Abstract
Type III secretion systems (T3SSs) are crucial for bacterial infections because they deliver effector proteins into host cells. The Escherichia coli type III secretion system 2 (ETT2) is present in the majority of E. coli strains, and although it is degenerate, ETT2 regulates bacterial virulence. An ATPase is essential for T3SS secretion, but the function of the ETT2 ATPase has not been demonstrated. Here, we show that EivC is homologous to the β subunit of F0F1 ATPases and it possesses ATPase activity. To investigate the effects of ETT2 ATPase EivC on the phenotype and virulence of avian pathogenic Escherichia coli (APEC), eivC mutant and complemented strains were constructed and characterized. Inactivation of eivC led to impaired flagella production and augmented fimbriae on the bacterial surface, and, consequently, reduced bacterial motility. In addition, the eivC mutant strain exhibited attenuated virulence in ducks, diminished serum resistance, reduced survival in macrophage cells and in ducks, upregulated fimbrial gene expression, and downregulated flagellar and virulence gene expression. The expression of the inflammatory cytokines interleukin (IL)-1β and IL-8 were increased in HD-11 macrophages infected with the eivC mutant strain, compared with the wild-type strain. These virulence-related phenotypes were restored by genetic complementation. These findings demonstrate that ETT2 ATPase EivC is involved in the motility and pathogenicity of APEC.
Keywords: avian pathogenic E. coli, ATPase, EivC, motility, virulence
Introduction
A number of Gram-negative pathogens, such as Escherichia coli (E. coli), Salmonella, Yersinia, and Pseudomonas utilize type III secretion systems (T3SSs) to deliver effector proteins into eukaryotic host cells and to assemble flagella, which facilitate infections and bacterial motility (Mota and Cornelis, 2005; Diepold and Armitage, 2015). The structure of the T3SS apparatus, known as an injectisome, is evolutionarily conserved among pathogenic T3SSs and flagella, which share a similar membrane-spanning basal body. T3SSs have an extracellular, needle-like projection connecting to the basal body, which serves as a channel for the translocation of effector proteins. These bacterial effector proteins interact and subvert the innate immunity of host cells, thereby facilitating bacterial growth and survival, which subsequently lead to diseases (Cornelis and Van Gijsegem, 2000; Coburn et al., 2007). In contrast, the basal body of flagella, which is the key bacterial organelle required for motility, is linked to an extracellular hook and a flagellar filament (Minamino et al., 2008; Li and Sourjik, 2011).
ATPases, which are thought to provide the energy for the secretion process (Paul et al., 2008), are an essential and highly conserved component of all T3SSs. Thus, ATPases are essential for T3SS function and bacterial virulence. Several T3SS ATPases from animal and plant pathogenic bacteria have been identified and characterized, including EscN of E. coli (Andrade et al., 2007), InvC and SscN of Salmonella (Eichelberg et al., 1994; Akeda and Galan, 2004; Yoshida et al., 2014), YscN of Yersinia (Woestyn et al., 1994; Blaylock et al., 2006), and HrcN of Xanthomonas (Lorenz and Buttner, 2009), and they show significant amino acid sequence homology to the catalytic β subunit of F0F1 ATPases and are able to hydrolyze ATP (Zarivach et al., 2007). Previous evidence indicated that ATP hydrolysis by T3SS ATPases is required for the dissociation of effector proteins from their cognate chaperones. In addition, it has been shown that ATPases are involved in the unfolding of effector proteins before their secretion (Minamino and MacNab, 2000; Jouihri et al., 2003; Akeda and Galan, 2005; Pallen et al., 2006).
Most bacteria contain only one T3SS, but some pathogens harbor multiple T3SSs, which function independently in different aspects of pathogenesis (Moest and Meresse, 2013). Intestinal pathogenic E. coli, such as enteropathogenic E. coli (EPEC), and enterohemorrhagic E. coli (EHEC), use the locus of enterocyte effacement (LEE)-encoded T3SS to cause attaching/effacing lesions and diarrheal disease (Franzin and Sircili, 2015). A second T3SS, named E. coli type III secretion system 2 (ETT2), was identified by sequencing the genome of an EHEC O157 strain (Hayashi et al., 2001). The intact ETT2 cluster contains at least 35 open reading frames, 19 of which are homologous to those of Salmonella pathogenicity island 1 (Hansen-Wester and Hensel, 2001; Hayashi et al., 2001). ETT2 is known to be present, in whole or in part, in the majority of E. coli strains. It is most frequent in intestinal pathogenic E. coli strains, but of low prevalence in uropathogenic E. coli (UPEC) strains (Miyazaki et al., 2002; Prager et al., 2004; Ren et al., 2004; Cheng et al., 2012; Zhou et al., 2014). Our previous study demonstrated that ETT2 is more widely distributed in avian pathogenic E. coli (APEC) isolates and exhibits more ETT2 isoforms compared with human extraintestinal pathogenic E. coli (ExPEC), such as UPEC and newborn meningitis E. coli (NMEC; Wang et al., 2016). A sequence analysis indicated that there are gene deletions and premature stop codons in the ETT2 locus of most E. coli strains, which probably abolish the functions of ETT2. However, ETT2 still plays a role in regulating bacterial virulence (Zhang et al., 2004; Ideses et al., 2005; Sheikh et al., 2006; Yao et al., 2009). However, the role of ETT2 genes in the pathogenicity of APEC infection is less clear.
In the present study, we investigated the effects of the ETT2 ATPase EivC on the phenotype and virulence of APEC. Like other T3SS ATPases, EivC is homologous to the β subunit of F0F1 ATPases and it possesses ATPase activity. Our results verified that EivC is involved in motility, serum resistance, and intra-macrophage survival of APEC, suggesting that EivC is involved in the pathogenicity of APEC.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Strains and plasmids used in this study are listed in Table 1. APEC strain APCE94 was isolated from a chicken with clinical septicemia symptoms of colibacillosis in Jiangsu, China, and it can cause severe colibacillosis symptoms in chickens and ducks. This APEC strain is well-characterized; thus, it was used for infection studies, mutant construction, and functional assays (Wang et al., 2016). All E. coli strains were grown in Luria-Bertani (LB) medium at 37°C with aeration. When necessary, the LB medium was supplemented with ampicillin (Amp, 100 μg/mL) and chloramphenicol (Cm, 30 μg/mL), unless otherwise specified.
Table 1.
Bacterial strains and plasmids used in this study.
| Strains or plasmids | Characteristics | Reference |
|---|---|---|
| Strains | ||
| APCE94 | APEC wild-type strain, O78 serotype | This study |
| APCE94ΔeivC | eivC deletion mutant in APCE94 | This study |
| APCE94CΔeivC | APCE94ΔeivC with plasmid pSTV28-eivC | This study |
| DH5α | F-, Δ(lacZYA-argF)U169, recA1, endA1, hsdR17 (rk-, mk+), phoA, supE44, λ- | TIANGEN |
| BL21 (DE3) | F-, ompT, hsdS (rB- mB-) gal, dcm (DE3) | TIANGEN |
| Plasmids | ||
| pET28a(+) | Kan, F1 origin, His tag | Novagen |
| pET28a-eivC | pET28a (+) carrying eivC gene | This study |
| pSTV28 | Cm, lacZ | Takara |
| pSTV28-eivC | pSTV28 derivative harboring eivC gene | This study |
| pKD46 | Amp; expresses λ red recombinase | Datsenko and Wanner, 2000 |
| pKD3 | Cm gene, template plasmid | Datsenko and Wanner, 2000 |
| pCP20 | Cm, Amp, yeast Flp recombinase gene, FLP | Datsenko and Wanner, 2000 |
Expression and Purification of a Histidine-Tagged EivC Fusion Protein (His-EivC)
The open reading frame of APCE94 eivC was amplified and cloned into pET28a, yielding plasmid pET28a-eivC. The His-EivC fusion protein was expressed in E. coli BL21 (DE3) cells by induction with 1 mM isopropyl-β-D-thiogalactopyranoside. Protein purification was performed using a HisTrap high-performance column (GE Healthcare, Shanghai, China) according to the manufacturer’s guidelines. The final protein concentration was determined by the Bradford method using a SmartSpec 3000 spectrophotometer (Bio-Rad, Shanghai, China).
ATPase Activity Assays
ATPase activity of the His-EivC fusion protein was determined using an ATPase/GTPase activity assay kit (Sigma, Shanghai, China) that measures the amount of inorganic phosphate (Pi) generated during hydrolysis, according to the manufacturer’s instructions. ATPase activity was expressed as 1 μmol phosphate released by 1 mg protein per min. All experiments were performed in triplicate.
Construction of Mutant and Complementation Strains
An isogenic mutant strain APCE94ΔeivC was constructed based on the lambda Red recombinase system (Datsenko and Wanner, 2000). Briefly, the eivC gene was replaced with a chloramphenicol resistance cassette, which was polymerase chain reaction (PCR)-amplified from plasmid pKD3. Then, the chloramphenicol resistance cassette was cured by plasmid pCP20. The mutant strain APCE94ΔeivC was confirmed by PCR and sequencing. Promoter prediction analyses were conducted with prediction program tools available at http://www.fruitfly.org/seq_tools/promoter. A complementation strain was generated by cloning the eivC gene and its predicted promoter into plasmid pSTV28 using the primer pair eivCCo-F and eivCCo-R (Table 2). The resulting plasmid, pSTV28-eivC, was transformed into APCE94ΔeivC to generate the complementation strain APCE94CΔeivC.
Table 2.
Primers used in this study.
| Primers | Sequence (5′ to 3′)a | Target genes | Products size |
|---|---|---|---|
| eivCEx-F | CGCGGATCCATGTCTTGCGATATGGAGC | eivC | 1317 bp |
| eivCEx-R | CCCAAGCTTCTTAACAATTCGTTCAAGCTCTT | eivC | |
| eivCMu-UF | AGAAGTTAACCCTACTGGACCC | Upstream region of eivC | 767 bp |
| eivCMu-UR | GAAGCAGCTCCAGCCTACACTTGCCAGCTTGCATAAATTT | eivC | |
| eivCMu-CF | AAATTTATGCAAGCTGGCAAGTGTAGGCTGGAGCTGCTTC | pKD3 | 1014 bp |
| eivCMu-CR | CTGATAAGTCGATTAACTT CATATGAATATCCTCCTTAG | pKD3 | |
| eivCMu-DF | CTAAGGAGGATATTCATATGAAGTTAATCGACTTATCAG | eivC | 671 bp |
| eivCMu-DR | GGATTCCTGTCAACCTAGAGAG | Downstream region of eivC | |
| C1 | TTATACGCAAGGCGACAAGG | pKD3 | |
| C2 | GATCTTCCGTCACAGGTAGG | pKD3 | |
| eivCup-F | GGCTGATAGCAATTCGCC | Upstream region of eivC | 3003 bp |
| eivCdown-R | CTGCGGTCCATTTATTTTGT | Downstream region of eivC | |
| eivCCo-F | GCGGAATTCGCCTTAGCATTATGGAGCCC | Upstream region of eivC | 2031 bp |
| eivCCo-R | CGCGGATCCGCTGTTGAACTATTTCCTTGTTAA | Downstream region of eivC | |
| dnaE RT-F | ATGTCGGAGGCGTAAGGCT | dnaE | 181 bp |
| dnaE RT-R | TCCAGGGCGTCAGTAAACAA | dnaE | |
| motA RT-F | GCTTTCTAATCTGAACGGTTACGC | motA | 96 bp |
| motA RT-R | TCCAGCTCAATAAACGACGGAC | motA | |
| flgB RT-F | GCAGCAAACATCGCCAATGCC | flgB | 101 bp |
| flgB RT-R | GTTGCATCCCGTCCACGTT | flgB | |
| flgD RT-F | GCACAAATCAGCACGGTCAGCG | flgD | 122 bp |
| flgD RT-R | ATCATCACGCCGTGACCGATCA | flgD | |
| flgF RT-F | ACGTCGCGTTGCAGCAGGAT | flgF | 128 bp |
| flgF RT-R | TATCACCGGATGCCCCTGAA | flgF | |
| fliC RT-F | CCAGTAGCTCCGTTAATCCCT | fliC | 132 bp |
| fliC RT-R | AGTTTTGCTCAGTACACCGGAA | fliC | |
| fimC RT-F | GCCGATGGTGTAAAGGATGG | fimC | 127 bp |
| fimC RT-R | AACTTTCCCGATCCTGTGGC | fimC | |
| ompA RT-F | GCTGAGCCTGGGTGTTTCCT | ompA | 171 bp |
| ompA RT-R | TCCAGAGCAGCCTGACCTTC | ompA | |
| aatA RT-F | CCGTACCCGTGTCGCTGTTAC | aatA | 98 bp |
| aatA RT-R | CAGCATTATCAGCATTGCCACT | aatA | |
| iss RT-F | CCGACAGCAGTAACACCAAAGG | iss | 105 bp |
| iss RT-R | TTCTGCACCGCCAACAAATT | iss | |
| tsh RT-F | GCACGAACTGGGAAGTATGGA | tsh | 118 bp |
| tsh RT-R | GGCATAGAAACCACCACCCC | tsh | |
| fyuA RT-F | TTGGCGACCAGGGTAAGAGC | fyuA | 145 bp |
| fyuA RT-R | AGACCCGCAGTAGGCACGAT | fyuA | |
| iucD RT-F | GCTGGGTAGCAGACGGATAT | iucD | 95 bp |
| iucD RT-R | GCATCACTGCCGATTCTTTA | iucD | |
| chβactin-F | GAGAAATTGTGCGTGACATCA | β-actin | 152 bp |
| chβactin-R | CCTGAACCTCTCATTGCCA | β-actin | |
| chIL1-F | TGGGCATCAAGGGCTACA | IL-1 | 224 bp |
| chIL1-R | TCGGGTTGGTTGGTGATG | IL-1 | |
| chIL8-F | TTGGAAGCCACTTCAGTCAGAC | IL-8 | 120 bp |
| chIL8-R | GGAGCAGGAGGAATTACCAGTT | IL-8 |
aRestriction sites are underlined.
Growth Curves and Motility Assays
To detect the effect of EivC on the growth rate of APEC, the growth kinetics of the APCE94, APCE94ΔeivC, and APCE94CΔeivC strains were determined in LB medium as described previously (Wang et al., 2014). Briefly, bacteria were incubated at 37°C with shaking, and the optical density of each strain was monitored at 1 h intervals by spectrophotometry.
A motility assay was performed as described previously (Han et al., 2015). Briefly, bacterial cultures were stabbed onto LB soft agar motility plates (0.5% agar) and incubated at 37°C. Bacterial motility haloes were measured after a 12 h incubation.
Transmission Electron Microscopy (TEM)
Morphological changes of the APCE94, APCE94ΔeivC, and APCE94CΔeivC strains were determined by transmission electron microscopy (TEM). Briefly, each strain was grown on LB agar plates and suspended and washed with phosphate-buffered saline (PBS). Then, the bacterial pellets were negatively stained using 2% phosphotungstic acid (Sigma). Finally, the stained bacteria were deposited on a carbon-coated grid, followed by observation under a FEI T12 transmission electron microscope (FEI, Ltd, Hillsboro, OR, USA).
Serum Bactericidal Assay
A bactericidal assay was performed in 96-well plates as described previously (Wang et al., 2014). Briefly, specific-pathogen-free (SPF) chicken sera were diluted to 12.5, 25, and 50% in PBS. Bacteria were incubated with different diluted sera or heat-inactivated sera (negative control) at 37°C for 30 min. Then, the bacteria were enumerated by plating on LB agar plates.
Bacterial Adhesion and Invasion Assays
Bacterial adhesion and invasion assays were performed as described previously (Wang et al., 2011). Chicken embryo fibroblast DF-1 cell monolayers were washed with Dulbecco’s modified Eagle’s medium (DMEM) without fetal bovine serum, and infected with bacteria at a multiplicity of infection (MOI) of 100 for 2 h at 37°C under 5% CO2. After washing with PBS, the cells were lysed with 0.5% Triton X-100, and bacteria were counted by plating on LB agar plates. For invasion assays, bacterial infection of cell cultures was performed as described for the bacterial adhesion assays. After a 1 h incubation, cells were washed and treated subsequently with DMEM containing gentamicin (100 μg/mL) for 1 h to kill extracellular bacteria. Then, the monolayers were washed and lysed with 0.5% Triton X-100, and invasive bacteria were enumerated by plating on LB agar plates. DH5α or DF-1 cells only were used as controls in all experiments. Assays were performed three times in triplicate.
Bacterial Intracellular Survival Assays
Bacterial intracellular survival was monitored as described previously (Wang et al., 2014). Briefly, avian HD-11 cells were infected with bacteria at a MOI of 100 for 1 h, as described for the invasion assays. Extracellular bacteria were killed with DMEM containing gentamicin (100 μg/mL) for 1 h, and intracellular bacteria were released, and they were defined as the initial number of invasive bacteria. Then, cells were grown in DMEM containing 10 μg/mL gentamicin for an additional 6, 12, or 24 h and lysed. Intracellular survival was expressed as the change (n-fold) in the bacterial number at a given time point relative to the initial number of invasive bacteria. This assay was performed three times in triplicate.
Animal Infection Experiments
All animal experiments were conducted in strict accordance with the Guidelines on the Humane Treatment of Laboratory Animals (Ministry of Science and Technology of the People’s Republic of China, Policy No. 2006 398) and were approved by the Institutional Animal Care and Use Committee at the Shanghai Veterinary Research Institute (permit No: Shvri-Po-0243).
The virulence of the APCE94, APCE94ΔeivC, and APCE94CΔeivC strains to ducks was determined. APEC strains were grown to the exponential phase and collected, washed twice in PBS, and then adjusted to the appropriate doses. Groups of eight 7-day-old ducks were inoculated intra-tracheally with 108 colony-forming units (CFUs) of bacteria, or with PBS as negative controls. Mortality was monitored daily until 7 days post-infection.
Bacterial colonization was determined during systemic infections, as described previously (Wang et al., 2011, 2014). Briefly, groups of eight 7-day-old ducks were infected intra-tracheally with a bacterial suspension containing 108 CFUs. At 24 h post-infection, ducks were bled, euthanized, and dissected. Bacterial loads in the blood were counted by plating onto LB agar plates. The liver, spleen, and lung were collected, weighed, and homogenized. The homogenates were diluted and plated onto LB agar to determine the bacterial numbers.
Quantitative Real-Time Reverse Transcription PCR (qRT-PCR)
Expression of bacterial virulence genes in the APCE94, APCE94ΔeivC, and APCE94CΔeivC strains was investigated by quantitative real-time reverse transcription PCR (qRT-PCR) as described previously (Wang et al., 2011). In addition, expression of interleukin (IL)-1β and IL-8 genes in HD-11 cells infected with the APCE94, APCE94ΔeivC, or APCE94CΔeivC strains was also investigated. In brief, total RNA was isolated from bacterial cultures or bacteria-infected HD-11 cells using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA). Contaminating DNA was removed from the samples with RNase-free DNase I (TaKaRa, Dalian, China). cDNA synthesis was performed using the PrimeScript® RT reagent kit (TaKaRa) according to the manufacturer’s protocol. qRT-PCR was performed using SYBR® Premix Ex TaqTM (TaKaRa) and gene-specific primers (Table 2). Relative gene expression was normalized to the expression of the housekeeping gene dnaE or β-actin via the ΔΔCT method. The PCR efficiency (>90%) for each of the genes was verified via standard dilution curves. The assay was performed in duplicate and repeated three times.
Statistical Analyses
Statistical analyses were conducted using the GraphPad Software package (GraphPad Software, La Jolla, CA, USA). One-way analysis of variance (ANOVA) was used to analyze motility and invasion assay data, and two-way ANOVA was performed to analyze the survival assays and qRT-PCR results. The animal infection data were analyzed using the non-parametric Mann–Whitney U-test. The mean values are shown in the figures. Statistical significance was established at p < 0.05.
Results
Sequence Analysis of the ETT2 ATPase EivC in APCE94
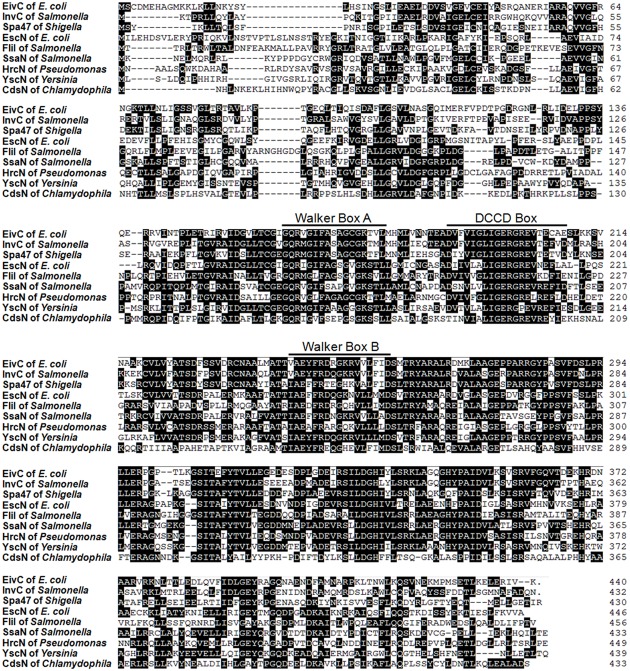
Our previous study showed that APCE94 harbors a type B ETT2 cluster. There is a 4.99-kb gene deletion in the eiv operon, which truncates the eivA and eivF genes (Wang et al., 2016). An amino acid sequence analysis indicated that EivC contains a glycine-rich region (Walker box A) and a nucleotide-binding protein region (Walker box B), which was the best conserved motif of the F0F1 ATPase (Zarivach et al., 2007). Moreover, a dicyclohexylcarbodiimide (DCCD) box was also present around a conserved glutamic acid residue, which reacted with DCCD in other ATP-binding proteins (Figure 1). These data suggest that the eivC gene likely encodes a F0F1 ATPase.
FIGURE 1.
Sequence analysis of ETT2 and EivC. Amino acid sequence comparisons of EivC with other T3SS ATPases. The locations of predicted Walker boxes A and B and a dicyclohexylcarbodiimide-binding site (DCCD box) within the amino acid sequence of EivC are indicated.
ATPase Activity of EivC
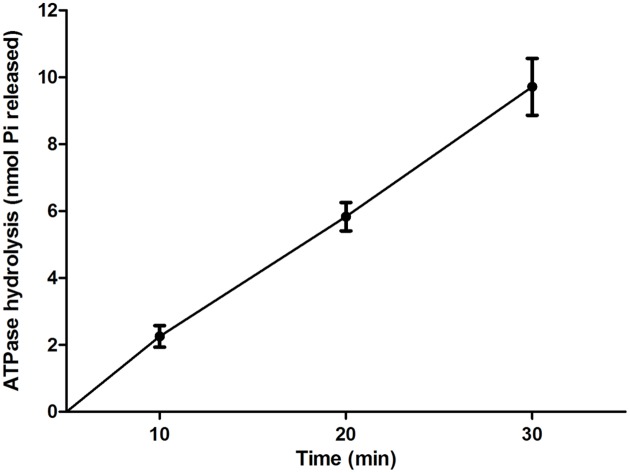
To determine whether EivC possess ATPase activity, a His-EivC fusion protein was purified and tested for its ability to hydrolyze ATP. The purified His-EivC fusion protein showed ATPase activity, with phosphate release rate of 0.28 ± 0.08 μmol/min/mg (Figure 2). This activity is comparable to that of the homologous ATPase InvC of Salmonella enterica serovar Typhimurium (Eichelberg et al., 1994; Akeda and Galan, 2004).
FIGURE 2.
ATPase activity of EivC. The His-EivC fusion protein was purified by nickel-affinity chromatography. ATP hydrolysis by the purified His-EivC fusion protein was determined using a malachite green assay over a period of 30 min. Error bars indicate standard deviations.
Inactivation of EivC Reduces the Motility of APCE94
The eivC mutant and complementation strains were successfully constructed using the lambda Red recombination system and plasmid pSTV28. No significant differences were observed in the generation times among the APCE94, APCE94ΔeivC, and APCE94CΔeivC strains during growth in LB medium (data not shown). After curing the chloramphenicol resistance cassette from the eivC gene, essential ribosomal binding sequences for the translation of downstream genes remained. Therefore, deletion of the eivC gene did not have a polar effect on the expression of downstream genes in ETT2 (data not shown).
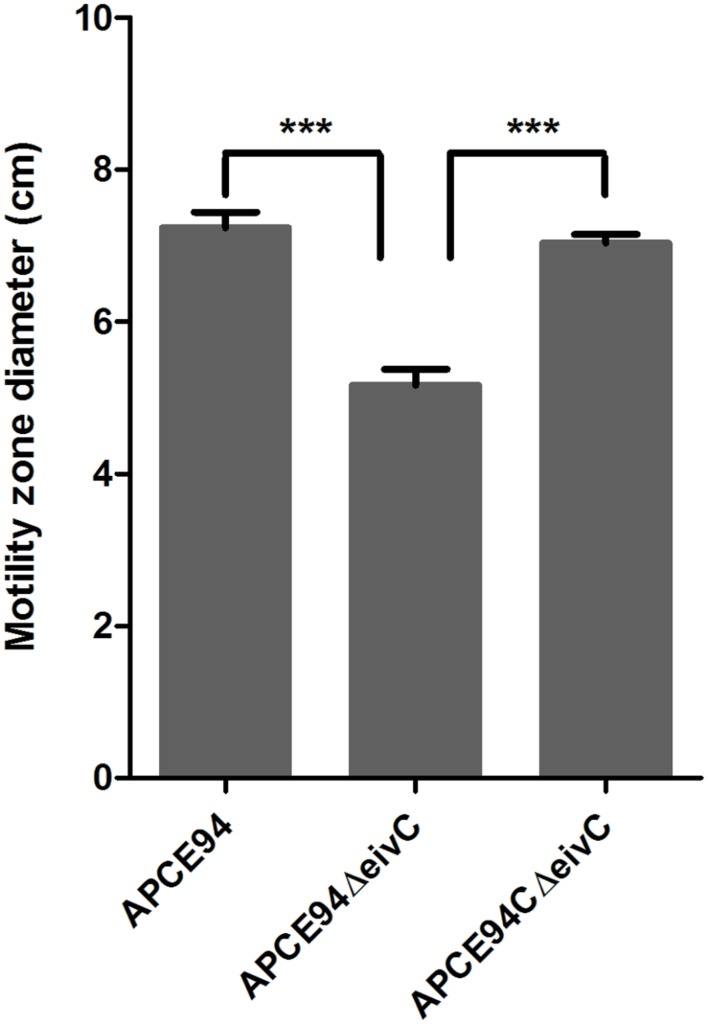
The F0F1 ATPase is similar to the FliI ATPase, which is an essential component of the bacterial flagellar motor that is required for bacterial motility (Minamino et al., 2008). Thus, we incubated the APCE94, APCE94ΔeivC, and APCE94CΔeivC strains on semisolid LB agar plates to examine whether EivC contributes to bacterial motility. The results showed that the diameter of the halo of the APCE94ΔeivC strain was significantly smaller than that of the APCE94 strain (p < 0.001), whereas the diameter of the halo of the APCE94CΔeivC strain was almost equal to that of the APCE94 strain (Figure 3). Therefore, deletion of eivC reduced the motility of APCE94.
FIGURE 3.
Effects of EivC on bacterial motility. Mutant strain APCE94ΔeivC exhibited a smaller motility diameter than the wild-type strain APCE94 and the complementation strain APCE94CΔeivC. Bars represent the average motility diameters of triplicate motility plates. Error bars represent the standard deviation. One-way ANOVA was used for the motility analysis (∗∗∗p < 0.001).
Knockout of eivC Leads to Fewer Flagella and Augmented Fimbriae
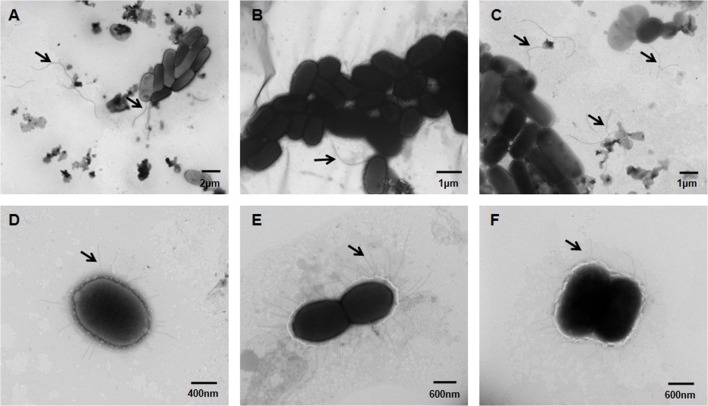
The influence of eivC on the bacterial morphological structure was examined by TEM. The results revealed remarkable differences in the flagella and fimbriae between the APCE94 and APCE94ΔeivC strains. The APCE94ΔeivC strain produced fewer flagella (including attached and broken flagella) than the APCE94 and APCE94CΔeivC strains (Figures 4A–C). In contrast, numerous long fimbriae surrounding the bacteria were observed in the APCE94ΔeivC strain, compared with that of the APCE94 strain. The fimbrial structures of the APCE94CΔeivC strain were similar to those of the APCE94 strain (Figures 4D–F). According to these results, deletion of eivC leads to impaired flagella production and augmented fimbriae on the bacterial surface.
FIGURE 4.
Transmission electron micrographs of APEC strains. Bacteria were grown overnight on LB agar, and then harvested, fixed, and negatively stained with 2% phosphotungstic acid. Numerous long flagella were observed on the surfaces of the wild-type strain APCE94 (A) and the complementation strain APCE94CΔeivC (C). Few flagella were observed on the mutant strain APCE94ΔeivC (B). Augmented fimbriae surrounding the bacteria were observed on the mutant strain APCE94ΔeivC (E), compared with those of the wild-type strain APCE94 (D) and the complementation strain APCE94CΔeivC (F).
EivC Is Required for the Serum Resistance of APCE94
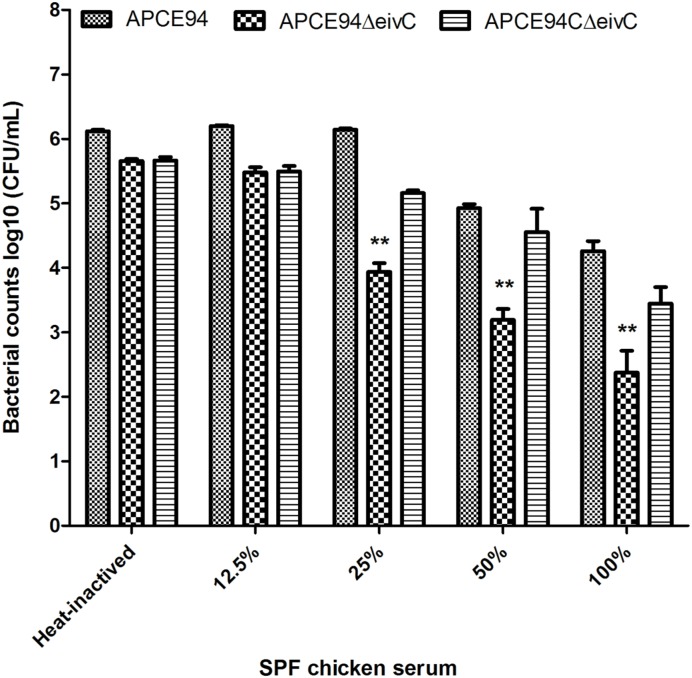
During colonization and survival in a host, resistance to serum killing is necessary for APEC pathogenicity (La Ragione and Woodward, 2002; Mellata et al., 2003). The serum bactericidal assay showed that the survival of the APCE94ΔeivC strain in SPF chicken serum was significantly reduced compared with that of the APCE94 strain (p < 0.01). Furthermore, the resistance to serum killing was restored in the APCE94CΔeivC strain (Figure 5). These observations indicate that EivC is required for the serum resistance of APCE94.
FIGURE 5.
Bacterial resistance to SPF chicken serum. The APCE94, APCE94ΔeivC, and APCE94CΔeivC strains were incubated with SPF chicken sera at different dilutions at 37°C for 30 min, and the surviving bacteria were enumerated. This data revealed that the mutant strain APCE94ΔeivC was significantly and highly susceptible to SPF chicken sera. A two-way ANOVA was performed for the survival assays (∗∗p < 0.01).
EivC Facilitates the Intracellular Survival of APCE94 in Avian HD-11 Cells
To investigate whether eivC affects bacterial adhesion and invasion capacities, the avian cell line DF-1 was infected with the APCE94, APCE94ΔeivC, or APCE94CΔeivC strains. Although the APCE94ΔeivC strain showed increased adherence to, and invasion of, DF-1 cells, these values were not significantly different from those of the APCE94 strain (data not shown). These results suggest that EivC does not affect the capacity of APEC to adhere to and invade into DF-1 cells.
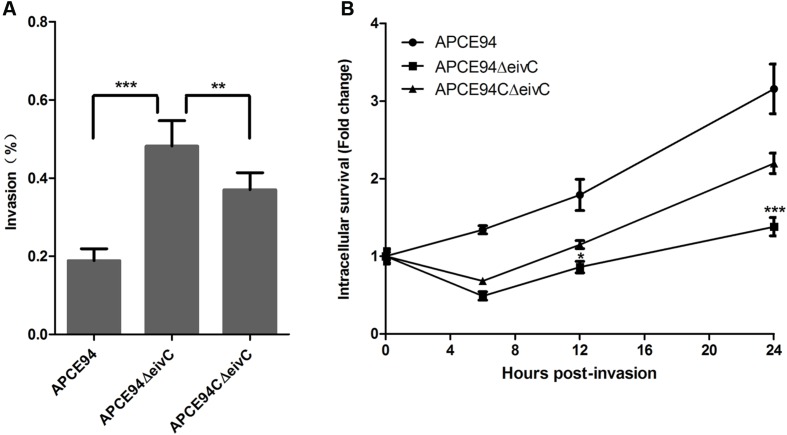
To determine whether eivC influences the intracellular survival and replication of APEC, the intracellular survival of the APCE94 strains in HD-11 cells was assessed at 0, 6, 12, and 24 h post-invasion. Unlike the case of DF-1 cells, the APCE94ΔeivC strain exhibited significantly increased invasiveness of avian HD-11 cells, compared with the APCE94 strains (p < 0.001). However, the intracellular survival of the APCE94ΔeivC strain in avian HD-11 cells was significantly lower than that of the APCE94 strain at all time points tested (p < 0.05). Complementation with eivC partially restored the invasion and intracellular survival capacities (Figure 6). These findings indicate that EivC contributes to the intracellular survival of APEC in chicken macrophages.
FIGURE 6.
Bacterial invasion and intracellular survival assays. (A) Invasion assay with avian HD-11 cells. The mutant strain APCE94ΔeivC showed significantly increased invasion to HD-11 cells, compared with the wild-type strain APCE94 and the complementation strain APCE94CΔeivC. Values are averages of three independent experiments. Error bars indicate standard deviations. Statistical significance was assessed using one-way ANOVA (∗∗p < 0.01; ∗∗∗p < 0.001). (B) Intracellular survival assay. Inactivation of EivC decreased its intracellular survival within avian HD-11 cells compared with APCE94, which was expressed as the fold change in bacterial numbers at 6, 12, and 24 h relative to the number of initial invasive bacteria. A two-way ANOVA was performed for the survival assays (∗p < 0.05; ∗∗∗p < 0.001).
EivC Contributes to Bacterial Colonization and Virulence during Systemic Infection In vivo
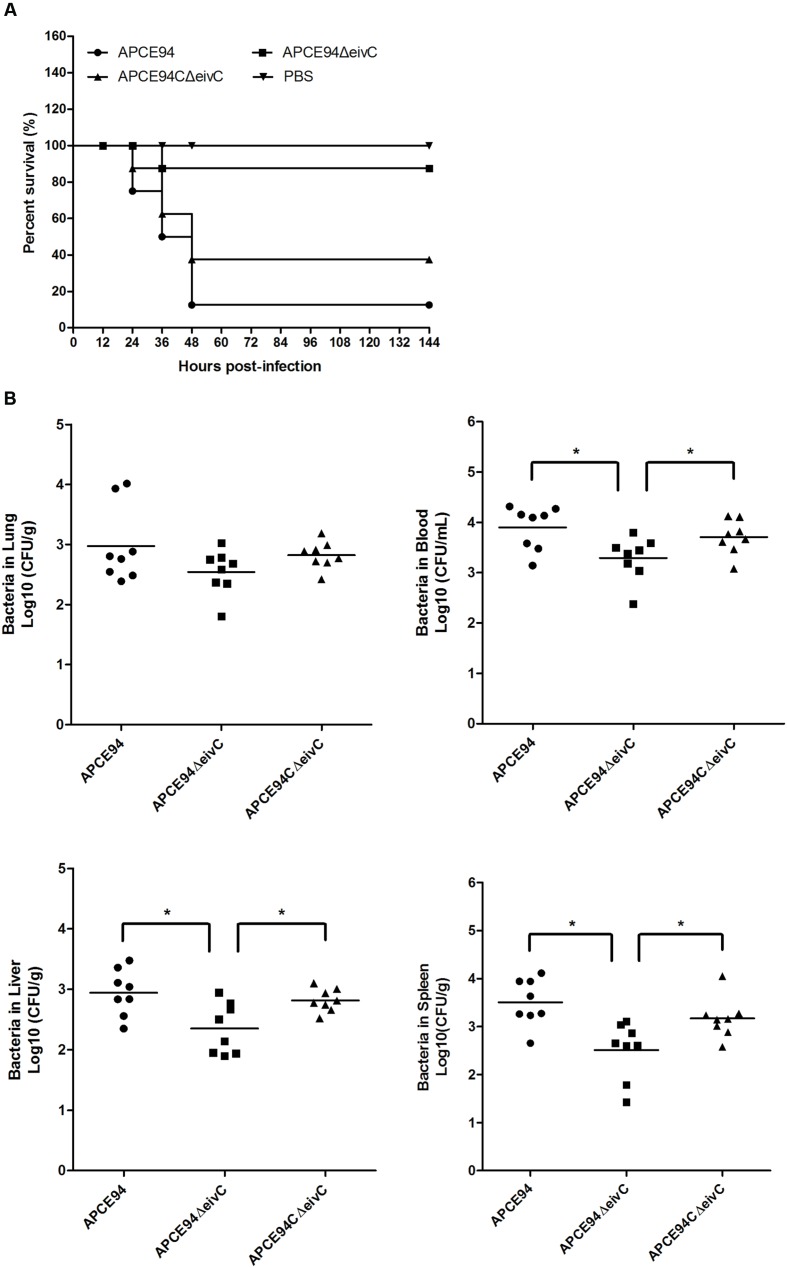
To investigate the effect of EivC on bacterial pathogenicity, the virulence of the APCE94, APCE94ΔeivC, and APCE94CΔeivC strains was compared using the duck model. As shown in Figure 7A, the mortality rate following infection with the APCE94ΔeivC strain (12.5%, 1/8) was significantly lower than that after infection with the APCE94 strain (87.5%, 7/8). The mortality rate following infection with the APCE94CΔeivC strain was restored to 62.5% (5/8). These results provide evidence that EivC contributes to the virulence of APEC.
FIGURE 7.
Animal systemic infection experiments. (A) Determination of bacterial virulence. Seven-day-old ducks were inoculated intra-tracheally with the APCE94, APCE94ΔeivC, or APCE94CΔeivC strains at 108 colony-forming units (CFUs). Ducks injected with PBS were used as negative controls. Survival was monitored until 7 days post-infection. (B) Bacterial colonization and proliferation in ducks. Groups of eight 7-day-old ducks were intra-tracheally infected with 108 CFUs of bacteria. Bacteria were recovered from the lung, blood, liver and spleen at 24 h post-infection. A non-parametric Mann–Whitney U-test was conducted to determine statistical significance (∗p < 0.05).
To measure the influence of eivC on APEC colonization in vivo, a systemic infection experiment was performed to assess bacterial proliferation in ducks. The bacterial loads in the lung, blood, liver, and spleen were determined at 24 h post-infection. The results showed a significant reduction in the number of recovered APCE94ΔeivC bacteria in the blood, liver, and spleen, compared with that of the APCE94 strain (p < 0.05), while the APCE94CΔeivC strain exhibited a higher bacterial colonization capacity. There were no significant differences in the colonization of the lungs among the APCE94ΔeivC, APCE94, and APCE94CΔeivC strains (p > 0.05; Figure 7B). These results indicate that EivC is involved in colonization and survival during infection in vivo.
Expression Profiling of Flagellar, Fimbrial, and Virulence Genes
To further explore the effect of eivC on the phenotypes and virulence of APEC, the expression of fimbrial, flagellar, and virulence genes in the APCE94, APCE94ΔeivC, and APCE94CΔeivC strains was analyzed by qRT-PCR. The transcription levels of the flagellar genes motA, flgB, flgD, flgF, and fliC, the type 1 fimbriae gene fimC, and the virulence genes ompA, aatA, iss, tsh, iucD, fyuA, and iucD in the APCE94ΔeivC strain were quantified as the fold-change relative to those of the APCE94 strain. As shown in Figure 8, the mRNA levels of the flgB, flgD, flgF, iss, and tsh genes were significantly downregulated in the APCE94ΔeivC compared with those in the APCE94 strain (p < 0.05; p < 0.001). By contrast, fimC and aatA expression was significantly upregulated in the APCE94ΔeivC strain (p < 0.001). The expression levels of these genes were restored in the APCE94CΔeivC strain. The qRT-PCR results are consistent with the phenotypes and virulence levels observed in this study.
FIGURE 8.
Quantification of virulence gene expressions. Quantitative real-time PCR (qRT-PCR) was performed to measure the expression levels of the flagellar genes motA, flgB, flgD, flgF, and fliC, the type 1 fimbrial gene fimC, and the virulence genes ompA, aatA, iss, tsh, fyuA, iucD in the APCE94, APCE94ΔeivC, and APCE94CΔeivC strains. Data were normalized to the housekeeping gene dnaE. The results are shown as relative expression ratios compared to expression in the wild-type strain APCE94. Statistical significance was assessed using two-way ANOVA (∗p < 0.05; ∗∗∗p < 0.001).
Determination of Cytokine Expression in APEC-Infected Avian HD-11 Cells
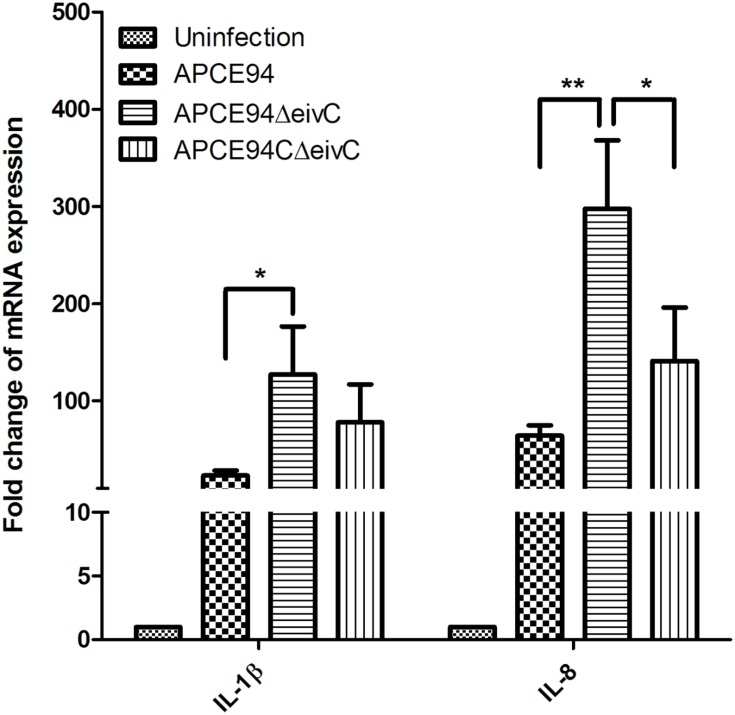
It is known that pathogen infection can stimulate the host inflammatory response (Raymond et al., 2013). Thus, the expression of the inflammatory cytokines IL-1β and IL-8 in APEC-infected avian HD-11 cells was assessed at 3 h post-invasion by qRT-PCR. The results showed that the expression of IL-1β and IL-8 was significantly upregulated in APEC-infected cells, compared with those in uninfected cells. Moreover, the IL-1β and IL-8 expression levels were significantly increased in HD-11 cells infected the APCE94ΔeivC strain, compared with those in HD-11 cells infected with the APCE94 strain (p < 0.05 and p < 0.01, respectively). The expression levels of IL-1β and IL-8 were partially restored in the APCE94CΔeivC strain (Figure 9).
FIGURE 9.
Cytokine expressions in APEC-infected avian HD-11 cells. Avian HD-11 cells were infected with the APCE94, APCE94ΔeivC, or APCE94CΔeivC strains. The mRNA levels of IL-1β and IL-8 in bacteria-infected HD-11 cells were analyzed at 3 h post-invasion by qRT-PCR. Data were normalized to the β-actin gene. Samples were calibrated to gene expression in uninfected HD-11 cells. Expression of IL-1β and IL-8 was significantly upregulated in cells infected with the APCE94ΔeivC strain, compared with those infected with the APCE94 strain. Statistical significance was assessed by two-way ANOVA (∗p < 0.05; ∗∗p < 0.01).
Discussion
Pathogenic E. coli are remarkably diverse, and they cause a wide range of diseases, ranging from gastroenteritis and diarrhea to extraintestinal infections, such as neonatal meningitis, urinary tract infections, pneumonia, and septicemia. Although the virulence factors and pathogenic mechanisms vary, pathogenic E. coli share some virulence strategies, such as a T3SS. ETT2 is known to be present, in whole or in part, in the majority of E. coli strains. Our previous study verified that the prevalence of ETT2 in APEC, similar to intestinal pathogenic E. coli, was markedly higher than that in human ExPEC (UPEC and NMEC); thus, it might be a potential risk to human health (Wang et al., 2016).
ATPases have been thought to be the key energizers of T3SSs, and their roles have been studied intensively (Paul et al., 2008). In this study, we determined the effects of the putative ETT2 ATPase EivC on the phenotype and virulence of APEC. Similar to other T3SS ATPases, EivC is homologous to the β subunit of F0F1 ATPases and contains the conserved Walker boxes A and B. We demonstrated that EivC possesses ATPase activity, hydrolyzing ATP at a rate of 0.28 ± 0.08 μmol/min/mg in this study. This ATP hydrolysis rate is similar to those of other T3SS ATPase family members, such as InvC (Eichelberg et al., 1994; Akeda and Galan, 2004), EscN (Andrade et al., 2007), SsaN (Yoshida et al., 2014), FliI (Fan and Macnab, 1996), YsaN (Chatterjee et al., 2013), HrcN (Lorenz and Buttner, 2009), and CdsN (Stone et al., 2008). Therefore, the functions of EivC in motility, serum resistance, and virulence in vitro and in vivo were evaluated.
T3SS ATPases are responsible for the export of flagellar- and virulence-related effector proteins (Cornelis and Van Gijsegem, 2000; Mota and Cornelis, 2005; Diepold and Wagner, 2014). Flagella are organelles whose main function is cell motility. Our results showed that inactivation of eivC decreased the production of flagella on the bacterial surface, thus reducing bacterial motility. However, type 1 fimbriae were augmented on the bacterial surface of the APCE94ΔeivC strain. Similarly, previous evidence indicated that inactivation of ETT2 affects the bacterial surface properties of septicemic E. coli (Ideses et al., 2005). The qRT-PCR analysis revealed that the APCE94ΔeivC strain exhibited downregulated flagellar gene (flgB, flgD, flgF) expression and upregulated fimbriae gene (fimC) expression. Moreover, previous evidence indicated that constitutive expression of type 1 fimbriae dramatically reduced the motility and flagellin expression of UPEC and E. coli K-12 MG1655 (Lane et al., 2007). Therefore, the increased fimbria expression in the APCE94ΔeivC strain might suppress flagellar gene expression, resulting in diminished motility.
T3SS ATPases are virulence factors, playing crucial roles in the pathogenicity of different pathogenic bacteria (Eichelberg et al., 1994; Fan and Macnab, 1996; Akeda and Galan, 2004; Blaylock et al., 2006; Andrade et al., 2007; Stone et al., 2008; Chatterjee et al., 2013). Here, we demonstrated that the ETT2 ATPase EivC is essential for APEC virulence. Inactivation of ATPase EivC led to significantly attenuated virulence, including impaired serum resistance, reduced intracellular survival in HD-11 cells, and diminished colonization in ducks. These defects were partially restored by complementation with eivC. Colonization and invasion play important roles during APEC infection. Fimbriae are known to contribute to adhesion to, and invasion of, host cells by bacteria (Luthje and Brauner, 2014). Previous studies showed that mutations of virulence genes (including ibeA, as well as those encoding the type VI secretion system, and the BarA-UvrY two component system) lead to a defect in expression of type 1 fimbriae, which may account for the diminished adhesion, invasion, and virulence of APEC (Herren et al., 2006; de Pace et al., 2010; Wang et al., 2011). Similarly, our study indicated that deletion of eivC increased the number and expression of type 1 fimbriae. It might be responsible for the increased invasiveness of HD-11 cells by the APCE94ΔeivC strain. However, the adherence to, and invasion of, DF-1 cells, as well as colonization of the lungs, did not differ significantly among the wild-type, mutant, and complementation strains. As well-known, in addition to colonization and invasion capacities, a number of virulence factors are also involved in the progress of infection, and these factors might be responsible for this phenomenon.
Avian pathogenic Escherichia coli initially infects poultry via the respiratory tract, and then it spreads systemically throughout the whole body. Thus, survival in serum is an important virulence parameter for APEC infection. There is a correlation between resistance to serum bactericidal effects and the capacity of APEC strains to cause septicemia and mortality (La Ragione and Woodward, 2002; Mellata et al., 2003). In addition, the capacity to survive within macrophages prevents the elimination of APEC by the host immune response. Bacterial surface components, such as lipopolysaccharide (LPS), capsules, as well as increased production of serum survival proteins, play important roles in resistance to serum and other factors present in macrophages. The bacteria surface structural alteration of the APCE94ΔeivC strain may contribute to reduced fitness within serum or a host. To determine whether the LPS of the APCE94ΔeivC strain was defective, LPS was purified from the APEC strains and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining. However, no changes of the LPS profile were found among the mutant strain APCE94ΔeivC, the wild-type strain APCE94, and the complementation strain APCE94CΔeivC. However, the qRT-PCR results demonstrated that the transcriptional levels of virulence genes involved in serum resistance (iss; Foley et al., 2000), and a gene encoding an autotransporter protein (tsh; Dozois et al., 2000), were significantly decreased in the APCE94ΔeivC strain, which might be a reason for the serum resistance defects, as well as the reduced colonization and proliferation capacities and virulence in vivo. Indeed, it has been discovered that ETT2 can affect the expression of virulence genes outside the ETT2 cluster, and it can indirectly regulate the virulence of intestinal pathogenic E. coli (Zhang et al., 2004; Sheikh et al., 2006). Similarly, previous evidence has shown that ETT2, although degenerate, is involved in serum survival, invasion, and intracellular survival, which are necessary for the full virulence of septicemic E. coli and NMEC (Ideses et al., 2005; Yao et al., 2009). However, the underlying mechanisms are unclear.
Bacteria utilize T3SSs to deliver effector proteins into host cells, which play various roles in invasion, intracellular survival, and subversion of innate immune responses. During infection, bacteria and their components can induce strong inflammatory and immune responses. Nevertheless, pathogenic bacteria are able to efficiently modulate host inflammatory and immune responses, thereby facilitating their proliferation and infections (Figueira and Holden, 2012; Raymond et al., 2013). Pathogens utilize multiple different mechanisms to disrupt the inflammatory response, including nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and mitogen-activated protein kinase (MAPK) signaling pathways. In EHEC and EPEC, the LEE effector proteins NleE, NleB, NleC, NleH, and Tir cooperatively interact with different proteins of the NF-κB signaling pathway, and subsequently, prevent the production of the inflammatory cytokine IL-8 (Nadler et al., 2010; Vossenkamper et al., 2010; Yen et al., 2010; Ruchaud-Sparagano et al., 2011; Pham et al., 2012; Gao et al., 2013). In addition, the MAPK signaling pathway is targeted by the effectors Tir and NleD to inhibit the inflammatory response and cytokine production (Baruch et al., 2011; Sham et al., 2011). A number of T3SS effector proteins, which also play roles in the inhibition of the inflammatory response, have been identified in Salmonella, Shigella, and Yersinia (Haraga and Miller, 2003; Zurawski et al., 2009; Paquette et al., 2012; Sanada et al., 2012), whereas the degenerate ETT2 is thought to be a non-functional secretion system because no effector proteins were identified in laboratory conditions following treatment with known inducers of T3SSs in other bacteria (Ren et al., 2004). T3SSs are generally activated by interactions with host cells, which might be a reason for the failure to identify effector proteins. Our results showed that the ETT2 ATPase EivC suppressed the expression of the inflammatory cytokines IL-1β and IL-8, and facilitated bacterial infection and virulence. APEC might utilize unidentified ETT2 effector proteins or other bacterial surface components to interfere with the host inflammatory response. However, the exact mechanisms by which the ETT2 interferes with the inflammatory response remain unknown. Such mechanisms need to be investigated in the future to help us prevent poultry colibacillosis and potential human infections (Kline et al., 2012; Marshall and Finlay, 2014).
In summary, we characterized the functions of the putative ETT2 ATPase EivC in an APEC strain. EivC is homologous to the β subunit of F0F1 ATPases and it possesses ATPase activity. EivC was crucial for bacterial flagellation and motility. In addition, ATPase EivC was involved in serum resistance, intra-macrophage survival, proliferation in vivo, and subversion of host inflammatory response.
Author Contributions
SW planned the experiments. SW, XL, XX, DY, DW, and YS conducted the experiments. SW, XL, XH, MT, CD, DP, and SY analyzed and discussed the experimental results. SW and SY wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by funds from the National Natural Science Foundation of China (grant no. 31572523 and 31370045), the Chinese Special Fund for Agro-scientific Research in the Public Interest (grant no 201303044), and the Shanghai Key Project on Agricultural Development through Science and Technology (grant no. 2016HNG4-1).
References
- Akeda Y., Galan J. E. (2004). Genetic analysis of the Salmonella enterica type III secretion-associated ATPase InvC defines discrete functional domains. J. Bacteriol. 186 2402–2412. 10.1128/JB.186.8.2402-2412.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akeda Y., Galan J. E. (2005). Chaperone release and unfolding of substrates in type III secretion. Nature 437 911–915. 10.1038/nature03992 [DOI] [PubMed] [Google Scholar]
- Andrade A., Pardo J. P., Espinosa N., Perez-Hernandez G., Gonzalez-Pedrajo B. (2007). Enzymatic characterization of the enteropathogenic Escherichia coli type III secretion ATPase EscN. Arch. Biochem. Biophys. 468 121–127. 10.1016/j.abb.2007.09.020 [DOI] [PubMed] [Google Scholar]
- Baruch K., Gur-Arie L., Nadler C., Koby S., Yerushalmi G., Ben-Neriah Y., et al. (2011). Metalloprotease type III effectors that specifically cleave JNK and NF-kappaB. EMBO J. 30 221–231. 10.1038/emboj.2010.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaylock B., Riordan K. E., Missiakas D. M., Schneewind O. (2006). Characterization of the Yersinia enterocolitica type III secretion ATPase YscN and its regulator, YscL. J. Bacteriol. 188 3525–3534. 10.1128/JB.188.10.3525-3534.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee R., Halder P. K., Datta S. (2013). Identification and molecular characterization of YsaL (Ye3555): a novel negative regulator of YsaN ATPase in type three secretion system of enteropathogenic bacteria Yersinia enterocolitica. PLoS ONE 8:e75028 10.1371/journal.pone.0075028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D., Zhu S., Su Z., Zuo W., Lu H. (2012). Prevalence and isoforms of the pathogenicity island ETT2 among Escherichia coli isolates from colibacillosis in pigs and mastitis in cows. Curr. Microbiol. 64 43–49. 10.1007/s00284-011-0032-0 [DOI] [PubMed] [Google Scholar]
- Coburn B., Sekirov I., Finlay B. B. (2007). Type III secretion systems and disease. Clin. Microbiol. Rev. 20 535–549. 10.1128/CMR.00013-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G. R., Van Gijsegem F. (2000). Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54 735–774. 10.1146/annurev.micro.54.1.735 [DOI] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pace F., Nakazato G., Pacheco A., de Paiva J. B., Sperandio V., da Silveira W. D. (2010). The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect. Immun. 78 4990–4998. 10.1128/IAI.00531-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A., Armitage J. P. (2015). Type III secretion systems: the bacterial flagellum and the injectisome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370:1679 10.1098/rstb.2015.0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A., Wagner S. (2014). Assembly of the bacterial type III secretion machinery. FEMS Microbiol. Rev. 38 802–822. 10.1111/1574-6976.12061 [DOI] [PubMed] [Google Scholar]
- Dozois C. M., Dho-Moulin M., Bree A., Fairbrother J. M., Desautels C., Curtiss R., III (2000). Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 68 4145–4154. 10.1128/IAI.68.7.4145-4154.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberg K., Ginocchio C. C., Galan J. E. (1994). Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J. Bacteriol. 176 4501–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F., Macnab R. M. (1996). Enzymatic characterization of FliI. An ATPase involved in flagellar assembly in Salmonella typhimurium. J. Biol. Chem. 271 31981–31988. 10.1074/jbc.271.50.31981 [DOI] [PubMed] [Google Scholar]
- Figueira R., Holden D. W. (2012). Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology 158(Pt. 5), 1147–1161. 10.1099/mic.0.058115-0 [DOI] [PubMed] [Google Scholar]
- Foley S. L., Horne S. M., Giddings C. W., Robinson M., Nolan L. K. (2000). Iss from a virulent avian Escherichia coli. Avian Dis. 44 185–191. 10.2307/1592523 [DOI] [PubMed] [Google Scholar]
- Franzin F. M., Sircili M. P. (2015). Locus of enterocyte effacement: a pathogenicity island involved in the virulence of enteropathogenic and enterohemorragic Escherichia coli subjected to a complex network of gene regulation. Biomed. Res. Int. 2015:534738 10.1155/2015/534738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Wang X., Pham T. H., Feuerbacher L. A., Lubos M. L., Huang M., et al. (2013). NleB, a bacterial effector with glycosyltransferase activity, targets GAPDH function to inhibit NF-kappaB activation. Cell Host Microbe 13 87–99. 10.1016/j.chom.2012.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Bai H., Tu J., Yang L., Xu D., Wang S., et al. (2015). Deletion of luxS further attenuates the virulence of the avian pathogenic Escherichia coli aroA mutant. Microb. Pathog. 88 39–47. 10.1016/j.micpath.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Hansen-Wester I., Hensel M. (2001). Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3 549–559. 10.1016/S1286-4579(01)01411-3 [DOI] [PubMed] [Google Scholar]
- Haraga A., Miller S. I. (2003). A Salmonella enterica serovar typhimurium translocated leucine-rich repeat effector protein inhibits NF-kappa B-dependent gene expression. Infect. Immun. 71 4052–4058. 10.1128/IAI.71.7.4052-4058.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Makino K., Ohnishi M., Kurokawa K., Ishii K., Yokoyama K., et al. (2001). Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8 11–22. 10.1093/dnares/8.1.11 [DOI] [PubMed] [Google Scholar]
- Herren C. D., Mitra A., Palaniyandi S. K., Coleman A., Elankumaran S., Mukhopadhyay S. (2006). The BarA-UvrY two-component system regulates virulence in avian pathogenic Escherichia coli O78:K80:H9. Infect. Immun. 74 4900–4909. 10.1128/IAI.00412-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideses D., Gophna U., Paitan Y., Chaudhuri R. R., Pallen M. J., Ron E. Z. (2005). A degenerate type III secretion system from septicemic Escherichia coli contributes to pathogenesis. J. Bacteriol. 187 8164–8171. 10.1128/JB.187.23.8164-8171.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouihri N., Sory M. P., Page A. L., Gounon P., Parsot C., Allaoui A. (2003). MxiK and MxiN interact with the Spa47 ATPase and are required for transit of the needle components MxiH and MxiI, but not of Ipa proteins, through the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 49 755–767. 10.1046/j.1365-2958.2003.03590.x [DOI] [PubMed] [Google Scholar]
- Kline T., Felise H. B., Sanowar S., Miller S. I. (2012). The type III secretion system as a source of novel antibacterial drug targets. Curr. Drug Targets 13 338–351. 10.2174/138945012799424642 [DOI] [PubMed] [Google Scholar]
- La Ragione R. M., Woodward M. J. (2002). Virulence factors of Escherichia coli serotypes associated with avian colisepticaemia. Res. Vet. Sci. 73 27–35. 10.1016/S0034-5288(02)00075-9 [DOI] [PubMed] [Google Scholar]
- Lane M. C., Simms A. N., Mobley H. L. (2007). complex interplay between type 1 fimbrial expression and flagellum-mediated motility of uropathogenic Escherichia coli. J. Bacteriol. 189 5523–5533. 10.1128/JB.00434-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Sourjik V. (2011). Assembly and stability of flagellar motor in Escherichia coli. Mol. Microbiol. 80 886–899. 10.1111/j.1365-2958.2011.07557.x [DOI] [PubMed] [Google Scholar]
- Lorenz C., Buttner D. (2009). Functional characterization of the type III secretion ATPase HrcN from the plant pathogen Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 191 1414–1428. 10.1128/JB.01446-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthje P., Brauner A. (2014). Virulence factors of uropathogenic E. coli and their interaction with the host. Adv. Microb. Physiol. 65 337–372. 10.1016/bs.ampbs.2014.08.006 [DOI] [PubMed] [Google Scholar]
- Marshall N. C., Finlay B. B. (2014). Targeting the type III secretion system to treat bacterial infections. Expert Opin. Ther. Targets 18 137–152. 10.1517/14728222.2014.855199 [DOI] [PubMed] [Google Scholar]
- Mellata M., Dho-Moulin M., Dozois C. M., Curtiss R., III, Brown P. K., Arne P., et al. (2003). Role of virulence factors in resistance of avian pathogenic Escherichia coli to serum and in pathogenicity. Infect. Immun. 71 536–540. 10.1128/IAI.71.1.536-540.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T., Imada K., Namba K. (2008). Mechanisms of type III protein export for bacterial flagellar assembly. Mol. Biosyst. 4 1105–1115. 10.1039/b808065h [DOI] [PubMed] [Google Scholar]
- Minamino T., MacNab R. M. (2000). FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol. Microbiol. 37 1494–1503. 10.1046/j.1365-2958.2000.02106.x [DOI] [PubMed] [Google Scholar]
- Miyazaki J., Ba-Thein W., Kumao T., Akaza H., Hayashi H. (2002). Identification of a type III secretion system in uropathogenic Escherichia coli. FEMS Microbiol. Lett. 212 221–228. 10.1111/j.1574-6968.2002.tb11270.x [DOI] [PubMed] [Google Scholar]
- Moest T. P., Meresse S. (2013). Salmonella T3SSs: successful mission of the secret(ion) agents. Curr. Opin. Microbiol. 16 38–44. 10.1016/j.mib.2012.11.006 [DOI] [PubMed] [Google Scholar]
- Mota L. J., Cornelis G. R. (2005). The bacterial injection kit: type III secretion systems. Ann. Med. 37 234–249. 10.1080/07853890510037329 [DOI] [PubMed] [Google Scholar]
- Nadler C., Baruch K., Kobi S., Mills E., Haviv G., Farago M., et al. (2010). The type III secretion effector NleE inhibits NF-kappaB activation. PLoS Pathog. 6:e1000743 10.1371/journal.ppat.1000743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen M. J., Bailey C. M., Beatson S. A. (2006). Evolutionary links between FliH/YscL-like proteins from bacterial type III secretion systems and second-stalk components of the FoF1 and vacuolar ATPases. Protein Sci. 15 935–941. 10.1110/ps.051958806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette N., Conlon J., Sweet C., Rus F., Wilson L., Pereira A., et al. (2012). Serine/threonine acetylation of TGFbeta-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc. Natl. Acad. Sci. U.S.A. 109 12710–12715. 10.1073/pnas.1008203109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K., Erhardt M., Hirano T., Blair D. F., Hughes K. T. (2008). Energy source of flagellar type III secretion. Nature 451 489–492. 10.1038/nature06497 [DOI] [PubMed] [Google Scholar]
- Pham T. H., Gao X., Tsai K., Olsen R., Wan F., Hardwidge P. R. (2012). Functional differences and interactions between the Escherichia coli type III secretion system effectors NleH1 and NleH2. Infect. Immun. 80 2133–2140. 10.1128/IAI.06358-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager R., Bauerfeind R., Tietze E., Behrend J., Fruth A., Tschape H. (2004). Prevalence and deletion types of the pathogenicity island ETT2 among Escherichia coli strains from oedema disease and colibacillosis in pigs. Vet. Microbiol. 99 287–294. 10.1016/j.vetmic.2004.01.011 [DOI] [PubMed] [Google Scholar]
- Raymond B., Young J. C., Pallett M., Endres R. G., Clements A., Frankel G. (2013). Subversion of trafficking, apoptosis, and innate immunity by type III secretion system effectors. Trends Microbiol. 21 430–441. 10.1016/j.tim.2013.06.008 [DOI] [PubMed] [Google Scholar]
- Ren C. P., Chaudhuri R. R., Fivian A., Bailey C. M., Antonio M., Barnes W. M., et al. (2004). The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J. Bacteriol. 186 3547–3560. 10.1128/JB.186.11.3547-3560.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud-Sparagano M. H., Muhlen S., Dean P., Kenny B. (2011). The enteropathogenic E. coli (EPEC) Tir effector inhibits NF-kappaB activity by targeting TNFalpha receptor-associated factors. PLoS Pathog. 7:e1002414 10.1371/journal.ppat.1002414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada T., Kim M., Mimuro H., Suzuki M., Ogawa M., Oyama A., et al. (2012). The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature 483 623–626. 10.1038/nature10894 [DOI] [PubMed] [Google Scholar]
- Sham H. P., Shames S. R., Croxen M. A., Ma C., Chan J. M., Khan M. A., et al. (2011). Attaching and effacing bacterial effector NleC suppresses epithelial inflammatory responses by inhibiting NF-kappaB and p38 mitogen-activated protein kinase activation. Infect. Immun. 79 3552–3562. 10.1128/IAI.05033-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh J., Dudley E. G., Sui B., Tamboura B., Suleman A., Nataro J. P. (2006). EilA, a HilA-like regulator in enteroaggregative Escherichia coli. Mol. Microbiol. 61 338–350. 10.1111/j.1365-2958.2006.05234.x [DOI] [PubMed] [Google Scholar]
- Stone C. B., Johnson D. L., Bulir D. C., Gilchrist J. D., Mahony J. B. (2008). Characterization of the putative type III secretion ATPase CdsN (Cpn0707) of Chlamydophila pneumoniae. J. Bacteriol. 190 6580–6588. 10.1128/JB.00761-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossenkamper A., Marches O., Fairclough P. D., Warnes G., Stagg A. J., Lindsay J. O., et al. (2010). Inhibition of NF-kappaB signaling in human dendritic cells by the enteropathogenic Escherichia coli effector protein NleE. J. Immunol. 185 4118–4127. 10.4049/jimmunol.1000500 [DOI] [PubMed] [Google Scholar]
- Wang S., Dai J., Meng Q., Han X., Han Y., Zhao Y., et al. (2014). DotU expression is highly induced during in vivo infection and responsible for virulence and Hcp1 secretion in avian pathogenic Escherichia coli. Front. Microbiol. 5:588 10.3389/fmicb.2014.00588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Liu X., Xu X., Zhao Y., Yang D., Han X., et al. (2016). Escherichia coli type III secretion system 2 (ETT2) is widely distributed in avian pathogenic Escherichia coli isolates from Eastern China. Epidemiol. Infect. 10.1017/S0950268816000820 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Niu C., Shi Z., Xia Y., Yaqoob M., Dai J., et al. (2011). Effects of ibeA deletion on virulence and biofilm formation of avian pathogenic Escherichia coli. Infect. Immun. 79 279–287. 10.1128/IAI.00821-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woestyn S., Allaoui A., Wattiau P., Cornelis G. R. (1994). YscN, the putative energizer of the Yersinia Yop secretion machinery. J. Bacteriol. 176 1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Xie Y., Perace D., Zhong Y., Lu J., Tao J., et al. (2009). The type III secretion system is involved in the invasion and intracellular survival of Escherichia coli K1 in human brain microvascular endothelial cells. FEMS Microbiol. Lett. 300 18–24. 10.1111/j.1574-6968.2009.01763.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H., Ooka T., Iguchi A., Hayashi T., Sugimoto N., Tobe T. (2010). NleC, a type III secretion protease, compromises NF-kappaB activation by targeting p65/RelA. PLoS Pathog. 6:e1001231 10.1371/journal.ppat.1001231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Miki T., Ono S., Haneda T., Ito M., Okada N. (2014). Functional characterization of the type III secretion ATPase SsaN encoded by Salmonella pathogenicity island 2. PLoS ONE 9:e94347 10.1371/journal.pone.0094347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarivach R., Vuckovic M., Deng W., Finlay B. B., Strynadka N. C. (2007). Structural analysis of a prototypical ATPase from the type III secretion system. Nat. Struct. Mol. Biol. 14 131–137. 10.1038/nsmb1196 [DOI] [PubMed] [Google Scholar]
- Zhang L., Chaudhuri R. R., Constantinidou C., Hobman J. L., Patel M. D., Jones A. C., et al. (2004). Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 72 7282–7293. 10.1128/IAI.72.12.7282-7293.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Guo Z., Duan Q., Hardwidge P. R., Zhu G. (2014). Escherichia coli type III secretion system 2: a new kind of T3SS? Vet. Res. 45:32 10.1186/1297-9716-45-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski D. V., Mumy K. L., Faherty C. S., McCormick B. A., Maurelli A. T. (2009). Shigella flexneri type III secretion system effectors OspB and OspF target the nucleus to downregulate the host inflammatory response via interactions with retinoblastoma protein. Mol. Microbiol. 71 350–368. 10.1111/j.1365-2958.2008.06524.x [DOI] [PMC free article] [PubMed] [Google Scholar]