Abstract
Tooth agenesis is one of the most common developmental anomalies in humans. Oligodontia, a severe form of tooth agenesis, is genetically and phenotypically a heterogeneous condition. Although significant efforts have been made, the genetic etiology of dental agenesis remains largely unknown. In the present study, we performed whole-exome sequencing to identify the causative mutations in Chinese families in whom oligodontia segregates with dominant inheritance. We detected a heterozygous missense mutation (c.632G>A [p.Arg211Gln]) in WNT10B in all affected family members. By Sanger sequencing a cohort of 145 unrelated individuals with non-syndromic oligodontia, we identified three additional mutations (c.569C>G [p.Pro190Arg], c.786G>A [p.Trp262∗], and c.851T>G [p.Phe284Cys]). Interestingly, analysis of genotype-phenotype correlations revealed that mutations in WNT10B affect the development of permanent dentition, particularly the lateral incisors. Furthermore, a functional assay demonstrated that each of these mutants could not normally enhance the canonical Wnt signaling in HEPG2 epithelial cells, in which activity of the TOPFlash luciferase reporter was measured. Notably, these mutant WNT10B ligands could not efficiently induce endothelial differentiation of dental pulp stem cells. Our findings provide the identification of autosomal-dominant WNT10B mutations in individuals with oligodontia, which increases the spectrum of congenital tooth agenesis and suggests attenuated Wnt signaling in endothelial differentiation of dental pulp stem cells.
Main Text
Our recent survey in China showed that the prevalence of tooth agenesis was approximately 5.89% among 6,015 adolescent individuals between 10 and 26 years old.1 However, the occurrence of oligodontia is relatively rare and has been estimated to be approximately 0.25% in the Chinese population2 and 0.14%–0.30% in various populations of Western countries.3
Oligodontia (ICD-10 diagnosis code K00.0) is the agenesis of six or more permanent teeth (excluding third molars), whereas absence of fewer than six teeth is referred to as hypodontia. Mutations in at least eight genes, including WNT10A (wingless-type MMTV integration site family, member 10A [MIM: 606268]), PAX9 (paired box gene 9 [MIM: 167416]), EDA (ectodysplasin A [MIM: 300451]), MSX1 (muscle segment homeobox 1 [MIM: 142983]), AXIN2 (axis inhibition protein 2 [MIM: 604025]), EDARADD (edar-associated death domain [MIM: 606603]), IKBKG (inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma [MIM: 300248]), and KRT17 (keratin 17, type I [MIM: 148069]) in order of decreasing frequency of mutant alleles, have been identified in individuals with non-syndromic oligodontia.4 Recently, multiple mutations in LRP6 (low density lipoprotein receptor-related protein 6 [MIM: 603507]), encoding a co-receptor in the Wnt pathway, were also found to cause autosomal-dominant oligodontia.5
By Sanger sequencing of these oligodontia-associated genes, we have previously identified mutations in most of these genes in Chinese individuals with non-syndromic oligodontia.6, 7, 8, 9, 10 However, no mutations were detected in nearly half of the examined affected individuals, suggesting the existence of unidentified genetic etiologies. Presumably, additional crucial components in several defined canonical pathways (such as BMP, FGF, NF-κB, P53, PAX, and Wnt, which involve tooth development) could be the candidates.11 In the present study, we applied whole-exome sequencing (WES) and Sanger sequencing to identify potential genetic defects in a large cohort of individuals with oligodontia. We also explored the potential effects of the identified mutations on cellular differentiation of dental pulp stem cells (DPSCs).
Our previous study detected WNT10A variants in 15.8% (75/474) of unrelated individuals with one to three missing teeth and 51.6% (16/31) of individuals with four or more missing teeth.8 Four families who are affected by inherited oligodontia and in whom the WNT10A genetic test had failed to detect a mutation were examined by WES as previously described.12 Informed consent was obtained from each of the individuals or from their parents in the case of minors. This study and associated research protocols were approved by the ethics committees of Wenzhou Medical University and Peking University.
In family ZZWX-1 (affected by inherited non-syndromic oligodontia), WES and Sanger sequencing identified a previously undescribed frameshift deletion mutation (c.289_296del [p.Ile97Leufs∗217]) in PAX9 (GenBank: NM_006194.3) in the proband (II-1; Figure S1A) and the affected mother (I-2; Figure S1A). This mutation results in the agenesis of almost all molars and several incisors (selective tooth agenesis 3 [MIM: 604625]; Figures S1A–S1D). MutationTaster analysis predicted that it is a disease-causing mutation because more than 70% of the encoded amino acid sequence (97–341) is replaced by a fragment of 217 amino acids with unknown functions. Two frameshift deletions (c.59delC and c.230_242del13), compared to 27 missense and nonsense mutations in PAX9, were previously identified to cause tooth agenesis (HGMD).13 These mutations might disturb dental bud morphogenesis and thus result in tooth agenesis.14
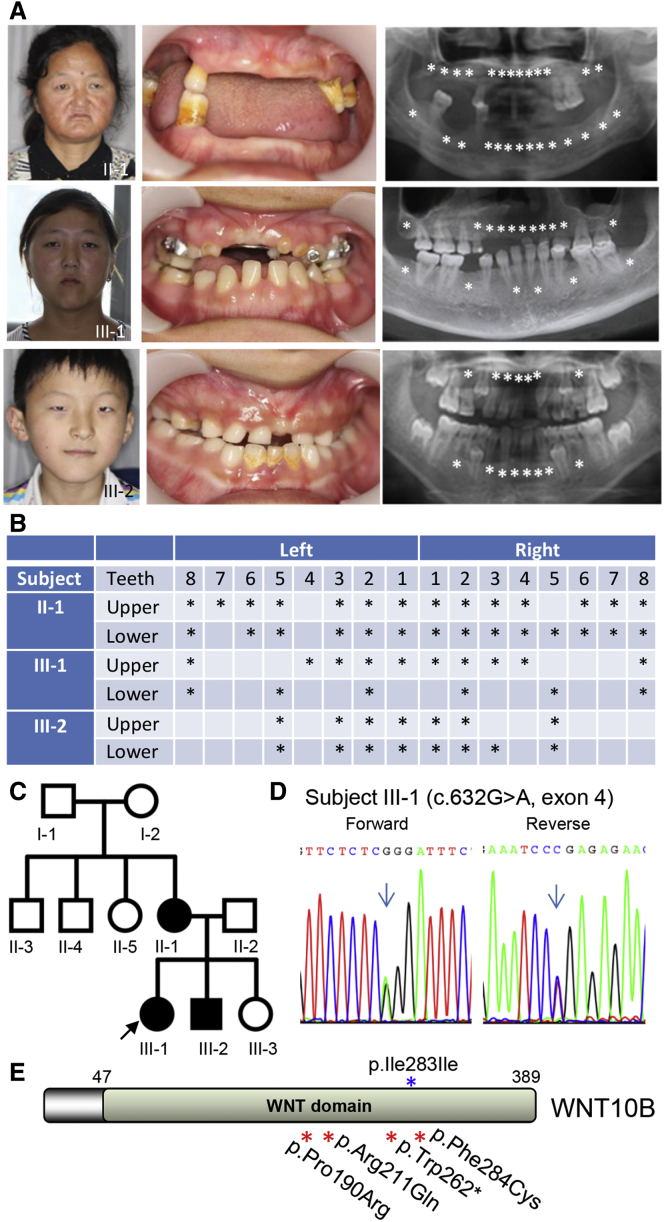
Because no mutations in known oligodontia-associated genes were found in the remaining families, we focused on the identification of mutations in other undetermined gene(s) by performing extensive bioinformatics analysis and analyzing pathways associated with tooth development. In family ZZYWL-2, three affected individuals were found to have autosomal-dominant congenital oligodontia (Figures 1A and 1B) accompanied by mild ectodermal dysplasia involving hair and eccrine sweat glands (Table S1). No obvious developmental abnormalities were seen in the ears, hands, or feet of the affected individuals (Figure S2).
Figure 1.
Autosomal-Dominant Oligodontia and Identification of the WNT10B Mutation
(A) Front views and panoramic radiographs of dentitions of three affected individuals (II-1, III-1, and III-2 in C). Missing teeth are denoted by asterisks.
(B) Schematic analysis shows the positions of missing teeth in each of the affected individuals. Missing teeth are denoted by asterisks.
(C) The ZZYWL-2 pedigree. Filled circles and squares represent affected female and male family members, respectively. Subjects I-1, I-2, II-3, II-4, and II-5 were not available for clinical evaluation or DNA analysis. WES and Sanger sequencing were performed for five family members (II-1, II-2, III-1, III-2, and III-3).
(D) Representative Sanger chromatograms show the heterozygous missense mutation c.632G>A, resulting in the p.Arg211Gln substitution.
(E) All four mutations and the rare variant c.849C>A (p.Ile283Ile) detected in the present study affect the middle region (amino acids 190–284) of WNT10B.
WES analyses of five members of the nuclear family, including three affected and two unaffected individuals (Figure 1C), generated an average of 56.34 Mb of aligned base reads per sample, of which 97.45% reads reached ≥10× coverage. A total of 38,125 single-nucleotide variants (SNVs; missense, nonsense, and splice-site mutations) and 1,378 indels (short coding insertions or deletions) were identified from the three affected individuals.
The pathogenic cause of the disorder was assumed to be the same heterozygous mutation in a single gene. No deleterious variant in known oligodontia-associated genes was found in this family. All known variants (present in normal family members, the Exome Aggregation Consortium [ExAC] Browser, or an in-house Han Chinese exome database containing 2,200 individuals at a minor allele frequency [MAF] > 0.0005) and non-deleterious SNVs determined by several commonly used algorithms (SIFT, PolyPhen-2, MutationTaster, and GERP++) were removed. Finally, a mutation predicted to be deleterious in WNT10B (Wnt family member 10B [MIM: 601906]), encoding a ligand for activation of canonical Wnt signaling, was prioritized for further analysis.
Using Sanger sequencing with specific primers (Table S2), we verified that the heterozygous mutation (c.632G>A) in WNT10B (GenBank: NM_003394.3) segregates with the affected individuals (chromatogram of proband III-1 is shown in Figure 1D), but not the unaffected family members. This missense mutation changed wild-type codon 211 from CGG to CAG, replacing arginine with glutamine (p.Arg211Gln) in the Wnt domain (Figure 1E) and thereby turning the positive charge of the side chain to neutral. One allele of the variant was present in 60,669 individuals in the ExAC Browser and also in the in-house exome database. It is worth noting that these exome databases are useful sources for variant classifications because they contain both normal and pathogenic or likely pathogenic variants in disease-associated genes.15
Multiple-sequence alignment of WNT10B showed that the residue Arg211 is evolutionarily conserved in all of the compared species and is also conserved in almost all members of the Wnt ligand family (Figures S3A and S3B). On the basis of 3D structural analysis of the WNT10B paralog in Xenopus,16 Arg211 was predicted to locate to helix D, which appears to interact with the cysteine-rich domain of Frizzled-8 (Figure S3C) and thus involve the binding affinity of the ligand with the domain. In addition, the c.632G>A (p.Arg211Gln) mutation was predicted to be disease causing by several major algorithms, including MutationTaster, PolyPhen-2 and GERP++.
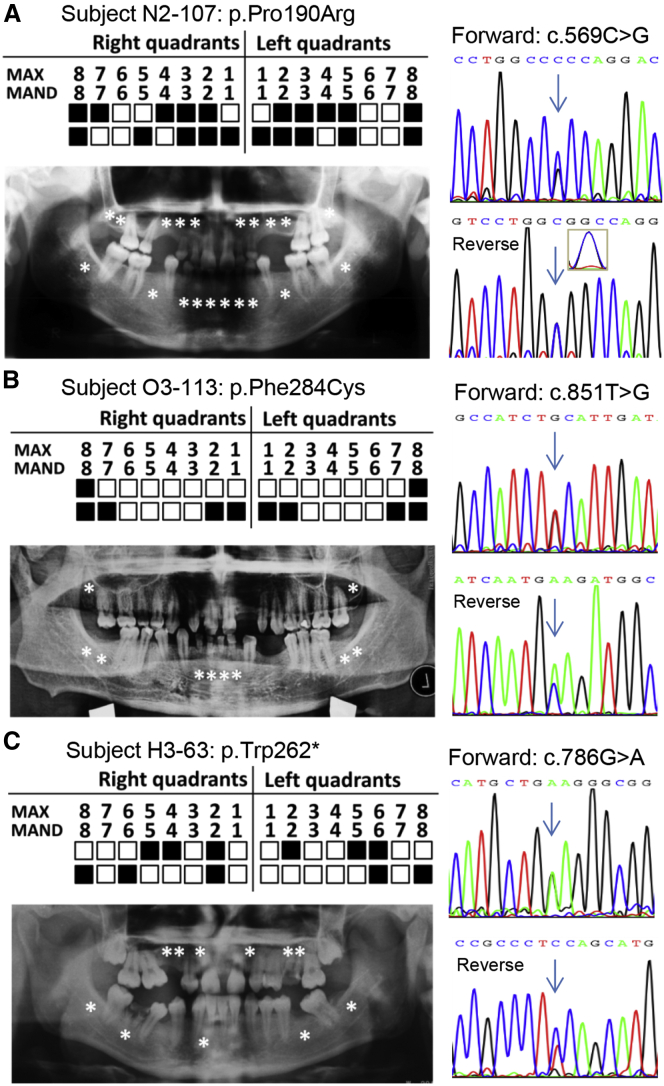
To evaluate the prevalence of the WNT10B mutations in unrelated individuals with oligodontia, we screened the whole coding region and the intron-exon boundaries of the gene (primer sequences are provided in Table S2) in a large cohort of 145 affected individuals who had not been screened for other candidate genes. As a result, we identified three additional mutations, including one nonsense and two missense mutations (Figure 1E and Figure 2).
Figure 2.
Genetic Screen of WNT10B Mutations in Unrelated Individuals with Oligodontia
(A) Subject N2-107. A panoramic radiograph (left) and the corresponding mutation in Sanger chromatograms (right) are shown. The positions of each of the missing teeth are indicated by asterisks in the radiograph or filled boxes in schematic maxillary (MAX) and lower mandible (MAND) locations. Also, double peaks of the heterozygous variant c.569C>G in a reverse Sanger chromatogram can be recognized in the amplified inset.
(B) A panoramic radiograph (left) and the mutation in Sanger chromatograms (right) of subject O3-113 are shown as in (A).
(C) A panoramic radiograph and the mutation in Sanger chromatograms of subject H3-63 are shown as in (A).
The c.569C>G (p.Pro190Arg) mutation, identified in subject N2-107 (Figure 2A), was predicted to be disease causing by MutationTaster because the residue Pro190 is evolutionarily conserved (Figure S4A). This variant was also seen in the ExAC Browser (4/121,292 alleles; MAF = 3.298e−5), but not in the in-house exome database. Another missense mutation, c.851T>G (p.Phe284Cys), identified in subject O3-113 (Figure 2B), was also conserved according to multiple-sequence alignment (Figure S4B) and predicted to be disease causing by several algorithms. Four alleles of c.851T>G were shown in the ExAC Browser (4/119,494 alleles; MAF = 3.347e−5), and one was in the in-house exome database (1/4,400 alleles; MAF = 0.00022).
Notably, a nonsense mutation (c.786G>A [p.Trp262∗]; Figure 2C) was identified in subject H3-63. This mutation was predicted to produce a truncated protein (missing 128 amino acids at the C terminus) and thereby disrupt the protein’s function. The residue Trp262 was found to be conserved in different species (Figure S4C). One allele of this variant was found in the ExAC Browser (1/118,298 alleles; MAF = 0.00011647), and none was present in the in-house exome database.
In addition, we detected a SNP (c.849C>A [p.Ile283Ile]) in four unrelated affected persons among 145 individuals (MAF = 0.0138; Figure S5) but only four such variants in 2,200 individuals from the in-house exome database (MAF = 0.0009), suggesting that this recurrent variant is associated with the oligodontia phenotype (p = 0.0003, exact binomial test). Also, this variant was predicted to be disease causing by MutationTaster (probability score = 1), e.g., phyloP and PhastCons scores were rated highly positive, reflecting an evolutionarily conserved nucleotide and flanking columns in genome sequences of 46 different species. Until recently, at least 50 human diseases have been associated with synonymous mutations affecting mRNA splicing, stability, and other properties.17 As an example, two synonymous mutations in catechol-O-methyltransferase (COMT [MIM: 116790]) were found to affect COMT mRNA stability and thus protein expression and enzymatic activity.18 Therefore, further studies are needed to investigate whether the synonymous SNP c.849C>A is able to cause changes in expression, conformation, or function of the encoded protein.
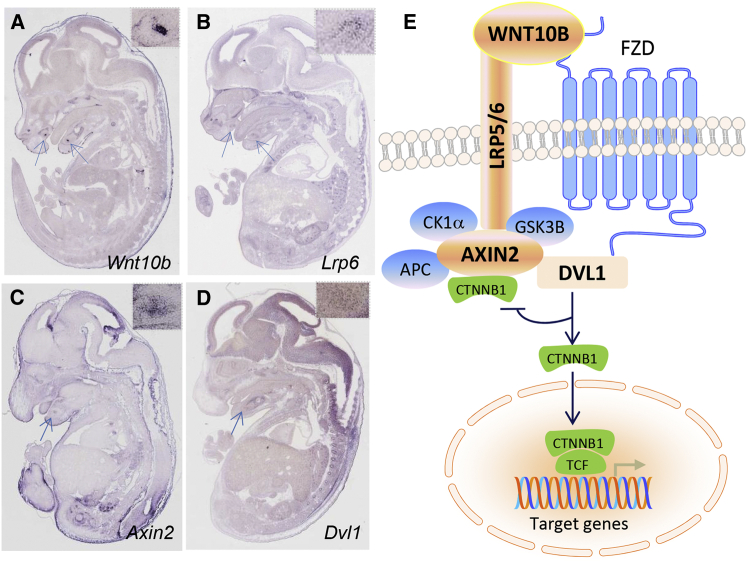
Murine Wnt10b expression was first detected at embryonic day 11.5 (E11.5), a stage showing a slight thickening of presumptive incisor epithelium,19, 20 which is the key feature of early tooth development. In later stages (i.e., the bud stage at E13.5, the cap stage at E14.5, and the early bell stage at E15.5), Wnt10b expression was gradually upregulated and extended distally from the midline in the incisor domain toward the molar primordia.19 To examine the temporal and spatial expression pattern of Wnt10b in whole-mount mouse embryos, we analyzed images of in situ hybridization from E11.5 to E15.5 (Allen Brain Atlas and Eurexpress). Compared to other embryonic tissues, the developing tooth field showed specific expression of Wnt10b, particularly high in the incisors at the E14.5 cap stage (Figure 3A). Because LRP5 and LRP6 are the receptors of WNT10B, the expression of Lrp6 partially overlapped that of Wnt10b (Figure 3B). Expression of two additional crucial components (Figures 3C and 3D), Axin2 and dishevelled 1 (Dvl1 [MIM: 601365]), which involve tooth agenesis in the canonical Wnt pathway (Figure 3E), was also detectable at the cap stage.
Figure 3.
Expression of Wnt10b in Embryonic Mice and the Wnt Pathway
(A) Wnt10b expression was detected in the developing tooth region by in situ hybridization, as indicated by arrows in the sagittal section of an E14.5 embryo (Eurexpress). The region of interest is shown in the inset.
(B) Lrp6 expression is shown as in (A).
(C) Analysis of Axin2 expression is shown as in (A).
(D) Expression of Dvl1 is shown as in (A). All expression data in Eurexpress are publically available; users can retrieve information tailored to their own needs.
(E) The binding of WNT10B ligand to a dual-receptor complex comprising the Wnt co-receptors LRP5 and LRP6 and one of the seven transmembrane receptors of the FZD family (e.g., Frizzled-8 [encoded by FZD8 (MIM: 606146)]) initiates Wnt-β-catenin signaling. AXIN2 moves to the LRP5-LRP6 tail at the membrane through its interaction with dishevelled (DVL1), which is recruited by FZD. Glycogen synthase kinase 3β (encoded by GSK3B [MIM: 605004]) is also included in this complex, which prevents phosphorylation of β-catenin (encoded by CTNNB1 [MIM: 116806]) and its proteosomal degradation. β-catenin is therefore accumulated in the cytoplasm and translocated into the nucleus, where it associates with members of the TCF and LEF transcription factor families to control transcription of target genes. In addition to WNT10A variants, variants in several Wnt-pathway components, including WNT10B, LRP6, AXIN2, and DVL1 (nodes in orange), have been found to cause tooth agenesis.
To investigate the effects of the WNT10B mutations identified here on Wnt signaling, we established four mutated WNT10B constructs by using the GeneArt SiteDirected Mutagenesis System (Thermo Fisher Scientific), specific primers (Table S2), and the template of wild-type WNT10B cDNA.21 To obtain the conditioned medium containing each of the WNT10B proteins from stable cells, we transfected COS-7 kidney fibroblast cells (CRL-1651, ATCC) with each of the plasmids by using Lipofectamine 2000 (Invitrogen) and selected stable transfectants in medium containing G418 (500 μg/ml, Invitrogen) for 4 weeks. Translational expression of these constructs was further confirmed by western blot (Figure S6). Wild-type WNT10B and each of the mutated WNT10B proteins were collected from the supernatant of the stable cells as described previously.22
Effects of WNT10B on canonical Wnt signaling can be measured by the TCF reporter system (TCF is encoded by lymphoid enhancer binding factor 1 [LEF1 (MIM: 153245)]; Figure 3E).23 Human hepatocellular carcinoma HEPG2 epithelial cells (HB-8065, ATCC), which have constitutive canonical Wnt signaling,24 were transfected with 2 μg of the plasmid pTOPFLASH luciferase reporter containing four TCF-responsive elements or with the pFOPFLASH luciferase reporter containing the dominant-negative TCF-binding sequence. The β-galactosidase (β-gal) expression plasmid (pCMV-β-gal) was co-transfected as an internal control. Four hours later, culture medium was replaced with the COS-7-supernatant-conditioned medium containing wild-type or mutant WNT10B. After 48 hr of incubation, the cells were lysed with the Dual-Luciferase Reporter Assay System (Promega) for measurement of luciferase activity and were normalized by β-gal levels.
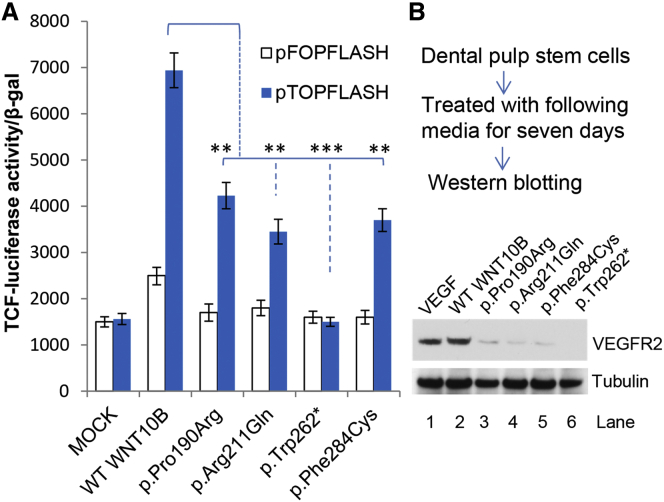
Further statistical analysis showed that the luciferase activity in each of the cells treated with mutant WNT10B supernatant was significantly lower than that in cells treated with wild-type WNT10B (Figure 4A). In particular, cells treated with p.Trp262∗ mutant supernatant did not show any luciferase activity, suggesting that the truncated protein failed to activate Wnt signaling.
Figure 4.
Functional Assessment of Mutant WNT10B in Wnt Signaling
(A) Effects of mutant WNT10B on luciferase activity. The Wnt signaling in HEPG2 cells was assessed with TCF reporter plasmids. HEPG2 cells were transiently transfected with either pTOPFLASH or pFOPFLASH and then cultured in no supernatant (Mock), wild-type WNT10B supernatant, or each of the four indicated mutant WNT10B supernatants. Error bars depict SDs; ∗∗p < 0.01, ∗∗∗p < 0.001. The results are from three independent experiments performed in triplicate.
(B) Impact of WNT10B on endothelial differentiation of DPSCs. DPSCs were isolated and cultured as described in our previous study.25 Cells were treated with medium conditioned with wild-type or mutant WNT10B (containing 50 ng/ml of each of the recombinant WNT10B proteins) for the purpose of inducing endothelial cell differentiation. Recombinant human VEGF-C (50 ng/ml, Sigma) served as a positive control. After 7 days, cells were lysed and prepared26 for western blots (ECL system, GE Healthcare Life Sciences) with anti-VEGFR2 polyclonal antibody (5 μg/mL, Invitrogen). The amount of tubulin, used as a loading control, was detected by monoclonal anti-α-tubulin antibody (1:500, Sigma).
In humans, WNT10B was significantly expressed in pulpal tissue (GEO Profile: 21100839) and DPSCs (GEO Profile: 111538043). Immunostaining and qRT-PCR showed that WNT10B was one of the key molecules involved in initiating epithelium invagination and epithelial-mesenchymal interaction when human DPSCs and oral epithelial cells were co-cultivated.27
Intriguingly, Wnt-β-catenin signaling was shown to inhibit rather than activate odontoblastic differentiation of DPSCs.28 In contrast, the WNT1-β-catenin pathway was recently found to determine the vasculogenic fate of post-natal DPSCs.26 This is consistent with the observation that blood capillaries and vascularization are detected by immunostaining and electron microscopy in the mouse embryonic molar mesenchyme.29
Considering the critical role of vascular endothelial cells, as well as odontoblasts, to the success of dental pulp tissue engineering, we were prompted to examine whether the mutated WNT10B affected differentiation of DPSCs.25 The cells were treated with conditioned media containing either wild-type or mutant WNT10B for 7 days and then harvested for western blot analysis. As a result, VEGF receptor 2 (encoded by KDR [MIM: 191306]), an endothelial differentiation marker of vasculogenesis, was activated by wild-type WNT10B, but not p.Trp262∗ WNT10B. Three media conditioned with missense mutant WNT10B could also activate VEGFR2, but to a lesser extent than media conditioned with wild-type WNT10B (Figure 4B). The impaired endothelial differentiation of the cultured DPSCs probably resulted from the p.Trp262∗ null allele and three hypomorphic alleles. However, whether these alleles have dominant-negative effects on endothelial differentiation cannot be entirely excluded.
Sequence analysis showed that WNT10B shares 60% identity and 72% similarity with WNT10A and also shares a similar localization pattern in the dental epithelium at the bud and cap stages, when tooth morphogenesis is first apparent.19 However, WNT10A expression gradually shifted to the underlying mesenchyme in later stages and was continuous in the process of odontoblast differentiation.30 We thus analyzed whether mutations in these two homolog Wnt ligands cause oligodontia at the same or at different positions. Genotype-phenotype analysis of individuals with WNT10A mutations by our previous study and others revealed that premolars were the most commonly missing teeth (62.5%–68.8%), whereas incisors were missing only 18.8%–31.3% of the time.8, 31, 32 In contrast, the most frequently missing permanent teeth in the affected individuals with WNT10B variants were the lateral incisors (83.3%; Table 1), whereas premolars were missing only 51.4% of the time, a pattern clearly different from the oligodontia patterns resulting from WNT10A mutations. Given that the most commonly affected teeth in individuals with hypodontia were mandibular second premolars, followed by maxillary lateral incisors, as reported in a recent meta-analysis,33 it is likely that there is an overlapping phenotype in some of the individuals with WNT10A and WNT10B mutations.
Table 1.
Analysis of Missing-Tooth Patterns in Individuals with WNT10B Variants
| Quadrant | Central Incisor (%) | Lateral Incisor (%) | Canine (%) | First Premolar (%) | Second Premolar (%) | First Molar (%) | Second Molar (%) |
|---|---|---|---|---|---|---|---|
| Maxillary | 8/18 (44.4) | 15/18 (83.3)a | 12/18 (66.7) | 9/18 (50.0) | 10/18 (55.6) | 4/18 (22.2) | 6/18 (33.3) |
| Mandibular | 12/18 (66.7) | 15/18 (83.3)a | 8/18 (44.4) | 4/18 (22.2) | 14/18 (77.8) | 4/18 (22.2) | 5/18 (27.8) |
The numerator is the number of missing teeth in the nine individuals, and the denominator is the total number of teeth that these nine individuals should have at each position. The third molar (wisdom tooth) is not included. The tooth number on the left side is combined with that on the right side because no significant difference, in terms of the number of missing teeth, was found at any of the corresponding positions.
The most frequently missing tooth positions.
The selective pattern in WNT10B mutants is also different from that of mutations in other oligodontia-associated genes, such as MSX1 (missing second premolar) and PAX9 (agenesis of molars), as described previously7, 10, 11, 34 and currently in the individuals with the p.Ile97Leufs∗217 variant (Figure S1). Regardless, protein-protein-interaction and genetic studies have shown crosstalk between PAX9 and the Wnt pathway (e.g., AXIN2) or MSX1 in dental tissues,11 which are essential for the establishment of the odontogenic potential of the mesenchyme. However, the underlying mechanism of different genotype-phenotype correlations observed here remains elusive.
Other significant homologs of human WNT10B, such as WNT4 (39% identity and 54% similarity) and WNT3A (38% identity and 52% similarity), were also found to be specifically expressed in the tooth developing region (Eurexpress). In particular, Wnt4 was found to stimulate mesenchymal expression of Msx1 during early tooth organogenesis.35 These observations could prompt further genetic screening of additional Wnt ligands in individuals with tooth agenesis.
It is worth noting that several homozygous mutations in WNT10B were previously detected in multiple individuals with a split-hand and/or split-foot malformation and other conditions (Table S3).36 However, no tooth agenesis was reported in those cases. One of the assumptions is that those mutations are either located at the 3′ end of the gene or derived from duplication events, and these are clearly different from the mutations we identified. Likewise, whereas heterozygous mutations in LRP6 were found to be responsible for oligodontia, homozygous deletions of Lrp6 in mice also resulted in limb malformation.37 Therefore, it is conceivable that mutations in other promising candidate genes in the canonical Wnt pathway (see the review by Baron and Kneissel)38—mutations that might show a phenocopying of the WNT10B mutants—could be identified in individuals with oligodontia or other anomalies involving ectodermal dysplasia.
In conclusion, our findings provide genetic and functional evidence that heterozygous mutations in WNT10B are a previously unidentified genetic cause of tooth agenesis. Our findings also reveal a potential genotype-phenotype correlation between WNT10B variants and the positions of missing teeth. Gene-disease pathway analysis suggests that more genetic defects in the canonical Wnt pathway, as well as other interacting pathways, could be identified from individuals with tooth agenesis. Further research of how the Wnt pathway regulates the decision between the odontoblastic fate and the vasculogenic fate of dental stem cells is warranted for shedding light on the pathogenesis of tooth agenesis and tooth regeneration and repair.
Acknowledgments
We are indebted to the individuals with tooth agenesis and their families for their participation in this study. We thank Dr. Curtis Harris’s lab for the wild-type WNT10B plasmid, Dr. DeeDee Smart’s lab for the pTOPFLASH and pCMV β-gal plasmids, and Qianzhi Shao for bioinformatics analysis. We wish to thank Drs. Yan Jin and Weidong Li for thoughtful suggestions. Research reported in this publication was supported by the Wenzhou Medical University Research Fund (to P.Y.), the Beijing Collaborative Innovative Research Center for Cardiovascular Diseases (PXM2014_014226_000002 to D.J.), and National Natural Science Foundation of China grant 81110114 (to Z.S.S.).
Published: June 16, 2016
Footnotes
Supplemental Data include six figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.05.012.
Contributor Information
Hailan Feng, Email: kqfenghl@bjmu.edu.cn.
Tao Cai, Email: tcai@mail.nih.gov.
Web Resources
Allen Brain Atlas, http://www.brain-map.org/
Eurexpress, http://www.eurexpress.org/ee/
ExAC Browser, http://exac.broadinstitute.org/
GEO Profiles, http://www.ncbi.nlm.nih.gov/geoprofiles
MutationTaster, http://www.mutationtaster.org/
OMIM, http://www.omim.org
Supplemental Data
References
- 1.Zhang J., Liu H.C., Lyu X., Shen G.H., Deng X.X., Li W.R., Zhang X.X., Feng H.L. Prevalence of tooth agenesis in adolescent Chinese populations with or without orthodontics. Chin. J. Dent. Res. 2015;18:59–65. [PubMed] [Google Scholar]
- 2.Feng H.L. [Prosthodontic treatment of congenital tooth agenesis I. The classicfication, prevalence and etiology of congenital tooth agenesis] Zhonghua Kou Qiang Yi Xue Za Zhi. 2011;46:54–57. [PubMed] [Google Scholar]
- 3.Dhanrajani P.J. Hypodontia: etiology, clinical features, and management. Quintessence Int. 2002;33:294–302. [PubMed] [Google Scholar]
- 4.Ruf S., Klimas D., Hönemann M., Jabir S. Genetic background of nonsyndromic oligodontia: a systematic review and meta-analysis. J. Orofac. Orthop. 2013;74:295–308. doi: 10.1007/s00056-013-0138-z. [DOI] [PubMed] [Google Scholar]
- 5.Massink M.P., Créton M.A., Spanevello F., Fennis W.M., Cune M.S., Savelberg S.M., Nijman I.J., Maurice M.M., van den Boogaard M.J., van Haaften G. Loss-of-Function Mutations in the WNT Co-receptor LRP6 Cause Autosomal-Dominant Oligodontia. Am. J. Hum. Genet. 2015;97:621–626. doi: 10.1016/j.ajhg.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H., Ding T., Zhan Y., Feng H. A Novel AXIN2 Missense Mutation Is Associated with Non-Syndromic Oligodontia. PLoS ONE. 2015;10:e0138221. doi: 10.1371/journal.pone.0138221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X.X., Wong S.W., Han D., Feng H.L. Simultaneous Occurence of an Autosomal Dominant Inherited MSX1 Mutation and an X-linked Recessive Inherited EDA Mutation in One Chinese Family with Non-syndromic Oligodontia. Chin. J. Dent. Res. 2015;18:229–234. doi: 10.3290/j.cjdr.a35147. [DOI] [PubMed] [Google Scholar]
- 8.Song S., Zhao R., He H., Zhang J., Feng H., Lin L. WNT10A variants are associated with non-syndromic tooth agenesis in the general population. Hum. Genet. 2014;133:117–124. doi: 10.1007/s00439-013-1360-x. [DOI] [PubMed] [Google Scholar]
- 9.Song S., Han D., Qu H., Gong Y., Wu H., Zhang X., Zhong N., Feng H. EDA gene mutations underlie non-syndromic oligodontia. J. Dent. Res. 2009;88:126–131. doi: 10.1177/0022034508328627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., Wu H., Wu J., Zhao H., Zhang X., Mues G., D’Souza R.N., Feng H., Kapadia H. Identification and functional analysis of two novel PAX9 mutations. Cells Tissues Organs (Print) 2009;189:80–87. doi: 10.1159/000151448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin W., Bian Z. The Gene Network Underlying Hypodontia. J. Dent. Res. 2015;94:878–885. doi: 10.1177/0022034515583999. [DOI] [PubMed] [Google Scholar]
- 12.Cai T., Yang L., Cai W., Guo S., Yu P., Li J., Hu X., Yan M., Shao Q., Jin Y. Dysplastic spondylolysis is caused by mutations in the diastrophic dysplasia sulfate transporter gene. Proc. Natl. Acad. Sci. USA. 2015;112:8064–8069. doi: 10.1073/pnas.1502454112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stockton D.W., Das P., Goldenberg M., D’Souza R.N., Patel P.I. Mutation of PAX9 is associated with oligodontia. Nat. Genet. 2000;24:18–19. doi: 10.1038/71634. [DOI] [PubMed] [Google Scholar]
- 14.Thesleff I. Current understanding of the process of tooth formation: transfer from the laboratory to the clinic. Aust. Dent. J. 2014;59(Suppl 1):48–54. doi: 10.1111/adj.12102. [DOI] [PubMed] [Google Scholar]
- 15.Song W., Gardner S.A., Hovhannisyan H., Natalizio A., Weymouth K.S., Chen W., Thibodeau I., Bogdanova E., Letovsky S., Willis A., Nagan N. Exploring the landscape of pathogenic genetic variation in the ExAC population database: insights of relevance to variant classification. Genet. Med. 2015 doi: 10.1038/gim.2015.180. Published online December 17, 2015. [DOI] [PubMed] [Google Scholar]
- 16.Janda C.Y., Waghray D., Levin A.M., Thomas C., Garcia K.C. Structural basis of Wnt recognition by Frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauna Z.E., Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 18.Nackley A.G., Shabalina S.A., Tchivileva I.E., Satterfield K., Korchynskyi O., Makarov S.S., Maixner W., Diatchenko L. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 19.Dassule H.R., McMahon A.P. Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev. Biol. 1998;202:215–227. doi: 10.1006/dbio.1998.8992. [DOI] [PubMed] [Google Scholar]
- 20.Sarkar L., Sharpe P.T. Expression of Wnt signalling pathway genes during tooth development. Mech. Dev. 1999;85:197–200. doi: 10.1016/s0925-4773(99)00095-7. [DOI] [PubMed] [Google Scholar]
- 21.Yoshikawa H., Matsubara K., Zhou X., Okamura S., Kubo T., Murase Y., Shikauchi Y., Esteller M., Herman J.G., Wei Wang X., Harris C.C. WNT10B functional dualism: beta-catenin/Tcf-dependent growth promotion or independent suppression with deregulated expression in cancer. Mol. Biol. Cell. 2007;18:4292–4303. doi: 10.1091/mbc.E06-10-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouji Y., Yoshikawa M., Shiroi A., Ishizaka S. Wnt-10b promotes differentiation of skin epithelial cells in vitro. Biochem. Biophys. Res. Commun. 2006;342:28–35. doi: 10.1016/j.bbrc.2006.01.104. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen P., Lee S., Lorang-Leins D., Trepel J., Smart D.K. SIRT2 interacts with β-catenin to inhibit Wnt signaling output in response to radiation-induced stress. Mol. Cancer Res. 2014;12:1244–1253. doi: 10.1158/1541-7786.MCR-14-0223-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibata T., Aburatani H. Exploration of liver cancer genomes. Nat. Rev. Gastroenterol. Hepatol. 2014;11:340–349. doi: 10.1038/nrgastro.2014.6. [DOI] [PubMed] [Google Scholar]
- 25.Gronthos S., Mankani M., Brahim J., Robey P.G., Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z., Nor F., Oh M., Cucco C., Shi S., Nör J.E. Wnt/β-catenin signaling determines the vasculogenic fate of post-natal mesenchymal stem cells. Stem Cells. 2016 doi: 10.1002/stem.2334. Published online February 11, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao L., Tsutsui T. Three-dimensional epithelial and mesenchymal cell co-cultures form early tooth epithelium invagination-like structures: expression patterns of relevant molecules. J. Cell. Biochem. 2012;113:1875–1885. doi: 10.1002/jcb.24056. [DOI] [PubMed] [Google Scholar]
- 28.Scheller E.L., Chang J., Wang C.Y. Wnt/beta-catenin inhibits dental pulp stem cell differentiation. J. Dent. Res. 2008;87:126–130. doi: 10.1177/154405910808700206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan G., Zhang L., Yang G., Yang J., Wan C., Zhang L., Song G., Chen S., Chen Z. The distribution and ultrastructure of the forming blood capillaries and the effect of apoptosis on vascularization in mouse embryonic molar mesenchyme. Cell Tissue Res. 2014;356:137–145. doi: 10.1007/s00441-013-1785-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamashiro T., Zheng L., Shitaku Y., Saito M., Tsubakimoto T., Takada K., Takano-Yamamoto T., Thesleff I. Wnt10a regulates dentin sialophosphoprotein mRNA expression and possibly links odontoblast differentiation and tooth morphogenesis. Differentiation. 2007;75:452–462. doi: 10.1111/j.1432-0436.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- 31.Bohring A., Stamm T., Spaich C., Haase C., Spree K., Hehr U., Hoffmann M., Ledig S., Sel S., Wieacker P., Röpke A. WNT10A mutations are a frequent cause of a broad spectrum of ectodermal dysplasias with sex-biased manifestation pattern in heterozygotes. Am. J. Hum. Genet. 2009;85:97–105. doi: 10.1016/j.ajhg.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arzoo P.S., Klar J., Bergendal B., Norderyd J., Dahl N. WNT10A mutations account for ¼ of population-based isolated oligodontia and show phenotypic correlations. Am. J. Med. Genet. A. 2014;164A:353–359. doi: 10.1002/ajmg.a.36243. [DOI] [PubMed] [Google Scholar]
- 33.Khalaf K., Miskelly J., Voge E., Macfarlane T.V. Prevalence of hypodontia and associated factors: a systematic review and meta-analysis. J. Orthod. 2014;41:299–316. doi: 10.1179/1465313314Y.0000000116. [DOI] [PubMed] [Google Scholar]
- 34.Xuan K., Jin F., Liu Y.L., Yuan L.T., Wen L.Y., Yang F.S., Wang X.J., Wang G.H., Jin Y. Identification of a novel missense mutation of MSX1 gene in Chinese family with autosomal-dominant oligodontia. Arch. Oral Biol. 2008;53:773–779. doi: 10.1016/j.archoralbio.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 35.Kettunen P., Løes S., Furmanek T., Fjeld K., Kvinnsland I.H., Behar O., Yagi T., Fujisawa H., Vainio S., Taniguchi M., Luukko K. Coordination of trigeminal axon navigation and patterning with tooth organ formation: epithelial-mesenchymal interactions, and epithelial Wnt4 and Tgfbeta1 regulate semaphorin 3a expression in the dental mesenchyme. Development. 2005;132:323–334. doi: 10.1242/dev.01541. [DOI] [PubMed] [Google Scholar]
- 36.Wend P., Wend K., Krum S.A., Miranda-Carboni G.A. The role of WNT10B in physiology and disease. Acta Physiol. (Oxf.) 2012;204:34–51. doi: 10.1111/j.1748-1716.2011.02296.x. [DOI] [PubMed] [Google Scholar]
- 37.Kelly O.G., Pinson K.I., Skarnes W.C. The Wnt co-receptors Lrp5 and Lrp6 are essential for gastrulation in mice. Development. 2004;131:2803–2815. doi: 10.1242/dev.01137. [DOI] [PubMed] [Google Scholar]
- 38.Baron R., Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat. Med. 2013;19:179–192. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.