Abstract
Genome-wide association studies (GWASs) have been successful in detecting variants correlated with phenotypes of clinical interest. However, the power to detect these variants depends on the number of individuals whose phenotypes are collected, and for phenotypes that are difficult to collect, the sample size might be insufficient to achieve the desired statistical power. The phenotype of interest is often difficult to collect, whereas surrogate phenotypes or related phenotypes are easier to collect and have already been collected in very large samples. This paper demonstrates how we take advantage of these additional related phenotypes to impute the phenotype of interest or target phenotype and then perform association analysis. Our approach leverages the correlation structure between phenotypes to perform the imputation. The correlation structure can be estimated from a smaller complete dataset for which both the target and related phenotypes have been collected. Under some assumptions, the statistical power can be computed analytically given the correlation structure of the phenotypes used in imputation. In addition, our method can impute the summary statistic of the target phenotype as a weighted linear combination of the summary statistics of related phenotypes. Thus, our method is applicable to datasets for which we have access only to summary statistics and not to the raw genotypes. We illustrate our approach by analyzing associated loci to triglycerides (TGs), body mass index (BMI), and systolic blood pressure (SBP) in the Northern Finland Birth Cohort dataset.
Introduction
Genome-wide association studies (GWASs) are conducted by collecting genotypes and phenotypes from a set of individuals. This is followed by a series of statistical tests to identify variants that are significantly associated with the phenotype. Recently, the sample size for GWASs has increased to tens of thousands or hundreds of thousands. These large studies have discovered hundreds of new variants involved in multiple common diseases.1, 2 Most of these variants have very small effect sizes, which emphatically supports the message that the larger the association study the better it fares in discovering associations.
Unfortunately, some phenotypes are either logistically difficult or very expensive to collect. For these phenotypes, it is impractical to perform GWASs with tens of thousands or hundreds of thousands of individuals with these phenotypes. Examples of these phenotypes include ones that require (1) obtaining an inaccessible tissue such as brain expression, (2) using a complex intervention such as a response to diet, or (3) re-contacting individuals simply because they were unmeasured in the original cohort. Investigators are often unable to collect samples that are large enough to discover variants with small effect sizes for these phenotypes. As a result, it is unlikely that GWASs will be effectively conducted on these phenotypes.
One approach to increase power for GWASs on a phenotype that is hard to collect is to utilize an intermediate or proxy phenotype that is correlated to the target phenotype of interest. In this approach, one intermediate or proxy phenotype, which is highly correlated and easily collectable, is collected and then a GWAS is performed on the intermediate phenotype in order to detect associated signals. For example, triglyceride levels can be collected as a proxy for obtaining information about metabolic diseases. This approach is known as intermediate phenotype analysis.3, 4
One way to interpret the intermediate phenotype analysis is to consider the target phenotype as missing data and use the intermediate phenotype as inferring the missing data. This connection to missing data analysis motivates the following intuition. In missing data analyses, it is well known that utilizing multiple sources of information can be more effective than using a single source of information, which has been shown in machine learning5, 6, 7, 8, 9 and genetics.10, 11, 12 This motivates an intuition that utilizing multiple phenotypes together as proxies for a trait can lead to better performance. This is the basis of our approach.
In this paper, we propose an approach called phenotype imputation that allows one to perform a GWAS on a phenotype that is difficult to collect. Our approach leverages the correlation structure between multiple phenotypes to impute the uncollected phenotype. Specifically, we estimate the correlation structure from a complete dataset that includes all phenotypes. The conditional distribution based on the multivariate normal (MVN) statistical framework is used to impute the uncollected phenotype in an incomplete dataset. Our imputation approach utilizes only phenotypic information and not genetic information; therefore, imputed phenotypes can be subsequently used for association testing without incurring data re-use. We provide an optimal meta-analysis strategy for situations where the final GWAS will include the complete and incomplete datasets. This strategy combines association results from the collected phenotype and imputed phenotype while accounting for imputation uncertainties. Moreover, we demonstrate that we can analytically calculate the statistical power of an association test using an imputed phenotype, which can be helpful for study design purposes. In addition, we show that the summary statistics of the imputed phenotype can be approximated by a weighted linear combination of summary statistics for the proxy phenotypes. This result makes our method applicable to datasets where we have access only to the summary statistics and not to the raw genotypes and phenotypes.
We show the effectiveness of our proposed approach by applying it to the Northern Finland Birth Cohort (NFBC) data.13 Imputing the triglyceride (TG), body mass index (BMI), and systolic blood pressure (SBP) phenotypes enable us to recover most of the significantly associated loci in the original data at the nominal significance level. This shows that even though the imputed phenotype might not provide sufficient power for discovery purposes due to imputation uncertainties, it can effectively be used for replication purposes.
Material and Methods
A Standard Genome-wide Association Study
Initially, we describe the standard GWAS framework for testing genetic effects on quantitative phenotypes. SNPs are the most common form of genetic variation; therefore, we consider SNPs throughout this paper. However, the frameworks can be generalized to other types of variants. Suppose that we collect genotypes of m SNPs and quantitative phenotypes for n individuals. Let Y indicate a matrix of phenotypic values where yk is a (n × 1) vector for the kth phenotype. Let yjk be the phenotypic value of the jth individual for the kth phenotype and gji = {0,1,2} be the minor allele count of the jth individual at the ith SNP. Let pi indicate the frequency of ith variant in the population. The derivations are simplified by standardizing the minor allele counts for each SNP to have a mean of 0 and a variance of 1, such that represents the standardized value of gji. Let xi be the (n × 1) vector of standardized minor allele counts at the ith SNP, where 1T xi = 0 and . We assume Fisher’s polygenic model where the phenotype and the genotype follow normal distributions. Under the additive model, each SNP contributes linearly toward the phenotype:
| (Equation 1) |
where μk is the phenotypic mean for the kth phenotype, 1 is a (n × 1) vector of all ones, and βik is the effect of the ith SNP toward the kth phenotype. is the environment and measurement errors where I is an identity matrix. We additionally assume that the phenotypes are standardized so that their means are 0 and their variances are 1.
In a standard GWAS, we consider one SNP and one phenotype at a time. We omit SNP index below (e.g., instead of xi, we use x) for notation clarity. The following model is used to test each SNP:
| (Equation 2) |
Equation 2 is different from Equation 1 in that it omits the effects of the other SNPs, which can manifest as background genetic effects. This was the motivation for using mixed model14, 15, 16, 17 in the situations where sample data have population structures. Equation 2 leads us to least square solutions, and , where “hat” over parameters denotes estimated values. is the residual error that is used to compute the standard error .18, 19, 20, 21 Note that the estimated effect size is equal to the correlation between the standardized minor allele counts and the standardized phenotypic values, = cor. If the sample size is large enough, follows a normal distribution with the mean equal to the true effect size βk. Thus, we can define a normally distributed association statistic as . Under the null hypothesis of no association (βk = 0), the statistic sk follows the standard normal distribution. Under the alternative hypothesis of true association, the statistic sk follows a normal distribution with non-centrality parameter (NCP) :14, 15, 20, 22
| (Equation 3) |
To reject the null hypothesis of no association, given the significance threshold α, we compute the p value, which is the probability that the observed statistic sk will be equally or more extreme under the null hypothesis, and determine that the association is significant if this probability is less than the significance threshold α (e.g., α = 5 × 10−8 in GWASs). Equivalently, we reject the null hypothesis when Φ(sk) < αs/2 or Φ(sk) > 1 − αs/2, where Φ(.) indicates the cumulative density function of the standard normal distribution.
The statistical power is the probability of detecting an association in a situation where an association is present with a certain effect size.22, 23, 24, 25 Intuitively, power measures the probability that the truly associated variants will be discovered. Statistical power depends on both the effect size and the number of individuals in the study; therefore, power estimates can guide the choice of study size as well as provide expectations for which effect sizes can and can not be discovered. Power is estimated as
| (Equation 4) |
which is a function of the effect size βk, its standard error σk, the number of individuals n, and the significance threshold α.
Phenotype Imputation
Phenotype Imputation Method
We consider two phenotype datasets in which we collected phenotypes from n1 and n2 individuals, respectively. Let Y(1) and Y(2) be matrices of phenotypic values of size and , and and be vectors of phenotypic values for the kth phenotype in the first and second datasets, respectively. We use to indicate phenotypes excluding the phenotype. Thus, and are row vectors of size for the jth individual phenotypes excluding the phenotype in Y(1) and Y(2), respectively.
We assume that the phenotypic values follow a multivariate normal distribution. In the Discussion, we explore the case where this assumption is violated. If we assume that each phenotype is standardized to a mean of 0 and variance of 1, then we model the joint distribution of multiple phenotypes as
This can be represented more compactly with a block matrix:
where is a row vector for the first phenotypic values for the jth individual obtained from Y(1) and is the same vector in column format. Let indicate the correlation between the two phenotypes k1 and k2, and let denote a vector of correlations between and the phenotypes in Y(1) excluding the phenotype. is a covariance matrix between the phenotypes in Y(1) excluding the phenotype.
Using the above joint distribution, we condition on phenotypes to compute the distribution of phenotypic values for the jth individual for the phenotype. This distribution is computed as follows:
| (Equation 5) |
We assume that the phenotype is not collected in the second study in the phenotype imputation problem. Let be the imputed phenotypic values for the uncollected phenotype. We assume that the correlation between any pair of phenotypes is the same in two datasets Y(1) and Y(2). As a result, the above joint distribution in Equation 5 holds for Y(2). Thus, we can perform a similar conditional analysis. The conditional distribution is computed as follows:
| (Equation 6) |
The method for imputing the missing phenotype for a particular individual j uses the mean of the conditional distribution as shown in Equation 6, , as our prediction. A more compact formula to impute the phenotype for all the individuals in the dataset Y(2) is as follows:
| (Equation 7) |
Equation 7 shows that the imputed phenotype is a linear weighted combination of other collected phenotypes. Thus, if our multivariate normal assumption holds, the imputed phenotype will also follow a normal distribution.
We utilized the imputed phenotype in the association study to compute the association statistic of the imputed phenotype as the ratio between the estimated effect size for the imputed phenotype and its standard error. The association statistic is:
| (Equation 8) |
where , , and are the estimated effect size, standard error, and residual error, respectively, as computed from the imputed values of the phenotype. Given a sufficiently large sample size, this statistic will follow a normal distribution. It will follow under the null hypothesis of no association to imputed phenotype.
Noisy Measurement Model
We introduce a model that is closely related to our phenotype imputation method. Under this model, called noisy measurement model (NMM), our method has interesting optimal properties that are related to the weighted sum of statistics approach. However, NMM is not a requirement for our method to work.
Under NMM, we assume that the phenotype has the main genetic effect, whereas other phenotypes can be modeled as the phenotype plus noise. We consider the other phenotypes as noisy measurements of the phenotype . Under this model, the pleiotropic genetic effects to other phenotypes are driven by the main genetic effect to phenotype . As a result, the observed genetic effect to each of the phenotypes cannot be greater than the genetic effect to phenotype . Generally, this can be a strict assumption, but considering our situation where only phenotype is missing, this can be a reasonable assumption; if the genetic effect is greater in phenotype , then it makes more sense to model the main effect driven by phenotype k. An analysis of the collected phenotype k data alone would be optimal, and we do not even need to perform phenotype imputation.
Specifically, we describe NMM as
| (Equation 9) |
where uk is “noise” in the measurement. We assume that the noise follows a normal distribution with mean zero and variance . We further assume that the noise is independent of genotypes. The denominator was formulated to standardize the phenotype .
Let be the correlation between and . It is straightforward to show that
Thus, we can re-write Equation 9 such as
| (Equation 10) |
An important property of NMM is that if NMM holds, then the strength of the effect of the variant on phenotype k is approximately the strength of the effect of the variant on phenotype times the correlation between the two phenotypes. That is, if , then approximately . This can be shown as
where we further assume that the residual errors are similar for two phenotypes ; this holds true if the genetic effects are small. A similar relationship arises when considering the statistics of two SNPs in linkage disequilibrium (LD) and the correlation between the two SNPs is r. Others have shown that the ratio between the NCPs of two statistics is the same as r.26, 27, 28, 29 This is similar to NMM in the sense that a causal SNP drives the genetic effect, and the proxy SNP can be thought of as a noisy measurement of the causal SNP due to LD.
Power of Phenotype Imputation
If NMM describes truth, it is possible to analytically calculate the power of our phenotype imputation method. We consider the situation that the variant we are testing under NMM is associated with the phenotype with NCP of . The NCP of the association statistic for the kth phenotype on the same variant is where is the correlation between the phenotypes k and . Here, instead of considering the correlation between the phenotype and another phenotype k, we consider the correlation between the phenotype and the imputed phenotype of .
The covariance of the imputed and true phenotype is:
| (Equation 11) |
We know that the variance of is 1, because we have already standardized the phenotypes. We compute the variance of the imputed phenotype as follows:
| (Equation 12) |
If we utilize the covariance between the imputed and true phenotypes and the variance of phenotypes, we can compute the correlation as follows:
| (Equation 13) |
Under NMM, each phenotype is modeled as a standardized linear combination of phenotype and noise. Imputed phenotype is also a linear combination of those phenotypes; thus, we can consider the imputed phenotype as a new phenotype where we can apply NMM. That is, we can consider the imputed phenotype as a noisy version of the true phenotype. Then, by the property of NMM,
| (Equation 14) |
We obtained NCP of the statistic for the imputed phenotype; therefore, we can analytically calculate power of our phenotype imputation using Equation 4.
It should be noted that the following quantity will have a mean of 0:
| (Equation 15) |
The variance of is computed as follows:
Our results evaluate this quantity in real dataset to determine whether our imputation method works as expected.
Relation to Optimal Linear Combinations of Marginal Statistics
The result of phenotype imputation is a weighted linear combination of the observed phenotypes. We show that under NMM, phenotype imputation is the “optimal” weighted combination of the phenotypes in terms of statistical power. Let be a vector of association statistics computed for the first phenotypes, . Under NMM, given that the NCP of the uncollected phenotype is , we have . We calculate the association statistic of the imputed phenotype as a linear combination of weighted statistics computed for the phenotypes. Let indicate the vector of weights where wi is the weight corresponding to the ith phenotype marginal statistics, then we have following formula:
| (Equation 16) |
Using the above formula and the fact the variance of the associated statistic is 1, we have:
It has been shown that power is maximized when we maximize the NCP.30 Thus, we find the set of weights that maximizes . Let and w′ = Aw, then our maximization problem reduces to following optimization:
If we let and use the Cauchy-Schwarz inequality, we get the following:
The optimal value for w′ is Θ and the maximum NCP is as follows:
This is exactly the NCP obtained from the previous section. Moreover, the optimal value for w is , which is the same vector of weights used in the previous section. This is the justification for Equation 14 above.
Interestingly, this result indicates that we can use Equation 16 and the optimal weights, which are obtained in this section, to estimate the marginal statistics of the imputed phenotype as weighted linear combinations of observed marginal statistics from other phenotypes. Thus, given the observed marginal statistics of the first phenotypes and the pairwise phenotype correlations, we can compute the estimated marginal statistics. Our method does not need to have access the raw genotypes and phenotypes. This makes our method applicable to datasets where we have access only to the summary statistics.
We note that for any vector of weights, including the ones utilized in imputation, the type I error rates are controlled. The reason is that if the variant we are testing is not associated with the phenotype, λ = 0, then the NCP of the imputed statistic for that variant is zero.
Optimal Meta-analysis Strategy for Combining Imputed and Observed Values
We use the phenotype imputation to fill the values of the phenotype for individuals whose phenotypic values are missing. We then want to obtain an association statistic for the combined dataset, including the imputed and observed phenotypes. However, our imputation is not always accurate; thus, it is suboptimal to use combined observed and imputed data without distinguishing between them. We propose to compute the association statistics by performing statistical tests on the collected phenotype and imputed phenotype separately. Then, we perform a fixed-effect meta-analysis to combine the two statistics.
We use Ym and Yc to indicate the missing and collected phenotypes, respectively. We compute the association statistic of each set separately. The association statistic for the collected phenotype is computed as where λc is the NCP of the phenotype and nc is the number of individuals whose phenotypic values are collected for this phenotype. We use Equation 14 to compute the Z-score for the imputed phenotype as where nm is the number of individuals whose phenotypic values are missing for this phenotype.
We combine the two statistics using the fixed-effects meta-analysis. The fixed-effects meta-analysis association statistic, sFE, is computed as , where wc and wm are computed such that the meta-analysis association statistic is maximized.31, 32 Other studies31, 33 show that the optimal weights are computed as and . Thus, we have:
| (Equation 17) |
We can use Equation 17 to combine the statistics computed for the collected and imputed phenotypes as a joint association statistic.
Polygenic Model
We described the properties of our method under NMM. However, NMM is a simple model and might not always hold true. We introduce a more complex model, which explicitly models both the genetic and environmental correlations in phenotypes. We suggest a strategy that is optimized for this model and show that the new strategy is equivalent to our standard strategy under some simplifying assumptions.
Let indicate the vector of true effect sizes of a given variant toward all phenotypes where βj is the effect size for the jth phenotype. Let E be a matrix which models the errors. We consider a multi-phenotype setting, where we perform a joint testing of a variant for all the phenotypes:
where vec() is an operator that converts a matrix to vector by stacking columns of matrix and is an operator that performs Kronecker product between two matrices.
This multi-phenotype setting enables us to model the genetic and environmental correlations. Let ρij and ξij indicate the genetic and environment correlations, respectively, between ith and jth phenotype. Let denote the genetic variance of phenotype i and denote the error variance of phenotype i. The true vector of effect sizes are assumed to follow a MVN in the multi-phenotype polygenic model,15, 34, 35, 36 such that
| (Equation 18) |
where is the proportion that the variant contributes to the genetic variance.15, 34, 35, 36 We assumed that is the same for all phenotypes. We define a variance matrix that encodes the environmental correlations in a similar manner as follows:
If we use the polygenic model, we have and where K is the kinship matrix that represents the genetic relatedness between individuals. We use the following matrix, V, that encodes the covariance for all pairs of phenotypes:
Let indicate the vector of estimated effect sizes for all the phenotypes for a given variant. Using the mixed model we have and . Let ψij be the ith row and jth column element of ψ. We can obtain marginal statistics for all the phenotypes by standardizing . Let indicate a vector of marginal statistics. The joint distribution of statistics follows a MVN where Λ is the vector of NCPs.
| (Equation 19) |
If we use Equation 18, we can assume a prior distribution for effect size of the single SNP that we test, such as . NCP is an expectation of marginal statistic, which is standardized effect size. Therefore, prior distribution for β gives us prior distribution for NCP.
| (Equation 20) |
where H is a matrix and hij is the ith row and jth column of matrix H. We have . We can utilize block matrix notation where and is a matrix of prior distribution for NCP of all the phenotypes excluding the phenotype.
In summary, we have and . We assume that the NCP for the phenotype is . Conditioned on this, the NCPs of the phenotypes excluding the phenotype is as follows:
| (Equation 21) |
As a result, the marginal statistics of all the phenotypes excluding the phenotype is as follows:
| (Equation 22) |
The equation above can be simplified by setting the to the mean of Equation 21. This assumption implies that the marginal statistics of all the phenotypes excluding, the phenotype is as follows:
| (Equation 23) |
Similarly, we consider that the imputed marginal statistics are a weighted linear combination of all the marginal statistics that maximizes the power. If we use Cauchy-Schwartz inequality, we can show that the maximum NCP of will be . The maximum NCP is achieved when the weights of the marginal statistics are . Therefore, we have successfully derived the weighted combination of marginal statistics that are optimized for the polygenic model.
Relation between Polygenic Model and Noisy Measurement Model
We show that under some simplifying assumptions, the method for the polygenic model is equivalent to the standard method for NMM. We make two assumptions. First, the pairwise genetic and environment correlations are equal (e.g., ρij = ξij) and the individuals are sufficiently unrelated so that we can approximate K with I. The second assumption implies that we have no population structure. Based on these two assumptions, we can simplify V, as follows:
| (Equation 24) |
where for any phenotypes as we standardized the phenotypes. Recall that we defined R as a phenotypic correlation matrix. Thus, . As a result, we have . Given the NCP for the phenotype is , then the NCPs of all the phenotype excluding the phenotype will have a distribution with mean equal to . Similar to previous section, if we fix NCP to its mean value for simplification, the method converges to the standard approach based on NMM. If we consider the two assumptions discussed above, then the result implies that our approach for the multi-phenotype polygenic model is equivalent to the standard strategy for NMM.
Avoiding Over-fitting
The number of phenotypes is large ( is large) in some datasets, such as eQTL datasets; thus, we have the risk of over-fitting, which occurs in a method where the number of parameters is large. These methods usually do not generalize, but it produces very high accuracy in the training dataset and very low accuracy in the test dataset. One way to avoid over-fitting is to add a sparsity prior, such as the Laplace prior,37 which reduces the linear regression to LASSO.38 The LASSO setting allows imputing of the phenotype, while utilizing few phenotypes to avoid over-fitting. Another solution is to select the most informative phenotypes and then apply our method. For example, we can pick the top ten phenotypes based on their correlation with the target phenotype. We use only these ten phenotypes in our method.
Handling Missing Data
Our method can handle missing data in the target dataset by performing imputation with only the available phenotypes for each individual. Some of the individuals will have more accurate imputation, because they utilize more phenotypes to perform the imputation. We have developed an optimal approach for performing an association test utilizing these differing degrees of quality of phenotype imputation, which we explain in Appendix A.
Adjusting for Covariates
In a typical GWAS, we usually adjust for the non-genetic factors that influence the phenotype, such as sex, age, study design, and known clinical covariates. Covariate adjustment reduces the spurious association signals in a study. Given that we have p covariates, we need to adjust for them by extending Equation 1. Thus, the polygenic model used to handle covariates for the kth phenotype is as follows:
| (Equation 25) |
where is the ith covariate and γik is the effect of that covariate toward the kth phenotype. Moreover, to perform the single SNP association test instead of using Equation 2, we need to adjust for the covariates. We use the following model for the single SNP association test:
| (Equation 26) |
There are two possible ways to adjust for covariates for phenotype imputation. The first is to impute the phenotype and then use Equation 26 for association testing. This testing is similar to testing collected phenotypes and adjusting for covariates. The second possible way is to regress out the covariates from all the collected phenotypes to generate new phenotypes where the covariates are removed. Then, we use our imputation method to impute the uncollected phenotype using the phenotypes where the covariates are regressed out. We can use Equation 2 to perform association testing.
Results
Overview of Phenotype Imputation
In phenotype imputation, we consider two datasets (D1 and D2) in which multiple phenotypes are collected along with genetic information to perform a GWAS. In the first dataset (D1), we collect the target phenotype and the related phenotypes. In the second dataset (D2), the related phenotypes have been collected for all of the individuals but the target phenotype has not been collected. These datasets are used to predict the uncollected target phenotype in the second dataset (D2) by leveraging the correlation structure between the additional phenotypes and the target phenotype. The first dataset (D1) is used to approximate this correlation structure. GWAS is performed after imputing the target phenotype to discover genetic variants that are significantly associated with the imputed target phenotype.
This framework allows for the estimation of the relative power of imputation compared to the power if the phenotype was collected in the sample. Intuitively, the power loss depends on how close the imputed phenotypes are to the true phenotypes. The correlation between the imputed and true phenotypes is defined as rimp, which can be estimated from the first dataset. This provides an idea of how well the imputation will perform in the target dataset. Under some additional assumptions, which we refer to as the noisy measurement model (NMM), the power in the imputed study with n individuals is equivalent to the power of a complete study where individuals are collected (see Material and Methods for the detailed derivation). The number of individuals that contribute toward the power of a statistical test for a phenotype is defined as the effective number of individuals. For example, we can impute triglyceride (TG) levels in the NFBC dataset13 using high-density lipoproteins (HDL), low-density lipoproteins (LDL), and systolic blood pressure (SBP) with a correlation of 0.5. As a result, in a study where HDL, LDL, and SBP were collected for 8,000 individuals, the power of GWAS on the imputed TG is equivalent to performing GWAS in 2,000 individuals where TG has been collected.
Phenotype Imputation Controls Type I Error
We simulated datasets for multiple phenotypes under the null model where the variant we are testing has no effect (effect size of zero) toward the target phenotype. We computed the type I error under five different significance thresholds: 0.05, 0.01, 0.005, 5 × 10−6, and 5 × 10−8. We generated 100,000,000 simulated datasets that consist of 1,000 individuals. The type I error rates for our imputation method were 0.049, 0.0099, 0.00489, 4.90 × 10−6, and 4.89 × 10−8 for the significance thresholds of 0.05, 0.01, 0.005, 5 × 10−6, and 5 × 10−8, respectively. This indicates that the type I error is correctly controlled in our imputation method. The Northern Finland Birth Cohort dataset13 was used to show that the type I error is controlled (see Figure S1). We plot the Q-Q plot of the Z-score for the imputed triglyceride (TG) phenotype from the Finland dataset. There is no inflation in the Q-Qplot as shown in Figure S1.
Phenotype Imputation on Northern Finland Birth Cohort
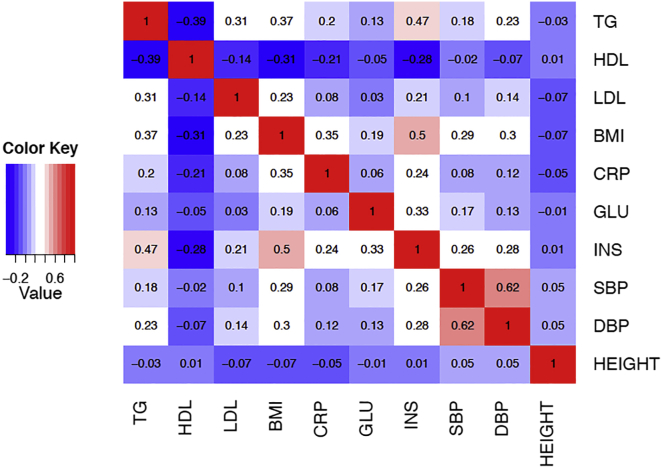
The Northern Finland Birth Cohort (NFBC) dataset13 was used to assess the performance of our method. The NFBC dataset consists of 10 phenotypes collected from 5,327 individuals. The 10 phenotypes are triglycerides (TG), high-density lipoproteins (HDL), low-density lipoproteins (LDL), glucose (GLU), insulin (INS), body mass index (BMI), C-reactive protein (CRP) as a measure of inflammation, systolic blood pressure (SBP), diastolic blood pressure (DBP), and height. The genotype data consists of 331,476 SNPs. Figure 1 shows the pairwise correlations between each pair of phenotypes. The correlation coefficients between the phenotypes in this data are between 0.01 and 0.62. SBP and DBP are the two phenotypes that show the highest correlation.
Figure 1.
The Pairwise Correlation between Each Phenotype Pair in the NFBC Dataset
We considered the possibility of imputing each of these ten phenotypes using the other nine phenotypes. First, the corresponding value of rimp was computed (Table S1). In order to evaluate our method, we are interested in the scenario where rimp is high and higher than the highest pairwise correlation. The TG, INS, DBP, BMI, and SBP phenotypes satisfied these criteria. INS and DBP do not have any significant associated variants; therefore, TG, BMI, and SBP phenotypes were the focus of the evaluation.
For our experiments, we assume that TG, BMI, and SBP phenotypes were collected for only 500 individuals to be used as a training dataset to estimate the correlation structure between phenotypes. The TG, BMI, and SBP phenotypic values were masked in the rest of the individuals and they were used only when the imputation accuracy was measured. The 500 individuals were used to compute the correlation structure between the phenotypes. Our method was used to impute the TG, BMI, and SBP phenotypes for the other individuals.
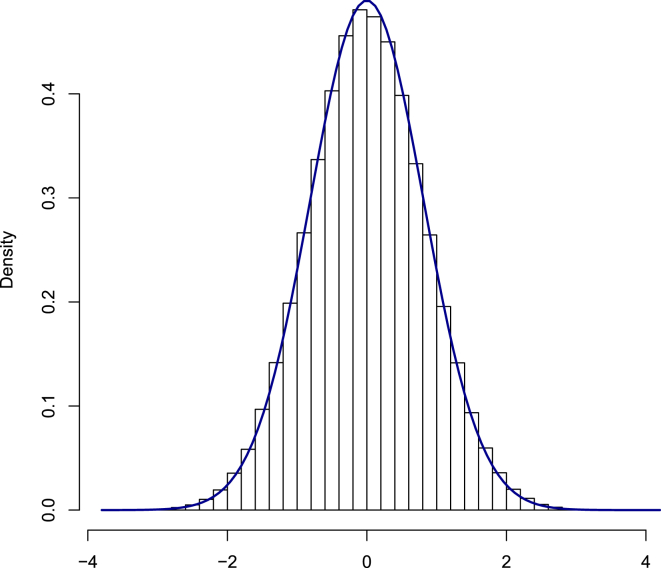
The correlation between the imputed phenotype and the true TG phenotypes was rimp = 0.58. Our estimate of this correlation from the training data was . This correlation coefficient and the size of the data resulted in an effective number of ∼1,620 (0.582 × (5,327 − 500) = 1,623) individuals. Therefore, we did not expect to see any significant loci in our imputed data. However, the size of the data was sufficient to observe an effect in a replication study. An association analysis was performed, using EMMAX14 on the imputed phenotypes, along with the original TG phenotypes for comparison. Table 1 shows the estimated effect size (β), standard error of the estimated effect size (se(β)), Z-scores, and p values. The result in Table 1 indicates that when EMMAX14 was run on the original TG phenotype in the test dataset, then seven loci passed our significance threshold of 5 × 10−6. When EMMAX14 was run on the imputed phenotypes for these seven loci, then most of these loci (six out of seven) passed the replication significance threshold of at least 0.05. Therefore, it appears that for most variants, phenotype imputation power was equivalent to collecting individuals. Surprisingly, the test statistic (Z-score) for the imputed phenotype of all variants, other than rs1260326, was close to rimp times the test statistic (Z-score) at the actual variant (Table 1). Two statistical values are defined as close when the difference between the two values is less than one standard deviation (SD = 1). This is exactly the result we expect under NMM. We also expect that if the assumption holds, the distribution of the statistic on the imputed data minus rimp times the statistic on the original data (last column of Table 1) over the whole data will follow a distribution with mean of 0 and variance of as described in the Material and Methods. In Figures 2, S3, and S4, we show that this is the case for the TG, BMI, and SBP phenotypes, respectively. These data demonstrate that although NMM is a simple model, NMM describes these datasets effectively. These results also show that performing a GWAS on the imputed phenotype has enough power to identify most of the associated loci that are significant when it is performed on the original phenotype.
Table 1.
Comparison between the Association Test on the Real Test Data for TG, BMI, and SBP Phenotypes and the Imputed Test Data in the NFBC Data
| Phenotype | rsID |
Real Test Dataa |
Imputed Test Data |
-∗ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | se(β) | Z-Score (Zreal) | p value | β | se(β) | Z-Score (Zimp) | p Value | |||
| TG | rs3923037 | 0.074 | 0.0149 | 4.96 | 7.14 × 10−7 | 0.0224 | 0.0083 | 2.700 | 0.006 | 0.17 |
| rs6728178 | 0.076 | 0.0149 | 5.10 | 3.45 × 10−7 | 0.0267 | 0.0083 | 3.209 | 0.001 | 0.24 | |
| rs6754295 | 0.074 | 0.0149 | 4.94 | 7.91 × 10−7 | 0.0266 | 0.0083 | 3.197 | 0.001 | 0.32 | |
| rs676210 | 0.0752 | 0.0149 | 5.01 | 5.38 × 10−7 | 0.0250 | 0.0083 | 2.996 | 0.002 | 0.084 | |
| rs673548 | 0.0762 | 0.0149 | 5.08 | 3.81 × 10−7 | 0.02530 | 0.0083 | 3.031 | 0.002 | 0.08 | |
| rs1260326 | −0.0807 | 0.0150 | −5.37 | 8.15 × 10−8 | −0.004 | 0.0084 | −0.534 | 0.59 | 2.58 | |
| rs10096633 | 0.0819 | 0.0147 | 5.55 | 3.00 × 10−8 | 0.0191 | 0.0082 | 2.324 | 0.02 | 0.79 | |
| BMI | rs987237 | −0.074 | 0.0150 | −4.97 | 6.63 × 10−7 | −0.037 | 0.00929 | −4.07 | 4.62 × 10−5 | 0.93 |
| rs11759809 | −0.074 | 0.0150 | −4.95 | 7.35 × 10−7 | −0.036 | 0.00931 | −3.96 | 7.43 × 10−5 | 0.84 | |
| SBP | rs782586 | 0.074 | 0.0149 | 4.96 | 7.43 × 10−7 | 0.036 | 0.01016 | 3.50 | 0.00047 | 0.37 |
| rs782588 | 0.074 | 0.0149 | 4.94 | 8.14 × 10−7 | 0.035 | 0.01014 | 3.43 | 0.00061 | 0.32 | |
| rs782602 | 0.075 | 0.0150 | 5.01 | 5.53 × 10−7 | 0.034 | 0.01016 | 3.39 | 0.00071 | 0.23 | |
| rs2627759 | 0.070 | 0.0150 | 4.65 | 3.44 × 10−6 | 0.032 | 0.01016 | 3.12 | 0.00183 | 0.19 | |
| rs10486523 | −0.073 | 0.0145 | −4.98 | 6.62 × 10−7 | −0.031 | 0.00999 | −3.08 | 0.00207 | 0.06 | |
| rs9791555 | −0.073 | 0.0145 | −4.97 | 6.79 × 10−7 | −0.031 | 0.00999 | −3.07 | 0.00214 | 0.06 | |
| rs7799346 | −0.073 | 0.0145 | −4.98 | 6.52 × 10−7 | −0.030 | 0.00999 | −3.04 | 0.00235 | 0.09 | |
| rs6976779 | 0.069 | 0.0146 | 4.71 | 2.59 × 10−6 | 0.039 | 0.01000 | 3.94 | 0.00008 | 0.97 | |
| rs2846572 | −0.067 | 0.0145 | −4.62 | 3.94 × 10−6 | −0.031 | 0.00998 | −3.10 | 0.00194 | 0.19 | |
Zimp and Zreal are the test statistics (Z-score) obtained from the imputed and original datasets, respectively. The last column is the difference between the imputed test statistics and the analytical test statistics.
The real test data is obtained from the NFBC data by removing the 500 individuals who are assumed to be missing in our experiment.
Figure 2.
Difference between the Imputed Marginal Statistics and Analytical Marginal Statistics for TG Phenotype
Imputed marginal statistics are obtained from the association between the genotype and the imputed phenotype. Analytical marginal statistics are equal to the marginal statistics computed on the true target phenotype scaled by rimp. The blue curve is the normal distribution with a mean of 0 and a variance of 1 − . This histogram indicates that the difference follows a normal distribution (mean 0 and variance 1 − ). Thus, for most null variants the NMM assumption holds.
A further investigation was performed on rs1260326, whose imputed Z-score was not close to the expected value. Table S2 shows the EMMAX14 results for rs1260326 on all of the phenotypes in the NFBC data. We observe that in the original data this SNP is significant only for the TG phenotype. Thus, the effect sizes of this SNP for multiple phenotypes are not well modeled by the overall phenotypic correlation. Therefore, our method, and any other approaches that use proxy phenotypes, will have limited performance in detecting such a locus.
Phenotype Imputation on Hybrid Mouse Diversity Panel
Our method was also applied to the Hybrid Mouse Diversity Panel (HMDP) collected in the Bennett et al. study,39 which consisted of 25 phenotypes, 894 animals, and 98 strains. We imputed body fat (BF) mass, which we considered to be the target phenotype, by using metabolic phenotypes (HDL, TG, TC, UC, FFA, and GLU) as the related phenotypes. The BF phenotype was measured by nuclear magnetic resonance (NMR). It was assumed that the BF phenotype was collected for only 200 animals, which was used as a training dataset to compute the pairwise correlations (see Figure S6). The correlation between the imputed phenotype and the true BF phenotype was rimp = 0.4. We performed experiments similar to those performed on the TG phenotype for the NFBC dataset. Table 2 indicates the significant SNPs, which passed our significant threshold of 0.05 for both imputed and real test datasets. These results are similar to the NFBC dataset. For all of the variants, the test statistic (Z-score) for the imputed phenotype is close to rimp times the test statistic (Z-score) at the actual variant (Table 2, last column).
Table 2.
Comparison between the Association Test for BF Phenotype on the Real Test Data and the Imputed Test Data in the HMDP
| rsID |
Real Test Data |
Imputed Test Data |
- 0.4 ∗ | ||||||
|---|---|---|---|---|---|---|---|---|---|
| β | se(β) | Z-Score (Zreal) | p Value | β | se(β) | Z-score (Zimp) | p Value | ||
| rs38946050 | −0.247 | 0.05887 | −4.200 | 3.04 × 10−5 | −0.093 | 0.03220 | −2.891 | 0.003 | 1.211 |
| rs37558901 | −0.163 | 0.03803 | −4.286 | 2.09 × 10−5 | −0.051 | 0.0209 | −2.448 | 0.01 | 0.733 |
| rs27178379 | −0.185 | 0.04433 | −4.176 | 3.36 × 10−5 | −0.055 | 0.02435 | −2.275 | 0.02 | 0.604 |
| rs50810977 | −0.163 | 0.03803 | −4.286 | 2.09 × 10−5 | −0.051 | 0.0209 | −2.448 | 0.01 | 0.733 |
| rs51148868 | −0.185 | 0.04433 | −4.176 | 3.36 × 10−5 | −0.055 | 0.02435 | −2.275 | 0.02 | 0.604 |
| rs32339557 | −0.163 | 0.03803 | −4.286 | 2.09 × 10−5 | −0.051 | 0.02093 | −2.448 | 0.01 | 0.733 |
| rs51646366 | −0.163 | 0.03803 | −4.286 | 2.09 × 10−5 | −0.051 | 0.02093 | −2.448 | 0.01 | 0.733 |
| rs31560659 | −0.163 | 0.03803 | −4.286 | 2.09 × 10−5 | −0.051 | 0.02093 | −2.448 | 0.01 | 0.733 |
| rs50923350 | −0.163 | 0.03803 | −4.286 | 2.09 × 10−5 | −0.051 | 0.02093 | −2.448 | 0.01 | 0.733 |
| rs37193394 | −0.205 | 0.04742 | −4.331 | 1.72 × 10−5 | −0.056 | 0.02599 | −2.161 | 0.03 | 0.428 |
| rs26890141 | −0.185 | 0.04433 | −4.1769 | 3.36 × 10−5 | −0.055 | 0.02435 | −2.275 | 0.02 | 0.604 |
| rs46913800 | −0.185 | 0.04433 | −4.1769 | 3.36 × 10−5 | −0.055 | 0.02435 | −2.275 | 0.02 | 0.604 |
| rs38214662 | −0.163 | 0.03803 | −4.2867 | 2.09 × 10−5 | −0.051 | 0.02093 | −2.448 | 0.01 | 0.733 |
| rs47384543 | −0.185 | 0.04433 | −4.1769 | 3.36 × 10−5 | −0.055 | 0.02435 | −2.275 | 0.02 | 0.604 |
| rs51585751 | −0.163 | 0.03803 | −4.2867 | 2.09 × 10−5 | −0.051 | 0.02093 | −2.448 | 0.01 | 0.733 |
| rs29268223 | −0.185 | 0.04433 | −4.1769 | 3.36 × 10−5 | −0.055 | 0.02435 | −2.275 | 0.02 | 0.604 |
Evaluating Imputation Power by Simulation
We evaluated the power of phenotype imputation through simulations. We removed the phenotype of interest from the dataset and applied phenotype imputation to predict its value and measure the corresponding association power after imputation. In order to robustly measure this power, we randomized the individuals from whom we removed the phenotype values.
Specifically, we performed the following simulation procedure. A locus that had a significant association was considered. First, we computed the number of individuals that were needed to remove their phenotypic values to obtain a statistical power of 50% for that locus. Let k indicate the number of individuals obtained from this step. The second step required random selection of k individuals and consideration of the phenotypic values for these k individuals that were missing. Our imputation model was used to impute the phenotypic values of these k individuals. An association test on the complete dataset was performed. The second step was repeated 10,000 times in order to compute the statistical power. The statistical power was computed as the number of times that the computed association statistic value was significant (with p < 10−6). A power increase greater than 50% was expected if the imputation was working; therefore, it was used as the reference for statistical power before imputation. The value of k was computed by randomly removing phenotypes of k individuals for 10,000 simulations. The value of k was checked by determining whether the number of simulations, where the association statistics is significant (with p < 10−6), equaled 5,000 (50% of total simulations that corresponded to a statistical power of 50%). The TG, BMI, and SBP phenotypes from the NFBC data were used to perform the power simulation. The power gained by imputing the missing phenotype was 8%–33% (Table 3).
Table 3.
Measuring Power of Imputation by Simulation in the NFBC Data
| Phenotype | rsID | Power after Imputation | Power before Imputation | Absolute Power Gain |
|---|---|---|---|---|
| TG | rs673548 | 83.59% | 50% | 33.59% |
| rs10096633 | 62.16% | 50% | 12.16% | |
| rs3923037 | 63.74% | 50% | 13.74% | |
| rs6728178 | 80.97% | 50% | 30.97% | |
| rs6754295 | 76.40% | 50% | 26.40% | |
| rs676210 | 82.16% | 50% | 32.16% | |
| BMI | rs987237 | 63.12% | 50% | 13.12% |
| rs11759809 | 61.33% | 50% | 11.33% | |
| SBP | rs782586 | 82.52% | 50% | 32.52% |
| rs782588 | 81.72% | 50% | 31.72% | |
| rs782602 | 81.99% | 50% | 31.99% | |
| rs2627759 | 74.05% | 50% | 24.05% | |
| rs9791555 | 58.77% | 50% | 8.77% | |
| rs7799346 | 58.63% | 50% | 8.63% |
The Material and Methods section provides an optimal weight for combining imputed and observed summary statistics in a fixed effect meta-analysis. This process is beneficial when we have access to the summary statistics. The simulation process described above was used. The k individuals were randomly selected to mask them as individuals with missing phenotypes. The summary statistics (sc) were computed for individuals whose phenotypic values were observed. The missing phenotypes were imputed and the summary statistics were computed for individuals whose phenotypic values were missing. There were two options for combining these statistics. The first option uses Equation 17 to combine the computed summary statistics in an optimal way. This option is referred to as imputation-based fixed-effect meta-analysis. The second option applies fixed-effect meta-analysis with typical fixed-effect meta-analysis weights. In this case, we use and . This option is called general fixed-effect meta-analysis. We lost power when we used the second option where the weights were not optimal (see Table 4). The first option, which is optimal, was compared to the previous simulations, where the imputed and observed phenotypic values were combined to compute the summary statistics. The data showed a small difference between them. We used the TG phenotype from the NFBC dataset for these experiments.
Table 4.
The Optimal Meta-analysis Strategy to Combine Summary Statistics for Imputed and Observed Phenotype Achieves Maximum Power
| rsID | Imputation-Based Fixed-effect Meta-analysis Power | General Fixed-Effect Meta-analysis Power |
|---|---|---|
| rs673548 | 83.56% | 82.30% |
| rs10096633 | 62.14% | 45% |
| rs3923037 | 63.65% | 60.86% |
| rs6728178 | 80.96% | 80.00% |
| rs6754295 | 75.49% | 74.31% |
| rs676210 | 82.01% | 80.85% |
Imputation-based fixed-effect meta-analysis uses the optimal weights that are shown in Equation 17. General fixed-effect meta-analysis uses the typical fixed-effect meta-analysis weights where the weight for each study is the square root of the number of samples in the study.
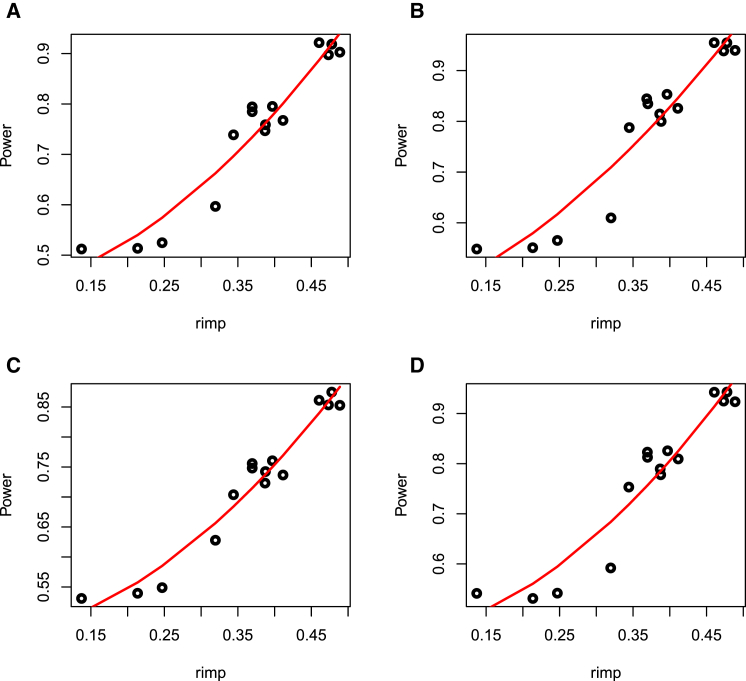
The statistical power of imputation depends on rimp, which is the correlation between the imputed and true phenotype (see Figure 3). We considered imputing the TG phenotype using HDL, LDL, CRP, and GLU phenotypes. There were 24 − 1 = 15 possible combinations for these four phenotypes to impute the TG phenotype (excluding one combination that refers to a case where none of the four phenotypes are used for imputation). The rimp and the statistical power for a given variant for each combination of phenotypes was computed. The black circle in Figure 3 indicates 1 of the 15 possible combinations for imputing TG phenotype. The x axis is the computed rimp for a given combination of phenotypes, and the y axis is the computed statistical power. The red curve indicates a second order polynomial that is fitted to the black circles. We observe that the statistical power increases as we increase the value of rimp (see Figure 3). Two factors increase rimp. The first factor is the number of phenotypes that satisfies the NMM assumption. As we use more phenotypes that satisfy the NMM assumption in our imputation method, we can increase rimp that result in increases of power. The second factor is the correlation between phenotypes that are used to impute target phenotype. As we use more correlated phenotypes, we can increase rimp that result in increases of power.
Figure 3.
An Increase of rimp Increases the Statistical Power
The x axis is the rimp and the y axis is computed power. Shown are the effect of rimp on the power of imputing the TG phenotype for rs6728178 (A), rs673548 (B), rs6754295 (C), and rs676210 (D). The TG phenotype in the NFBC data was imputed using HDL, LDL, CRP, and GLU phenotypes. The black circle indicates the rimp and the statistical power for a combination of four phenotypes to impute TG for one variant. The red curve indicates a second order polynomial that is fitted to the black circles.
Utilizing Simulation Data to Validate Our Model
In the Material and Methods section, we show that the rimp, which is the correlation between imputed and true phenotype, is equal to . One of the phenotypes was imputed by utilizing any combination of the remaining nine phenotypes. There are 29 − 1 possible combinations for these nine phenotypes to impute the desired phenotype in the NFBC dataset. The computed difference between rimp and is small (see Figure S5). The rimp was computed as a correlation between the imputed and true phenotypes. This experiment was performed for all the nine phenotypes (TG, HDL, LDL, BMI, CRP, GLU, INS, SBP, and DBP) in the NFBC dataset.
Next, the difference between the computed association statistics for imputed phenotype and the analytical association statistics were obtained from Equation 14. We simulated phenotypes for 1,000, 5,000, and 10,000 individuals and we considered three, four, five, and six phenotypes in each simulation. Multi-phenotypes were simulated utilizing the matrix-variate, as previous reported.15, 34, 35, 36 We run each of the simulations for 10,000 times and our result is the average of 10,000 runs (Table S3).
Discussion
We propose a method for resolving the problem of phenotype imputation. The primary advantage of our framework is that it increases the power of GWASs on phenotypes that are difficult to collect. Analytical power computation is provided that allows investigators to determine the benefit of the imputation for a given dataset prospectively. Another advantage of this method is that it allows the use of summary statistics when the raw genotypes are not available.
Our model assumes that the phenotypes follow a normal distribution. This assumption is widely accepted in the GWAS community.14, 15, 20 When the phenotypes are not normal, one possible solution is to transform the phenotypes to follow a normal. We applied inverse normal transformation to the data, a procedure that is heavily used in many studies.40, 41, 42 We verified that when all of the phenotypes in the NFBC data were transformed, the phenotypes as a set follow a multivariate normal distribution (see Figure S2). Another possible way to deal with non-normal phenotypes is to use the weighted combination of statistics approach. Asymptotically, the multivariate central limit theorem applies if the datasets are large enough and the statistics themselves will follow a multivariate normal distribution. Thus, using a weighted combination of Z-scores will control the type I error, but its optimal properties might not be guaranteed for non-normal phenotypes.
Our framework is closely related to the noisy measurement model (NMM) in that both the power calculation and the connection to weighted combination of statistics are based on NMM. In Material and Methods, we showed that we can assume a more complex polygenic model. NMM is equivalent to a polygenic model where we assume that the genetic correlation is the same as the environmental correlation. We also developed a weighted combination of statistics approach for situations where this is not the case; it is optimized for the polygenic model. This approach might show a better performance if we have an accurate estimate of genetic and environmental correlations. However, estimating genetic correlations using SNP data often requires thousands of individuals. On the other hand, the phenotypic correlations can be accurately measured relatively easily from a much smaller set of individuals. Therefore, we expect that our standard solution based on phenotypic correlation and NMM will be a practical solution for situations where the size of the complete dataset is small. Moreover, our analysis is based on real data, which shows that NMM is a reasonable model for most loci that we evaluated.
An implicit assumption of our approach is that we expect that we can borrow information of a target phenotype from the proxy phenotypes. We assume that there will be pleiotropy between phenotypes that are reflected in correlations. If this is not the case, such as the TG-associated locus (rs1260326), then the power to detect such a locus using other phenotypes is considerably limited. Note that this is not the limitation of only our method, but can be a limitation of any possible approaches that depend on proxy phenotypes. Nevertheless, our NFBC analysis shows that such a situation is relatively rare (one out of seven loci) compared to the situations where our method was effective.
It is worth mentioning that phenotype imputation has some similarities to phenotype prediction. In phenotype prediction, one typically predicts phenotypes based on available genetic information. One of the widely used methods for phenotype prediction is BLUP (best linear unbiased prediction).43 Phenotype prediction is an active research area, and various approaches have been proposed to solve this problem efficiently.44, 45 The main difference between phenotype prediction and phenotype imputation lies in the main goal of the approaches. The main goal of phenotype prediction is to have a method that predicts the phenotypic values as close as possible to the true value using the genetic data and possibly using other phenotypes. However, in phenotype imputation, the goal is to impute the phenotypic values using other phenotypes such that we can recover the associated signals if we have collected the imputed phenotype. Therefore, we cannot use the genetic data for phenotype imputation. If the genetic data in our imputation are used, we would not be able to perform genetic association, because the genetic data would be used twice (once in imputation and once again in the GWAS).
Phenotype imputation is analogous to genotype imputation in several ways.46, 47, 48, 49, 50 Genotype imputation involves imputing the missing genotypes. As in phenotype imputation, if we use one tagged variant in the genotype imputation to impute the missing variant, we lack sufficient power when we perform a GWAS on the imputed genotype. However, if we use a panel of reference individuals and multiple variants, we can achieve higher power. This is similar to our phenotype imputation where utilization of multiple phenotypes will achieve higher power than only one phenotype. These similarities are the reasons we use the name “phenotype imputation” for this problem.
Our method controls type I errors even in situations where there are systematic differences between the reference (first dataset) and target (second dataset) datasets. Power will be affected, but our method will not report false positives.
We acknowledge the fact that more sophisticated machine learning can be utilized, including techniques such as support vector machines (SVM),51 LASSO,38 Elastic-net,52 and supervised PCA53 to solve the phenotype imputation problem and improve the imputation power. Moreover, these methods do not make any assumption on the distribution of collected phenotypes. However, these methods are designed for general missing data problems and do not utilize the genetic data. A recent multiple imputation method54 was proposed that incorporates the genetic similarity (kinship) between individuals to perform phenotype imputation. This method performs better than generalized machine learning methods described above. However, all of these methods require access to individuals’ raw data, which is not possible in most cases. One the main advantages of our method is that we can perform imputation using available summary statistics. In addition, we provide an analytical power calculation for our method, although performing analytical power computation is not easy for other methods.
Our approach allows us to know the exact distribution of the imputed phenotype due to our parametric assumptions. We can directly use the mean value of this distribution as the imputed value. Furthermore, we utilize the variance of the missing phenotype in our analysis of the statistical power. If we use a more sophisticated machine learning method for the imputation, as mentioned above, then we can use multiple imputation techniques8, 55 to obtain the confidence intervals for the imputation.
Acknowledgments
F.H., E.Y.K., M.B., and E.E. are supported by National Science Foundation grants 0513612, 0731455, 0729049, 0916676, 1065276, 1302448, 1320589, and 1331176 and NIH grants K25-HL080079, U01-DA024417, P01-HL30568, P01-HL28481, R01-GM083198, R01-ES021801, R01-MH101782, and R01-ES022282. B.H. is supported by a grant (2016-708) from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea. S.M. and C.V. are supported by NIH grant R01-GM083198-01A1. E.E. is supported in part by the NIH BD2K award U54EB020403. We acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691).
Published: June 9, 2016
Footnotes
Supplemental Data include six figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.04.013.
Contributor Information
Buhm Han, Email: buhm.han@amc.seoul.kr.
Eleazar Eskin, Email: eeskin@cs.ucla.edu.
Appendix A. Phenotype Imputation for Cases Where Different Subsets of Phenotypes Are Missing
The Material and Methods section explains the method we use when the target phenotype is the only missing phenotype. Unfortunately, if the number of related phenotypes is large, then there are many individuals where one or more phenotypic values are missing. Let indicate a vector of size where each element of the vector has value of 0 or 1. Vector indicates which phenotypes are missing, excluding the target phenotype. The ith element of is one for the cases where the ith phenotype is missing. We refer to as one configuration of missing phenotypes in the second dataset. If we have phenotypes, then we have at most such configurations. Let indicate the set of all possible configurations, . Let indicate a new partition of the second dataset to a set of individuals which miss exactly the phenotypes denoted by configuration . We can easily extend our method to impute the target phenotype for those individuals, who belong to configuration by removing the phenotypes that are missing for these individuals. Thus, and are computed in a manner similar to the methods as mentioned in previous section, while we exclude the phenotypes that are missing for these individuals.
We apply Equations 7 and 14 to compute the imputed target phenotype and the imputed marginal statistics, respectively, for only those individuals utilizing the observed phenotypes. It is possible to have up to different configurations and up to different marginal statistics for each configuration. Let indicate the imputed marginal statistics for the configuration . Then, we compute the total marginal statistics by applying the fixed-effect meta-analysis as shown in previous section. Thus, we have:
| (Equation A1) |
where wi is the optimal weight for the marginal statistics for the configuration . This is proportional to the correlation between the imputed target phenotypic values and the true uncollected phenotypic values for all the individuals in configuration .
Web Resources
PhenIMP, http://genetics.cs.ucla.edu/phenIMP
Supplemental Data
References
- 1.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., MAGIC investigators. GIANT Consortium Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schunkert H., König I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F.R., Barbalic M., Gieger C., Cardiogenics. CARDIoGRAM Consortium Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer-Lindenberg A., Weinberger D.R. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat. Rev. Neurosci. 2006;7:818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- 4.Gordon T., Castelli W.P., Hjortland M.C., Kannel W.B., Dawber T.R. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am. J. Med. 1977;62:707–714. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- 5.Little R.J.A., Rubin D.B. Wiley-Blackwell; 2002. Statistical Analysis with Missing Data. [Google Scholar]
- 6.Allison P.D. Missing data: Quantitative applications in the social sciences. Br. J. Math. Stat. Psychol. 2002;55:193–196. [Google Scholar]
- 7.Ghosh S. Statistical analysis with missing data. Technometrics. 1988;30 455–455. [Google Scholar]
- 8.Rubin D.B. Inference and missing data. Biometrika. 1976;63:581–592. [Google Scholar]
- 9.Sterne J.A., White I.R., Carlin J.B., Spratt M., Royston P., Kenward M.G., Wood A.M., Carpenter J.R. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ. 2009;338:b2393. doi: 10.1136/bmj.b2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bobb J.F., Scharfstein D.O., Daniels M.J., Collins F.S., Kelada S. Multiple imputation of missing phenotype data for QTL mapping. Stat. Appl. Genet. Mol. Biol. 2011;10:29. doi: 10.2202/1544-6115.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaitsiakhovich T., Drichel D., Angisch M., Becker T., Herold C., Lacour A. Analysis of the progression of systolic blood pressure using imputation of missing phenotype values. BMC Proc. 2014;8(Suppl 1):S83. doi: 10.1186/1753-6561-8-S1-S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balise R.R., Chen Y., Dite G., Felberg A., Sun L., Ziogas A., Whittemore A.S. Imputation of missing ages in pedigree data. Hum. Hered. 2007;63:168–174. doi: 10.1159/000099829. [DOI] [PubMed] [Google Scholar]
- 13.Sabatti C., Service S.K., Hartikainen A.-L., Pouta A., Ripatti S., Brodsky J., Jones C.G., Zaitlen N.A., Varilo T., Kaakinen M. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.-Y.Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X., Carbonetto P., Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013;9:e1003264. doi: 10.1371/journal.pgen.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lippert C., Listgarten J., Liu Y., Kadie C.M., Davidson R.I., Heckerman D. FaST linear mixed models for genome-wide association studies. Nat. Methods. 2011;8:833–835. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- 17.Listgarten J., Lippert C., Kadie C.M., Davidson R.I., Eskin E., Heckerman D. Improved linear mixed models for genome-wide association studies. Nat. Methods. 2012;9:525–526. doi: 10.1038/nmeth.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCulloch C., Searle S., Neuhaus J. Wiley; 2011. Generalized, Linear, and Mixed Models. Wiley Series in Probability and Statistics. [Google Scholar]
- 19.Han B., Kang H.M., Eskin E. Rapid and accurate multiple testing correction and power estimation for millions of correlated markers. PLoS Genet. 2009;5:e1000456. doi: 10.1371/journal.pgen.1000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hormozdiari F., Kostem E., Kang E.Y., Pasaniuc B., Eskin E. Identifying causal variants at loci with multiple signals of association. Genetics. 2014;198:497–508. doi: 10.1534/genetics.114.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hormozdiari F., Kichaev G., Yang W.-Y., Pasaniuc B., Eskin E. Identification of causal genes for complex traits. Bioinformatics. 2015;31:i206–i213. doi: 10.1093/bioinformatics/btv240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han B., Hackel B.M., Eskin E. Postassociation cleaning using linkage disequilibrium information. Genet. Epidemiol. 2011;35:1–10. doi: 10.1002/gepi.20544. [DOI] [PubMed] [Google Scholar]
- 23.Spencer C.C.A., Su Z., Donnelly P., Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5:e1000477. doi: 10.1371/journal.pgen.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohashi J., Tokunaga K. The power of genome-wide association studies of complex disease genes: statistical limitations of indirect approaches using SNP markers. J. Hum. Genet. 2001;46:478–482. doi: 10.1007/s100380170048. [DOI] [PubMed] [Google Scholar]
- 25.Stranger B.E., Stahl E.A., Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunning A.M., Durocher F., Healey C.S., Teare M.D., McBride S.E., Carlomagno F., Xu C.F., Dawson E., Rhodes S., Ueda S. The extent of linkage disequilibrium in four populations with distinct demographic histories. Am. J. Hum. Genet. 2000;67:1544–1554. doi: 10.1086/316906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- 28.Pritchard J.K., Przeworski M. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abecasis G.R., Noguchi E., Heinzmann A., Traherne J.A., Bhattacharyya S., Leaves N.I., Anderson G.G., Zhang Y., Lench N.J., Carey A. Extent and distribution of linkage disequilibrium in three genomic regions. Am. J. Hum. Genet. 2001;68:191–197. doi: 10.1086/316944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eskin E. Increasing power in association studies by using linkage disequilibrium structure and molecular function as prior information. Genome Res. 2008;18:653–660. doi: 10.1101/gr.072785.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Bakker P.I.W., Ferreira M.A.R., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17(R2):R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willer C.J., Speliotes E.K., Loos R.J.F., Li S., Lindgren C.M., Heid I.M., Berndt S.I., Elliott A.L., Jackson A.U., Lamina C., Wellcome Trust Case Control Consortium. Genetic Investigation of ANthropometric Traits Consortium Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaitlen N., Eskin E. Imputation aware meta-analysis of genome-wide association studies. Genet. Epidemiol. 2010;34:537–542. doi: 10.1002/gepi.20507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J.J., Cho M.H., Lange C., Lutz S., Silverman E.K., Laird N.M. Integrating multiple correlated phenotypes for genetic association analysis by maximizing heritability. Hum. Hered. 2015;79:93–104. doi: 10.1159/000381641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X., Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods. 2014;11:407–409. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furlotte N.A., Eskin E. Efficient multiple-trait association and estimation of genetic correlation using the matrix-variate linear mixed model. Genetics. 2015;200:59–68. doi: 10.1534/genetics.114.171447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park T., Casella G. The bayesian lasso. J. Am. Stat. Assoc. 2008;103:681–686. [Google Scholar]
- 38.Tibshirani R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B. 1996;58:267–288. [Google Scholar]
- 39.Bennett B.J., Farber C.R., Orozco L., Kang H.M., Ghazalpour A., Siemers N., Neubauer M., Neuhaus I., Yordanova R., Guan B. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ardlie K.G., Deluca D.S., Segrè A.V., Sullivan T.J., Young T.R., Gelfand E.T., Trowbridge C.A., Maller J.B., Tukiainen T., Lek M., GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okada Y., Kubo M., Ohmiya H., Takahashi A., Kumasaka N., Hosono N., Maeda S., Wen W., Dorajoo R., Go M.J., GIANT consortium Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat. Genet. 2012;44:302–306. doi: 10.1038/ng.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Speliotes E.K., Yerges-Armstrong L.M., Wu J., Hernaez R., Kim L.J., Palmer C.D., Gudnason V., Eiriksdottir G., Garcia M.E., Launer L.J., NASH CRN. GIANT Consortium. MAGIC Investigators. GOLD Consortium Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henderson C.R. Sire evaluation and genetic trends. J. Anim. Sci. 1973;1973:10–41. [Google Scholar]
- 44.Meuwissen T., Goddard M. Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics. 2010;185:623–631. doi: 10.1534/genetics.110.116590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ober U., Ayroles J.F., Stone E.A., Richards S., Zhu D., Gibbs R.A., Stricker C., Gianola D., Schlather M., Mackay T.F., Simianer H. Using whole-genome sequence data to predict quantitative trait phenotypes in Drosophila melanogaster. PLoS Genet. 2012;8:e1002685. doi: 10.1371/journal.pgen.1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Browning S.R. Missing data imputation and haplotype phase inference for genome-wide association studies. Hum. Genet. 2008;124:439–450. doi: 10.1007/s00439-008-0568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marchini J., Howie B. Comparing algorithms for genotype imputation. Am. J. Hum. Genet. 2008;83:535–539. doi: 10.1016/j.ajhg.2008.09.007. author reply 539–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cortes C., Vapnik V. Support-vector networks. Mach. Learn. 1995;20:273–297. [Google Scholar]
- 52.Zou H., Hastie T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Series B Stat. Methodol. 2005;67:301–320. [Google Scholar]
- 53.Bair E., Hastie T., Paul D., Tibshirani R. Prediction by supervised principal components. J. Am. Stat. Assoc. 2006;101:119–137. [Google Scholar]
- 54.Dahl A., Iotchkova V., Baud A., Johansson Å., Gyllensten U., Soranzo N., Mott R., Kranis A., Marchini J. A multiple-phenotype imputation method for genetic studies. Nat. Genet. 2016;48:466–472. doi: 10.1038/ng.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubin D.B. Volume 81. John Wiley & Sons; 2004. (Multiple Imputation for Nonresponse in Surveys). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.