Abstract
Background
The ginsenoside Rb1 (Rb1) is the most abundant compound in the root of Panax ginseng. Recent studies have shown that Rb1 has a neuroprotective effect. However, the mechanisms underlying this effect are still unknown.
Methods
We used stable isotope labeling with amino acids in cell culture, combined with quantitative mass spectrometry, to explore a potential protective mechanism of Rb1 in β-amyloid-treated neuronal cells.
Results
A total of 1,231 proteins were commonly identified from three replicate experiments. Among these, 40 proteins were significantly changed in response to Rb1 pretreatment in β-amyloid-treated neuronal cells. Analysis of the functional enrichments and protein interactions of altered proteins revealed that actin cytoskeleton proteins might be linked to the regulatory mechanisms of Rb1. The CAP1, CAPZB, TOMM40, and DSTN proteins showed potential as molecular target proteins for the functional contribution of Rb1 in Alzheimer's disease (AD).
Conclusion
Our proteomic data may provide new insights into the protective mechanisms of Rb1 in AD.
Keywords: actin skeleton, Alzheimer's disease, β-amyloid, ginsenoside Rb1, mass spectrometry
1. Introduction
The root of Panax ginseng (Ginseng) has been used in traditional oriental medicine to improve health for more than a thousand years in Asia. A number of studies have reported the neuroprotective effects of ginseng [1]. Cognitive behavior in patients with Alzheimer's disease (AD) was improved by ginseng powder [2]. Ginseng extract prevented the development of locomotion deficits in patients with Parkinson's disease [3].
The main bioactive components of ginseng are known as ginsenosides, which have been identified in > 30 species [4]. It has been reported that neuroprotective effects on central nervous system disorders and neuronal diseases can be attributed to the ginsenosides [5]. The effects of ginsenosides have been shown via increased cell survival, extension of neurite growth, and neuronal rescues both in vivo and in vitro [1]. Of these, ginsenoside Rb1 (Rb1) has been reported to be the primary ginsenoside responsible for the neuroprotective effects of neurodegenerative diseases [6]. Hippocampal neurons were protected by Rb1 against either ischemia or glutamate-induced neuronal diseases [7]. Recently, several studies have reported the protective effects of Rb1 against AD. Rb1 improved AD by increasing brain-derived neurotrophic factor and decreasing Tau protein [8] and protected neuronal cells from injury with β-amyloid (Aβ) treatment [9], [10]. Additionally, Rb1 demonstrated anti-neuroinflammation effects in a rat model of AD [11].
In the past decade, many studies using state-of-the-art technologies have tried to understand the molecular mechanisms and to find biomarkers for the early diagnosis and treatment of AD [12]. In particular, proteomic studies, which provide powerful tools to identify the dynamic expression of proteins in biological samples, have been used to identify the molecular pathways involved in neuropathogenesis. From these studies, a number of potential target proteins have been identified for AD [13], [14], [15]. Recently, these proteomic studies have attempted to investigate the molecular effects of ginsenosides in cancer, smooth muscle cells, and diabetes [16], [17], [18]. However, even though a number of studies examining the protective effects of Rb1 are ongoing, our understanding of the regulatory mechanisms of Rb1 in AD is still lacking.
We performed a mass spectrometry (MS)-based proteomics experiment using stable isotope labeling with amino acids in cell culture (SILAC) to identify any proteins that are significantly altered by the neuroprotective effects of Rb1 in Aβ-treated neuronal cells. By following this approach, our data provide several new candidate proteins involved in the protective mechanisms of Rb1 and offer new insights into the potential molecular mechanisms of Rb1 in AD.
2. Materials and methods
2.1. SILAC
SILAC experiments were carried out as previously described [19]. In brief, SH-SY5Y cells were grown for at least five cell divisions in either “light media” containing 12C6-Arg and 12C6, 14N2-Lys or “heavy media” containing 13C6-Arg and 13C6, 15N2-Lys supplemented with 10% dialyzed fetal bovine serum (Invitrogen, New York, NY, USA), 50 IU/mL penicillin, and 50 mg/mL streptomycin. The labeled cells were pretreated with (light media) or without (heavy media) 100μM Rb1 for 24 h and then exposed to 25μM Aβ25–35 (Sigma-Aldrich, St. Louis, MO, USA) for 24 h. The cells were then lysed in buffer containing 1% Triton X-100, 150mM NaCl, 1mM EDTA, 50mM Tris–HCl (pH 8.0), 1mM sodium orthovanadate, 5mM NaF, 5mM sodium pyrophosphate, 1mM phenylmethylsulfonyl fluoride (PMSF), aprotinin (1.5 μg/mL), antipain (10 μg/mL), leupeptin (10 μg/mL), and benzamidine (0.1 mg/mL). The lysates were centrifuged at 160,000g and mixed at a 1:1 ratio according to their protein concentration. The combined protein lysates were separated via 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie Brilliant Blue G-250 (Bio-Rad, Hercules, CA, USA).
2.2. In-gel digestion and MS analysis
Each gel was sliced into 10 bands of equal size, destained with 50% acetonitrile (ACN) in 25mM ammonium bicarbonate and dried in a speed vacuum concentrator. Dried gel pieces were rehydrated using 25mM ammonium bicarbonate (pH 8.0) containing 50 ng of trypsin and incubated at 37°C for 16–24 h. Tryptic peptide mixtures were extracted with 50% ACN in 5% formic acid (FA) and dried in a speed vacuum concentrator. Extracted peptides were analyzed using the Agilent HPLC-Chip/TOF MS system with the Agilent 1260 nano-LC system, HPLC-Chip-cube MS interface, and a 6530 QTOF single quadrupole-TOF mass spectrometer (Agilent Technologies, Santa Clara, CA, USA). The dried peptides were resuspended in 2% ACN/0.1% FA and concentrated on a large-capacity HPLC Chip incorporated with an enrichment column (9 mm, 75 μm I.D., 160 nL) and a reverse-phase column (15 cm, 76 μm I.D., packed with Zorbax 300SB-C18 5-μm resin). The peptides were separated by a 70-min gradient of 3–45% buffer B (buffer A contained 0.1% FA and buffer B contained 90% ACN/0.1% FA) at a flow rate of 300 nL/min. The MS and MS/MS data were acquired in the positive ion mode and data stored centroid mode. The chip spray voltage was set at 1850 V and maintained under chip conditions. The drying gas temperature was set at 325°C with a flow rate of 3.5 L/min. A medium isolation (4 m/z) window was used for precursor isolation. A collision energy with a slope of 3.7 V/100 Da and an offset of 2.5 V was used for fragmentation. Additionally, whereas the MS data were acquired over a mass range of 300–3,000 m/z, the MS/MS data were acquired over a 50–2500 m/z mass range. Reference mass correction was performed using a reference mass of 922. Precursors were set in an exclusion list for 0.5 min after two MS/MS spectra. The elution profiles of the light and heavy peptides were isolated and quantified based on the area of each peptide peak, and the abundance ratio was calculated based on these areas by Xpress. Database searches were performed with a peptide mass tolerance of 20 ppm, an MS/MS tolerance of 0.5 Da, and a strict tryptic specificity (cleavage after lysine and arginine) allowing one missed cleavage site; carbamidomethylation of Cys was set as a fixed modification, whereas methione oxidation (M) was considered a variable modification.
Quantitative protein ratios were determined by the average levels of quantified peptides.
2.3. Bioinformatics analysis
Gene Ontology (GO) distribution analysis was performed using the DAVID database. Analysis of the protein–protein interaction networks was carried out using the STRING database Cytoscape plugin [20].
2.4. Immunoblotting
Protein lysates were separated by 7.5% SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). The membranes were blocked with 4% skim milk and then incubated with anti-PARP (Cell Signaling, Danvers, MA, USA) and anti-Mortalin (NeuroMab, Davis, CA, USA), followed by incubation with horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G at room temperature. The proteins were visualized using enhanced chemiluminescence (ECL).
3. Results and discussion
3.1. SILAC-based proteomic analysis
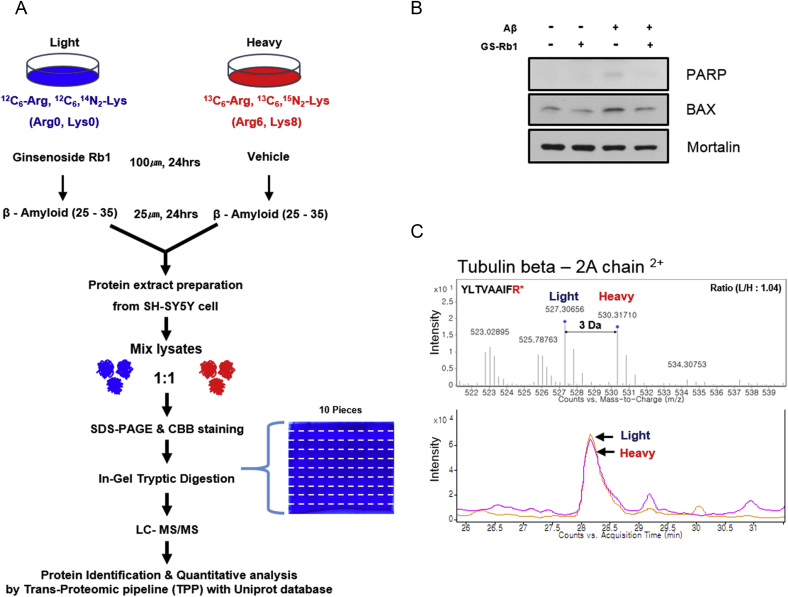
To explore the potential protective mechanisms of Rb1 in AD, we performed a large-scale proteomic analysis using SILAC combined with nano-LC tandem mass spectrometry (nano-LC-MS/MS) to investigate proteins expressed differentially owing to pretreatment with Rb1 in Aβ-treated SH-SY5Y cells. Cells grown in “light” media containing 12C6-Arg and 12C6,14N2-Lys were exposed to Aβ after pre-treatment with Rb1, whereas the cells grown in “heavy” media containing 13C6-Arg and 13C6, 15N2-Lys were treated with Aβ after pretreatment with vehicle (Fig. 1A). As seen in previous studies [21], Aβ treatment resulted in PARP-1 cleavage and increased Bax levels, which is a marker of apoptotic cells. These changes were prevented by Rb1 pretreatment (Fig. 1B). These results showed that Aβ treatment efficiently induced neurotoxicity, and Rb1 prevented Aβ-induced neurotoxicity in SH-SY5Y cells. Analysis of the monoisotopic peaks from our SILAC experiments indicated an expected 1:1: ratio between Rb1 pretreated and control samples, as illustrated in Fig. 1C.
Fig. 1.
SILAC analysis of pretreatment with Rb1 in Aβ-induced neurocytotoxicity. (A) An overview of SILAC experiments. Cells were pretreated with or without 100μM Rb1 for 24 h and then exposed to 25μM Aβ for 24 h. Each cell was lysed, and equal amounts of proteins were combined and then separated by SDS-PAGE. The gel lane was divided into 10 regions and analyzed using nano-LC MS/MS as described in the Materials and methods section. (B) Immunoblot analysis of Rb1 during Aβ exposure in SH-SY5Y cells. Decreased PARP-1 cleavage and Bax were observed with Rb1 pretreatment. (C). Paired peptides of a tubulin beta-2A chain showed an approximate ratio of 1:1. SH-SY5Y cells were cultured in light media containing 12C6-Arg and 12C6, 14N2-Lys or heavy media containing 13C6-Arg and 13C6, 15N2-Lys. Equal amounts of protein concentration were combined at a 1:1 ratio and were identified and quantified by nano-LC MS/MS. Aβ, β-amyloid; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; SILAC, stable isotope labeling with amino acids in cell culture.
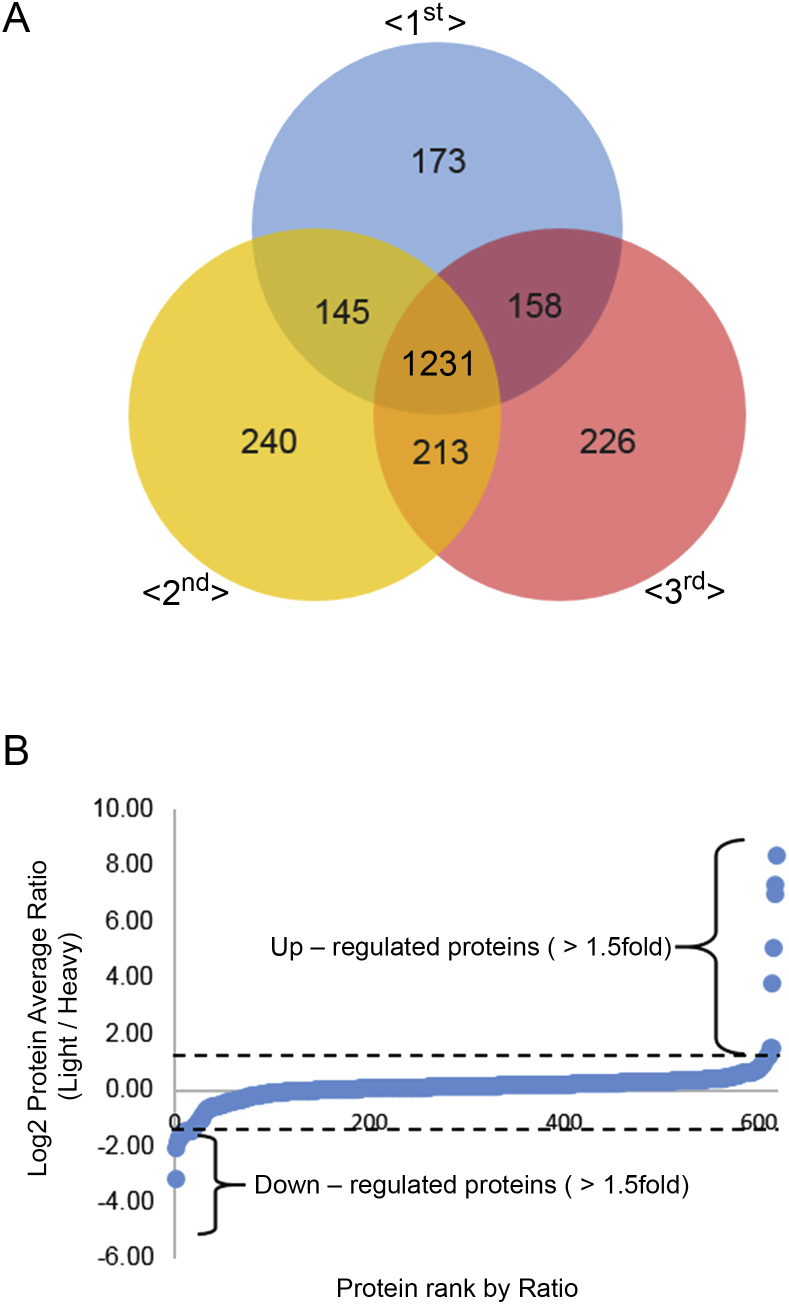
We performed three independent SILAC-based proteomic experiments and used protein sets with paired light- and heavy-labeled peptides found in the three replicate experiments. A total of 1707 proteins in the first dataset, 1829 proteins in the second dataset, and 1828 proteins in the third dataset were identified. Of these, 1231 proteins were commonly identified in all three independent datasets (Table S1; Fig. 2A). To detect the differentially expressed proteins, XPRESS was used to compare the extracted ion chromatography of the light- and heavy-labeled peptides from nano-LC-MS/MS [19]. A fold-change cutoff of > 1.5 was applied, and the commonly detected proteins were only used in three independent datasets for quantitative analysis. Forty of the proteins showed a significant difference of > 1.5-fold (Fig. 2B). Among these, 36 proteins were upregulated and four proteins were downregulated by pretreatment with Rb1 in Aβ-treated SH-SY5Y cells (Fig. 2B and Table 1).
Fig. 2.
Quantitative analysis of the proteins differentially expressed following Rb1 pretreatment of Aβ-induced neurotoxicity. (A) A Venn diagram demonstrating the overlap between the proteins identified in three replicates after Rb1 pretreatment in Aβ-induced neurotoxicity. A total of 1,231 proteins overlapped from the three replicates. (B) Distribution of protein expression levels by SILAC. Thirty-six proteins were significantly upregulated, and four proteins were significantly downregulated by > 1.5-fold. Aβ, β-amyloid; SILAC, stable isotope labeling with amino acids in cell culture.
Table 1.
List of proteins altered with a >1.5-fold change in three independent replicates
| Accession number1) | Gene name | Protein description | Log2 ratio (treat/control) |
|---|---|---|---|
| P16402 | HIST1H1D | Histone H1.3 | 7.32 |
| P17066 | HSPA6 | Heat shock 70 kDa protein 6 | 5.67 |
| P36776-2 | LONP1 | Isoform 2 of Lon protease homolog, mitochondrial | 3.82 |
| P31939-2 | ATIC | Isoform 2 of bifunctional purine biosynthesis protein PURH | 1.50 |
| O43809 | NUDT21 | Cleavage and polyadenylation specificity factor subunit 5 | 1.48 |
| P07951-2 | TPM2 | Isoform 2 of tropomyosin beta chain | 1.30 |
| P60981-2 | DSTN | Isoform 2 of destrin | 1.24 |
| O96008 | TOMM40 | Mitochondrial import receptor subunit TOM40 homolog | 1.22 |
| P10412 | HIST1H1E | Histone H1.4 | 1.21 |
| P16403 | HIST1H1C | Histone H1.2 | 1.17 |
| O14874-2 | BCKDK | Isoform 2 of {3-methyl-2-oxobutanoate dehydrogenase [lipoamide]} kinase, mitochondrial | 1.14 |
| P47914 | RPL29 | 60S ribosomal protein L29 | 1.04 |
| Q01518 | CAP1 | Adenylyl cyclase-associated protein 1 | 1.00 |
| Q12849-5 | GRSF1 | Isoform 2 of G-rich sequence factor 1 | 0.91 |
| P51570-2 | GALK1 | Isoform 2 of galactokinase | 0.90 |
| Q8NBX0 | SCCPDH | Saccharopine dehydrogenase-like oxidoreductase | 0.90 |
| P17980 | PSMC3 | 26S protease regulatory subunit 6A | 0.81 |
| P61221 | ABCE1 | ATP-binding cassette subfamily E member 1 | 0.73 |
| P46940 | IQGAP1 | Ras GTPase-activating-like protein IQGAP1 | 0.72 |
| P12955-2 | PEPD | Isoform 2 of Xaa-Pro dipeptidase | 0.71 |
| Q16795 | NDUFA9 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial | 0.67 |
| Q8NBU5-2 | ATAD1 | Isoform 2 of ATPase family AAA domain-containing protein 1 | 0.66 |
| P47756-2 | CAPZB | Isoform 2 of F-actin-capping protein subunit beta | 0.66 |
| Q5JTZ9 | AARS2 | Alanine–tRNA ligase, mitochondrial | 0.65 |
| Q9Y2V2 | CARHSP1 | Calcium-regulated heat stable protein 1 | 0.65 |
| P84103-2 | SRSF3 | Isoform 2 of serine/arginine-rich splicing factor 3 | 0.64 |
| Q9UJZ1-2 | STOML2 | Isoform 2 of stomatin-like protein 2, mitochondrial | 0.64 |
| P62314 | SNRPD1 | Small nuclear ribonucleoprotein Sm D1 | 0.64 |
| Q9NVP1 | DDX18 | ATP-dependent RNA helicase DDX18 | 0.63 |
| Q9Y266 | NUDC | Nuclear migration protein nudC | 0.63 |
| Q9NZI8 | IGF2BP1 | Insulin-like growth factor 2 mRNA-binding protein 1 | 0.63 |
| P62266 | RPS23 | 40S ribosomal protein S23 | 0.62 |
| Q15637-2 | SF1 | Isoform 2 of Splicing factor 1 | 0.62 |
| P12268 | IMPDH2 | Inosine-5′-monophosphate dehydrogenase 2 | 0.60 |
| Q9NXF1-2 | TEX10 | Isoform 2 of testis-expressed sequence 10 protein | 0.59 |
| P37108 | SRP14 | Signal recognition particle 14 kDa protein | −0.59 |
| P39748-2 | FEN1 | Isoform FENMIT of Flap endonuclease 1 | −0.66 |
| O60506-2 | SYNCRIP | Isoform 2 of heterogeneous nuclear ribonucleoprotein Q | −0.82 |
| Q8NBQ5 | HSD17B11 | Estradiol 17-beta-dehydrogenase 11 | −1.49 |
Accession numbers are from the Uniprot database; significantly different protein modulations (p < 0.05); fold change is calculated using Rb1 pretreated/control (unlabeled/labeled) ratios quantitated from integrated proteomics software. Ratios were obtained from n = 3.
3.2. Functional enrichment analysis of the changed proteins
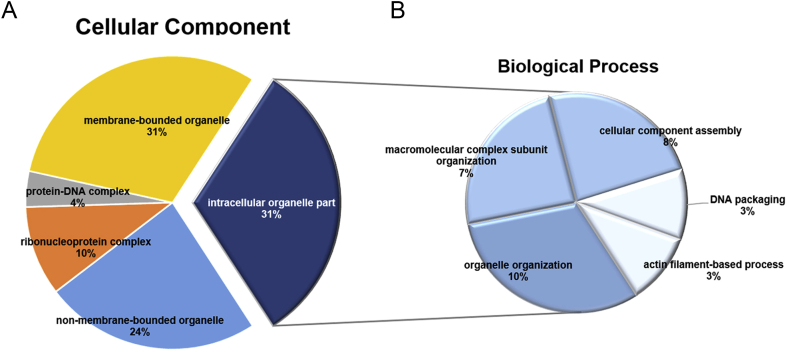
To investigate the functional roles of Rb1 in AD, the altered proteins were subjected to GO-based enrichment analysis using DAVID [22]. As shown in Fig. 3A, the cellular component term annotation reveals a major spectrum of cellular localizations involved in the protein–DNA complex, ribonucleoprotein complex, membrane-bound organelle, nonmembrane-bound organelle, and intracellular organelle part GO terms. To further understand the biological implications of the intracellular organelle part term-related proteins and investigate which have the best p values in the cellular component category (Table S2), enrichment was performed for different aspects of the biological process. The biological process category showed that a majority of proteins were associated with macromolecular complex subunit organization, cellular component assembly, DNA packing, actin filament-based process, and organelle organization, for which five subcategory terms were identified (Fig. 3B; Table S3).
Fig. 3.
Gene Ontology (GO) term annotation analysis of the proteins altered by Rb1. (A) The differentially expressed proteins are categorized by cellular components using GO annotation. A total of five subcategory terms are identified. (B) Intracellular organelle part, which has the most significant p value enrichment of the cellular component categories, is categorized by biological process according to GO annotation. The biological process category shows that most proteins were involved in macromolecular complex subunit organization, cellular component assembly, DNA packing, actin filament-based process, and organelle organization, for which five subcategory terms were identified.
3.3. Actin cytoskeleton
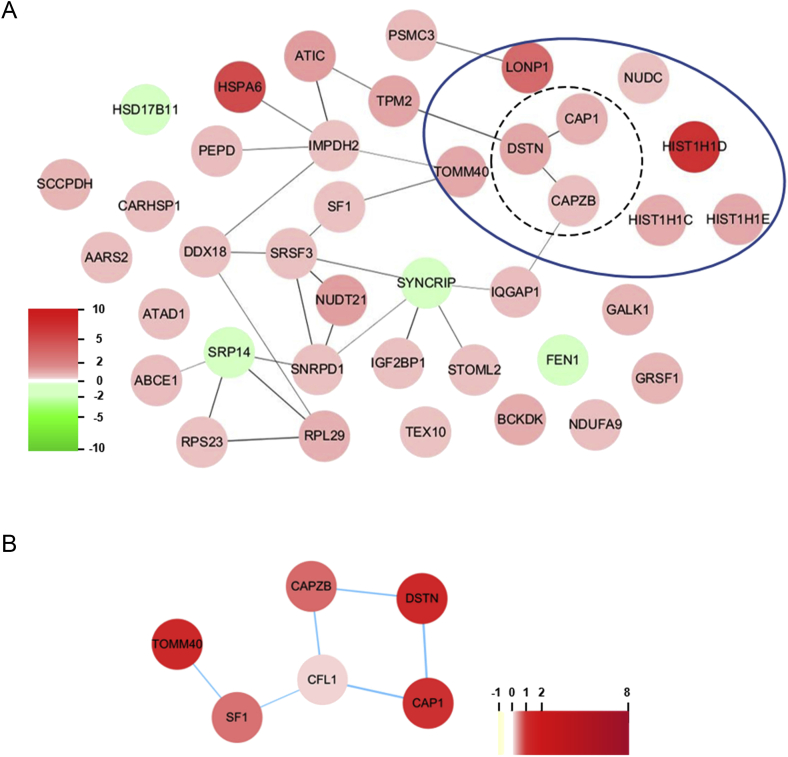
To investigate the regulatory mechanisms of Rb1 in AD, we performed an analysis of protein–protein interaction networks using the STRING database plugin Cytoscape [20]. We analyzed the proteins that were differentially expressed by Rb1 pretreatment. The subnetworks with upregulated proteins revealed a strong interaction network. Interestingly, consistent with our biological process category from the GO term analysis (Fig. 3B), the functional subnetwork associated actin binding and cytoskeletal protein binding was mainly clustered in the network list following the analysis of molecular function (Fig. 4A). We noted that cytoskeletal abnormalities induce neurodegenerative diseases such as AD and Parkinson's disease [23], [24].
Fig. 4.
Protein interaction network of the proteins altered by Rb1 in β-amyloid (Aβ)-induced neurotoxicity. (A) The proteins are grouped using Gene Ontology (GO) biological process terms (Fig. 3B). Protein interaction networks were analyzed using STRING database information and visualized by Cytoscape. The networks show the clustered proteins implicated in the actin cytoskeleton. The functionally significant proteins are clustered with GO biological process terms (blue, dashed line) and actin cytoskeleton (black, dotted line). (B) The analysis of the protein interaction network of cofilin-1 with the altered proteins was performed using STRING database information and visualized by Cytoscape. The node colors represent the expression levels of each protein (red, increase; green, decrease).
CAP1, CAPZB, and DSTN are known to be related to the actin cytoskeleton and have been reported to be associated with nervous system injury [23]. In particular, the actin cytoskeleton in association with these proteins has been recently reported to play a critical role in regulating AD [25], [26]. CAP1 is one of the main proteins that regulate actin dynamics [27], and it has been linked to a variety of human diseases [28], [29]. CAP1 controls actin filament turnover through the recycling of the cofilin-1 and actin proteins [30]. Knockdown of CAP1 regulates cell motility in cells [27]. The changes in CAP1 expression after sciatic nerve injury affect the motility and differentiation of Schwann cells [31]. Interestingly, it has been shown that CAP1 knockdown results in the aggregation and dephosphorylation of cofilin-1 in cells, similar to that seen in AD [27], [32], whereas the expression of CAP1 protein was dramatically increased in Rb1-pretreated samples (Table 1). Therefore, our results suggest that CAP1 may be one of candidate proteins related to the regulatory mechanism of Rb1 in AD. CAPZB, which is an actin cytoskeleton regulator, directly binds to the barbed end of F-actin and β-tubulin [33]. The interaction between CAPZB and β-tubulin exerts an effect on microtubule polymerization and is essential for growth cone morphology and neurite outgrowth [34]. The mRNA levels of CAPZB were increased in hippocampus CA1 pyramidal neurons at the mid stage of AD progression but decreased at the severe stage of AD [26]. However, little is known about the functional roles and protein expression of CAPZB in AD. DSTN is known to be an actin-depolymerizing factor [35]. Recent studies have demonstrated that DSTN plays an important role in human diseases such as cancer [36]. By contrast, the relevance of DSTN to AD is not yet clear. Additionally, previous studies have reported that DSTN binds to cofilin-1 and regulates its functions similar to CAP1 [35].
Interestingly, previous studies have reported that three proteins of actin filament-based processes from the GO term analysis are closely related to cofilin-1 [24]. Furthermore, a number of studies have shown the functional relevance between cofilin-1 and AD. In particular, aberrant cofilin-1 activity led to cognitive decline in AD, and its dephosphorylation without a change in protein expression was observed with age and in AD pathological conditions [37]. We were also able to identify cofilin-1 from our MS data set (Table S1). Cofilin-1 expression was not affected by Rb1 pretreatments or Aβ treatment, as similarly observed in previous studies [37]. This is in contrast to other actin cytoskeleton proteins, which were increased by Rb1 pretreatments prior to Aβ treatment (Fig. 4B). Thus, the protein–protein interactions of cofilin-1 were analyzed using our dataset. A total of five proteins from the STRING database were found to interact with cofilin-1. The SF1 and TOMM40 proteins were identified as well (Fig. 4B). Interestingly, a number of studies have suggested that TOMM40 is a biomarker for AD [38]. Additionally, previous studies showed that TOMM40 was dramatically decreased in whole blood from AD patients [39], whereas Rb1 pretreatment increased the expression of TOMM40 in Aβ-treated cells (Fig. 4B; Table S3). Moreover, a previous study showed that Aβ treatment of cortical neurons induced a dramatic perturbation of the neurotubule network with curly unparalleled segments, but Rb1 pretreatment preserved a normal neurotubule organization [40]. This result supports our data. Further studies are required to determine the mechanism by which Rb1 regulates neurotubule organization in Aβ-induced neurotoxicity. Taken together, our results suggest that Rb1 might play an important role in regulating actin cytoskeleton organization in AD.
In conclusion, we performed a comparative MS-based proteomic analysis using SILAC to investigate the potential protective mechanisms of Rb1 in AD. We identified a total of 1231 proteins from three independent samples. Among these proteins, 40 proteins showed significant fold changes after Rb1 pretreatment in Aβ-treated cells. Our bioinformatics analysis revealed the significance of actin cytoskeleton-related proteins for the protective mechanisms of Rb1 in AD. Therefore, CAP1, CAPZB, TOMM40, and DSTN proteins might be potential biomarkers and regulatory proteins of Rb1 pretreatment for AD protection. Additional studies are required to determine whether the proteins found in our study might be regulatory proteins for the protective mechanisms of Rb1 in AD.
Conflicts of interest
All authors have no conflicts of interest to declare.
Acknowledgments
This work was carried out with the support of the “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ00983503)” Rural Development Administration, Republic of Korea and Korea Basic Science Institute (KBSI) research grant G35110.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jgr.2015.09.004.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Radad K., Gille G., Liu L., Rausch W.D. Use of ginseng in medicine with emphasis on neurodegenerative disorders. J Pharmacol Sci. 2006;100:175–186. doi: 10.1254/jphs.crj05010x. [DOI] [PubMed] [Google Scholar]
- 2.Lee S.T., Chu K., Sim J.Y., Heo J.H., Kim M. Panax ginseng enhances cognitive performance in Alzheimer disease. Alzheimer Dis Assoc Disord. 2008;22:222–226. doi: 10.1097/WAD.0b013e31816c92e6. [DOI] [PubMed] [Google Scholar]
- 3.Van Kampen J.M., Baranowski D.B., Shaw C.A., Kay D.G. Panax ginseng is neuroprotective in a novel progressive model of Parkinson's disease. Exp Gerontol. 2014;50:95–105. doi: 10.1016/j.exger.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Liu C.X., Xiao P.G. Recent advances on ginseng research in China. J Ethnopharmacol. 1992;36:27–38. doi: 10.1016/0378-8741(92)90057-x. [DOI] [PubMed] [Google Scholar]
- 5.Liao B., Newmark H., Zhou R. Neuroprotective effects of ginseng total saponin and ginsenosides Rb1 and Rg1 on spinal cord neurons in vitro. Exp Neurol. 2002;173:224–234. doi: 10.1006/exnr.2001.7841. [DOI] [PubMed] [Google Scholar]
- 6.Yoshikawa T., Akiyoshi Y., Susumu T., Tokado H., Fukuzaki K., Nagata R., Samukawa K., Iwao H., Kito G. Ginsenoside Rb1 reduces neurodegeneration in the peri-infarct area of a thromboembolic stroke model in non-human primates. J Pharmacol Sci. 2008;107:32–40. doi: 10.1254/jphs.fp0071297. [DOI] [PubMed] [Google Scholar]
- 7.Lim J.H., Wen T.C., Matsuda S., Tanaka J., Maeda N., Peng H., Aburaya J., Ishihara K., Sakanaka M. Protection of ischemic hippocampal neurons by ginsenoside Rb1, a main ingredient of ginseng root. Neurosci Res. 1997;28:191–200. doi: 10.1016/s0168-0102(97)00041-2. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y., Feng Y., Fu Q., Li L. Panax notoginsenoside Rb1 ameliorates Alzheimer's disease by upregulating brain-derived neurotrophic factor and downregulating Tau protein expression. Exp Ther Med. 2013;6:826–830. doi: 10.3892/etm.2013.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie X., Wang H.T., Li C.L., Gao X.H., Ding J.L., Zhao H.H., Lu Y.L. Ginsenoside Rb1 protects PC12 cells against beta-amyloid-induced cell injury. Mol Med Rep. 2010;3:635–639. doi: 10.3892/mmr_00000308. [DOI] [PubMed] [Google Scholar]
- 10.Qian Y.H., Han H., Hu X.D., Shi L.L. Protective effect of ginsenoside Rb1 on beta-amyloid protein(1-42)-induced neurotoxicity in cortical neurons. Neurol Res. 2009;31:663–667. doi: 10.1179/174313209X385572. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Liu J., Zhang Z., Bi P., Qi Z., Zhang C. Anti-neuroinflammation effect of ginsenoside Rbl in a rat model of Alzheimer disease. Neurosci Lett. 2011;487:70–72. doi: 10.1016/j.neulet.2010.09.076. [DOI] [PubMed] [Google Scholar]
- 12.Han G., Sun J., Wang J., Bai Z., Song F., Lei H. Genomics in neurological disorders. Genomics Proteomics Bioinformatics. 2014;12:156–163. doi: 10.1016/j.gpb.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sowell R.A., Owen J.B., Butterfield D.A. Proteomics in animal models of Alzheimer's and Parkinson's diseases. Ageing Res Rev. 2009;8:1–17. doi: 10.1016/j.arr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sultana R., Butterfield D.A. Redox proteomics studies of in vivo amyloid beta-peptide animal models of Alzheimer's disease: insight into the role of oxidative stress. Proteomics Clin Appl. 2008;2:685–696. doi: 10.1002/prca.200780024. [DOI] [PubMed] [Google Scholar]
- 15.Swomley A.M., Forster S., Keeney J.T., Triplett J., Zhang Z., Sultana R., Butterfield D.A. Abeta, oxidative stress in Alzheimer disease: evidence based on proteomics studies. Biochim Biophys Acta. 2014;1842:1248–1257. doi: 10.1016/j.bbadis.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S.Y., Kim G.T., Roh S.H., Song J.S., Kim H.J., Hong S.S., Kwon S.W., Park J.H. Proteomic analysis of the anti-cancer effect of 20S-ginsenoside Rg3 in human colon cancer cell lines. Biosci Biotechnol Biochem. 2009;73:811–816. doi: 10.1271/bbb.80637. [DOI] [PubMed] [Google Scholar]
- 17.Cho W.C., Yip T.T., Chung W.S., Lee S.K., Leung A.W., Cheng C.H., Yue K.K.M. Altered expression of serum protein in ginsenoside Re-treated diabetic rats detected by SELDI-TOF MS. J Ethnopharmacol. 2006;108:272–279. doi: 10.1016/j.jep.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Ma Z.C., Gao Y., Wang Y.G., Tan H.L., Xiao C.R., Wang S.Q. Ginsenoside Rg1 inhibits proliferation of vascular smooth muscle cells stimulated by tumor necrosis factor-alpha. Acta Pharmacol Sin. 2006;27:1000–1006. doi: 10.1111/j.1745-7254.2006.00331.x. [DOI] [PubMed] [Google Scholar]
- 19.Choi J.W., Song M.Y., Park K.S. Quantitative proteomic analysis reveals mitochondrial protein changes in MPP(+)-induced neuronal cells. Mol Biosyst. 2014;10:1940–1947. doi: 10.1039/c4mb00026a. [DOI] [PubMed] [Google Scholar]
- 20.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abeti R., Abramov A.Y., Duchen M.R. Beta-amyloid activates PARP causing astrocytic metabolic failure and neuronal death. Brain. 2011;134:1658–1672. doi: 10.1093/brain/awr104. [DOI] [PubMed] [Google Scholar]
- 22.Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler R.W., Lane H.C. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMurray C.T. Neurodegeneration: diseases of the cytoskeleton? Cell Death Differ. 2000;7:861–865. doi: 10.1038/sj.cdd.4400764. [DOI] [PubMed] [Google Scholar]
- 24.Li H., Zhu Y.H., Chi C., Wu H.W., Guo J. Role of cytoskeleton in axonal regeneration after neurodegenerative diseases and CNS injury. Rev Neurosci. 2014;25:527–542. doi: 10.1515/revneuro-2013-0062. [DOI] [PubMed] [Google Scholar]
- 25.Penzes P., Vanleeuwen J.E. Impaired regulation of synaptic actin cytoskeleton in Alzheimer's disease. Brain Res Rev. 2011;67:184–192. doi: 10.1016/j.brainresrev.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao P.F., Davis D.A., Banigan M.G., Vanderburg C.R., Seshadri S., Delalle I. Modulators of cytoskeletal reorganization in CA1 hippocampal neurons show increased expression in patients at mid-stage Alzheimer's disease. PLoS One. 2010;5:e13337. doi: 10.1371/journal.pone.0013337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H., Ghai P., Wu H., Wang C., Field J., Zhou G.L. Mammalian adenylyl cyclase-associated protein 1 (CAP1) regulates cofilin function, the actin cytoskeleton, and cell adhesion. J Biol Chem. 2013;288:20966–20977. doi: 10.1074/jbc.M113.484535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamazaki K., Takamura M., Masugi Y., Mori T., Du W., Hibi T., Hiraoka N., Ohta T., Ohki M., Hirohashi S. Adenylate cyclase-associated protein 1 overexpressed in pancreatic cancers is involved in cancer cell motility. Lab Invest. 2009;89:425–432. doi: 10.1038/labinvest.2009.5. [DOI] [PubMed] [Google Scholar]
- 29.He J., Whelan S.A., Lu M., Shen D., Chung D.U., Saxton R.E., Faull K.F., Whitelegge J.P., Chang H.R. Proteomic-based biosignatures in breast cancer classification and prediction of therapeutic response. Int J Proteomics. 2011;2011:896476. doi: 10.1155/2011/896476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moriyama K., Yahara I. Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J Cell Sci. 2002;115:1591–1601. doi: 10.1242/jcs.115.8.1591. [DOI] [PubMed] [Google Scholar]
- 31.Zhu X., Yao L., Guo A., Li A., Sun H., Wang N., Liu H., Duan Z., Cao J. CAP1 was associated with actin and involved in Schwann cell differentiation and motility after sciatic nerve injury. J Mol Histol. 2014;45:337–348. doi: 10.1007/s10735-013-9554-z. [DOI] [PubMed] [Google Scholar]
- 32.Mitake S., Ojika K., Hirano A. Hirano bodies and Alzheimer's disease. Kaohsiung J Med Sci. 1997;13:10–18. [PubMed] [Google Scholar]
- 33.Delalle I., Pfleger C.M., Buff E., Lueras P., Hariharan I.K. Mutations in the Drosophila orthologs of the F-actin capping protein alpha- and beta-subunits cause actin accumulation and subsequent retinal degeneration. Genetics. 2005;171:1757–1765. doi: 10.1534/genetics.105.049213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis D.A., Wilson M.H., Giraud J., Xie Z., Tseng H.C., England C., Herscovitz H., Tsai L.H., Delalle I. Capzb2 interacts with beta-tubulin to regulate growth cone morphology and neurite outgrowth. PLoS Biol. 2009;7:e1000208. doi: 10.1371/journal.pbio.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian L., Liao M.F., Zhang L., Lu Q.S., Jing Z.P. A study of the expression and interaction of Destrin, cofilin, and LIMK in Debakey I type thoracic aortic dissection tissue. Scand J Clin Lab Invest. 2010;70:523–528. doi: 10.3109/00365513.2010.521572. [DOI] [PubMed] [Google Scholar]
- 36.Klose T., Abiatari I., Samkharadze T., De Oliveira T., Jager C., Kiladze M., Valkovska N., Friess H., Michalski C.W., Kleeff J. The actin binding protein destrin is associated with growth and perineural invasion of pancreatic cancer. Pancreatology. 2012;12:350–357. doi: 10.1016/j.pan.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 37.Barone E., Mosser S., Fraering P.C. Inactivation of brain Cofilin-1 by age, Alzheimer's disease and gamma-secretase. Biochim Biophys Acta. 2014;1842:2500–2509. doi: 10.1016/j.bbadis.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Elias-Sonnenschein L.S., Helisalmi S., Natunen T., Hall A., Paajanen T., Herukka S.K., Laitinen M., Remes A.M., Koivisto A.M., Mattila K.M. Genetic loci associated with Alzheimer's disease and cerebrospinal fluid biomarkers in a Finnish case-control cohort. PLoS One. 2013;8:e59676. doi: 10.1371/journal.pone.0059676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chong M.S., Goh L.K., Lim W.S., Chan M., Tay L., Chen G., Feng L., Ng T.P., Tan C.H., Lee T.S. Gene expression profiling of peripheral blood leukocytes shows consistent longitudinal downregulation of TOMM40 and upregulation of KIR2DL5A, PLOD1, and SLC2A8 among fast progressors in early Alzheimer's disease. J Alzheimers Dis. 2013;34:399–405. doi: 10.3233/JAD-121621. [DOI] [PubMed] [Google Scholar]
- 40.Chen X., Huang T., Zhang J., Song J., Chen L., Zhu Y. Involvement of calpain and p25 of CDK5 pathway in ginsenoside Rb1's attenuation of beta-amyloid peptide25–35-induced tau hyperphosphorylation in cortical neurons. Brain Res. 2008;1200:99–106. doi: 10.1016/j.brainres.2007.12.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.