Abstract
The transmission of Trypanosoma cruzi by vectors is confined to the Americas, and the infection circulates in at least two broadly defined transmission cycles occurring in domestic and sylvatic habitats. This study sought to detect and characterize infection by T. cruzi and other trypanosomes using PCR strategies in blood samples from free-ranging howler monkeys, Alouatta caraya, in the northeastern Argentina. Blood samples were collected at four sites with variable levels of habitat modification by human activity. PCR was conducted using primers for kinetoplast DNA, satellite DNA and ribosomal DNA of the trypanosomatid parasites. Ribosomal and satellite DNA fragments were sequenced to identify the trypanosomatid species and to characterize the discrete typing units (DTUs) of T. cruzi. Overall, 46% (50/109) of the howlers were positive according to the kDNA-PCR assay, but only 7 of the howlers were positive according to the SatDNA-PCR protocol. We sequenced the amplicons of the satellite DNA obtained from five specimens, and the sequences were 99% and 100% similar to T. cruzi. A sequence typical of DTU T. cruzi I was found in one howler monkey from the “remote” site, while sequences compatible with DTUs II, V, and VI were found in howlers from the “remote”, “rural” and “village” sites. We detected 96% positive samples for RibDNA-PCR, 9 of which were sequenced and displayed 99% identity with Trypanosoma minasense, while none showed identity with T. cruzi. The results demonstrated the presence of T. cruzi and a species closely related to T. minasense in blood samples from free-ranging A. caraya, belonging to different T. cruzi DTUs circulating in these howler monkey populations. The results obtained in this study could help evaluate the role of A. caraya as a reservoir of T. cruzi in regions where Chagas disease is hyper-endemic and where the human-wildlife interface is increasing.
Keywords: Trypanosoma cruzi, Alouatta caraya, Enzootic cycle, Howler monkey, TcI–TcVI DTUs, Trypanosoma minasense
Graphical abstract

Highlights
-
•
Trypanosome infection was assessed in wild howler monkeys from northern Argentina.
-
•
Seven out of 109 individuals were positive for Trypanosoma cruzi.
-
•
Satellite DNA sequencing were typical of T. cruzi I and II/V/VI.
-
•
Trypanosoma minasense was detected for first time in free-ranging Alouatta caraya.
-
•
Mixed infections by T. cruzi and T. minasense were observed in a group of animals.
1. Introduction
Chagas disease is the most important parasitic disease in Latin America, and as a result of infection of humans by the parasite Trypanosoma cruzi (Kinetoplastida: Trypanosomatidae), approximately eight million people are infected worldwide (World Health Organization, 2010). The transmission of this protozoan parasite by vectors is confined to the Americas and circulates in at least two broadly defined transmission cycles, occurring in domestic and sylvatic habitats. Infection with T. cruzi is a complex zoonosis, transmitted to vertebrate hosts by the feces of multiple blood-sucking triatomine species (Reduviidae: Triatominae) and sustained by more than 160 species of mammals belonging to 25 families in the Americas, with marsupials, edentates, and rodents being the most frequent sylvatic hosts. Chagas disease has emerged in regions previously considered to be relatively free of the disease, such as the Amazon basin, where mainly sylvatic, rather than domestic, vectors transmit the parasite, and local micro-epidemics of orally transmitted disease have been observed (World Health Organization, 2010). Infection in humans can also occur via blood transfusion, organ transplant, and vertical transmission from mother to offspring in endemic and non-endemic areas. Oral transmission has been observed in humans exposed to contaminated food (Nóbrega et al., 2009, Souza-Lima et al., 2013), which seems to be the main infection mechanism in wild mammals (Roque et al., 2008, Marcili et al., 2009, Rocha et al., 2013).
T. cruzi is currently classified into six discrete typing units (DTUs), T. cruzi I – T. cruzi VI (Tibayrenc, 2003, Zingales et al., 2009), defined as sets of stocks that are genetically more related to each other than to any other stock and that are identifiable by common genetic, molecular or immunological markers (Tibayrenc, 1998). The DTUs of T. cruzi are distributed differentially among triatomine insects, vertebrate host species and habitats in different geographical areas (Higo et al., 2004, Maffey et al., 2012). T. cruzi I occurs in many domestic cycles and mainly in sylvatic cycles throughout the Americas, involving opossums (genus Didelphis), which live in both arboreal and terrestrial sylvatic and peridomestic ecotopes (Bernabé et al., 2000, Yeo et al., 2005, Orozco et al., 2013). T. cruzi II, V and VI occur primarily in domestic cycles in Brazil and in the southern cone countries of South America (Yeo et al., 2005, Noireau et al., 2009). T. cruzi III has been detected in armadillos (Dasypus novemcinctus) throughout the Americas (Yeo et al., 2005, Lewis et al., 2009, Orozco et al., 2013), whereas T. cruzi IV has been isolated mostly from sylvatic mammals in the northern Amazon basin and in the United States (Bernabé et al., 2000, Yeo et al., 2005, Llewellyn et al., 2009, Marcili et al., 2009). Wild non-human primates appear to be associated naturally with T. cruzi I, T. cruzi II and T. cruzi IV in Atlantic and Amazonian forests (Lisboa et al., 2007, Da Silva et al., 2008, Marcili et al., 2009, Araújo et al., 2011, Lisboa et al., 2015).
In addition to two zoonotic trypanosome species, T. cruzi and T. rangeli, neotropical non-human primates, primarily species of the Cebidae family, can also be infected with T. (Megatrypanum) minasense (Chagas, 1908, Dunn et al., 1963, Hoare, 1972, Deane et al., 1974, De Resende et al., 1994, Ziccardi et al., 2000, Sato et al., 2008, Tenório et al., 2014). Regarding the genus Alouatta, trypomastigotes of T. minasense have been recorded by morphologic techniques in wild individuals of Alouatta palliata from Costa Rica (Chinchilla et al., 2005) and by molecular diagnosis in one captive A. caraya in the Centre for Wild Fauna (CCWF) from Brazil (Tenório et al., 2014). The highly pleomorphic nature of T. (Megatrypanum) minasense in the bloodstream requires accurate identification of the species based on not only morphology. Sato et al. (2008) detected T. minasense infection by molecular diagnosis in Saguinus midas exported to Japan as experimental laboratory animals. The first molecular phylogenetic characterization of T. minasense, showed that it is closely related to trypanosomes with T. theileri-like morphology (Sato et al., 2008).
The “Gran Chaco” ecoregion includes northern Argentina, Bolivia, Paraguay and southwestern Brazil, and it is a hyperendemic region for Chagas disease. In the southeastern limit of the “Gran Chaco”, known as humid Chaco, and in the neighboring savannas and gallery forests in northeastern Argentina, studies of the dynamics of T. cruzi transmission and infection characterization of T. cruzi have been scarce (Bar et al., 1999, Bar et al., 2010). In this region, some wild triatomine insects that are potential vectors of Chagas disease, such as Triatoma sordida and Psammolestes coreodes, colonize wild biotopes, such as palms, tree hollows and bird nests (Bar and Wisnivesky-Colli, 2001, Damborsky et al., 2001, Bar et al., 2010). Additionally, T. cruzi has been detected in T. sordida, suggesting that this triatomine species could play a role in the maintenance of the wild T. cruzi transmission cycle in northeastern Argentina (Bar and Wisnivesky-Colli, 2001).
Black and gold howler monkeys (Alouatta caraya) inhabit flooded forests in the island system of the Parana River and in the humid Chaco gallery forests east and west of the Parana River (Zunino and Kowalewski, 2008). Northern Argentina comprises the boundary of the southern distribution of A. caraya, which accounts for the largest proportion of biomass of any arboreal mammal (Brown and Zunino, 1994). Like other howlers, A. caraya often copes well with moderate deforestation (Bicca-Marques, 2003, Zunino et al., 2007). Habitat fragmentation involves the alteration of habitat, resulting in changes in the interaction between wildlife and humans, which can contribute to the proliferation of emergent zoonotic diseases (Nunn and Altizer, 2006, Gillespie et al., 2008). The available data on Trypanosoma infection in A. caraya have been scarce (Funayama and Barretto, 1970, Travi et al., 1982, Santa Cruz et al., 2000), and the limited sensitivity and specificity of the diagnostic methods used have hindered the level of detection. Aiming to assess the possible role of howler monkeys in the T. cruzi transmission cycle, this study sought to assess and characterize the prevalence of T. cruzi DTUs and of other trypanosomatid infections in populations of A. caraya living at sites with variable levels of contact with humans and with associated domestic animals.
2. Materials and methods
2.1. Study sites
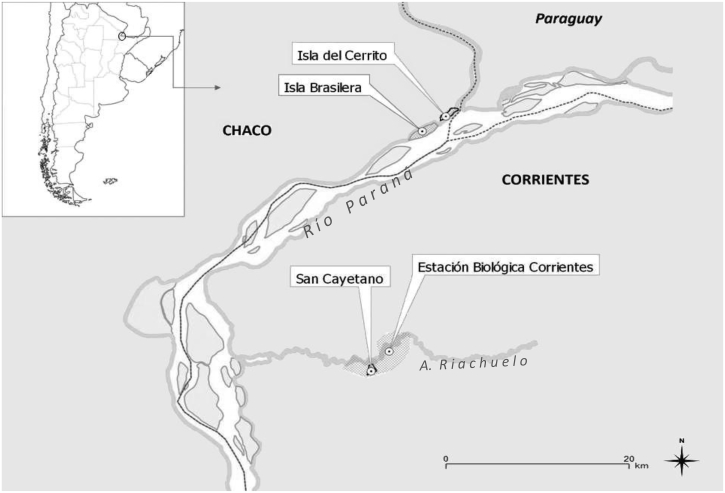
We collected blood samples from howlers at four sites that differed in their degrees of contact with humans: Isla Brasilera (IB) (27° 20′ S, 58° 40′ W) (“remote”); Estación Biológica Corrientes-Biological Field Station Corrientes (EBCo) (27° 30′ S, 58° 41′ W) (“rural”); Isla del Cerrito (IC) (27° 17′ S, 58° 37′ W) (“village”); and San Cayetano (SC) (27° 34′ S, 58° 42′ W) (“village”) (Fig. 1). Long-term studies of the ecology and behavior of A. caraya in these sites allowed us to localize and identify the selected studied howler groups. SC and IC are village sites where howler groups live in close proximity to family houses, with tree crowns that are contiguous with trees in the nearby forest fragments. IB is located 20 km north of EBCo and is an island characterized by a continuously flooded forest and little to no human contact. The island has an area of 290 ha, and 32–35 groups of primates have been identified. EBCo is a rural site characterized by a semi-deciduous forest, where at least 34 howler groups are under study in 24 forest fragments covering a total area of 3000 ha (Zunino et al., 2007, Kowalewski et al., 2010).
Fig. 1.
Location of study areas in Northeastern Argentina: San Cayetano (SC), Estación Biológica Corrientes (EBCo), Isla Brasilera (IB) and Isla del Cerrito (IC).
2.2. Capture of howler monkeys and sample collection
A total of 109 howler monkeys of different ages were captured and examined for trypanosome infections in different study areas (Table 2). Captures were conducted between July and August 2010. The animals were captured and immobilized to obtain blood samples, for which the assistance of at least two veterinarians was necessary. Chemical immobilization of the animals was performed with anesthetic darts (1 cc Pneu-dart type P) shot using an air rifle. Parenteral anesthesia was accomplished with xylazine chlorohydrate (Xilacina 20®, Richmond Vet Pharma, Buenos Aires, Argentina) (20 mg/ml), combined with midazolam (Midazolam®, Richmond Vet Pharma) (5 mg/ml), and ketamine (Ketonal 100®, Richmond Vet Pharma) (100 mg/ml), at the minimum dose appropriate for species and weight (Kreeger and Arnemo, 2007).
Table 2.
Capture of howler monkeys by study site, sex ratio and age categories.
| Type of environment | Number of groups | Number of individuals | Sex ratio male:female | Age categories |
|||
|---|---|---|---|---|---|---|---|
| Adult | Subadult | Young | |||||
| EBCo | Rural | 11 | 42 | 1.47 | 19 | 11 | 12 |
| IB | Remote | 15 | 49 | 1.58 | 33 | 7 | 9 |
| SC | Village | 5 | 16 | 1.67 | 14 | 1 | 1 |
| IC | Village | 1 | 2 | 1 | 2 | 0 | 0 |
The animals were captured from 11:00 h to 13:00 h and from 14:00 h to 16:00 h. During these time intervals, the temperature is warm and howlers are active at relatively low heights in the canopy. A sequence of procedures was applied to each howler: darting; physical examination and health assessment; and collection of blood samples, among others. Individuals in the groups were classified by sex/age categories, according to the classification of Rumiz (1990). We collected 1 ml of peripheral blood per individual from the femoral vein. After blood collection, we placed the blood samples in EDTA 0.5 M-containing tubes (1:1), which were stored at 4 °C for up to 72 h or at −20 °C until processing for molecular studies. After collecting the samples, the individuals were placed in an open cloth bag for recovery from anesthesia. Once the animals fully recovered, they were released at the point of capture and were monitored for several hours to be certain that they were unharmed and capable of a full range of locomotion behavior. On subsequent days, the animals were monitored to follow up their normal recuperation.
These procedures were successfully used in other research studies (Schmidt et al., 2007, Oklander et al., 2007). This study complied with the laws of Argentina and the University of Illinois guidelines for the ethical treatment of primates (IACUC protocol 09267).
2.3. Molecular detection of trypanosomatid infections
2.3.1. DNA extraction
DNA was isolated from 200 μl of blood using the High Pure PCR Template Preparation Kit (Roche Diagnostic Corp., Indiana, USA), following the instructions of the manufacturer. Prior to DNA extraction, 200 pg of linearized p-ZErO plasmid containing a sequence of Arabidopsis thaliana was added to each sample as an external control for DNA extraction and polymerase chain reaction (PCR) (Duffy et al., 2009).
2.3.2. DNA amplification
Different PCR strategies were applied, as follows. 1) Detection of T. cruzi infections: 1a) kDNA-PCR, amplification of the 330 bp fragment from the minicircle DNA of the kinetoplastid genome of T. cruzi using the primers “121” (5′-AAATAATGTACGGG(T/G)GAGATGCATGA-3′) and “122” (5′-GGTCGATTGGGGTTGGTGTAATATA-3′) and the cycling conditions reported by Burgos et al. (2007). 1b) Real-time SatDNA-PCR amplification of 166 bp fragments from specific satellite sequences of T. cruzi using the primers “cruzi 1” (5′-ASTCGGCTGATCGTTTTCGA-3′) and “cruzi 2” (5′-AATTCCTCCAAGCAGCGGATA-3′) and the probe “cruzi 3” (5′-CACACACTGGACACCAA-3′); the probe was labeled with 5_FAM (6-carboxyfluorescein) and 3_MGB (minor groove binder), and the cycling conditions were as reported by Duffy et al. (2013). PCR was performed using a Rotor Gene 3000 (Corbett Research, Sydney, Australia) Real Time thermocycler. Only positive kinetoplastid DNA-PCR (kDNA-PCR) samples were analyzed by this protocol. 2) Detection of trypanosomatid infections other than T. cruzi: RibDNA-PCR amplification was performed from the polymorphic D7 domain of the 24Sα rRNA genes using the primers “D75” (5′-GCAGATCTTGGTTGGCGTAG-3′) and “D76” (5′-GGTTCTCTGTTGCCCCTTTT-3′) and the cycling conditions reported in Schijman et al. (2006) (Table 1).
Table 1.
Summary of PCR protocols used for the detection of T. cruzi and others trypanosomes.
| Protocol | Target | Primers or probe | PCR | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| kDNA | Kinetoplast DNA | 121 122 |
C | 330 – T. cruzi 330 to 380 – T. rangeli |
Schijman et al., 2003; Burgos et al., 2007; Botero et al. 2010 |
| SatDNA | Satellite DNA | cruzi1 cruzi2 cruzi3 |
RT | 166 – T. cruzi |
Piron et al., 2007; Schijman et al., 2011; Duffy et al., 2013 |
| RibDNA | Ribosomal DNA | D75 D76 |
C | 270 – T. cruzi I 290 – T. cruzi II-V-VI 240 – T. rangeli |
Souto et al., 1999; Schijman et al., 2006; Burgos et al., 2007 |
C: Conventional PCR
RT: Real Time PCR
Each PCR run included an amplification reaction without DNA as a negative PCR mixture control and an amplification reaction with total DNA from the T. cruzi reference strain (T. cruzi VI CL-Brener) and/or from T. rangeli stock from Brazil (Cuba Cuba C. personal communication; Gurgel-Gonçalves et al., 2012) as positive controls. Up to 10 μl of the amplified product by conventional PCR (kDNA-PCR and RibDNA-PCR) was analyzed by 2% agarose gel electrophoresis in TBE buffer containing ethidium bromide (0.5 μg/ml), which was added previously to the gel.
2.3.3. Satellite and ribosomal DNA sequencing
PCR products were loaded on 2% low melting point agarose gels and were purified using a QIA quick® Gel Extraction Kit (Qiagen). Both satellite and ribosomal amplicons were sequenced with both forward and reverse primers, following the instructions of the processing site (Unidad de Genómica, Instituto de Biotecnología, Instituto Nacional de Tecnología Agropecuaria, Castelar, Provincia de Buenos Aires, Argentina).
2.4. Data analysis
The data were analyzed using Quantitative Parasitology (QP3.0) software (Rozsa et al., 2000) and InfoStat software, version 2014 (Di Rienzo et al., 2014). Ninety-five percent confidence intervals of prevalence rates were calculated, and the chi-square test was used to determine the differences in relative frequencies of positive samples among sex and age categories or among the study site categories.
3. Results
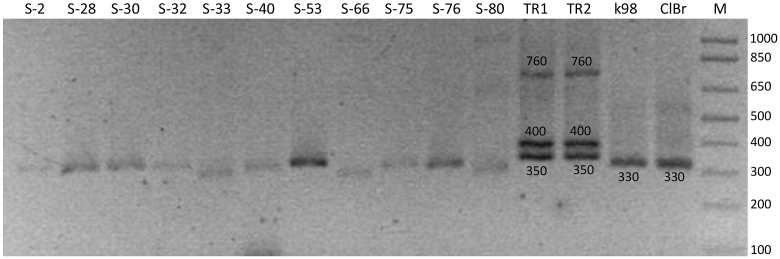
Fifty (46%) howlers were PCR positive according to kDNA-PCR (Table 3 and Fig. 2). There was no statistically significant difference between sexes (chi-square test: X2 = 0.44, degrees of freedom (df) = 1, p-value = 0.8) or between age categories (X2 = 0.46, df = 2, p-value = 0.5; respectively) for positive kDNA-PCR samples. In addition, we identified kDNA amplicons with slightly variable molecular weights between 300 and 330 bp (Fig. 3), which were very similar to the size of the T. cruzi DNA fragment used as a reference and slightly smaller than the T. rangeli amplicon used as a reference.
Table 3.
Infection prevalence determined by kDNA-PCR, RibDNA-PCR and SatDNA-PCR in wild howlers from Estación Biológica Corrientes (EBCo), Isla Brasilera (IB), San Cayetano (SC), and Isla del Cerrito (IC). Data presented as percentage, with 95% confidence intervals.
| Study site – PCR protocol | No. positive groups/No. examined groups | No. positive monkeys/No. examined monkeys | % Prevalence (IC 95%) |
|---|---|---|---|
| EBCo (rural) | |||
| kDNA | 8/11 | 19/42 | 45.2 (29.84–61.33) |
| RibDNA | 11/11 | 40/42 | 95.2 (83.83–99.42) |
| SatDNA | 3/8 | 3/19b | 15,8 (3.38–39.58) |
| IB (remote) | |||
| kDNA | 13/15 | 22/49 | 44.9 (30.66–59.77) |
| RibDNA | 15/15 | 48/49 | 98 (89.14–99.95) |
| SatDNA | 3/13 | 3/22b | 13.6 (2.90–34.92) |
| SC (village) | |||
| kDNA | 4/5 | 8/16 | 50 (24.65–75.35) |
| RibDNA | 5/5 | 15/16 | 93.8 (69.76–99.85) |
| SatDNA | 1/4 | 1/8b | 12.5 (0.31–52.66) |
| IC (village) | |||
| kDNA | 1/1 | 1/2 | a |
| RibDNA | 1/1 | 2/2 | a |
| SatDNA | 0/1 | 0/1 | a |
Prevalence was not estimated due to the very limited sample size.
Sat-DNA PCR was carried out only in kDNA-PCR-positive samples.
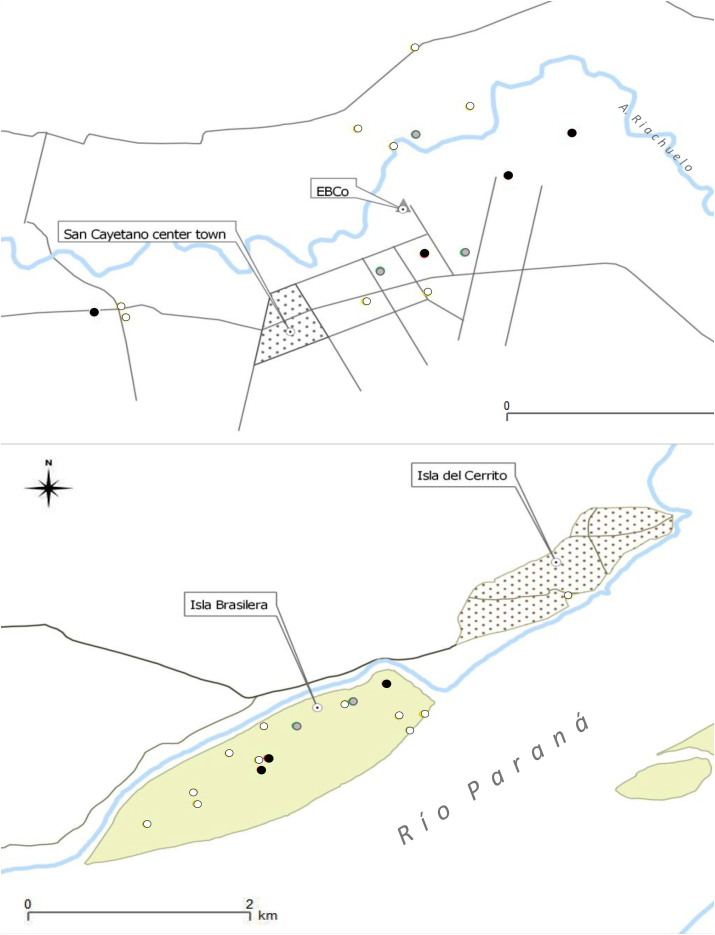
Fig. 2.
Study areas showing the locations of the sampled howler groups and the results of molecular analysis: groups with only RibDNA PCR-positive howler monkeys (gray circle); groups with RibDNA and kDNA-PCR-positive howler monkeys (white circle); groups with RibDNA, kDNA, and SatDNA-PCR-positive howler monkeys positive (black circle). A: Isla Brasilera (IB) and Isla del Cerrito (IC); B: San Cayetano (SC) and Estación Biológica de Corrientes (EBCo).
Fig. 3.
Size variation of amplified kDNA fragments revealed by electrophoresis and ethidium bromide staining. Samples are indicated by their ID numbers. Reference strains used as positive controls: TR1 and TR2: T. rangeli, k98: T. cruzi I, ClBr: T. cruzi VI (Cl Brener). M: 1 kb DNA molecular ladder. Fragment size is indicated in base pairs.
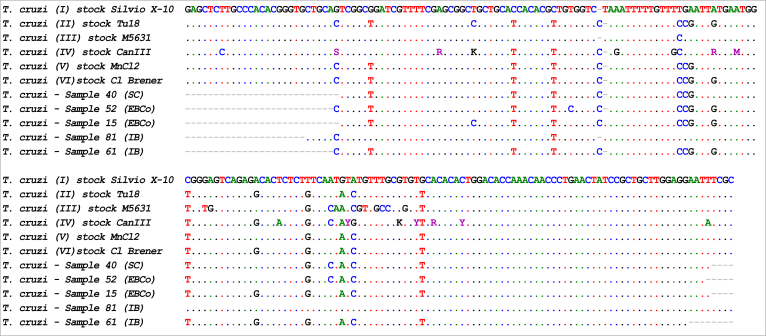
Using the Real Time SatDNA-PCR protocol, we detected infection with T. cruzi in seven howler monkeys (Table 3 and Fig. 2), which yielded kDNA fragments of 330 bp (six samples) and 300 bp (one sample). Sequencing was performed on five of seven SatDNA PCR-positive samples (S-15 and S-52 [EBCo], S-40 [SC], S-61 and S-81 [IB], all yielding kDNA fragments of 330 bp). The two remaining SatDNA PCR-positive samples did not yield sufficient material for purification and sequencing. A nucleotide identity of 99% with T. cruzi database reference sequences was obtained in all of the cases except for S-61, which had 100% sequence identity. The alignment of satellite sequences with those from representative strains of the different T. cruzi DTUs allowed for the characterization of T. cruzi in the SC and IB samples (Fig. 4). These sequence data are available in the GenBank database under the accession numbers KT369011 to KT369015. The sequence obtained in one howler from IB (S-81, accession number: KT369014) showed high similarity (99%) to those reported for T. cruzi strains, which are more related to the wild transmission cycle in Didelphis sp. In contrast, the satellite DNA sequences obtained from one howler from IB (S-61, accession number: KT369013) and from three monkeys from SC (S-15, S-52, and S-40, accession numbers: KT369011, KT369015, and KT369012, respectively) shared 100% and 99% identity with those previously reported for the TcII, TcV, and TcVI strains, which are associated with the domestic transmission cycle of T. cruzi.
Fig. 4.
Alignment of the DNA satellite repetitive sequences obtained from five samples of howlers (GenBank accession numbers KT369011 to KT369015) with the equivalent regions of reference stocks representative of the six DTUs, T. cruzi I – T. cruzi VI obtained from Genbank. DNA chromatograms were aligned using the Clustal W algorithm (Thompson et al., 1994) and were analyzed with BioEdit Sequence Alignment Editor software. The sequences were identified by a BLAST search against Genbank.
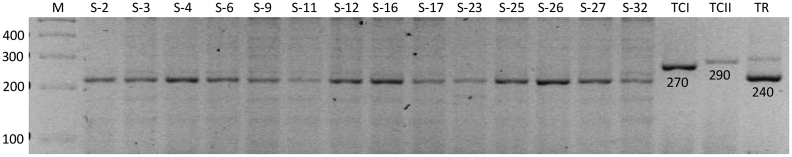
The RibDNA-PCR protocol detected 96% (105/109) of the positive samples (see Table 3). Identical fragments with sizes of 240 bp were obtained in all of the RibDNA-positive samples (Fig. 5). This fragment size was expected for T. rangeli. However, RibDNA sequences from 9 samples (5 from EBCo and 4 from IB) (GenBank accession number: KT369016), of which 3 were kDNA PCR-positive, displayed maximum nucleotide identity with the homologous gene of T. minasense when a BLAST search was performed against a reference sequence published in GenBank (Max. Ident.: 99.5%, accession number: AB362411, Sato et al., 2008). This finding suggested the presence of T. minasense or a very closely related species in the nine specimens, which yielded identical sequences. In addition, 49 of 105 specimens positive for RibDNA were also kDNA PCR-positive; therefore, these animals might have been co-infected with T. minasense and with T. cruzi or other trypanosomatid. It was not possible to obtain a culture of T. minasense to be used as a positive control to compare the sizes of DNA fragments obtained by RibDNA-PCR.
Fig. 5.
Agarose gel (2%) showing RibDNA-amplified fragments stained with ethidium bromide. Samples are indicated by their ID numbers. Reference strains were used as positive controls: TCI: T. cruzi I, TCII: T. cruzi II, and TR: T. rangeli. M: 1 kb DNA molecular ladder. Fragment size is indicated in base pairs.
There was no statistically significant difference among positive kDNA (X2 = 0.15, df = 3, p-value = 0,99), SatDNA (X2 = 0.23, df = 3, p-value = 0.97), and RibDNA-PCR (X2 = 0.89, df = 3, p-value = 0,83) samples among the four study sites.
4. Discussion
This study was the first to describe the molecular detection of T. cruzi infection in free-ranging howler monkeys from Argentina, confirming previous results based on morphological and serological techniques (Travi et al., 1982, Santa Cruz et al., 2000). We detected 46% of positive samples by amplification of the minicircle variable region of Trypanosoma sp. in all of the study sites, whereas only 6% (n = 7) of these samples were also positive according to SatDNA-PCR, confirming T. cruzi infection. This result was expected because SatDNA-PCR is less sensitive than kDNA-PCR, especially for infections with T. cruzi I or T. cruzi IV (Duffy et al., 2009, Schijman et al., 2011, Ramírez et al., 2015). Therefore, the low prevalence of T. cruzi infection obtained with SatDNA-PCR might have been due to a low parasite burden. Alternatively, several samples that yielded positive results for kDNA-PCR might have come from individuals infected by trypanosomatids other than T. cruzi, which were not distinguishable by the protocols implemented here. This possibility would be compatible with the variability of fragment sizes obtained by kDNA-PCR.
The prevalence of positive samples for the three diagnostic protocols (kDNA, SatDNA and RibDNA-PCR) were similar among the four study sites. However, we identified howler monkeys infected with different DTUs, as revealed by the SatDNA sequences. For instance, we recorded the occurrence of T. cruzi I in one monkey from IB (our “remote” site), consistent with the association of this DTU with the wild transmission cycle (Bernabé et al., 2000, Yeo et al., 2005, Orozco et al., 2013). We also found sequences from IB that shared 99–100% identity with those previously reported in DTUs II, V, and VI. These DTUs seem to be primarily associated with the domestic transmission cycle of T. cruzi (Yeo et al., 2005), although they have also been found in the wild (Noireau et al., 2009). Therefore, the presence of these DTUs in IB was not so unexpected. Monkeys from SC and EBCo were infected by strains associated with DTUs II, V, and VI. Unfortunately, the DTU identification was possible in only five monkeys (belonging to five groups and three sites), most likely due to low parasitic loads.
Palms, armadillo burrows, and the nests of birds and rodents are the specific ecotopes of most of the triatomines found in the surroundings of the study sites (Bar and Wisnivesky-Colli, 2001, Damborsky et al., 2001, Bar et al., 2010). Although the associations between vector and ecotope can vary, some species display a close relationship with one type of habitat (i.e., Psammolestes species with nests of Furnariidae), while others exhibit a great range of terrestrial and arboreal ecotopes (Gaunt and Miles, 2000, Noireau et al., 2009). We have observed cavities and bird nests that could be used by wild triatomines in trees in which A. caraya sleeps. Small mammal burrows were also seen at the bases of these sleeping site trees. Psammolestes coreodes associated with bird nests was found within of the home range of the studied howler groups (Martínez, unpublished data). Birds are refractory to T. cruzi infection but can be important sources of blood meals for triatomines. These triatomines, which are potential vectors of trypanosomes (Cruz-Guzmán et al., 2014), possibly feed from birds (Rabinovich et al., 2011) or small mammals that live in these nests, but the possibility that they fed on howler monkeys cannot be discounted.
In non-human primates, it is presumed that transmission of trypanosomes in enzootic cycles occurs mainly through the ingestion of infected triatomines (Da Silva et al., 2008, Roque et al., 2008, Marcili et al., 2009). The diet of howler monkeys primarily consists of leaves, fruits and flowers (Fernández, 2014, Dias and Rangel-Negrín, 2015), but they can ingest insects accidentally when eating fruits and leaves or when removing ectoparasites (grooming). More rarely, they might eat ants from old trees (Raño, personal communication). Therefore, howler monkeys could be infected in a number of different manners. Nevertheless, if the studied monkey population was already chronically infected, congenital (vertical) transmission from mother to offspring is possible (Eberhard and D’Alessandro, 1982).
Trypomastigotes of T. minasense were originally described by Chagas (1908) in the blood of a marmoset, Callithrix penicillate, from southeastern Brazil. This trypanosome is a widely distributed species detected in several neotropical non-human primates, such as A. palliata, Saimiri sciureus, Saguinus midas, and Callithrix penicillata (Dunn et al., 1963, Souza and Dawson, 1976, Ziccardi and Lourenço de Oliveira, 1997, Ziccardi et al., 2000, Chinchilla et al., 2005, Sato et al., 2008). The transmission route of T. minasense is unknown. Because most of the recorded primate hosts are primarily arboreal, Dunn et al. (1963) suggested that the vectors for T. minasense are probably arboreal or partially arboreal insects.
Our results showed identical rDNA amplicons that displayed nucleotide identity with the reference strain of T. minasense, which was determined for the first time by Sato et al. (2008). The percentage of identity (99.5%) was similar than that obtained between available 24S ribosomal sequences of two T. rangeli strains (“San Agustín“ and “SC58”: 99.58%; GenBank Accession Nos.: U73612, and KJ742907, respectively) and was greater than that obtained between T. cruzi stocks belonging to the same DTU (i.e., stocks “DM28” and “La Cruz” [TC I: 96.4%] [Souto et al. (1999) and Accession No.: L22334, respectively] and stocks “TC IIb” and “Y” [TC II: 92.7%] [Accession Nos.: GQ303145, and M28885, respectively]).
The samples that were kDNA-positive (46%) and their corresponding ribosomal sequences showed that identity with T. minasense was likely to indicate mixed infection with T. minasense/T. cruzi or with other Trypanosomatids that were not identified using our methods. T. minasense could have resulted in a 330 bp kDNA fragment; however, only 46% of the samples were positive according to kDNA-PCR. This finding suggests that the primers used for this protocol do not recognize the kDNA target in T. minasense. However, the lack of reference DNA from T. minasense precluded the confirmation of this possibility. In addition, in mixed infections, RibDNA-PCR yielded only T. minasense fragments but not T. cruzi-specific fragments (Schijman et al., 2006), perhaps because of a higher burden of T. minasense in mixed infection, favoring the amplification of the parasite DNA in a larger amount.
This study was the first to show the presence of T. minasense or a species closely related to T. minasense in wild howler monkeys. Tenório et al. (2014) found T. minasense in one captive individual of A. caraya that had 90% similarity with the homologous sequence referenced by Sato et al. (2008). The prevalence found in our study (96%) was much higher than that recorded in the updated bibliography of non-human primates; however, these reports were based on blood smear analyses, hemoculture and/or xenodiagnosis (De Resende et al., 1994, Ziccardi and Lourenço de Oliveira, 1997; Ziccardi et al., 2000). Our prevalence results showed similar values to those obtained by Sato et al. (2008), who detected T. minasense in 93% of wild-caught Saguinus midas individuals by PCR amplification. These high prevalence values detected were likely due to the greater sensitivity of molecular diagnostic methods.
Our current knowledge regarding the transmission process of T. minasense, the effects of the infection on non-human primates, and how it or other non-zoonotic trypanosomes interact with T. cruzi in a mixed infection is very scarce. The results of the present study enabled us to survey the biology of T. minasense in free-ranging non-human primate populations, as well as how it interacts with T. cruzi infection dynamically.
Parasitological surveys of non-human primates provide an important opportunity to better understand the epidemiology, transmission dynamics and emergence risk of various anthropozoonoses such as Chagas disease, which affect the human populations in most locations in the semiarid and humid Argentine Chaco region. The results obtained in this study will help evaluate the role of A. caraya as a reservoir of human trypanosomes in regions where Chagas disease is hyper-endemic and where the human-wildlife interface is increasing. Massive changes in land use, among other effects, usually result in an increasing probability of parasite cross-transmission as the contact between howlers and humans and associated domestic animals increases. Our current knowledge regarding the impact of T. cruzi on the health of free-ranging A. caraya populations remains minimal. Consequently, we believe that research into the epidemiology, transmission ecology, and clinical consequences of T. cruzi infections of A. caraya and other non-human primate populations will provide more accurate information about the dynamics of this disease in the wild-domestic interface.
Acknowledgements
We are grateful to Gabriel Zunino, Federico Pontón, Hernán Argibay, Rut Pernigotti, Ramón Martínez, Mariana Raño, Vanina Fernández, Malén Balbis, and the technician personnel from Maiztegui National Human Viral Disease Institute (INEVH) for their inestimable help in the fieldwork. A special thanks goes to Carolina Cura for her laboratory assistance and to Mariano Giombini for his comments on the manuscript and for English corrections. Our acknowledgments go to César Cuba Cuba for providing the reference strain of T. rangeli. This research was partially supported by Consejo Nacional de Investigaciones Científicas y Técnicas, “Agencia Nacional de Ciencia y Técnica” (ANPCyT) through PICT 2011-0207 and subsidies from “Administración Nacional de Laboratorios e Institutos de Salud” - FOCANLIS 2009–2010.
References
- Araújo C.A.C., Waniek P.J., Xavier S.C.C., Jansen A.M. Genotype variation of Trypanosoma cruzi isolates from different Brazilian biomes. Exp. Parasitol. 2011;127:308–312. doi: 10.1016/j.exppara.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Bar M.E., Wisnivesky-Colli C. Triatoma sordida stål 1859 (Hemiptera, Reduviidae: Triatominae) in palms of Northeastern Argentina. Mem. Inst. Oswaldo Cruz. 2001;96:895–899. doi: 10.1590/s0074-02762001000700002. [DOI] [PubMed] [Google Scholar]
- Bar M.E., Alvarez B.M., Oscherov E.B., Damborsky M.P., Jörg E.M. Contribution to knowledge of reservoirs of Trypanosoma cruzi (Chagas, 1909) in Corrientes province, Argentina. Rev. Soc. Bras. Med. Trop. 1999;32:271–276. doi: 10.1590/s0037-86821999000300008. [DOI] [PubMed] [Google Scholar]
- Bar M.E., Oscherov E.B., Damborsky M.P., Borda M. Epidemiología de la Tripanosomiasis americana en el norte de Corrientes. Medicina-Buenos Aires. 2010;70:133–138. [PubMed] [Google Scholar]
- Bernabé C., Brisse S., Tibayrenc M. Population structure and genetic typing of Trypanosoma cruzi, the agent of Chagas disease: a multilocus enzyme electrophoresis approach. Parasitology. 2000;120:513–526. doi: 10.1017/s0031182099005661. [DOI] [PubMed] [Google Scholar]
- Bicca-Marques J.C. How do howler monkeys cope with habitat fragmentation? In: Marsh L.K., editor. Primates in Fragments: Ecology and Conservation. Kluwer Academic/Plenum Publishers; New York: 2003. pp. 283–303. [Google Scholar]
- Botero A., Ortiz S., Muñoz S., Triana O., Solari A. Differentiation of Trypanosoma cruzi and Trypanosoma rangeli of Colombia using minicircle hybridization tests. Diagn. Micr. Infec. Dis. 2010;68:265–270. doi: 10.1016/j.diagmicrobio.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Brown A.D., Zunino G.E. Hábitat, densidad y problemas de conservación de los primates de Argentina. Vida Silv. Neotropical. 1994;3:30–40. [Google Scholar]
- Burgos J.M., Altcheh J., Bisio M., Duffy T., Valadares H.M.S., Seidenstein M.E., Piccinali R. Direct molecular profiling of minicircle signatures and lineages of Trypanosoma cruzi bloodstream populations causing congenital Chagas disease. Int. J. Parasitol. 2007;37:1319–1327. doi: 10.1016/j.ijpara.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Chagas C. Trypanosoma minasense: nota preliminar. Brazil-Medico. 1908;22:471. [Google Scholar]
- Chinchilla M., Troyo A., Guerrero O.M., Gutiérrez-Espeleta G.A., Sánchez R. Presencia de Trypanosoma minasense (Kinetoplastida: Trypanosomatidae) en Alouatta palliata (Primates: Cebidae) de Costa Rica. Parasitol. Latinoam. 2005;60:90–92. [Google Scholar]
- Santa Cruz A.C.M., Prieto O.H., Roux J.P., Patiño E.M., Borda J.T., Gómez L.G., Schiebler N. Comunicaciones Científicas y Tecnológicas, Universidad Nacional del Nordeste; 2000. Endo y ectoparasitosis en mono aullador (Alouatta caraya) (Humbolt, 1812), Mammalia, Cebidae. Informe preliminar; pp. 3–6. [Google Scholar]
- Cruz-Guzmán P.J., Morocoima A., Chique J.D., Ramonis-Quintero J., Toquero Uzcátegui M., Carrasco H.J. Psammolestes arthuri naturally infected with Trypanosoma cruzi found in sympatry with Rhodnius prolixus and Triatoma maculata on bird nests in Anzoátegui state, Venezuela. Saber Univ. Oriente Venezuela. 2014;26:428–440. [Google Scholar]
- Da Silva M.F., Naiff R.D., Marcili A., Gordo M., D’Affonseca Neto J.A., Naiff M.F., Franco A.M.R. Infection rates and genotypes of Trypanosoma rangeli and T. cruzi infecting free-ranging Saguinus bicolor (Callitrichidae), a critically endangered primate of the Amazon Rainforest. Acta Trop. 2008;107:168–173. doi: 10.1016/j.actatropica.2008.05.015. [DOI] [PubMed] [Google Scholar]
- Damborsky M.P., Bar M.E., Oscherov E.B. Detection of triatomines (Hemiptera: Reduviidae) in domiciliary and extra-domiciliary ecotopes. Corrientes, Argent. Cad. Saúde Pública, Río Jan. 2001;17:843–849. doi: 10.1590/s0102-311x2001000400018. [DOI] [PubMed] [Google Scholar]
- De Resende D.M., Pereira L.H., Lobo A. Long-term patency of blood parasites by Trypanosoma minasense and microfilariae in Callithrix penicillata marmosets (Primates, Callitrichidae), caught at the wild and maintained in captivity. Mem. Inst. Oswaldo Cruz. 1994;89:127–128. doi: 10.1590/s0074-02761994000100025. [DOI] [PubMed] [Google Scholar]
- Deane L.M., Da Silva J.E., Loures Filho L. Nycthemeral variation in the parasitaemia of Trypanosoma minasense in naturally infected marmosets of the genus Callithrix (primates, Callithricidae) Rev. Inst. Med. Trop. São Paulo. 1974;16:1–6. [PubMed] [Google Scholar]
- Di Rienzo J.A., Casanoves F., Balzarini M.G., Gonzalez L., Tablada M., Robledo C.W. Grupo InfoStat, FCA, Universidad Nacional de Córdoba; Argentina: 2014. InfoStat Versión 2014.http://www.infostat.com.ar [Google Scholar]
- Dias P.A.D., Rangel-Negrín A. Diets of howler monkeys. In: Kowalewski M.M., editor. Howler Monkeys. Springer; New York: 2015. pp. 21–56. [Google Scholar]
- Duffy T., Bisio M., Altcheh J., Burgos J.M., Diez M., Levin M.J., Favaloro R.R. Accurate real-time PCR strategy for monitoring bloodstream parasitic loads in Chagas disease Patients. PLoS Negl. Trop. Dis. 2009;3:e419. doi: 10.1371/journal.pntd.0000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy T., Cura C.I., Ramirez J.C., Abate T., Cayo N.M., Parrado R., Bello Z.D. Analytical performance of a multiplex Real-Time PCR assay using TaqMan probes for quantification of Trypanosoma cruzi satellite DNA in blood samples. PLoS Negl. Trop. Dis. 2013;7:e2000. doi: 10.1371/journal.pntd.0002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn F.L., Lambrecht F.L., Du Plessis R. Trypanosomes of South American monkeys and marmosets. Am. J. Trop. Med. Hyg. 1963;12:524–534. doi: 10.4269/ajtmh.1963.12.524. [DOI] [PubMed] [Google Scholar]
- Eberhard M., D’Alessandro A. Congenital Trypanosoma cruzi infection in a laboratory-born squirrel monkey, Saimiri sciureus. Am. J. Trop. Med. Hyg. 1982;31:931–933. doi: 10.4269/ajtmh.1982.31.931. [DOI] [PubMed] [Google Scholar]
- Fernández V.A. Universidad de Buenos Aires; Buenos Aires, Argentina: 2014. Ecología nutricional del mono aullador negro y dorado (Alouatta caraya) en el límite sur de su distribución. PhD Thesis. [Google Scholar]
- Funayama G.K., Barretto M.P. Estudos sobre reservatorios e vectores silvestres do Trypanosoma cruzi. XLII-Infeccão natural do simio, Alouatta caraya (Humboldt, 1812) pelo T. cruzi. Rev. Inst. Med. Trop. São Paulo. 1970;12:257–265. [PubMed] [Google Scholar]
- Gaunt M., Miles M. The ecotopes and evolution of triatomine bugs (Triatominae) and their associated trypanosomes. Mem. Inst. Oswaldo Cruz. 2000;95:557–565. doi: 10.1590/s0074-02762000000400019. [DOI] [PubMed] [Google Scholar]
- Gillespie T.R., Nunn C.L., Leendertz F.H. Integrative approaches to the study of primate infectious disease: implications for biodiversity conservation and global health. Am. J. Phys. Anthropol. Suppl. 2008;47:53–69. doi: 10.1002/ajpa.20949. [DOI] [PubMed] [Google Scholar]
- Gurgel-Gonçalves R., Cura C., Schijman A.G., Cuba Cuba C.A. Infestation of Mauritia flexuosa palms by triatomines (Hemiptera: Reduviidae), vectors of Trypanosoma cruzi and Trypanosoma rangeli in the Brazilian savanna. Acta Trop. 2012;121:105–111. doi: 10.1016/j.actatropica.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Higo H., Miura S., Horio M., Mimori T., Hamano S., Agatsuma T., Yanagi T. Genotypic variation among lineages of Trypanosoma cruzi and its geographic aspects. Parasitol. Int. 2004;53:337–344. doi: 10.1016/j.parint.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Hoare C.A. Blackwell Scientific Publication; Oxford. U.K.: 1972. The Trypanosomes of Mammals: a Zoological Monograph; p. 749. [Google Scholar]
- Kowalewski M.M., Salzer J.S., Deutsch J.C., Raño M., Kuhlenschmidt M.S., Gillespie T.R. Black and gold howler monkeys (Alouatta caraya) as sentinels of ecosystem health: patterns of zoonotic protozoa infection relative to degree of human-primate contact. Am. J. Primatol. 2010;73:75–83. doi: 10.1002/ajp.20803. [DOI] [PubMed] [Google Scholar]
- Kreeger T.J., Arnemo J.M. Vancouver, British Columbia, Sunquest, Canadá; 2007. Handbook of Wildlife Chemical Immobilization. [Google Scholar]
- Lewis M.D., Ma J., Yeo M., Carrasco H.J., Llewellyn M.S., Miles M.A. Genotyping of Trypanosoma cruzi: systematic selection of assays allowing rapid and accurate discrimination of all known lineages. Am. J. Trop. Med. Hyg. 2009;81:1041–1049. doi: 10.4269/ajtmh.2009.09-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisboa C.V., Pinho A.P., Monteiro R.V., Jansen A.M. Trypanosoma cruzi (kinetoplastida Trypanosomatidae): biological heterogeneity in the isolates derived from wild hosts. Exp. Parasitol. 2007;116:150–155. doi: 10.1016/j.exppara.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Lisboa C.V., Monteiro R.V., Martins A.F., Xavier S.C.D.C., Lima V.D.S., Jansen A.M. Infection with Trypanosoma cruzi TcII and TcI in free-ranging population of lion tamarins (Leontopithecus spp): an 11-year follow-up. Mem. Inst. Oswaldo Cruz. 2015;110:394–402. doi: 10.1590/0074-02760140400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn M.S., Lewis M.D., Acosta N., Yeo M., Carrasco H.J., Segovia M., Vargas J. Trypanosoma cruzi IIc: phylogenetic and phylogeographic insights from sequence and microsatellite analysis and potential impact on emergent Chagas disease. PLoS Negl. Trop. Dis. 2009;3:e510. doi: 10.1371/journal.pntd.0000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffey L., Cardinal M.V., Ordóñez-Krasnowski P.C., Lanati L.A., Lauricella M.A., Schijman A.G., Gürtler R.E. Direct molecular identification of Trypanosoma cruzi discrete typing units in domestic and peridomestic Triatoma infestans and Triatoma sordida from the Argentine Chaco. Parasitology. 2012;139:1570–1579. doi: 10.1017/S0031182012000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcili A., Valente V.C., Valente S.A., Junqueira A.C.V., Silva F.M.D., Pinto A.Y.D.N., Naiff R.D. Trypanosoma cruzi in Brazilian Amazonia: lineages TCI and TCIIa in wild primates, Rhodnius spp. and in humans with Chagas disease associated with oral transmission. Int. J. Parasitol. 2009;39:615–623. doi: 10.1016/j.ijpara.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Nóbrega A.A., Garcia M.H., Tatto E., Obara M.T., Costa E., Sobel J., Araujo W.N. Oral transmission of Chagas disease by consumption of açaí palm fruit, Brazil. Emerg. Infect. Dis. 2009;15:653–655. doi: 10.3201/eid1504.081450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireau F., Diosque P., Jansen A.M. Trypanosoma cruzi: adaptation to its vectors and its hosts. Vet. Res. 2009;40:26. doi: 10.1051/vetres/2009009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn C.L., Altizer S. Behavior, Ecology and Evolution. Oxford Series in Ecology and Evolution. Oxford University; New York: 2006. Infectious diseases in primates. [Google Scholar]
- Oklander L.I., Zunino G.E., Di Fiore A., Corach D. Isolation, characterization and evaluation of 11 autosomal STRs suitable for population studies in black and gold howler monkeys Alouatta caraya. Mol. Ecol. Notes. 2007;7:117–120. [Google Scholar]
- Orozco M.M., Enriquez G.F., Alvarado-Otegui J.A., Cardinal M.V., Schijman A.G., Kitron U., Gürtler R.E. New sylvatic hosts of Trypanosoma cruzi and their reservoir competence in the humid Chaco of Argentina: a longitudinal study. Am. J. Trop. Med. Hyg. 2013;88:872–882. doi: 10.4269/ajtmh.12-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piron M., Fisa R., Casamitjana N., López-Chejade P., Puig L., Vergés M., Gascón J. Development of a real-time PCR assay for Trypanosoma cruzi detection in blood samples. Acta Trop. 2007;103:195–200. doi: 10.1016/j.actatropica.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Rabinovich J.E., Kitron U.D., Obed Y., Yoshioka M., Gottdenker N., Chaves L.F.L. Ecological patterns of blood-feeding by kissing-bugs (Hemiptera: Reduviidae: Triatominae) Mem. Inst. Oswaldo Cruz. 2011;106:479–494. doi: 10.1590/s0074-02762011000400016. [DOI] [PubMed] [Google Scholar]
- Ramírez J.C., Cura C.I., Moreira O.C., Lages-Silva E., Juiz N., Velázquez E., Ramírez J.D. Analytical validation of quantitative real-time PCR methods for quantification of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. J. Mol. Diagn. 2015;17:605–615. doi: 10.1016/j.jmoldx.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha F.L., Roque A.L.R., de Lima J.S., Cheida C.C., Lemos F.G., de Azevedo F.C., Arrais R.C. Trypanosoma cruzi infection in neotropical wild carnivores (Mammalia: Carnivora): at the top of the T. cruzi transmission chain. PloS One. 2013;8:e67463. doi: 10.1371/journal.pone.0067463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque A.L.R., Xavier S.C.C., Da Rocha M.G., Duarte A.C.M., D’Andrea P.S., Jansen A.M. Trypanosoma cruzi transmission cycle among wild and domestic mammals in three areas of orally transmitted Chagas Disease outbreaks. Am. J. Trop. Med. Hyg. 2008;79:742–749. [PubMed] [Google Scholar]
- Rozsa L., Reiczigel J., Majoros G. Quantifying parasites in samples of hosts. J. Parasitol. 2000;86:228–232. doi: 10.1645/0022-3395(2000)086[0228:QPISOH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Rumiz D.I. Alouatta caraya: population density and demography in Northern Argentina. Am. J. Primatol. 1990;21:279–294. doi: 10.1002/ajp.1350210404. [DOI] [PubMed] [Google Scholar]
- Sato H., Leo N., Katakai Y., Takano J., Akari H., Nakamura S., Une Y. Prevalence and molecular phylogenetic characterization of Trypanosoma (Megatrypanum) minasense in the peripheral blood of small neotropical primates after a quarantine period. J. Parasitol. 2008;94:1128–1138. doi: 10.1645/GE-1513.1. [DOI] [PubMed] [Google Scholar]
- Schijman A.G., Altcheh J., Burgos J.M., Biancardi M., Bisio M., Levin M.J., Freilij H. Aetiological treatment of congenital Chagas’ disease diagnosed and monitored by the polymerase chain reaction. J. Antimicrob. Chemoth. 2003;52:441–449. doi: 10.1093/jac/dkg338. [DOI] [PubMed] [Google Scholar]
- Schijman A.G., Lauricella M.A., Marcet P.L., Duffy T., Cardinal M.V., Bisio M., Levin M.J. Differential detection of Blastocrithidia triatomae and Trypanosoma cruzi by amplification of 24salpha ribosomal RNA genes in faeces of sylvatic triatomine species from rural northwestern Argentina. Acta Trop. 2006;99:50–54. doi: 10.1016/j.actatropica.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Schijman A.G., Bisio M., Orellana L., Sued M., Duffy T., Mejia Jaramillo A.M., Cura C. International study to evaluate PCR methods for detection of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. PLoS. Negl. Trop. Dis. 2011;5:e931. doi: 10.1371/journal.pntd.0000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D.A., Kowalewski M.M., Ellersieck M.R. Serum nutritional profiles of free-ranging Alouatta caraya in Northern Argentina: Lipoproteins; amino acids; vitamins A, D, and E; carotenoids; and minerals. Int. J. Primatol. 2007;28:1093–1107. [Google Scholar]
- Souto R.P., Vargas N., Zingales B. Trypanosoma rangeli: discrimination from Trypanosoma cruzi based on a variable domain from the large subunit ribosomal RNA gene. Exp. Parasitol. 1999;91:306–314. doi: 10.1006/expr.1998.4380. [DOI] [PubMed] [Google Scholar]
- Souza O.E., Dawson G.A. Trypanosome infections in the marmosets (Saguinus geoffroyi) from the Panama Canal zone. Am. J. Trop. Med. Hyg. 1976;25:407–409. doi: 10.4269/ajtmh.1976.25.407. [DOI] [PubMed] [Google Scholar]
- Souza-Lima R.D.C.D., Vale Barbosa G., Rodrigues Coura J., Lima Arcanjo A.R., Da Silva Nascimento A., Barbosa Ferreira J.M.B., Magalhães L.K. Outbreak of acute Chagas disease associated with oral transmission in the Rio Negro region, Brazilian Amazon. Rev. Soc. Bras. Med. Trop. 2013;46:510–514. doi: 10.1590/0037-8682-1367-2013. [DOI] [PubMed] [Google Scholar]
- Tenório M.S., Oliveira e Sousa L., Alves-Martin M.F., Paixão M.S., Rodrigues M.V., Starke-Buzetti W.A., Araújo Junior J.P. Molecular identification of trypanosomatids in wild animals. Vet. Parasitol. 2014;203:203–206. doi: 10.1016/j.vetpar.2014.02.010. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence waiting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M. Genetic epidemiology of parasitic protozoa and other infectious agents: the need for an integrated approach. Int. J. Parasitol. 1998;28:85–104. doi: 10.1016/s0020-7519(97)00180-x. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M. Genetic subdivisions within Trypanosoma cruzi (Discrete Typing Units) and their relevance for molecular epidemiology and experimental evolution. Kinetoplastid Bio. Dis. 2003;2:12. doi: 10.1186/1475-9292-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travi B.L., Ruiz A.M., Colillas O.J., Segura E.L. Trypanosomiasis en monos neotropicales. Medicina-Buenos Aires. 1982;42:55–60. [PubMed] [Google Scholar]
- World Health Organization . Sixty-Third World Health Assembly; 2010. Chagas Disease: Control and Elimination. [Google Scholar]
- Yeo M., Acosta N., Llewellyn M., Sánchez H., Adamson S., Miles G.A.J., López E. Origins of Chagas disease: Didelphis species are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. Int. J. Parasitol. 2005;35:225–233. doi: 10.1016/j.ijpara.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Ziccardi M., Lourenço de Oliveira R. The infection rates of trypanosomes in squirrel monkeys at two sites in the Brazilian Amazon. Mem. Inst. Oswaldo Cruz. 1997;92:465–470. doi: 10.1590/s0074-02761997000400003. [DOI] [PubMed] [Google Scholar]
- Ziccardi M., Lourenço de Oliveira R., Lainson R., de Oliveira Brígido M. do C., Pereira Carneiro Muniz J.A. Trypanosomes of non-human primates from the National Centre of primates, Ananindeua, State of Pará, Brazil. Mem. Inst. Oswaldo Cruz. 2000;95:157–159. doi: 10.1590/s0074-02762000000200004. [DOI] [PubMed] [Google Scholar]
- Zingales B., Andrade S.G., Briones M.R.S., Campbell D.A., Chiari E., Fernandes O., Guhl F. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem. Inst. Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- Zunino G.E., Kowalewski M.M. Primate research and conservation in northern Argentina: the field station Corrientes (Estación Biológica de Usos Múltiples - EBCo) Trop. Conserv. Sci. 2008;1:140–150. [Google Scholar]
- Zunino G.E., Kowalewski M.M., Oklander L.I., González V. Habitat fragmentation and population size of the black and gold howler monkey (Alouatta caraya) in a semideciduous forest in northern Argentina. Am. J. Primatol. 2007;69:966–975. doi: 10.1002/ajp.20389. [DOI] [PubMed] [Google Scholar]