Abstract
The Community Structure-Activity Resource (CSAR) benchmark exercise provides a unique opportunity for researchers to objectively evaluate the performance of protein-ligand docking methods. Patch-Surfer and PL-PatchSurfer, molecular surface-based methods for predicting binding ligands of proteins developed in our group, were tested on both CSAR 2013 and 2014 benchmark exercises in combination with an empirical scoring function-based method, AutoDock, while we only participated in CSAR 2013 using Patch-Surfer. The prediction results for Phase 1 task in CSAR 2013 showed that Patch-Surfer was able to rank all the four designed binding proteins within top ranks, outperforming AutoDock Vina. In Phase 2 of 2013, PL-PatchSurfer correctly selected the correct ligand pose for two target proteins. PL-PatchSurfer performed reasonably well in ranking ligands according to their binding affinity and in selecting near-native ligand poses in 2013 Phase 3 and 2014 Phase 1, respectively, although AutoDock Vina showed better performance. Lastly, in the 2014 Phase 2 exercise, the PL-PatchSurfer scores computed for ligands to target protein pairs correlated well with their pIC50 values, which was better or comparable to results by other participants. Overall, our methods showed fairly good performance in CSAR 2013 and 2014. Unique characteristics of the methods are discussed in comparison with AutoDock.
Keywords: Patch-Surfer, PL-PatchSurfer, 3D Zernike Descriptor, AutoDock Vina, docking, CSAR, score function, virtual screening, drug design, protein-ligand interaction
1. INTRODUCTION
Substantial progress has been made in the past two decades in developing virtual screening methods; however, developing accurate scoring functions for evaluating binding energy of ligand-protein interaction is still a challenging problem1, 2. A scoring function is aimed at not only identifying the correct docking pose of a ligand, but also differentiating the binding affinity between different small molecules.
Scoring functions can be classified into three different categories: molecular force fields3–7, statistical knowledge-based scoring functions8–12, and empirical scoring functions13–16. Force field scoring functions calculate potential energy between a protein and a small molecule, which usually contain several energy terms derived from the first principles of physics, such as van der Waals, electrostatic, and hydrogen-bond interactions. Solvent effects are often included in an energy term as well. In contrast, statistical knowledge-based scoring functions are derived from the frequency of observed interacting atomic pairs and other structural features in a database of known protein-ligand complexes. Using the Boltzmann relationship, the observed frequency can be used to compute the energy of the structural feature17. Knowledge-based scoring functions do not provide individual energetic contributions to protein-ligand interaction, but they provide an efficient and practical way of calculating the binding affinity of ligands. Empirical scoring functions, the last category, combine force-field-based energy terms, knowledge-based terms, and other physically meaningful terms. Typically, weighting factors associated to different energy terms are calibrated by training on a set of known protein-ligand complexes with known binding affinity.
Since there are many different kinds of scoring functions but none are sufficiently accurate and while remaining efficient, it is important for the community to have objective benchmarks to validate and compare existing methods. In the past years, Dr. Heather Carlson and her team at the University of Michigan have been leading an effort of providing experimental datasets of crystal structures and binding affinities for diverse protein-ligand complexes, which are referred to as CSAR (Community Structure-Activity Resource, http://www.csardock.org/)18–21. The 2013 CSAR benchmark exercise included selecting artificially designed proteins that bind to a ligand molecule, which made the exercise more interesting for the community. The 2014 CSAR exercise was to predict correct poses from sets of docking decoys and to rank-order compounds.
We participated and submitted predictions in CSAR 2013 using two methods of different types, Patch-Surfer22, 23 and AutoDock programs, AutoDock424 and AutoDock Vina25. Patch-Surfer, which is developed in our group, makes binding ligand prediction for a target pocket by searching similar known ligand binding pockets. The method uses a local surface patch representation of binding pockets, which facilitates correct identification of local surface similarity and increases the search speed. In this article, we further extend our submitted predictions by applying our newly developed protein-ligand virtual screening method called PL-PatchSurfer26–28, which predicts binding ligands for a query binding pocket by directly evaluating complementarity between the protein pocket and ligands. PL-PatchSurfer was benchmarked in CSAR2013 Phase 2, Phase 3, as well as CSAR2014 Phase 1 and 2.
Patch-Surfer performed very well in Phase 1 of CSAR 2013, being able to rank all four designed binding proteins that bind to a target ligand within top ranks. This performance was better than AutoDock Vina. In Phase 2 of 2013, PL-PatchSurfer correctly selected the correct ligand pose for two target proteins. PL-PatchSurfer performed reasonably well in ranking ligands according to their binding affinity in 2013 Phase 3 and in selecting near-native ligand docking pose in 2014 Phase 1, although AutoDock Vina showed better performance. In the 2014 Phase 2 exercise, the PL-PatchSurfer scores computed for ligands to target protein pairs correlated well with their pIC50 values, which was better or comparable to results by other participants.
Although Patch-Surfer and PL-PatchSurfer employ rather coarse-grained molecular surface-based representations of binding pockets and ligands, which are very different from conventional virtual screening methods, they showed fairly good performance in CSAR 2013 and 2014. At the same time, comparison with performance of AutoDock and results with other participants revealed weakness of the methods. Unique strengths of the Patch-Surfer and PL-PatchSurfer as well as weakness identified through the CSAR exercise are discussed.
2. METHODS
2.1 Data sets
CSAR 2013 was based on the experimental data of artificially designed ligand-binding proteins by the David Baker’s group of University of Washington29. In the first Phase, the organizers provided sequences of 16 designed proteins and a ligand molecule, a derivative of steroid digoxigenin. Its chemical structure in the SMILES representation is [C(C(=O)NCCC)O[C@H]1CC[C@]2([C@@H](C1)CC[C@@H]1[C@@H]2C[C@@H](O)[C@]2([C@]1(O)CC[C@@H]2[C@@H]1COC(=O)C1)C)C″)]. The participants were asked to predict which proteins bind to the ligand and also to rank the binding ability of the designed proteins. In the second phase, the organizers provided the structures of two proteins and a set of pre-generated docking ligand decoys and the participants were asked to score the provided poses and rank them. These two proteins were designed and produced by the Baker group from a putative isomerase (PDB ID: 1Z1S)29. In the third phase, the organizers provided one protein structure, which is one of the two designed proteins whose crystal structures have been solved by the Baker group (PDB ID: 4J9A. This PDB ID was reported after the 2013 CSAR exercise), and ten potential inhibitors. Participants were asked to predict the relative binding affinity of the ten different inhibitors to the protein and predict the best three poses of each ligand.
Although we did not participate in CSAR 2014 at the time of the exercise, in this work we tested PL-PatchSurfer on the benchmark datasets provided in its two phases. For Phase 1, similar to Phase 2 of CSAR 2013, the organizers provided pre-generated 200 docking poses for 22 protein-ligand complexes. The target proteins were coagulation factor Xa (FXa) (three datasets each with a different ligand molecule), Spleen tyrosine kinase (SYK) (five sets), and tRNA-methyltransferase (TRMD) (fourteen sets). The participants were asked to find the nearest native pose to the crystal structure. Phase 2 of CSAR 2014 was a ligand ranking problem of given congeneric ligand sets for the three proteins in Phase 1. The five ligand sets were given to the participants, three for FXa, one for SYK, and one for TRMD, and each ligand set consisted of 31–276 ligands in the SMILES string format.
2.2 Patch-Surfer
Patch-Surfer is a program for predicting a binding ligand for a query protein by comparing the shape and physicochemical properties of a potential binding pocket of the query to known pockets in a database22, 23. In Patch-Surfer, the pocket surface is represented by a set of overlapping local surface patches. A surface patch is characterized by four features: geometric shape, electrostatic potential, hydrophobicity, and visibility (concavity), each of which is described by three dimensional Zernike Descriptors (3DZD), which is a mathematical series expansion of a 3D function. Here, we briefly explain 3DZD. For more details, refer to the original papers30, 31. To describe a surface with 3DZD, a surface patch is considered as a three dimensional (3D) function, f(x), in the 3D space. To represent the geometric shape of a surface patch, the surface is mapped on a 3D grid, where 1s are placed for positions that are occupied by the surface and 0 otherwise. For the other properties, a 3D grid holds the property’s value at each position. The 3D grid with mapped values is considered as a 3D function, f(x). The function can be expanded into a series in terms of the Zernike-Canterakis basis:
| (1) |
where
| (2) |
In this Zernike-Canterakis basis, Rnl(r) is the radial function and is the spherical harmonics. m and l are integers that have ranges −1 < m < 1 and 0 ≤ 1 ≤ n. are called 3D Zernike moments and the 3DZD, Fnl, are calculated as norms of vectors Ωnl as shown in Equation 3. The norm gives rotational invariance to the descriptor.
| (3) |
To compare two pockets, similar patches from the two pockets are matched and a similarity score is computed, which reflects the similarity of the features of matched patches and the relative positions of corresponding patches in each pocket.
Patch-Surfer was originally designed to compare the similarity between two protein pockets. In Phase 1 of CSAR 2013 benchmark, we used Patch-Surfer in its original aim of comparing pockets. Two scoring terms, one for considering geometric shape similarity and another for comparing visibility (concavity) were used. In Phases 2 and 3, we modified the program so that it can compare the surface of a binding pocket with molecular surface of a small ligand molecule. For pocket-to-ligand comparison, we used scoring terms for shape and the electrostatic potential to quantify complementarity of the two properties of a pocket and a ligand molecule.
2.3. PL-PatchSurfer
We ran PL-PatchSurfer for the benchmark datasets of Phase 2 and 3 of CSAR2013 and Phase 1 and 2 of CSAR2014. PL-PatchSurfer searches complementarity between a receptor pocket and a ligand by surface-patch comparison between the molecules represented by 3DZD. Thus, while Patch-Surfer compares a query pocket against known ligand binding pockets, PL-PatchSurfer compares a query pocket to ligands. Complementarity of a pocket and a ligand surface patch pair is evaluated in terms of shape, electrostatic potential, hydrogen-bond donor and acceptor positions, hydrophobicity, and the relative position of the patch in the molecule. In addition to the five features, the overall score of a pocket and a ligand considers similarity of relative position of corresponding patches in each molecule. These terms were combined with weighting factors that were trained to maximize accuracy of virtual screening tests. To score given a ligand for a target pocket, a maximum of 50 3D structures of the ligand were generated from its SMILE representation using OMEGA32. Then, surface of each conformation of the ligands was generated and converted into 3DZD. The score of a ligand for a target protein was defined as the maximum score among the scores computed for all the conformations of the ligand. For more details, refer to the original paper26.
2.4 AutoDock programs
We used two versions of the AutoDock program, AutoDock424 and its subsequent version, Autdock Vina25. Although both of the programs were developed by the same group, the scoring functions and sampling methods are different. AutoDock4 uses Lamarkian Genetic algorithm and force-field based scoring function composed of van der Waals, Coulombic interaction, hydrogen bonding, solvation, and torsional entropy terms24, while AutoDock Vina uses Local Search and empirical scoring function with steric, hydrogen bonding, hydrophobic, and torsional entropy terms25. Weight parameters of both scoring functions that associate with these terms were calibrated using a set of protein-ligand complexes with known binding affinities. Although the same types of scoring terms were considered by the two programs, they have different implementations and thus performance can be different25. To run AutoDock4 and AutoDock Vina, input files of a target protein and a ligand were prepared with AutoDockTools (ADT) tools and Python scripts named prepare_ligand4.py and prepare_receptor4.py, which are associated with the AutoDock program. The binding pocket position in target protein was specified with the ADT molecular viewer. The parameters were kept at their default values.
For Phase 2 and Phase 3 in CSAR2013, we combined the scores from AutoDock Vina and Patch-Surfer, considering the complementary nature between the two. Because the scales of the two scores are different, we calculated the Z-score of each score and summed the two Z-scores as the final score.
3. RESULTS AND DISCUSSION
3.1 Phase 1 Results
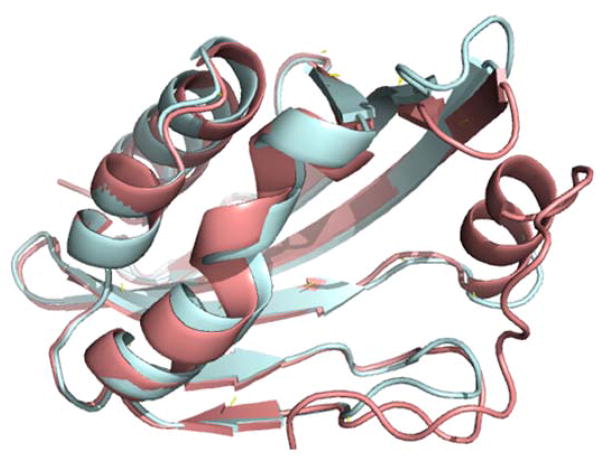
The task of the Phase 1 of the CSAR 2013 exercise was to identify proteins out of 16 artificially designed proteins that bind to a derivative of the steroid digoxigenin (SMILES of this molecule provided in the Dataset section). Since only amino acid sequences of the 16 proteins were provided, we needed to model 3D structures of the proteins. The modeling was performed using threading web servers, HHPred32 and LOMETS33, both of which build a structure model of a query protein based on a known protein structure that is used as a template. LOMETS takes a meta-server approach, which runs 10 independent prediction programs. Thus, by adding HHPred, we had 11 independent predictions for each target protein. Among the 11 structure models, we have selected the one which was built based on a template protein selected by the majority of the programs. The left side of Table 1 shows the templates identified by this procedure for building the target proteins and the sequence identity between each template to its target protein. The structure models were expected to be sufficiently accurate for the subsequent docking prediction because the sequence identities were all very high. Indeed, RMSD of Cα atoms of the model structure for DIG19 to its crystal structure that was revealed after Phase 1 experiment (PDB ID: 4J9A) was 0.602 Å (Fig. 1).
Table 1.
Template proteins used for building the target proteins in 2013 Phase 1 exercise.
| Target ID | High Sequence Identity Template a) | Low Sequence Identity Template b) | ||
|---|---|---|---|---|
| PDB ID a) | Sequence Identity (%) | PDB ID | Sequence Identity (%) | |
| DIG1 | 1GY7B | 86.9 | 1ZO2A | 39.3 |
| DIG2 | 1MVEA | 89.9 | 1CPNA | 27.5 |
| DIG3 | 1YNAA | 81.3 | 3AKQA | 54.7 |
| DIG4 | 3JUMB | 87.9 | 3FF0A | 48.3 |
| DIG5 | 1Z1SA | 91.5 | 1S5AA | 36.1 |
| DIG6 | 3CU3A | 80.0 | 4I4KA | 25.2 |
| DIG7 | 3GWRB | 86.2 | 3CNXA | 24.2 |
| DIG8 | 3HK4A | 78.1 | 5AIGA | 23.9 |
| DIG9 | 1I60A | 88.2 | 2ZVRA | 22.1 |
| DIG10 | 1Z1SA | 92.2 | 1S5AA | 35.3 |
| DIG12 | 2OWPA | 88.8 | 2RCDA | 46.5 |
| DIG13 | 2OX1A | 90.8 | 4CNNA | 19.6 |
| DIG14 | 3E5ZA | 92.0 | 3DR2A | 28.3 |
| DIG17 | 3CU3A | 85.0 | 4I4KA | 24.6 |
| DIG18 | 1Z1SA | 89.9 | 1S5AA | 35.3 |
| DIG19 | 1Z1SA | 86.0 | 1S5AA | 33.6 |
Templates with a high sequence identity to the target proteins.
Templates with a low sequence identity to the targets. The models were used in the post-analyses to investigate how the quality of homology models affects to the predicting ligand-binding proteins.
Figure 1.
Superposition between the model and the crystal structure of DIG19 (PDBID: 4J9A). The crystal structure is showed in cyan and the modeled structure is shown in pink. The RMSD between the model and the crystal structure is 0.602 Å.
For further post-analysis, we have also constructed a model for each target protein using a template structure that has a lower sequence identity to the target (the right columns in Table 1). Later in this section we investigate how the model quality affects the accuracy of selecting targets that bind to the ligand.
We applied Patch-Surfer to the homology models to determine which proteins have a binding pocket that is the most similar to known steroid binding pockets. The reference steroid binding pockets were identified by keyword searches on the PDB website. Two entries of steroid binding proteins were found, 1HDC (20β-hydroxysteorid dehydrogenase) and 3UP0 (nuclear receptor DAF-12). 1HDC has bound carbenoxolone (PDB ID: CBO) in the crystal structure while 3UP0 has bound (5β, 14β, 17α, 25S)-3-oxocholest-7-en-26-oic acid (PDB ID: D7S). Among these two PDB entries, we decided to use 1HDC as the reference because its ligand, CBO, has 5 ring structures, which is consistent with the target ligand.
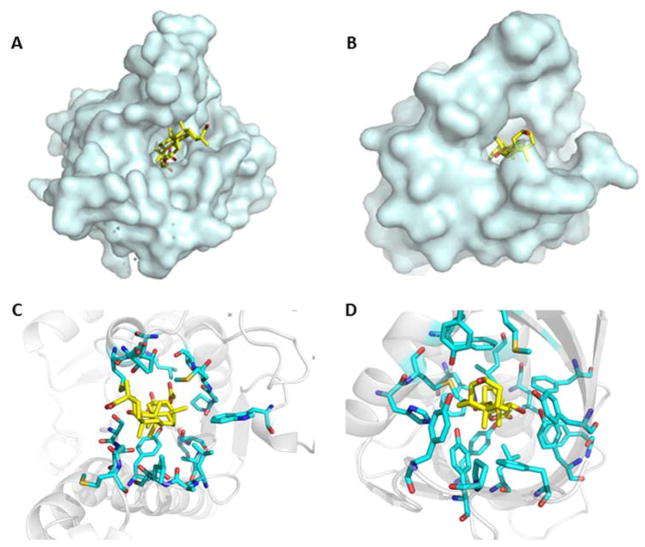
Table 2 shows the rank of the 16 proteins based on the Patch-Surfer score, which quantifies the similarity of the pockets in the targets and the steroid binding pocket of 1HDC. Since the Patch-Surfer score is meaningful in ranking proteins relative to each other but does not provide an absolute indication of ligand binding, we decided to submit the eight top-ranked target proteins as binders. They are, DIG5, DIG8, DIG19, DIG10, DIG18, DIG9, DIG2, and DIG3. The Patch-Surfer’s prediction was very successful in ranking all three positive proteins, whose binding affinity to DOG was confirmed by experiments to be in the μM range, within the top four ranks. Shown in Figure 2 are the crystal structures of 1HDC and the designed protein (DIG19) with bound ligands, which show that the ligands have similar binding mode in the two pockets. In both proteins, the ligands bind vertically in the pockets and the hydrophobic ring structures in the ligands (the picene ring and the phenanthren ring for the ligands of 1hdc and DIG19, respectively) bind to a hydrophobic core of the pockets.
Table 2.
Rank of predicted binding affinity of the target designed proteins using Patch-Surfer.
| Rank | Target protein | Patch-Surfer Score a) | Binding or not |
|---|---|---|---|
| 1 | DIG5 | 0.592 | Yes (205μM) |
| 2 | DIG8 | 0.653 | No |
| 3 | DIG19 | 0.684 | Yes (541 pM) |
| 4 | DIG10 | 0.686 | Yes (8.9 μM) |
| 5 | DIG18 | 0.688 | Yes b) |
| 6 | DIG9 | 0.702 | No |
| 7 | DIG2 | 0.711 | No |
| 8 | DIG3 | 0.723 | No |
| 9 | DIG13 | 0.724 | No |
| 10 | DIG6 | 0.729 | No |
| 11 | DIG7 | 0.752 | No |
| 12 | DIG4 | 0.785 | No |
| 13 | DIG12 | 0.789 | No |
| 14 | DIG17 | 0.802 | No |
| 15 | DIG14 | 0.847 | No |
| 16 | DIG1 | 0.963 | No |
Eight targets in bold (DIG5 to DIG3 in the table) were predicted to bind the target ligand.
A small Patch-Surfer score indicates that the putative binding pocket is similar to the reference steroid binding pocket.
The binding affinity of this protein is not available because it was not reported in the paper by Tinberg et al. 29.
Figure 2.
The crystal structures of 1HDC and the designed protein DIG19 (PDB ID: 4J9A). Ligands are shown in yellow. The ligands of 1HDC and DIG19 are carbenoxolone and digoxigenin, respectively. A, B: the overall structure of 1HDC and DIG19, respectively. C, D: Binding sites of 1HDC and DIG19, respectively.
For Phase 1 of CSAR 2013 exercise, there were 16 predictions submitted. Among them, only four of them, including Patch-Surfer, correctly ranked all four binding proteins within top 5 ranks. Thus, Patch-Surfer was successful relative to the other participants in the benchmark exercise.
Besides the prediction by Patch-Surfer (Table 2), we also submitted a separate prediction in parallel that used a combined score of Patch-Surfer and four modes of AutoDock. Three scores are computed from AutoDock: one by flexible docking by AutoDock Vina, which explores various conformations of a ligand that give the lowest energy, another score by the Vina rigid docking mode, which treats a ligand as a rigid molecule, and lastly a score by the AutoDock4 flexible docking mode. With Patch-Surfer, we computed two scores, the pocket similarity score for the binding pocket of each target and either of 1HDC or 3UP0. Thus, in total, we had five different scores. These five scores were then normalized by computing a Z-score, and the sum of the five Z-scores was used to obtain the final rank of the target proteins (Table 3). It turned out that this prediction was worse than the Patch-Surfer’s prediction in Table 2, not being able to select the best binder, DIG19, within top half of the targets.
Table 3.
Rank of predicted binding affinity of the designed proteins using the combined score.
| Rank | Target protein | Total Z-score | Binding or not |
|---|---|---|---|
| 1 | DIG5 | −4.21 | Yes (205uM) |
| 2 | DIG18 | −2.71 | Yes |
| 3 | DIG10 | −2.61 | Yes (8.9 uM) |
| 4 | DIG2 | −2.30 | No |
| 5 | DIG3 | −2.21 | No |
| 6 | DIG8 | −1.95 | No |
| 7 | DIG4 | −1.47 | No |
| 8 | DIG17 | −1.27 | No |
| 9 | DIG6 | −0.83 | No |
| 10 | DIG13 | −0.65 | No |
| 11 | DIG14 | −0.59 | No |
| 12 | DIG19 | 0.48 | Yes (541pM) |
| 13 | DIG9 | 2.03 | No |
| 14 | DIG7 | 2.51 | No |
| 15 | DIG12 | 2.93 | No |
| 16 | DIG1 | 12.87 | No |
Target proteins in bold (the top eight targets) were predicted to bind to the target ligand.
After CSAR2013, we extended the analysis in two directions to further understand the performance of Patch-Surfer. First, we made homology models of the designed proteins using templates with a lower sequence identity and examined how prediction results are affected by the quality of the models. Second, we used a variety of ligand binding pockets as the reference to investigate how they influence prediction results.
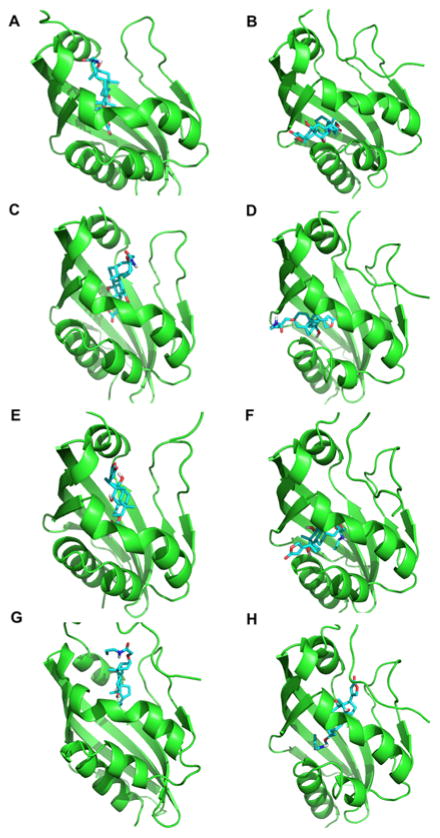
The right columns in Table 1 show template structures with a lower sequence identity used to build homology models for the first part of the extended analysis. These templates were identified by HHpred and the models were generated using MODELLER34 based on the templates. The structural difference of the models with close and distant templates is not large, on average, RMSD between them was 2.17 Å (Table 4, the rightmost column). The RMSD is even smaller, 0.98 Å, for the models of ligand binding proteins, DIG5, DIG10, DIG18, and DIG19. However, this difference of models made substantial difference in the prediction results (Table 4). Among the four ligand binding proteins, only DIG5 was ranked within the top while the other three proteins were below the half of the rank. By performing docking the target ligand, the derivative of digoxigenin, to the structure models, it turned out that this difference was enough to make very different binding modes of the ligand to the proteins (Fig. 3). The models built with high sequence identity templates (Fig. 3A, C, E, G) have one larger pocket that is consistent with the reference steroid binding protein, 1HDC. On the other hand, the corresponding pockets in the models built on lower sequence identity templates are smaller, which caused the positions of bound ligand the other side of the helix in the middle of the structures. These results suggest that, when computational protein models are used in ligand-binding prediction, their quality is critical to prediction results.
Table 4.
Rank of predicted binding affinity of the target designed proteins using structure models with a lower quality.
| Rank | Target protein | Patch-Surfer Score | Binding or not | RMSD (Å) a) |
|---|---|---|---|---|
| 1 | DIG5 | 0.592 | Yes (205uM) | 1.02 |
| 2 | DIG13 | 0.614 | No | 3.31 |
| 3 | DIG14 | 0.626 | No | 0.86 |
| 4 | DIG9 | 0.645 | No | 2.80 |
| 5 | DIG6 | 0.657 | No | 4.83 |
| 6 | DIG4 | 0.675 | No | 0.36 |
| 7 | DIG7 | 0.680 | No | 1.86 |
| 8 | DIG8 | 0.688 | No | 3.35 |
| 9 | DIG18 | 0.689 | Yes | 0.93 |
| 10 | DIG2 | 0.696 | No | 3.83 |
| 11 | DIG3 | 0.734 | No | 0.60 |
| 12 | DIG10 | 0.742 | Yes (8.9uM) | 1.00 |
| 13 | DIG12 | 0.742 | No | 0.48 |
| 14 | DIG17 | 0.767 | No | 7.67 |
| 15 | DIG19 | 0.768 | Yes (541pM) | 0.97 |
| 16 | DIG1 | 0.988 | No | 0.91 |
Figure 3.
Predicted binding mode using models built on high and low sequence identity templates. At each row, the structure on the left/right is a model based on the high/low sequence identity template. The target ligand was docked using the AutoDock Vina in the flexible docking mode. The lowest energy structures are shown. A, B: DIG5; C, D: DIG10; E, F: DIG18; G, H: DIG19.
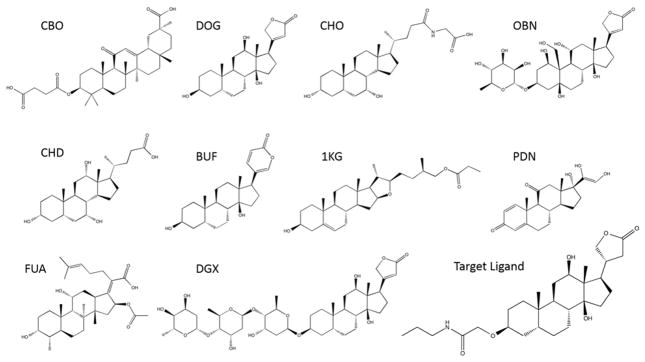
For the second post-analysis, we examined how different reference pockets affect the Patch-Surfer results. In addition to 1HDC we originally used as the reference pocket, we newly selected nine more protein-ligand complexes as the references. These additional compounds were identified by the SIMCOMP webserver35 using the target ligand as the query molecule. Using Patch-Surfer, a reference pocket was compared with binding pockets of the structure models of the 16 target proteins built based on the high sequence identity templates, and the target proteins were ranked according to the Patch-Surfer pocket similarity scores. The nine binding pockets and their bound ligands are listed in Table 5 with five distance (i.e. dissimilarity) scores to the target ligand. Their two-dimensional (2D) structures are shown in Figure 4. All of the compounds but CBO share the four ring structure, cyclopenta[a]phenanthrene, with the target ligand (bottom right corner in Figure 4). The five compound distances measure different aspects of compounds, and thus their distances are not necessarily consistent. Zernike (3D Zernike descriptors) compares global surface shape of molecules28, 31, 36. SIMCOMP evaluates 2D graph similarity of molecules35. LIGSIFT compares molecules by overlapping Gaussian distributions that represent global shape of the molecules. We used two options of LIGSIFT, one that consider the shape only (LIGSIFT_SHP) and the other that also consider chemical nature of molecules (LIGSIFT_CHEM)37. The last one is the Tanimoto coefficient computed with the Open Babel software38, which indicates the fraction of common fingerprints of molecules.
Table 5.
Distance (dissimilarity) of bound ligands of ten different reference pockets and predictive accuracy using the references in the post-analysis for 2013 Phase 1.
| PDB code | Ligand | Distance of the Ligand to Target Ligand a) | Prediction Results b) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Zernike | SIMCOMP | LS_SHP | LS_CHEM | Babel | AUC | Top4 Acc | Top6 Acc | Top8 Acc | ||
| 1hdc | CBO | 0.698 | 0.727 | 0.0122 | 0.0246 | 0.587 | 0.938 | 0.75 | 1.00 | 1.00 |
| 1lke | DOG | 0.546 | 0.375 | 0.0075 | 0.0133 | 0.287 | 0.417 | 0.25 | 0.25 | 0.25 |
| 2b04 | CHO | 0.678 | 0.459 | 0.0405 | 0.0725 | 0.578 | 0.646 | 0.25 | 0.50 | 0.75 |
| 3a3y | OBN | 0.739 | 0.440 | 0.0131 | 0.0254 | 0.431 | 0.688 | 0.50 | 0.75 | 0.75 |
| 3aqi | CHD | 0.578 | 0.460 | 0.0160 | 0.0286 | 0.687 | 0.875 | 0.75 | 0.75 | 0.75 |
| 4res | BUF | 0.590 | 0.277 | 0.0117 | 0.0203 | 0.863 | 0.312 | 0.25 | 0.25 | 0.25 |
| 4jch | 1KG | 0.568 | 0.650 | 0.0220 | 0.0409 | 0.571 | 0.646 | 0.25 | 0.50 | 0.75 |
| 2q1v | PDN | 0.624 | 0.589 | 0.0197 | 0.0341 | 0.660 | 0.438 | 0.25 | 0.50 | 0.50 |
| 1q23 | FUA | 0.697 | 0.724 | 0.0444 | 0.0884 | 0.553 | 0.604 | 0.50 | 0.50 | 0.50 |
| 3b0w | DGX | 0.519 | 0.517 | 0.0198 | 0.0485 | 0.441 | 0.542 | 0.25 | 0.50 | 0.50 |
Distance (dissimilarity) of the bound ligand in each PDB entry to the actual compound, the derivative of digoxigenin, was computed using five compound comparison methods. For the SIMCOMP score and the Tanimoto Coefficient computed with Babel, the original value was subtracted from 1.0 to make all the distance values consistently smaller for more similar ligands.
AUC, the Area Under the Receiver Operator Characteristic Curve. Top 4, 6, 8 accuracies compute the fraction of the four binding proteins (DIG5, DIG10, DIG18, DIG19) within top ranks.
Figure 4.
Bound ligands of the ten reference pockets used in Table 5.
On the right side of Table 5, Patch-Surfer’s predictive accuracies computed using each of the ten reference binding pockets were shown. The accuracies are represented in terms of Area Under the Receiver Operator Characteristics Curve (AUC), Top 4, Top 6, and Top 8 accuracies. Top 4, 6, 8 accuracies show the ratio of the four designed proteins that bind the target ligand (DIG5, DIG10, DIG18, and DIG19) ranked within each respective rank. Using 1HDC as the reference pocket performed best, with an AUC value and Top 4, 6, 8 accuracies of 0.938, 0.75, 1.00, and 1.00, respectively. The second and the third well performed pockets were 3AQI and 3A3Y. Interestingly, the bound ligands of these three proteins, 1HDC, 3AQI, and 3A3Y, are not particularly similar to the target ligand, according to the five compound distance measures. In Table 6, we further computed Pearson’s correlation coefficient between compound distance measures and accuracy measures. Global shape difference measured with 3D Zernike descriptors (3DZD) and SIMCOMP showed relatively large correlation to the accuracy measures, but overall the correlations were not substantially high between those ligand distance measures and the predictive accuracies. The reason of the highest correlation to the accuracies with the Zernike distance might be because Patch-Surfer uses the 3DZD for describing binding pocket surface properties, although the way 3DZD is used for the ligand distance measure and for Patch-Surfer is different. The former uses it for representing global shape of ligands while the latter uses it for representing segmented pocket surface regions.
Table 6.
Pearson’s correlation coefficients between reference ligand dissimilarity to the target ligand and predictive accuracies of Patch-Surfer in CSAR 2013 Phase 1.
| AUC | Top 4 Acc | Top 6 Acc | Top 8 Acc | |
|---|---|---|---|---|
| Zernike | 0.400 | 0.433 | 0.532 | 0.504 |
| SIMCOMP | 0.516 | 0.360 | 0.534 | 0.559 |
| LIGSIFT_SHP | 0.067 | −0.110 | −0.070 | 0.131 |
| LIGSIFT_CHEM | 0.075 | −0.097 | −0.049 | 0.117 |
| Babel | −0.047 | 0.115 | 0.008 | 0.020 |
The two post analyses revealed that the original choices of the templates and the reference pocket we made were very appropriate. The analyses also confirm that the qualities of homology models are very important in ligand binding prediction as also reported by previous works39–41.
3.2. 2013 Phase 2 Results
In Phase 2 of CSAR 2013 exercise, the organizers provided structures of two proteins, DIG18 and DIG20 (PDB ID: 4J8T), as well as 200 pre-generated ligand poses of the target ligand, for both of the proteins. Participants were asked to score those 200 ligand poses to identify the correct pose of the ligand. We submitted two predictions for this phase, one by using AutoDock Vina in the score only mode and the other one by using the consensus score between Vina and Patch-Surfer. Moreover, we ran PL-PatchSurfer for this exercise as a post-analysis.
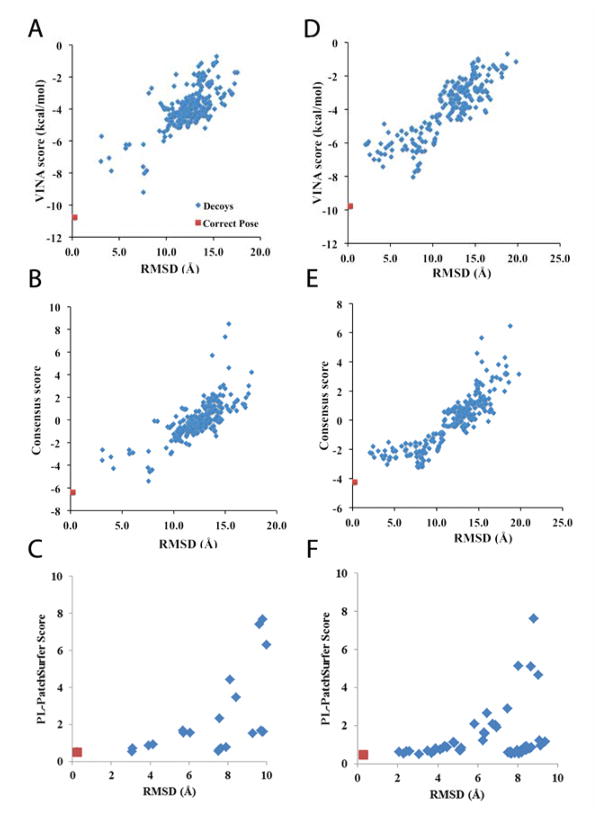
All the three methods successfully selected the correct pose of the ligand with the lowest score among all the pre-generated poses. Figure 5 shows the distribution of the scores and the RMSD of the 200 ligand poses of DIG18 (Fig. 5A, B, C) and DIG20 (Fig. 5D, E, F). Spearman’s rank coefficients between the Vina results and the consensus results were 0.642 and 0.886 for DIG18 and DIG20, respectively, which indicate that the ranks by the two scores are different but correlated. To run PL-PatchSurfer, we used the binding pocket of an engineered lipocalin protein structure (PDB ID: 1LKE), which was co-crystallized with digoxigenin (DOG), instead of using the homology models prepared for Phase 1. Ligand poses that have an RMSD of 10 Å or higher to the reference binding pose of 1LKE were not scored because they were obviously dissimilar to the reference pose.
Figure 5.
Score distributions relative to the RMSD of ligand poses. The correct pose is shown in red and the other poses are in blue. A, B, C are for DIG18 and D, E, F are results for DIG20. A and D showed results using the Vina score, B and E used the consensus score, and C and F used PL-PatchSurfer. In C and F, the ligand poses that have an RMSD of 10 Å or higher to the reference binding pose of 1LKE were not scored because they were obviously dissimilar to the reference.
3.3. 2013 Phase 3 Results
In Phase 3 of CSAR 2013 exercise, the organizer provided the 3D structure of one of the artificially designed proteins, DIG19 (PDB ID: 4J9A), and ten different small molecules. Participants were asked to find the correct binding pose of each ligand and its corresponding binding affinity. We used Vina to generate a set of ligand poses and used the Vina score and the consensus of Vina and Patch-Surfer scores to rank the poses. AutoDock Vina was run in the flexible docking mode. For the most cases, Vina generated nine poses (Table S1 in Supporting Information). In addition to the submitted predictions, we newly ran PL-PatchSurfer as a post analysis of this exercise.
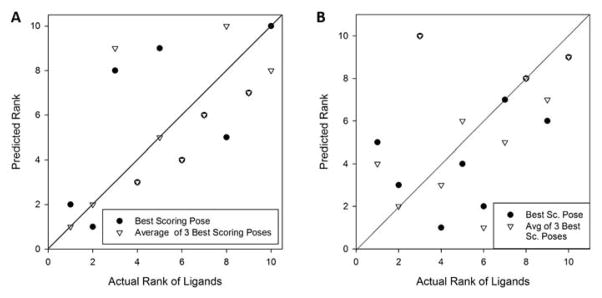
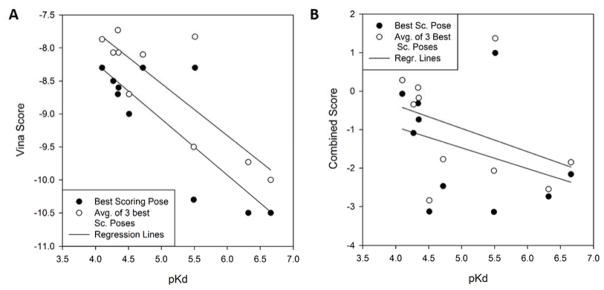
Figure 6A shows how the Vina score predicted the rank of the ligands in the order of their binding affinity. The ligands were ranked in two ways: First, we ranked them based on the Vina score of the lowest energy conformation among those which were generated (circles in Figure 6A). Next, we ranked the ligand using the average of the top three lowest energies among the generated conformations (triangles). Overall predicted ranks showed reasonable agreement with the experimental results. The two highest affinity ligands (the left bottom corner) were ranked correctly when the average score of the three best poses was considered and also selected by the best pose energy as the two best binders but with reversed rank. Moreover, the ligand with the lowest affinity (the right upper corner) was selected correctly when considering the best pose energy. The correlation coefficient between the best Vina score (filled circles) and the experimental pKd was 0.819, while it was 0.834 when the average score of the three best poses was considered (empty circles) (Fig. 7A).
Figure 6.
Correct and predicted rank of the ten ligands in CSAR 2013 Phase 3. A: Ranks of ligands were predicted by AutoDock Vina scores. B: Prediction of the rank was performed by the combined scores of Vina and Patch-Surfer. Filled circles, ranking of the ligands based on the lowest energy (score) among the constructed conformations for the ligands. Triangles, ranking based on the average of the lowest three energies (scores) among the constructed conformations for the ligands.
Figure 7.
The correlation between the scores and experimentally determined pKd in 2013 Phase 3. Filled circles, for each ligand, its lowest energy (score) among the constructed conformations was plotted. Empty circles, the average of the three lowest energies (scores) among the constructed conformations was plotted for each ligand. Lines are linear regression between pKd and the scores. A: Correlation with the Vina score of the ligands. B: Correlation with the combined score of Vina and Patch-Surfer.
Next, in Figure 6B and 7B, we examined how the combined score of Vina and Patch-Surfer works for ranking and predicting binding affinity of ligands. The results were not as good as using the Vina score only (Figs. 6A and 7A). The correlation coefficients were −0.344 and −0.394, when the best score (filled circles) and the average of the three best scores (empty circles) were used, respectively.
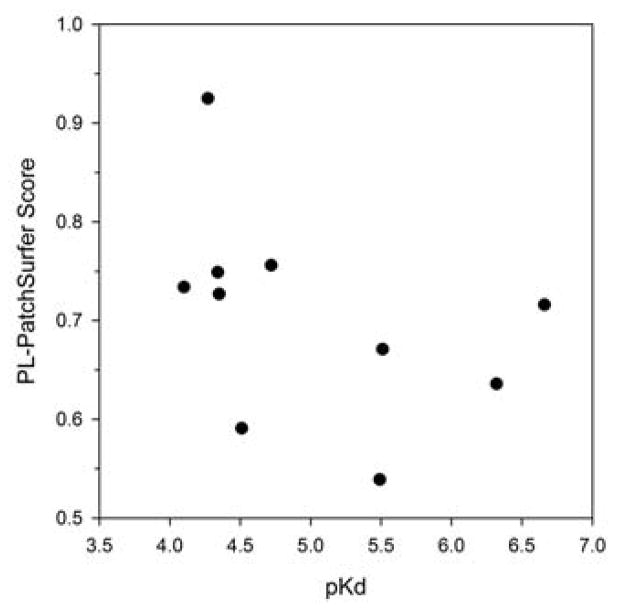
At last, PL-PatchSurfer’s results on this benchmark are shown in Figure 8. Pearson’s correlation coefficient between the PL-PatchSurfer score and pKd is Phase is −0.41. Although this is not a strong correlation, it shows that the program could discriminate non-binders from active compounds, as it is originally designed. The PL-PatchSurfer’s correlation is not as good as that of AutoDock Vina but better than the combined score of Vina and Patch-Surfer.
Figure 8.
The correlation between the PL-PatchSurfer score and pKd in 2013 Phase 3.
According to the CSAR organizer’s paper42, our results using Vina is the 6th among 27 predictions submitted to this phase, a larger correlation (0.819 and 0.834), while the results with the combined score and PL-PatchSurfer were among the lower ranks.
3.4. Results for Phase 1 of CSAR 2014
We did not submit our predictions for CSAR 2014, but here we report results of PL-PatchSurfer we newly ran on the exercise datasets of Phase 1 and 2. The dataset of Phase 1 contained 22 protein-ligand pairs with 200 pre-generated decoys for each ligand. The target proteins were FXa, SYK, and TRMD. Participants were asked to score the decoy poses for each protein-ligand pair. To apply PL-PatchSurfer in this phase, weighting factors of the PL-PatchSurfer scoring function were trained for each of the three target proteins using five crystal structures each for the target proteins (FXa: 2PR3, 2VVV, 2VWO, 2WYG, 3CEN; SYK: 1XBB, 3TUC, 4FYO, 4PV0, 4PX6; TRMD: 1P9P, 4MCB, 4MCC, 4MCD, 4YVJ). The ligands of these crystal structures were not the same as the target ligands of this exercise. Using the crystal structures, decoy conformations of the cognate ligand for each target protein were generated by DOCK643. The average number of the generated decoys were 763.4, 701.2, and 259.0 for FXa, SYK, and TRMD, respectively. Using the decoys, the weights were trained to maximize the Pearson’s correlation coefficients between RMSD of the docked ligand and the PL-PatchSurfer score of the decoys.
Table 7 summarizes the results of PL-PatchSurfer’s prediction. PL-PatchSurfer was able to select the nearest native (i.e. correct) pose as the top 1 choice for two out of three FXa decoy sets, three out of five SYK decoy sets. As for TRMD decoy sets, although the top 1 rank was correct only for one out of fourteen set sets, the top 1 rank was within 2 Å RMSD from the nearest-native binding pose for additional nine cases. When Top 3 ranks were considered, the correct pose was selected for the all three FXa decoy sets, and four out of five SYK sets, and five TMSD sets. If the consideration was further extended to top 10 ranks, correct poses were selected for all but three decoy sets. Although these results by PL-PatchSurfer is not as high as AutoDock Vina we have also applied (Table S2 in Supporting Information), it shows that PL-PatchSurfer is able to predict a near native pose within a top rank.
Table 7.
PL-PatchSurfer’s results of CSAR 2014 Phase 1.
| Target | Top 1 a) | Top 3 | Top 5 | Top 10 |
|---|---|---|---|---|
| 01_FXA_gtc101 | X b) | X | X | X |
| 02_FXA_gtc398 | 1.630 | X | X | X |
| 03_FXA_gtc401 | X | X | X | X |
| 04_SYK_gtc224 | 2.053 | 1.550 | 1.550 | 1.550 |
| 05_SYK_gtc225 | X | X | X | X |
| 06_SYK_gtc233 | 2.370 | X | X | X |
| 07_SYK_gtc249 | X | X | X | X |
| 08_SYK_gtc250 | X | X | X | X |
| 09_TRMD_gtc445 | 3.214 | 3.096 | X | X |
| 10_TRMD_gtc446 | 3.181 | 3.095 | X | X |
| 11_TRMD_gtc447 | 1.772 | 1.772 | 1.772 | X |
| 12_TRMD_gtc448 | 2.564 | 2.564 | 2.541 | X |
| 13_TRMD_gtc451 | 1.614 | X | X | X |
| 14_TRMD_gtc452 | 1.746 | 1.477 | 1.477 | 1.477 |
| 15_TRMD_gtc453 | 1.501 | 1.501 | X | X |
| 16_TRMD_gtc456 | 3.920 | X | X | X |
| 17_TRMD_gtc457 | 1.536 | X | X | X |
| 18_TRMD_gtc458 | 1.602 | 1.602 | X | X |
| 19_TRMD_gtc459 | 1.721 | 1.710 | 1.495 | X |
| 20_TRMD_gtc460 | 1.442 | X | X | X |
| 21_TRMD_gtc464 | X | X | X | X |
| 22_TRMD_gtc465 | 1.919 | 1.919 | 1.919 | 1.919 |
The lowest RMSD (Å) within top 1, 3, 5, 10 were reported.
X shows that the best pose (the pose that is nearest to the native) was selected within the specified top hits. The values are RMSD from the best pose (Å).
3.5. 2014 Phase 2 results
The exercise for Phase 2 of CSAR2014 was to rank the ligands in the give ligand library in terms of their pIC50 values (three sets for FXa, one set for SYK, and one set for TRMD). The numbers of ligands in the library are 45, 67, and 51 for the three sets for FXa, 31 for TRMD, and 276 for SYK. We used PL-PatchSurfer to compute the scores for the all ligands and compared the scores with provided pIC50 values by computing the Pearson’s correlation coefficients (Table 8). Moderate correlation coefficients of −0.590 and −0.671 were observed for the dataset 1 of FXa and TRMD, respectively. Correlation was weak for the other cases. However, when compared with results of other participants44–46 that are available at the time of writing (Table 8), PL-PatchSurfer was the best among the other available prediction results for FXa dataset 1 and competitive for TRMD.
Table 8.
Pearson’s correlation coefficients between PL-PatchSurfer score and pIC50.
| Protein-Ligand Dataset | PL-PatchSurfer | N.-Govindan et al. 47 | Kumar et al.44 | Yan et al.48 | Hogues et al.45 | Baumgartner et al.49 | Martiny et al.46 | |
|---|---|---|---|---|---|---|---|---|
| FXa | Set 1 | −0.590 | −0.26/−0.43 | 0.15/0.44/0.30/0.09 | 0.263/0.139 | 0.10/0.00/0.00/0.00 | 0.039 | |
| Set 2 | −0.094 | −0.16/−0.14 | 0.135 | −0.11/0.08/0.14/0.19 | 0.088/0.057 | 0.35/0.30/0.44/0.36 | 0.077 | |
| Set 3 | −0.139 | −0.22/−0.16 | −0.18/−0.24/−0.18/0.11 | 0.019/0.091 | 0.33/0.28/0.48/0.40 | 0.126 | ||
| SYK | −0.244 | −0.38/−0.38 | 0.784 | 0.31/0.53/0.59/0.33 | 0.120/0.127 | 0.62/0.62/0.10/0.25 | 0.265 | |
| TRMD | −0.671 | −0.61/−0.56 | 0.179 | 0.82/0.25/0.24/0.65 | 0.514/0.058 | 0.69/0.75/0.73/0.82 | 0.591 | |
The correlations of Nedumpully-Govidan et al. were taken from Figure 4 of their paper47. Kumar et al. 44 were taken from Table 2 of their paper. For FXa, they provided an overall correlation for the three sets. Yan et al. are from Table V of their paper48. The first three values are IT-Score and its variations and the last one is score by AutoDock Vina. The values for Hogues et al. 45 are from Table 3 of their paper. Values of Baumgartner et al. 49 were computed from Figure 6 of their paper. Values of Matiny et al.46 were computed from R2 values in Table 3 of their paper. Since it was not clear if their score has a positive or negative correlation to pIC50, we chose to put positive values.
4. Conclusions and Discussions
The CSAR benchmark exercise provided a unique opportunity for researchers who develop or use protein-ligand docking methods to objectively evaluate the performance of such methods. We participated in all three phases of CSAR 2013 and submitted our predictions. In the submitted predictions, we used Patch-Surfer in combination with AutoDock. Moreover, in this work we have further used PL-PatchSurfer to complete exercises provided in CSAR 2014. Patch-Surfer and PL-PatchSurfer are both for predicting binding ligands for a query pocket in a protein surface but achieve in complementary ways: The former is designed to compare a query pocket against a database of known ligand binding pockets while the latter compares molecular surface of ligands to a query pocket.
It was our pleasant surprise that Patch-Surfer performed well in 2013 Phase 1, even better than AutoDock, in selecting designed proteins that bind to the target ligand. PL-PatchSurfer performed well in 2013 Phase 2 in identifying correct binding pose of ligands and in 2014 Phase 2 in terms of correlation to pIC50. In 2014 Phase 2, PL-PatchSurfer performed the best among other available participants’ results in one of the datasets (FXa dataset 1) and also better or comparable in another set (TRMD). On the other hand, PL-PatchSurfer showed weakness in 2013 Phase 3 and 2014 Phase 1, whose aims were ranking of ligands and ligand binding poses, respectively, which probably needed more detailed atomic detailed energy evaluation than PL-PatchSurfer is equipped with. The surface-based coarse-grained molecular representation used in PL-PatchSurfer seemed not work well in these two exercises, however, the coarse-grained representation can be an advantage in certain situations, including virtual screening for binding pockets in apo form as we showed in our recent study27. Thus, it is important to use the methods for appropriate purposes for their algorithms, knowing their characteristics and when they show their strengths.
Supplementary Material
Acknowledgments
The authors thank Lenna Peterson for proofreading the manuscript. This work was partly supported by the National Institute of General Medical Sciences of the National Institutes of Health (R01GM097528) and the National Science Foundation (IIS1319551, DBI1262189, IOS1127027) and The National Natural Science Foundation of China (21403002).
Footnotes
The number of poses for each ligand generated by Autodock Vina in Phase 3 CSAR 2013 exercise; Autodock Vina scores for protein-ligand pairs of CSAR 2014 Phase 1 using two different atom charge assignments; Vina score distributions with the two different atom charges on the 22 target-ligand dataset in Phase 1 of CSAR 2014.
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49(20):5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 2.Cheng T, Li X, Li Y, Liu Z, Wang R. Comparative assessment of scoring functions on a diverse test set. J Chem Inf Model. 2009;49(4):1079–1093. doi: 10.1021/ci9000053. [DOI] [PubMed] [Google Scholar]
- 3.Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput -Aided Mol Des. 2001;15(5):411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 4.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christen M, Hunenberger PH, Bakowies D, Baron R, Burgi R, Geerke DP, Heinz TN, Kastenholz MA, Krautler V, Oostenbrink C, Peter C, Trzesniak D, van Gunsteren WF. The GROMOS software for biomolecular simulation: GROMOS05. J Comput Chem. 2005;26(16):1719–1751. doi: 10.1002/jcc.20303. [DOI] [PubMed] [Google Scholar]
- 7.Tirado-Rives J, Jorgensen WL. Viability of molecular modeling with pentium-based PCs. J Comput Chem. 1996;17(11):1385–1386. doi: 10.1002/(SICI)1096-987X(199608)17:11<1385::AID-JCC11>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 8.Grinter SZ, Zou X. A Bayesian statistical approach of improving knowledge-based scoring functions for protein-ligand interactions. J Comput Chem. 2014;35(12):932–943. doi: 10.1002/jcc.23579. [DOI] [PubMed] [Google Scholar]
- 9.Huang SY, Zou X. An iterative knowledge-based scoring function to predict protein-ligand interactions: I. Derivation of interaction potentials. J Comput Chem. 2006;27(15):1866–1875. doi: 10.1002/jcc.20504. [DOI] [PubMed] [Google Scholar]
- 10.Muegge I, Martin YC. A general and fast scoring function for protein-ligand interactions: a simplified potential approach. J Med Chem. 1999;42(5):791–804. doi: 10.1021/jm980536j. [DOI] [PubMed] [Google Scholar]
- 11.Gohlke H, Hendlich M, Klebe G. Knowledge-based scoring function to predict protein-ligand interactions. J Mol Biol. 2000;295(2):337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- 12.Mooij WT, Verdonk ML. General and targeted statistical potentials for protein-ligand interactions. Proteins. 2005;61(2):272–287. doi: 10.1002/prot.20588. [DOI] [PubMed] [Google Scholar]
- 13.Bohm HJ. The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J Comput -Aided Mol Des. 1994;8(3):243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- 14.Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput -Aided Mol Des. 2002;16(1):11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 15.Korb O, Stutzle T, Exner TE. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J Chem Inf Model. 2009;49(1):84–96. doi: 10.1021/ci800298z. [DOI] [PubMed] [Google Scholar]
- 16.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput -Aided Mol Des. 1997;11(5):425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 17.Sippl MJ. Knowledge-based potentials for proteins. Curr Opin Struct Biol. 1995;5(2):229–235. doi: 10.1016/0959-440x(95)80081-6. [DOI] [PubMed] [Google Scholar]
- 18.Dunbar JB, Jr, Smith RD, Damm-Ganamet KL, Ahmed A, Esposito EX, Delproposto J, Chinnaswamy K, Kang YN, Kubish G, Gestwicki JE, Stuckey JA, Carlson HA. CSAR data set release 2012: ligands affinities complexes and docking decoys. J Chem Inf Model. 2013;53(8):1842–1852. doi: 10.1021/ci4000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damm-Ganamet KL, Smith RD, Dunbar JB, Jr, Stuckey JA, Carlson HA. CSAR benchmark exercise 2011–2012: evaluation of results from docking and relative ranking of blinded congeneric series. J Chem Inf Model. 2013;53(8):1853–1870. doi: 10.1021/ci400025f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith RD, Dunbar JB, Jr, Ung PM, Esposito EX, Yang CY, Wang S, Carlson HA. CSAR benchmark exercise of 2010: combined evaluation across all submitted scoring functions. J Chem Inf Model. 2011;51(9):2115–2131. doi: 10.1021/ci200269q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunbar JB, Jr, Smith RD, Yang CY, Ung PM, Lexa KW, Khazanov NA, Stuckey JA, Wang S, Carlson HA. CSAR benchmark exercise of 2010: selection of the protein-ligand complexes. J Chem Inf Model. 2011;51(9):2036–2046. doi: 10.1021/ci200082t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu X, Xiong Y, Kihara D. Large-scale binding ligand prediction by improved patch-based method Patch-Surfer2. 0. Bioinformatics. 2015;31(5):707–713. doi: 10.1093/bioinformatics/btu724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sael L, Kihara D. Detecting local ligand-binding site similarity in nonhomologous proteins by surface patch comparison. Proteins. 2012;80(4):1177–1195. doi: 10.1002/prot.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu B, Zhu X, Monroe L, Bures MG, Kihara D. PL-PatchSurfer: A Novel Molecular Local Surface-Based Method for Exploring Protein-Ligand Interactions. Int J Mol Sci. 2014;15(9):15122–15145. doi: 10.3390/ijms150915122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin WH, Bures MG, Kihara D. PatchSurfers: Two Methods for Local Molecular Property-Based Binding Ligand Prediction. Methods. 2015 doi: 10.1016/j.ymeth.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin WH, Zhu X, Bures MG, Kihara D. Three-Dimensional Compound Comparison Methods and Their Application in Drug Discovery. Molecules. 2015;20(7):12841–12862. doi: 10.3390/molecules200712841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tinberg CE, Khare SD, Dou J, Doyle L, Nelson JW, Schena A, Jankowski W, Kalodimos CG, Johnsson K, Stoddard BL, Baker D. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501(7466):212–216. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canterakis N. 3D Zernike moments and Zernike affine invariants for 3D image analysis and recognition. Proc.11th Scandinavian Conference on Image Analysis; 1999; pp. 85–93. [Google Scholar]
- 31.Sael L, Li B, La D, Fang Y, Ramani K, Rustamov R, Kihara D. Fast protein tertiary structure retrieval based on global surface shape similarity. Proteins. 2008;72(4):1259–1273. doi: 10.1002/prot.22030. [DOI] [PubMed] [Google Scholar]
- 32.Hawkins PC, Skillman AG, Warren GL, Ellingson BA, Stahl MT. Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J Chem Inf Model. 2010;50(4):572–584. doi: 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35(10):3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 35.Hattori M, Tanaka N, Kanehisa M, Goto S. SIMCOMP/SUBCOMP: chemical structure search servers for network analyses. Nucleic Acids Res. 2010;38(Web Server issue):W652–W656. doi: 10.1093/nar/gkq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venkatraman V, Chakravarthy PR, Kihara D. Application of 3D Zernike descriptors to shape-based ligand similarity searching. J Cheminform. 2009;1:19. doi: 10.1186/1758-2946-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy A, Skolnick J. LIGSIFT: an open-source tool for ligand structural alignment and virtual screening. Bioinformatics. 2015;31(4):539–544. doi: 10.1093/bioinformatics/btu692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michino M, Abola E, Brooks CL, 3rd, Dixon JS, Moult J, Stevens RC. Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat Rev Drug Discov. 2009;8(6):455–463. doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bordogna A, Pandini A, Bonati L. Predicting the accuracy of protein-ligand docking on homology models. J Comput Chem. 2011;32(1):81–98. doi: 10.1002/jcc.21601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kufareva I, Katritch V, Stevens RC, Abagyan R. Advances in GPCR modeling evaluated by the GPCR Dock 2013 assessment: meeting new challenges. Structure. 2014;22(8):1120–1139. doi: 10.1016/j.str.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith RD, Damm-Ganamet KL, Dunbar JB, Jr, Ahmed A, Chinnaswamy K, Delproposto JE, Kubish GM, Tinberg CE, Khare SD, Dou J, Doyle L, Stuckey JA, Baker D, Carlson HA. CSAR Benchmark Exercise 2013: Evaluation of Results from a Combined Computational Protein Design, Docking, and Scoring/Ranking Challenge. J Chem Inf Model. 2015 doi: 10.1021/acs.jcim.5b00387. Article ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen WJ, Balius TE, Mukherjee S, Brozell SR, Moustakas DT, Lang PT, Case DA, Kuntz ID, Rizzo RC. DOCK 6: Impact of new features and current docking performance. J Comput Chem. 2015;36(15):1132–1156. doi: 10.1002/jcc.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar A, Zhang KY. Application of Shape Similarity in Pose Selection and Virtual Screening in CSARdock2014 Exercise. J Chem Inf Model. 2015 doi: 10.1021/acs.jcim.5b00279. Article ASAP. [DOI] [PubMed] [Google Scholar]
- 45.Hogues H, Sulea T, Purisima EO. Evaluation of the Wilma-SIE Virtual Screening Method in Community Structure-Activity Resource 2013 and 2014 Blind Challenges. J Chem Inf Model. 2015 doi: 10.1021/acs.jcim.5b00278. Article ASAP. [DOI] [PubMed] [Google Scholar]
- 46.Martiny VY, Martz F, Selwa E, Iorga BI. Blind Pose Prediction, Scoring, and Affinity Ranking of the CSAR 2014 Dataset. J Chem Inf Model. 2015 doi: 10.1021/acs.jcim.5b00337. Article ASAP. [DOI] [PubMed] [Google Scholar]
- 47.Nedumpully-Govindan P, Jemec DB, Ding F. CSAR Benchmark of Flexible MedusaDock in Affinity Prediction and Nativelike Binding Pose Selection. J Chem Inf Model. 2015 doi: 10.1021/acs.jcim.5b00303. Article ASAP. [DOI] [PubMed] [Google Scholar]
- 48.Yan C, Grinter SZ, Merideth BR, Ma Z, Zou X. Iterative Knowledge-Based Scoring Functions Derived from Rigid and Flexible Decoy Structures: Evaluation with the 2013 and 2014 CSAR Benchmarks. J Chem Inf Model. 2015 doi: 10.1021/acs.jcim.5b00504. Article ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baumgartner MP, Camacho CJ. Choosing the Optimal Rigid Receptor for Docking and Scoring in the CSAR 2013/2014 Experiment. J Chem Inf Model. 2015 doi: 10.1021/acs.jcim.5b00338. Article ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.