical activity of the novel BMI-1 inhibitor PTC-209 in acute myeloid leukemia: implications for leukemia therapy. Cancer Sci. 2015;106(12):1705–1713. doi: 10.1111/cas.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawa M, Yamamoto K, Yokozawa T, et al. BMI-1 is highly expressed in M0-subtype acute myeloid leukemia. Int J Hematol. 2005;82(1):42–47. doi: 10.1532/IJH97.05013. [DOI] [PubMed] [Google Scholar]
- 23.Song LB, Li J, Liao WT, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest. 2009;119(12):3626–3636. doi: 10.1172/JCI39374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conde-Perez A, Gros G, Longvert C, et al. A caveolin-dependent and PI3K/AKT-independent role of PTEN in beta-catenin transcriptional activity. Nat Commun. 2015;6:8093. doi: 10.1038/ncomms9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang XY, Zhang C, Cai JB, et al. Comprehensive multiple molecular profile of epithelial mesenchymal transition in intrahepatic cholangiocarcinoma patients. PLoS One. 2014;9(5):e96860. doi: 10.1371/journal.pone.0096860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oishi N, Kumar MR, Roessler S, et al. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology. 2012;56(5):1792–1803. doi: 10.1002/hep.25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto K, Tajima H, Nakanuma S, et al. Angiotensin II enhances epithelial-to-mesenchymal transition through the interaction between activated hepatic stellate cells and the stromal cell-derived factor-1/CXCR4 axis in intrahepatic cholangiocarcinoma. Int J Oncol. 2012;41(2):573–582. doi: 10.3892/ijo.2012.1499. [DOI] [PubMed] [Google Scholar]
- 28.Yao X, Wang X, Wang Z, et al. Clinicopathological and prognostic significance of epithelial mesenchymal transition-related protein expression in intrahepatic cholangiocarcinoma. Onco Targets Ther. 2012;5:255–261. doi: 10.2147/OTT.S36213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu MJ, Choi JH. Epithelial-mesenchymal transition phenotypes are associated with patient survival in intrahepatic cholangiocarcinoma. J Clin Pathol. 2014;67(3):229–234. doi: 10.1136/jclinpath-2013-201806. [DOI] [PubMed] [Google Scholar]