Abstract
Platelet production, maintenance, and clearance are tightly controlled processes indicative of platelets’ important roles in hemostasis and thrombosis. Platelets are common targets for primary and secondary prevention of several conditions. They are monitored clinically by complete blood counts, specifically with measurements of platelet count (PLT) and mean platelet volume (MPV). Identifying genetic effects on PLT and MPV can provide mechanistic insights into platelet biology and their role in disease. Therefore, we formed the Blood Cell Consortium (BCX) to perform a large-scale meta-analysis of Exomechip association results for PLT and MPV in 157,293 and 57,617 individuals, respectively. Using the low-frequency/rare coding variant-enriched Exomechip genotyping array, we sought to identify genetic variants associated with PLT and MPV. In addition to confirming 47 known PLT and 20 known MPV associations, we identified 32 PLT and 18 MPV associations not previously observed in the literature across the allele frequency spectrum, including rare large effect (FCER1A), low-frequency (IQGAP2, MAP1A, LY75), and common (ZMIZ2, SMG6, PEAR1, ARFGAP3/PACSIN2) variants. Several variants associated with PLT/MPV (PEAR1, MRVI1, PTGES3) were also associated with platelet reactivity. In concurrent BCX analyses, there was overlap of platelet-associated variants with red (MAP1A, TMPRSS6, ZMIZ2) and white (PEAR1, ZMIZ2, LY75) blood cell traits, suggesting common regulatory pathways with shared genetic architecture among these hematopoietic lineages. Our large-scale Exomechip analyses identified previously undocumented associations with platelet traits and further indicate that several complex quantitative hematological, lipid, and cardiovascular traits share genetic factors.
Introduction
The number and size of circulating blood cells are determined by multiple genetic and environmental factors, and abnormal values are a common manifestation of human disease. The three major cell types—red blood cells (RBCs), white blood cells (WBCs), and platelets—have distinct biological roles, with platelets serving as important mediators of hemostasis and wound healing. Platelet count (PLT) and mean platelet volume (MPV), a measure of platelet size, are clinical blood tests that are used to screen for and diagnose disease. A number of well-described rare genetic disorders, including Bernard-Soulier syndrome (MIM: 231200), Glanzmann thrombasthenia (MIM: 273800), and Wiskott-Aldrich syndrome (MIM: 301000), as well as common conditions such as acute infection are characterized by abnormalities in the number, size, and/or reactivity of circulating blood platelets. MPV has also been reported to be an independent risk factor for myocardial infarction (MI) in population-based studies.1 Accordingly, anti-platelet medications including aspirin and ADP/P2Y12 receptor blockers such as clopidogrel and GIIb/IIIa inhibitors that reduce platelet reactivity are commonly used in the primary and secondary prevention of several cardiovascular conditions, including stroke and MI.2, 3 Investigating the biological mechanisms that govern platelet number (PLT) and size (MPV) can provide insights into platelet development and clearance and has the potential to enhance our understanding of human diseases.
Genome-wide association studies (GWASs) have successfully identified numerous loci where variants are associated with PLT and MPV.4, 5, 6, 7, 8, 9, 10, 11, 12, 13 To date, the largest GWAS of PLT (n = 66,867) and MPV (n = 30,194) identified 68 distinct loci.8 Subsequent functional experiments of several identified genes, including ARHGEF3 (MIM: 612115), DNM3 (MIM: 611445), JMJD1C (MIM: 604503), and TPM1 (MIM: 191010), demonstrated their roles in hematopoiesis and megakaryopoesis,8, 14 as well as the potential of human genetic association methods to identify genetic factors that functionally contribute to platelet biology and dysfunction in disease.
Despite these successes, much of the heritability of these traits remains unexplained.15 GWASs of PLT and MPV have largely focused on more common (minor allele frequency [MAF] > 0.05) genetic variation, with many of the associated markers located in intronic or intergenic regions. The examination of rare (MAF < 0.01) and low-frequency (MAF: 0.01–0.05) variants, particularly those in protein coding regions, has the potential to identify previously unidentified causal variants. Indeed, recent studies reaching sample sizes of 31,340 individuals have identified rare to low-frequency coding variants associated with PLT in MPL (MIM: 159530), CD36 (MIM: 173510), and JAK2 (MIM: 147796), among others.16, 17 Studies with larger sample size are needed to further characterize the contribution of rare and low-frequency genetic variation to PLT and MPV.
To conduct such a study of platelet-related traits, we formed the Blood Cell Consortium (BCX) to perform a large-scale meta-analysis of Exomechip association results of blood cell traits. In this report, we describe results from a meta-analysis of Exomechip association data in 157,293 and 57,617 participants for PLT and MPV, respectively. The Exomechip is a custom genotyping array enriched for rare to low-frequency coding variants; in addition, the Exomechip contains a scaffold of nonsynonymous variants and common SNPs obtained from the NHGRI GWAS catalog of complex disorders and traits. With increased sample size and use of the Exomechip, our goal was to identify rare, low-frequency, and common variants associated with PLT and MPV.
Material and Methods
Study Participants
The Blood Cell Consortium (BCX) was formed to identify genetic variants associated with blood cell traits using the Exomechip genotyping array. As the BCX is interested in the genetics of common hematological measures, our collaborative group is divided into three main working groups: RBC, WBC, and platelet.18, 19 For the platelet working group, our sample is comprised of 157,293 participants from 26 discovery and replication cohorts of five ancestries: European (EA), African American (AA), Hispanic, East Asian, and South Asian. Detailed descriptions of the participating cohorts are provided in the Tables S1–S4. All participants provided informed consent, and all protocols were approved by the participating studies’ respective institutional review boards. In the platelet working group, we analyzed two traits: PLT (× 109/L of whole blood) and MPV (fL) (Table S3).
Genotyping and Quality Control
Each participating study used one of the following Exomechip genotyping arrays: Illumina ExomeChip v.1.0, Illumina ExomeChip v.1.1_A, Illumina ExomeChip-12 v.1.1, Illumina ExomeChip-12 v.1.2, Affymetrix Axiom Biobank Plus GSKBB1, or Illumina HumanOmniExpress ExomeChip (Table S2). Genotypes were called using either (1) a combination of the Illumina GenomeStudio and zCall software or (2) the Exomechip joint calling plan developed by the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium (Table S2).20 Standard quality-control criteria were applied by each study. Exclusion criteria included: (1) sample call rates, (2) excess heterozygosity rate, (3) Hardy-Weinberg equilibrium p values < 1 × 10−6, and (4) sex mismatch. Additionally, ancestry was confirmed through principal components or multi-dimensional scaling analyses using linkage disequilibrium (LD) pruned markers (r2 < 0.2) with MAF > 1%. Scatterplots anchored using the 1000 Genomes Project populations were visually inspected and ancestry outliers excluded. We included only autosomal and X chromosome variants. All remaining variants (including monomorphic variants) were aligned to the forward strand and alleles checked to ensure that the correct reference allele was specified. We performed study-specific quality control on each trait association results using EasyQC.21 We plotted variant allele frequencies from each study against ethnicity-specific reference population allele frequencies to identify allele frequency deviations and presence of flipped alleles. After all quality-control procedures, each study generated an indexed variant call file (VCF) for subsequent analyses that was checked for allele alignment using the checkVCF package.
Association Analysis
To assess the association between the blood cell traits and Exomechip variants in the BCX, we considered blood cell traits measured in standard peripheral complete blood counts. When possible, we excluded individuals with blood cancer, leukemia, lymphoma, bone marrow transplant, congenital or hereditary anemia, HIV, end-stage kidney disease, dialysis, splenectomy, and cirrhosis, and those with extreme measurements of platelet traits. We also excluded individuals on erythropoietin treatment as well as those on chemotherapy. Additionally, we excluded women who were pregnant and individuals with acute medical illness at the time of complete blood count.
For platelet traits, we used raw values of PLT (×109/L) and MPV (fL). In each participating study, residuals for PLT and MPV were first calculated from linear regression models that adjusted for age, age2, sex, study center (where applicable), and principal components of genotype data. We then transformed these residuals using the rank-based inverse normal transformation. To confirm proper implementation of this transformation in each cohort, a scatterplot of the median standard error versus study-specific sample size was visually inspected for deviations using EasyQC.21 Autosomal and X chromosome variants were then tested for association with each blood cell trait using either RvTests or RAREMETALWORKER. Within individual cohorts, we performed analyses in ancestry-stratified groups: EA, AA, Hispanic, East Asian, and South Asian. Both statistical packages generate single variant association score summary statistics, variance-covariance matrices containing LD relationships between variants within a 1 MB window, and variant-specific parameters including MAF, chromosome, position, strand, genotype call rate, and Hardy-Weinberg equilibrium p values.
Discovery Association Meta-analysis
We performed ancestry-stratified (EA and AA) and combined all ancestry (All) meta-analyses of single variant association results using the Cochran-Mantel-Haenszel approach implemented in RareMETALS.22 In the multi-ancestry meta-analyses (All), we combined individuals of EA, AA, Hispanic, South Asian, and East Asian ancestries. We included variants in the meta-analysis if the genotype call rate was ≥95%. For palindromic variants (i.e., A/T and C/G variants), we compared allele frequencies taken across the entire consortium in order to detect flipped alleles. We kept variants with an allele frequency difference < 0.30 or < 0.60 for race-specific (EA and AA) or combined all ancestry analyses, respectively.21 Heterogeneity metrics (I2 and heterogeneity p value) were calculated using METAL.23 Using single-variant score statistics and variance-covariance matrices of LD estimates, we performed two types of gene-based tests: (1) variable threshold (VT) burden test with greatest power when all rare variants in a gene are associated consistently with a trait24 and (2) sequence kernel association test (SKAT)25 with better power than the burden approach when rare variants in a gene have heterogeneous effects. For all gene-based tests, we considered only missense, nonsense, and splice site single-nucleotide variants (SNVs) with MAF ≤ 1%. Similar to the single variant meta-analyses, gene-based results were generated for each major ancestry group (EA and AA) and for the combined multi-ancestry (All) samples.
Conditional Analysis
To identify independent signals around significant associations, we performed stepwise conditional analyses conditioning on the most significant single variant in a 1 MB window in RareMETALS. This procedure was repeated until there was no additional SNP significantly associated with phenotype in each region, defined as a p value that accounts for the number of markers tested in each ancestry group. For discovery and conditional single variant analyses, the threshold was: AA p < 3.03 × 10−7, EA p < 2.59 × 10−7, and All p < 2.20 × 10−7. For gene-based tests, the significance threshold accounted for the number of genes tested: AA p < 2.91 × 10−6, EA p < 2.90 × 10−6, and All p < 2.94 × 10−6. In regions like chromosome 12q24 with known extended LD structure spanning more than 1 MB, we performed a stepwise conditional analysis in GCTA using the Montreal Heart Institute Biobank cohort to disentangle seven independent PLT-associated SNVs (Table S9),26 conditioning on the most significant variant in the region.
Replication Meta-analysis
We attempted to replicate PLT and MPV associations with independent SNVs that reached significance levels in six independent cohorts (Figure 1, Table S4). Single variant association results of the six independent cohorts were combined in RareMETALS. Contributing replication cohorts adhered to identical quality control and association analysis procedures described previously for the discovery phase. We combined results in EA (PLT n = 19,939, MPV n = 15,519) and All (PLT n = 35,436, MPV n = 16,088) ethnicity groupings (Table S4). The results of discovery and replication phases were further combined using fixed effects inverse variance weighted meta-analysis in METAL.23
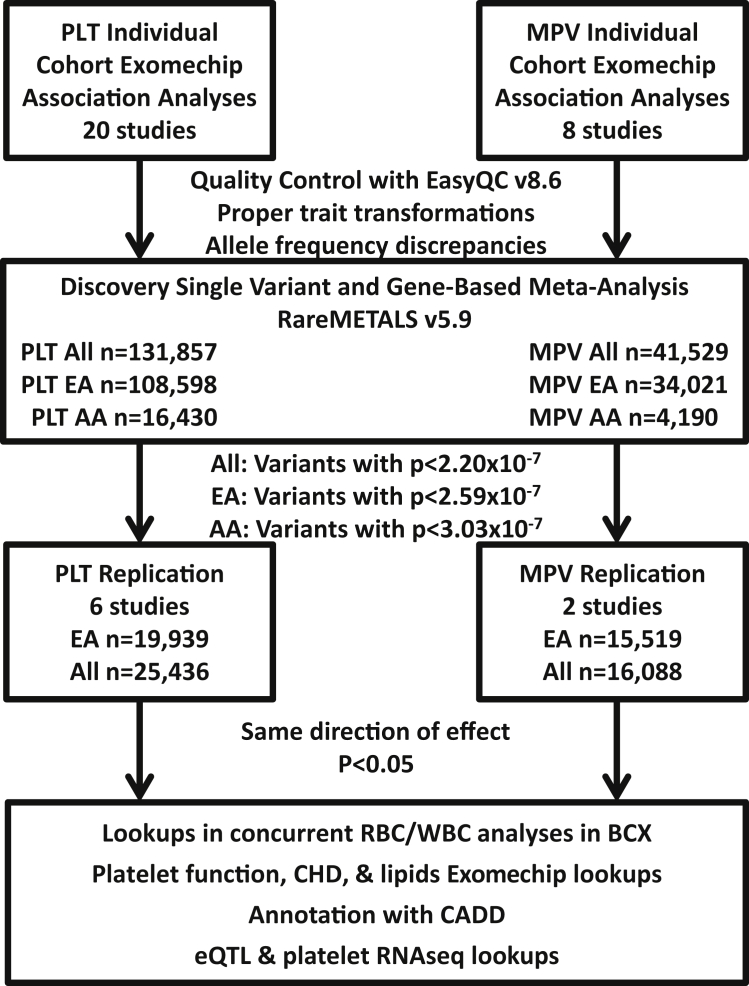
Figure 1.
Study Design and Flow
Individual study-level association analyses were performed using RareMetalWorker or RVTests. To perform quality control of individual study association results, we used EasyQC v.8.6 to ensure proper trait transformations, to assess allele frequency discrepancies, and to evaluate other metrics. We then combined results in meta-analysis with RareMETALS v5.9 in three groups: African ancestry (AA), European ancestry (EA), and combined all five (AA, EA, Hispanic-Latino, East Asian, South Asian) ancestries (All). Independent variants identified by conditional analysis in RareMETALS with a p value less than the threshold corrected for multiple testing (All, p < 2.20 × 10−7; EA, p < 2.59 × 10−7; AA, p < 3.03 × 10−7) were carried forward for replication. Markers showed replication if they had p < 0.05 in the same direction of effect in the replication analyses. Associated markers were further annotated using various resources: (1) concurrent BCX Exomechip analyses of RBC and WBC traits, (2) on-going Exomechip analyses of platelet aggregation, quantitative lipids, and coronary heart disease (CHD) traits, (3) severity prediction by CADD, (4) an internal database of reported eQTL results, and (5) platelet RNA-seq data.
Platelet Function Exomechip
Two BCX cohorts, GeneSTAR and the Framingham Heart Study (FHS), measured platelet aggregation in a subset of genotyped participants. Platelet aggregation measures are described in detail elsewhere and briefly below (Table S18).27 Both studies isolated platelet-rich plasma from fasting blood samples and measured platelet aggregation after addition of agonists using a four-channel light transmission aggregometer (Bio/Data Corporation). FHS (Offspring Exam 5) tested aggregation for periods of 4 min after administration of ADP (0.05, 0.1, 0.5, 1.0, 3.0, 5.0, 10.0, and 15.0 μM) and 5 min after administration of epinephrine (0.01, 0.03, 0.05, 0.1, 0.5, 1.0, 3.0, 5.0, and 10.0 μM), as well as lag time(s) to aggregation with 190 μg/mL calf skin-derived type I collagen (Bio/Data Corporation). Threshold concentrations (EC50) were determined as the minimal concentration of agonist required to produce a >50% aggregation. The maximal aggregation response (% aggregation) was also determined for each participant at each concentration tested. GeneSTAR recorded maximal aggregation (% aggregation) for periods of 5 min after ADP (2.0 and 10.0 μM) and 5 min after epinephrine administration (2.0 and 10.0 μM), as well as lag time(s) to aggregation with equine tendon-derived type I collagen (1, 2, 5, and 10 μg/mL). Exomechip genotyping, quality control, and association analyses adhered to methods described previously for PLT and MPV analysis. We queried independent SNVs associated with PLT (n = 79) and/or MPV (n = 38) in these platelet aggregation association results and report platelet aggregation associations with p < 0.001.
Further Variant Annotation
In addition to primary analyses completed in this investigation, we took advantage of several existing resources to annotate our associated SNVs. Associated variants were cross-referenced with Combined Annotation Dependent Depletion (CADD) scores for Exomechip.28 The Global Lipids Genetics Consortium (GLGC), the CARDIoGRAM Exome Consortium, and Myocardial Infarction Genetics Consortium have each performed independent Exomechip analysis of lipids traits and coronary heart disease (CHD).29, 30 The CHD phenotype reflected a composite endpoint that included MI, CHD, coronary artery bypass graft, and hospitalized angina, among others.29 Similar to the platelet aggregation lookups, we queried our list of PLT- and/or MPV-associated SNVs against their Exomechip association results for lipids and CHD. We report lipid and CHD associations with p < 0.0001. From a curated collection of more than 100 separate expression quantitative trait loci (eQTL) datasets, we conducted a more focused query of whether platelet loci were also associated with transcript expression in blood, arterial, and adipose-related tissues. A general overview of a subset of >50 eQTL studies has been published (Supplemental Data).31 Separately, we queried transcripts in loci corresponding to previously unreported associated variants and/or marginally associated variants showing further evidence of association in our replication analyses to assess their platelet expression levels using the largest platelet RNA-seq dataset to date (n = 32 patients with MI).32
Results
Discovery Meta-Analysis
In our discovery phase, we performed a meta-analysis of the associations of 246,925 single-nucleotide variants (SNVs) with PLT and MPV in 131,857 and 41,529 individuals, respectively (Figures 1, S1, and S2; Tables S1–S4). After the initial meta-analyses, we ran conditional analyses to identify independent loci and found 79 independent PLT and 38 independent MPV SNVs (Tables 1, 2, and S5–S8). One association, rs12692566 in LY75-CD302, with PLT in EA did not surpass the initial discovery statistical significance threshold but surpassed the threshold when conditioned on nearby rs78446341 (p = 2.48 × 10−7). There were no associations unique to the AA ancestry group, which had a limited sample size (Tables S10 and S11). Single variant meta-analysis results for each ancestry grouping that met our significance thresholds are available in the Supplement (Tables S10 and S11). Additionally, full discovery meta-analysis results are available online (Web Resources).
Table 1.
Previously Unreported Associations with PLT
| rsID | Ref/Alt | Function | Gene |
European Ancestry (EA) |
Combined All Ancestry (All) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Discovery (n = 108,598) |
Replication (n = 19,939) |
Combined |
Discovery (n = 131,857) |
Replication (n = 25,436) |
Combined |
||||||||||
| EAF | Beta | p Value | Beta | p Value | p Value | EAF | Beta | p Value | Beta | p Value | p Value | ||||
| rs3091242 | C/T | intron | TMEM50A | 0.54 | −0.026 | 9.68 × 10−8 | −0.017 | 0.124 | 3.85 × 10−8 | 0.50 | −0.02 | 1.03 × 10−5 | −0.0084 | 0.390 | 1.24 × 10−5 |
| rs12566888 | G/T | intron | PEAR1∗ | 0.094 | 0.040 | 1.42 × 10−7 | 0.061 | 1.26 × 10−3 | 1.17 × 10−9 | 0.16 | 0.034 | 2.09 × 10−8 | 0.047 | 4.31 × 10−4 | 5.71 × 10−11 |
| rs200731779 | C/G | missense | FCER1A | 1.5 × 10−5 | −2.96 | 2.48 × 10−7 | NA | NA | 2.48 × 10−7 | 1.2 × 10−5 | −2.96 | 2.48 × 10−7 | NA | NA | 2.48 × 10−7 |
| rs6734238 | A/G | intergenic | IL1F10/IL1RN | 0.41 | 0.022 | 9.55 × 10−6 | 0.0075 | 0.487 | 1.64 × 10−5 | 0.41 | 0.026 | 7.19 × 10−9 | 0.015 | 0.117 | 3.77 × 10−9 |
| rs12692566a | C/A | missense | LY75-CD302∗ | 0.82 | −0.029 | 9.19 × 10−7 | −0.042 | 2.50 × 10−3 | 1.23 × 10−8 | 0.83 | −0.026 | 2.27 × 10−6 | −0.05 | 7.84 × 10−5 | 3.65 × 10−9 |
| rs78446341 | G/A | missense | LY75-CD302∗ | 0.02 | 0.092 | 4.16 × 10−9 | 0.14 | 5.01 × 10−5 | 1.98 × 10−12 | 0.018 | 0.094 | 3.06 × 10−10 | 0.13 | 9.23 × 10−5 | 1.97 × 10−13 |
| rs56106611b | T/G | missense | KALRN∗ | 0.012 | 0.11 | 3.51 × 10−8 | 0.11 | 7.14 × 10−3 | 8.51 × 10−10 | 0.01 | 0.11 | 8.59 × 10−8 | 0.11 | 7.37 × 10−3 | 2.14 × 10−9 |
| rs1470579 | A/C | intron | IGF2BP2 | 0.32 | −0.028 | 1.08 × 10−7 | −0.0073 | 0.562 | 2.82 × 10−7 | 0.38 | −0.023 | 6.07 × 10−7 | −0.012 | 0.272 | 5.15 × 10−7 |
| rs1126673 | C/T | ncRNA | LOC100507053 | 0.69 | 0.026 | 6.38 × 10−8 | 0.019 | 9.63 × 10−2 | 1.81 × 10−8 | 0.71 | 0.025 | 1.87 × 10−8 | 0.014 | 0.168 | 1.12 × 10−8 |
| rs1473247b | T/C | intron | RNF145∗ | 0.27 | −0.029 | 3.01 × 10−8 | −0.022 | 8.32 × 10−2 | 7.28 × 10−9 | 0.32 | −0.026 | 1.32 × 10−8 | −0.025 | 1.85 × 10−2 | 7.66 × 10−10 |
| rs2256183 | A/G | intron | MICA | 0.56 | 0.03 | 6.78 × 10−7 | −0.022 | 0.104 | 2.60 × 10−6 | 0.59 | 0.028 | 2.13 × 10−7 | 0.011 | 0.389 | 3.20 × 10−7 |
| rs1050331 | T/G | 3′ UTR | ZMIZ2 | 0.47 | 0.037 | 1.32 × 10−15 | 0.036 | 5.80 × 10−4 | 3.28 × 10−18 | 0.48 | 0.035 | 3.09 × 10−17 | 0.031 | 8.80 × 10−4 | 1.26 × 10−19 |
| rs755109 | T/C | intron | HEMGN | 0.37 | 0.028 | 2.87 × 10−9 | 0.039 | 6.84 × 10−4 | 1.17 × 10−11 | 0.34 | 0.028 | 9.03 × 10−11 | 0.044 | 2.18 × 10−5 | 2.59 × 10−14 |
| rs2068888 | G/A | nearGene-3 | EXOC6 | 0.45 | −0.023 | 2.81 × 10−7 | −0.012 | 0.266 | 2.47 × 10−7 | 0.44 | −0.022 | 1.19 × 10−7 | −0.012 | 0.212 | 8.61 × 10−8 |
| rs3794153 | C/G | missense | ST5 | 0.45 | −0.027 | 7.28 × 10−9 | −0.026 | 1.53 × 10−2 | 3.57 × 10−10 | 0.40 | −0.027 | 2.19 × 10−9 | −0.023 | 2.47 × 10−2 | 1.74 × 10−10 |
| rs174583 | C/T | intron | FADS2 | 0.34 | 0.031 | 8.79 × 10−9 | 0.048 | 1.22 × 10−4 | 1.03 × 10−11 | 0.34 | 0.028 | 4.72 × 10−9 | 0.042 | 1.10 × 10−4 | 4.42 × 10−12 |
| rs45535039 | T/C | 3′ UTR | CCDC153 | 0.28 | 0.04 | 4.02 × 10−10 | 0.071 | 5.31 × 10−2 | 8.48 × 10−11 | 0.28 | 0.04 | 2.5 × 10−12 | 0.056 | 8.56 × 10−2 | 6.25 × 10−13 |
| rs11616188 | G/A | nearGene3 | LTBR | 0.42 | −0.025 | 1.26 × 10−8 | −0.031 | 3.59 × 10−3 | 1.81 × 10−10 | 0.37 | −0.025 | 7.57 × 10−9 | −0.033 | 1.07 × 10−3 | 4.20 × 10−11 |
| rs10506328b | A/C | intron | NFE2 | 0.64 | 0.033 | 5.63 × 10−11 | 0.06 | 5.88 × 10−8 | 2.01 × 10−16 | 0.69 | 0.038 | 3.79 × 10−15 | 0.059 | 2.33 × 10−8 | 2.73 × 10−21 |
| rs2279574 | C/A | missense | DUSP6 | 0.54 | −0.023 | 2.47 × 10−7 | −0.0082 | 0.442 | 4.28 × 10−7 | 0.50 | −0.021 | 1.57 × 10−7 | −0.006 | 0.531 | 4.04 × 10−7 |
| rs61745424 | G/A | missense | CUX2 | 0.025 | −0.064 | 2.36 × 10−6 | −0.085 | 6.79 × 10−3 | 6.49 × 10−8 | 0.023 | −0.068 | 1.37 × 10−7 | −0.073 | 1.43 × 10−2 | 6.30 × 10−9 |
| rs2784521 | A/G | nearGene-5 | DDHD1 | 0.83 | 0.025 | 1.62 × 10−5 | 0.0096 | 0.486 | 2.24 × 10−5 | 0.76 | 0.028 | 2.92 × 10−8 | 0.01 | 0.363 | 5.56 × 10−8 |
| rs55707100 | C/T | missense | MAP1A∗ | 0.032 | 0.095 | 7.03 × 10−14 | 0.073 | 3.87 × 10−2 | 9.53 × 10−15 | 0.028 | 0.092 | 6.85 × 10−14 | 0.082 | 1.62 × 10−2 | 3.77 × 10−15 |
| rs10852932 | G/T | intron | SMG6∗ | 0.36 | −0.024 | 1.82 × 10−6 | −0.042 | 8.93 × 10−4 | 1.42 × 10−8 | 0.39 | −0.025 | 4.79 × 10−8 | −0.036 | 6.99 × 10−4 | 2.15 × 10−10 |
| rs76066357 | G/C | missense | ITGA2B∗ | 0.014 | −0.17 | 6.92 × 10−16 | −0.19 | 2.88 × 10−5 | 1.05 × 10−19 | 0.013 | −0.16 | 1.92 × 10−15 | −0.18 | 6.00 × 10−5 | 5.78 × 10−19 |
| rs1801689 | A/C | missense | APOH∗ | 0.036 | 0.083 | 6.34 × 10−12 | 0.13 | 2.44 × 10−5 | 1.82 × 10−15 | 0.032 | 0.090 | 8.64 × 10−15 | 0.12 | 2.03 × 10−5 | 1.57 × 10−18 |
| rs892055 | A/G | missense | RASGRP4 | 0.34 | 0.029 | 5.30 × 10−10 | 0.018 | 9.87 × 10−2 | 2.01 × 10−10 | 0.38 | 0.025 | 3.49 × 10−9 | 0.017 | 8.13 × 10−2 | 9.96 × 10−10 |
| rs3865444 | C/A | 5′ UTR | CD33∗ | 0.32 | −0.026 | 1.11 × 10−6 | −0.034 | 2.52 × 10−3 | 1.27 × 10−8 | 0.29 | −0.026 | 2.10 × 10−7 | −0.032 | 3.03 × 10−3 | 2.59 × 10−9 |
| rs6136489b | T/G | intergenic | SIRPA∗ | 0.34 | −0.033 | 8.69 × 10−13 | −0.028 | 1.24 × 10−2 | 4.00 × 10−14 | 0.39 | −0.030 | 1.8 × 10−12 | −0.024 | 1.30 × 10−2 | 8.78 × 10−14 |
| rs855791 | A/G | missense | TMPRSS6∗ | 0.56 | −0.031 | 3.96 × 10−11 | −0.017 | 0.130 | 2.34 × 10−11 | 0.60 | −0.029 | 2.34 × 10−11 | −0.022 | 3.52 × 10−2 | 2.97 × 10−12 |
| rs1018448 | A/C | missense | ARFGAP3 | 0.54 | −0.028 | 4.02 × 10−10 | −0.0053 | 0.618 | 2.62 × 10−9 | 0.59 | −0.025 | 1.55 × 10−9 | −0.0065 | 0.515 | 6.13 × 10−9 |
| rs738409 | C/G | missense | PNPLA3∗ | 0.23 | −0.042 | 1.49 × 10−14 | −0.042 | 1.75 × 10−3 | 1.03 × 10−16 | 0.22 | −0.044 | 1.33 × 10−18 | −0.038 | 1.61 × 10−3 | 9.73 × 10−21 |
We show variants in previously unreported loci (n = 32) and retained after conditional analyses in European ancestry (EA) (p < 2.59 × 10−7) and all ancestry (All) (p < 2.20 × 10−7) analyses. Associations in African ancestry (AA) had previously been reported in the literature (Table S10). Asterisks (∗) indicate variants (20/32) showing evidence of replication (p < 0.05, same direction of effect). If multiple genes/transcripts were annotated to a variant, the transcript most expressed in Eicher et al.32 (Table S22) was selected. Full results and annotations are available in Table S5. Abbreviations are as follows: PLT, platelet count; MPV, mean platelet volume; REF, reference allele; ALT, alternate allele; EAF, effect allele frequency.
Surpasses significance threshold after conditioning on rs78446341 (p = 2.48 × 10−7).7
Previous association with MPV.
Table 2.
Previously Unreported Associations with MPV
| rsID | Ref/Alt | Function | Gene |
European Ancestry (EA) |
Combined All Ancestry (All) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Discovery (n = 34,021) |
Replication (n = 15,519) |
Combined |
Discovery (n = 41,529) |
Replication (n = 16,088) |
Combined |
||||||||||
| EAF | Beta | p Value | Beta | p Value | p Value | EAF | Beta | p Value | Beta | p Value | p Value | ||||
| rs6687605 | T/C | missense | LDLRAP1∗ | 0.53 | 0.046 | 8.27 × 10−12 | 0.025 | 3.74 × 10−2 | 1.80 × 10−9 | 0.51 | 0.046 | 9.92 × 10−11 | 0.024 | 3.58 × 10−2 | 3.80 × 10−11 |
| rs56043070a | G/A | splice | GCSAML∗ | 0.069 | 0.094 | 1.30 × 10−9 | 0.19 | 4.48 × 10−16 | 1.12 × 10−21 | 0.064 | 0.092 | 2.25 × 10−10 | 0.19 | 3.66 × 10−16 | 2.42 × 10−22 |
| rs1339847a | G/A | missense | TRIM58 | 0.10 | −0.10 | 1.47 × 10−13 | −0.037 | 5.44 × 10−2 | 9.31 × 10−13 | 0.10 | −0.11 | 2.18 × 10−17 | −0.032 | 9.77 × 10−2 | 1.06 × 10−15 |
| rs34968964a | G/C | missense | IQGAP2 | 0.0049 | 0.32 | 7.65 × 10−9 | 0.12 | 9.18 × 10−2 | 1.99 × 10−8 | 0.004 | 0.32 | 2.11 × 10−9 | 0.11 | 0.106 | 8.18 × 10−9 |
| rs34950321a | C/T | missense | IQGAP2∗ | 0.018 | 0.18 | 7.80 × 10−10 | 0.14 | 1.49 × 10−3 | 6.03 × 10−12 | 0.016 | 0.17 | 2.61 × 10−9 | 0.14 | 1.59 × 10−3 | 1.86 × 10−11 |
| rs34592828a | G/A | missense | IQGAP2∗ | 0.037 | 0.22 | 1.72 × 10−27 | 0.16 | 2.73 × 10−9 | 1.61 × 10−34 | 0.032 | 0.23 | 1.68 × 10−31 | 0.16 | 2.95 × 10−9 | 2.98 × 10−38 |
| rs1012899a | G/A | missense | LRRC16A | 0.77 | 0.051 | 1.40 × 10−7 | 0.012 | 0.417 | 1.24 × 10−6 | 0.77 | 0.042 | 1.32 × 10−6 | 0.016 | 0.273 | 2.50 × 10−6 |
| rs664370 | A/G | missense | PXT1∗ | 0.30 | −0.034 | 8.03 × 10−5 | −0.025 | 5.61 × 10−2 | 1.39 × 10−5 | 0.35 | −0.042 | 5.77 × 10−8 | −0.028 | 2.78 × 10−2 | 7.23 × 10−9 |
| rs2343596a | C/A | intron | ZFPM2 | 0.31 | 0.062 | 2.02 × 10−13 | 0.012 | 0.357 | 3.32 × 10−11 | 0.38 | 0.052 | 1.59 × 10−11 | 0.012 | 0.339 | 4.35 × 10−10 |
| rs55895668a | T/C | missense | PLEC | 0.43 | −0.042 | 5.94 × 10−7 | −0.013 | 0.350 | 2.19 × 10−6 | 0.47 | −0.041 | 1.23 × 10−7 | −0.011 | 0.409 | 5.97 × 10−7 |
| rs4909945 | T/C | missense | MRVI1∗ | 0.68 | −0.048 | 1.25 × 10−8 | −0.035 | 8.41 × 10−3 | 5.19 × 10−10 | 0.71 | −0.041 | 3.96 × 10−7 | −0.035 | 7.42 × 10−3 | 1.06 × 10−8 |
| rs11125 | A/T | missense | LGALS3 | 0.078 | −0.091 | 1.55 × 10−8 | −0.037 | 0.117 | 2.76 × 10−8 | 0.07 | −0.09 | 4.22 × 10−9 | −0.037 | 0.117 | 7.21 × 10−9 |
| rs2010875a | C/T | missense | PLEKHO2∗ | 0.14 | −0.076 | 1.33 × 10−7 | −0.042 | 1.62 × 10−2 | 2.10 × 10−8 | 0.15 | −0.063 | 3.01 × 10−7 | −0.042 | 1.62 × 10−2 | 2.43 × 10−8 |
| rs10512472a | T/C | missense | SLFN14∗ | 0.18 | −0.059 | 1.37 × 10−8 | −0.059 | 1.96 × 10−4 | 1.12 × 10−11 | 0.18 | −0.058 | 3.15 × 10−10 | −0.059 | 1.20 × 10−4 | 1.67 × 10−13 |
| rs35385129 | C/A | missense | PVR∗ | 0.16 | −0.058 | 6.24 × 10−8 | −0.044 | 7.36 × 10−3 | 2.01 × 10−9 | 0.15 | −0.055 | 3.00 × 10−8 | −0.043 | 7.13 × 10−3 | 8.79 × 10−10 |
| rs2243603 | C/G | missense | SIRPB1 | 0.77 | 0.044 | 5.89 × 10−6 | 0.077 | 0.167 | 2.62 × 10−6 | 0.79 | 0.049 | 4.58 × 10−8 | 0.088 | 7.78 × 10−2 | 1.25 × 10−8 |
| rs1018448 | A/C | missense | ARFGAP3∗ | 0.55 | 0.056 | 1.13 × 10−12 | 0.051 | 1.78 × 10−5 | 1.04 × 10−16 | 0.60 | 0.055 | 1.52 × 10−13 | 0.05 | 2.16 × 10−5 | 1.68 × 10−17 |
| rs1997715 | G/A | 3′ UTR | ZXDB∗ | 0.26 | 0.048 | 1.93 × 10−9 | 0.084 | 5.83 × 10−2 | 4.26 × 10−10 | 0.35 | 0.04 | 4.58 × 10−8 | 0.08 | 3.99 × 10−2 | 8.88 × 10−9 |
We show variants in previously unreported MPV loci (n = 18) and retained after conditional analyses in European ancestry (EA) (p < 2.59 × 10−7) and all ancestry (All) (p < 2.20 × 10−7) analyses. Associations in African ancestry (AA) had previously been reported in the literature (Table S11). Asterisk (∗) indicates variants (11/18) that showed evidence of replication (p < 0.05, same direction of effect). If multiple genes/transcripts were annotated to a variant, the transcript more expressed in Eicher et al.32 (Table S22) was selected. Full results and annotations are available in Table S7. Abbreviations are as follows: MPV, mean platelet volume; PLT, platelet count; REF, reference allele; ALT, alternate allele; EAF, effect allele frequency.
Previous association with PLT.
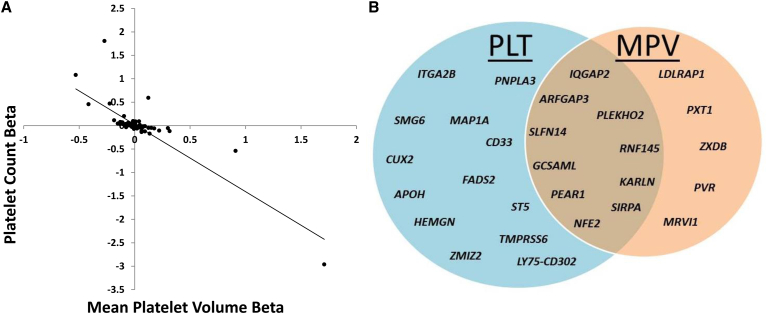
Of these independently associated single variants, 32 PLT and 18 MPV variants were in loci not previously reported (Tables 1 and 2). Of these 32 PLT loci, 4 had previously been identified as MPV loci (Table 1), and 10 of the 18 MPV loci had previously been identified with PLT (Table 2).8, 9, 17 Of the independent loci in our study, 23 SNVs showed association with both PLT and MPV (Table 3, Figure 2). All but one (rs6136489 intergenic to SIRPA [MIM: 602461] and LOC727993) had opposite directions of effect for PLT and MPV. Additionally, the observed effect sizes for PLT and MPV displayed strong negative correlations (Figure 2), indicative of the strong negative correlation between these traits.
Table 3.
Variants Associated with Both PLT and MPV
| rsID | Gene | PLT | MPV |
|---|---|---|---|
| rs12566888 | PEAR1 | ↑ | ↓ |
| rs1668873 | TMCC2 | ↑ | ↓ |
| rs56043070 | GCSAML | ↓ | ↑ |
| rs12485738 | ARHGEF3 | ↑ | ↓ |
| rs56106611 | KALRN | ↑ | ↓ |
| rs34592828 | IQGAP2 | ↓ | ↑ |
| rs1012899 | LRRC16A | ↓ | ↑ |
| rs342293 | PIK3CG | ↓ | ↑ |
| rs2343596 | ZFPM2 | ↓ | ↑ |
| rs10761731 | JMJD1C | ↑ | ↓ |
| rs11602954 | BET1L | ↑ | ↓ |
| rs10506328 | NFE2 | ↑ | ↓ |
| rs2958154 | PTGES3 | ↓ | ↑ |
| rs7961894 | WDR66 | ↓ | ↑ |
| rs1465788 | ZFP36L1 | ↑ | ↓ |
| rs2297067 | EXOC3L4 | ↑ | ↓ |
| rs2138852 | TAOK1 | ↓ | ↑ |
| rs10512472 | SLFN14 | ↑ | ↓ |
| rs11082304 | CABLES1 | ↓ | ↑ |
| rs6136489∗ | SIRPA/LOC727993 | ↓ | ↓ |
| rs41303899 | TUBB1 | ↓ | ↑ |
| rs6070697 | TUBB1 | ↑ | ↓ |
| rs1018448 | ARFGAP3 | ↓ | ↑ |
All variants listed here showed association with both PLT and MPV in the opposite direction of effect as indicated by the arrows, except for rs6136489 (denoted by asterisk), which showed association with decreased PLT and decreased MPV. Abbreviations are as follows: PLT, platelet count; MPV, mean platelet volume.
Figure 2.
Shared PLT and MPV Genetic Associations
(A) Comparing PLT and MPV effects sizes (r = −0.84) in European ancestry (EA) analyses of all identified SNVs identified (n = 124). Examined SNPs include all those from Tables 1, 2, S5–S9, S14, and S15.
(B) 56 independent SNVs showed association to PLT only, and 15 independent SNVs were associated with MPV only. 23 independent SNVs were associated with both PLT and MPV. Named genes indicate that the association was not previously reported in the literature.
Associated variants ranged in allele frequency and included rare, low-frequency, and common SNVs. Most of the previously unreported associations were with common variants (PLT n = 25, MPV n = 15), although associations with low-frequency (PLT n = 6, MPV n = 2) and rare (PLT n = 1, MPV n = 1) variants were observed. Rare (PLT n = 6, MPV n = 1) SNVs associated with PLT and MPV had larger effects compared to common and low-frequency SNVs (Tables 1, 2, and S5–S8). A large majority of associated SNVs did not exhibit heterogeneous effects; however, one previously unreported association with MRVI1 and a few known associated loci (e.g., MYL2/SH2B3/ATXN2, ARHGEF3, WDR66/HPD, and JAK2) did show moderate to substantial heterogeneity across discovery studies (Table S23). Gene-based tests of missense, nonsense, and splice-site rare variants that found significant results largely reflected rare and low-frequency single variant results, with variants in TUBB1 (MIM: 612901), JAK2, LY75 (MIM: 604524), IQGAP2 (MIM: 605401), and FCER1A (MIM: 147140) showing associations (Tables S12 and S13).
Replication Meta-Analysis
We attempted to replicate our associations in six independent cohorts (PLT n = 25,436, MPV n = 16,088) (Figure 1, Table S4). Of the loci not previously associated, 20/32 PLT and 11/18 MPV variants showed evidence of replication with p < 0.05 and the same direction of effect (Tables 1 and 2). In addition to the significant SNVs in our discovery analysis, we carried forward 13 PLT and 10 MPV sub-threshold variants that approached discovery significance thresholds with p values ranging from 2.47 × 10−7 to 1.99 × 10−6 (Tables S14 and S15). Of these, 7/13 PLT and 4/10 MPV showed associations in same direction of effect with p < 0.05 and surpassed significance thresholds when discovery and replication results were combined (Tables S14 and S15).
Intersection with Other Cardiovascular and Blood Traits
The BCX also completed analyses of RBC and WBC traits, so we cross-referenced our list of PLT- and MPV-associated SNVs with the results of the other blood cell traits.18, 19 Of our replicated platelet loci previously unreported in the literature, six SNVs in TMPRSS6 (MIM: 609862), MAP1A (MIM: 600178), PNPLA3 (MIM: 609567), FADS2 (MIM: 606149), TMEM50A (MIM: 605348), and ZMIZ2 (MIM: 611196) showed association with RBC-related traits (p < 0.0001) (Table 4). Similarly, five replicated platelet SNVs previously unreported in the literature in PEAR1 (MIM: 610278), CD33 (MIM: 159590), SIRPA, ZMIZ2, and LY75 showed association with WBC-related traits (p < 0.0001) (Table 4). To explore possible shared genetic associations of platelet size/number with platelet reactivity, we examined the association of PLT/MPV-associated SNVs with platelet reactivity to collagen, epinephrine, and ADP in GeneSTAR and FHS. Eight SNVs associated with PLT and/or MPV were also associated with platelet reactivity (p < 0.001) (Tables 5, S18, and S19). The most strongly associated SNVs were located in genes implicated with platelet reactivity in prior GWASs, including PEAR1, MRVI1 (MIM: 604673), JMJD1C, and PIK3CG (MIM: 601232).27 However, we did observe new suggestive relationships between platelet reactivity and SNVs in PTGES (MIM: 607061), LINC00523, and RASGRP4 (MIM: 607320) (Table 5).
Table 4.
Intersection of Platelet-Associated Variants with RBC and WBC Traits
| SNP | MarkerName | Gene | PLT | Trait | Other Blood Cell |
|---|---|---|---|---|---|
| rs855791 | chr22: 37,462,936 | TMPRSS6 | ↓ | MCH, MCV, HGB MCHC, HCT | ↑ |
| rs855791 | chr22: 37,462,936 | TMPRSS6 | ↓ | RDW | ↓ |
| rs55707100 | chr15: 43,820,717 | MAP1A | ↑ | HGB, MCH, HCT, MCHC | ↓ |
| rs174583 | chr11: 61,609,750 | FADS2 | ↑ | RDW | ↓ |
| rs174583 | chr11: 61,609,750 | FADS2 | ↑ | HGB, RBC, HCT, MCHC | ↑ |
| rs738409 | chr22: 44,324,727 | PNPLA3 | ↓ | HCT, HGB | ↑ |
| rs3091242 | chr1: 25,674,785 | TMEM50A | ↓ | RDW | ↑ |
| rs1050331 | chr7: 44,808,091 | ZMIZ2 | ↑ | MCH, MCV | ↓ |
| rs1050331 | chr7: 44,808,091 | ZMIZ2 | ↑ | WBC | ↑ |
| rs6734238a | chr2: 113,841,030 | IL1F10/IL1RN | ↑ | MCH | ↓ |
| rs6734238a | chr2: 113,841,030 | IL1F10/IL1RN | ↑ | WBC, NEU | ↑ |
| rs12566888 | chr1: 156,869,047 | PEAR1 | ↑ | WBC, NEU, MON | ↓ |
| rs3865444 | chr19: 51,727,962 | CD33 | ↓ | WBC | ↓ |
| rs6136489 | chr20: 1,923,734 | SIRPA/LOC727993 | ↓ | WBC, LYM | ↓ |
| rs2256183a | chr6: 31,380,529 | MICA | ↑ | BAS | ↑ |
| rs12692566 | chr2: 160,676,427 | LY75-CD302 | ↓ | WBC | ↓ |
We cross-referenced novel variants associated with platelet count (PLT) and/or mean platelet volume (MPV) in RBC and WBC association analyses in the Blood Cell Consortium (BCX). Here, we show RBC/WBC-associated platelet variants with p < 0.0001. Full details of RBC/WBC associations are shown in Tables S16 and S17. Arrows denote direction of effect for the platelet and other blood cell trait(s). Abbreviations are as follows: BCX, Blood Cell Consortium; RBC, red blood cell; WBC, white blood cell; PLT, platelet count; MCH, mean corpuscular hemoglobin; MCV, mean corpuscular volume; HGB, hemoglobin; MCHC, mean corpuscular hemoglobin concentration; HCT, hematocrit; RDW, red blood cell distribution width; PLT, platelet count; NEU, neutrophil; MON, monocyte; LYM, lymphocyte; BAS, basophil.
Marker not replicated in platelet analyses.
Table 5.
Overlap of Associations of Platelet Count and Mean Platelet Volume Variants with Platelet Reactivity
| rsID | Gene | PLT | MPV | Agonist(s)a | Direction of Effectsb |
|---|---|---|---|---|---|
| rs12566886 | PEAR1 | ↑ | ↓ | epi, ADP, collagen | ↓↓↓ |
| rs10761731 | JMJD1C | ↑ | ↓ | epi, ADP | ↑↑ |
| rs12355784 | JMJD1C | ↑ | ns | epi | ↑ |
| rs342293 | PIK3CG | ↓ | ↑ | epi | ↓ |
| rs4909945 | MRVI1 | ns | ↓ | epi, ADP | ↓↓ |
| rs2958154 | PTGES3 | ↓ | ↑ | collagen | ↑ |
| rs12883126 | LINC00523 | ↑ | ns | epi | ↑ |
| rs892055 | RASGRP4 | ↑ | ns | epi | ↓ |
Variants were examined using platelet reactivity phenotypes (Table S18) in GeneSTAR and the Framingham Heart Study (FHS). Arrows denote direction of effect for platelet count (PLT), mean platelet volume (MPV), and platelet reactivity (p < 0.001). Multiple arrows refer to direction for respective agonist for platelet reactivity. Detailed association results for platelet reactivity are given in Table S19. Abbreviations are as follows: PLT, platelet count; MPV, mean platelet volume; ns, not significant (p > 0.05); epi, epinephrine.
Platelet reactivity associations with p < 0.001.
Collagen measurements reflect lag time to aggregation, so direction of effect has been flipped to denote a negative direction of effect as less reactive and positive direction of effect as more reactive.
In addition to examining possibly shared genetic associations with blood cell-specific traits, we queried our list of associated platelet SNVs against independent Exomechip genotyping efforts in lipids and CHD by the GLGC, CARDIoGRAM Exome Consortium, and Myocardial Infarction Genetics Consortium Exomechip studies.29, 30 Numerous platelet-associated SNVs (n = 37), including those in GCKR (MIM: 600842), FADS1 (MIM: 606148), FADS2, MAP1A, APOH (MIM: 138700), and JMJD1C, showed association with one or more lipids traits (p < 0.0001) (Table S20). Far fewer (n = 4; MYL2 [MIM: 160781], SH2B3 [MIM: 605093], BRAP [MIM: 604986], APOH) showed association with CHD (p < 0.0001) (Table S20).
Annotation of Associated Variants
We used various resources to annotate our platelet-associated variants. First, we used CADD to predict the putative functional severity of associated variants.28 As expected, rare and low-frequency coding SNVs were predicted to be more severe than common, non-coding variation (Tables 1, 2, and S5–S8). To assess potential impact on gene expression, we queried our list of platelet-associated SNVs against a collection of results from existing eQTL datasets.31 Many (n = 67) platelet-associated SNVs were also associated with gene expression in blood, arterial, or adipose tissues (Table S21). These included the reported trans-eQTL rs12485738 in ARHGEF3 with several platelet-related transcript targets (e.g., GP1BA, GP6, ITGA2B, MPL, TUBB1, and VWF),33 as well as eQTLs in newly identified PLT/MPV loci (e.g., rs1018448 with ARFGAP3/PACSIN2, rs1050331 with ZMIZ2, and rs174546 with FADS1/FADS2/TMEM258 expression). Using platelet RNA-seq data from 32 subjects with MI, we found that almost all of the genes closest to our previously unreported associated SNVs or marginal SNVs with evidence of replication were expressed in platelets, indicating the feasibility of potential functional roles in the relevant target cell type (Table S22).32
Discussion
Here, we present a large-scale meta-analysis of Exomechip association data with two clinical platelet measurements, PLT and MPV. By combining Exomechip association results in 157,293 and 57,617 participants, respectively, we detected numerous associations with rare, low-frequency, and common variants. There was substantial overlap of our platelet associations with concurrent Exomechip association findings for RBC and WBC traits, indicating shared genetic influence on regulatory and functional mechanisms among the three different blood cell lineages.18, 19 More surprisingly, we observed shared associations of platelet and lipids loci. The identification of shared blood cell and lipids associations as well as identifying genes with entirely new associations reveals candidates for further examination in order to elucidate the mechanisms underlying platelet development and function.
Using Exomechip to Identify Previously Unreported Genetic Associations
Using the Exomechip that has an emphasis on rare and infrequent coding variation, we found associations with variants that ranged from common to rare in allele frequency. We attempted to replicate independent associations, although our replication cohorts were underpowered to associations of rare variants. To inform our replication criteria, we conducted a power analysis by using a sample size of 20,000 and considering multiple combinations of allele frequencies and effect sizes. Based on allele frequency and effect size, our most difficult to replicate variant was rs56106611 (MAF = 0.012, Beta = 0.11). However, we still had approximately 80% power to detect this association in the replication stage. Despite this, replication of extremely rare variants remains a challenge. For example, there were associations with rare coding variants with large effect sizes in FCER1A, MPL, JAK2, SH2B3, TUBB1, and IQGAP2.16, 17 The overall effect size of these rare variants must be validated in independent studies. The PLT-associated and predicted deleterious variant rs200731779 in FCER1A (p.Leu114Val) had a large effect (β = −2.96) in discovery analyses, but could not be replicated in available samples due to its extremely rare allele frequency (MAF = 1.48 × 10−5 in EA). The affected amino acid is extracellularly positioned near the interface of two Ig-like domains, an area of the protein critical for FC-IgE interaction as shown through its crystal structure, biochemical data, and mutagenesis studies.34, 35, 36, 37 Other variants in FCER1A, a subunit of the allergy response IgE receptor and basophil differentiation factor, have previously been associated with IgE levels and monocyte counts.38, 39 Increased platelet activation has been postulated to contribute to or be a consequence of allergic and inflammatory responses.40 Our association of rare deleterious variation in FCER1A to reduced PLT provides a further link between platelet biology and allergy response.
Although SNVs in IQGAP2 have previously been associated with PLT, we detected independent IQGAP2 low-frequency and rare missense variants associated with increased MPV (Table 2, Figures S3 and S4).8, 17 Located proximal to thrombin receptor F2R (MIM: 187930), IQGAP2 functions in the cytoskeletal dynamics in response to thrombin-induced platelet aggregation.41 We did not observe IQGAP2 associations with platelet aggregation, which may be due to the rare/low-frequency nature of the SNVs and the absence of thrombin-induced aggregation data in the available cohorts. Nonetheless, the associations of rare and low-frequency variants in IQGAP2 further strengthen its contribution to platelet biology. In addition to IQGAP2, we observed other low-frequency associations, including nonsynonymous coding variants in ITGA2B (MIM: 607759), LY75, MAP1A, and APOH. The SNV rs76066357 in ITGA2B, a gene implicated in Glanzmann’s thrombasthenia (MIM: 273800), was associated with decreased PLT (Table 1). Moreover, ITGA2B codes for the platelet glycoprotein alpha-IIb, which is part of the target receptor of GIIb/IIIa inhibitors (e.g., eptifibatide and abciximab) used in the acute management of acute coronary syndromes. Although ClinVar lists rs76066357 as pathogenic (ID: 216944) with limited evidence, rs76066357 is a non-rare, predicted benign variant that contributes to population variability in PLT in our study as opposed to a severe Mendelian disorder of platelet reactivity.42 Previous studies do suggest a potential role for variants in ITGA2B and ITGB3 (MIM: 173470) leading to thrombocytopenia as well as abnormalities in platelet reactivity.43
In addition to rare and low-frequency variant associations, we detected previously unreported associations for PLT and MPV at 25 and 15 common loci, respectively. For example, a common missense SNV rs1018489 in ARFGAP3 (MIM: 612439) showed association with decreased PLT and increased MPV. This variant is an eQTL for both ARFGAP3 and neighboring gene PACSIN2 (MIM: 604960) in blood tissues (Table S21, Figures S5 and S6). Although the possible role of the androgen receptor (AR) gene target and cellular secretory factor ARFGAP3 is unknown in platelets,44, 45, 46 PACSIN2 functions in the formation of the megakaryocyte demarcation membrane system during platelet production through interactions with FlnA.47 Genetic variation that influences PACSIN2 expression may hinder the formation of the megakaryocyte demarcation membrane system and lead to the production of fewer but larger and potentially more reactive platelets. We also observed several other novel associations with common variants, including those in SMG6 (MIM: 610963), a mediator of embryonic stem cell differentiation through nonsense-mediated decay, and LY75, an endocytotic immunity-related receptor highly expressed on dendritic cells where it is involved in recognition of apoptotic and necrotic cells.48, 49, 50
Overlap with Other Platelet and Blood Cell Traits
There was substantial overlap of variants associated with both PLT and MPV (n = 23) as well as a strong negative correlation in effect sizes, consistent with the documented negative correlation between the two traits in population studies (Figure 2).51 Only rs6136489, a reported eQTL for SIRPA, showed the same direction of effect for both PLT and MPV. SIRPA directly interacts with CD47, and SIRPA/CD47 signaling plays an important role in platelet clearance and the etiology of immune thrombocytopenia purpura.52, 53, 54 Knockout Sirpa mice exhibit thrombocytopenia phenotypes, although they have similar MPV to control animals.54 How genetic variation in SIRPA influences MPV in addition to its demonstrated contribution to PLT remains to be characterized. In addition to shared associations of PLT and MPV, there was overlap in the parallel Exomechip analyses of platelet reactivity. Largely mirroring results from previous GWASs, markers within PEAR1, JMJD1C, PIK3CG, and MRVI1 showed the strongest associations with PLT/MPV and platelet reactivity.27, 55, 56, 57 Other PLT/MPV-associated markers in PTGES3, LINC00523, and RASGRP4 showed marginal associations. Notably, PTGES3 is linked to prostaglandin synthesis and the RasGRP family has been shown to have functional roles in blood cells including in platelet adhesion.58 The association of platelet reactivity genes, particularly PEAR1 and MRVI1, with PLT/MPV further supports a biological relationship between processes that control platelet function, megakaryopoiesis, and clearance.51, 59, 60 However, these large-scale association analyses are unable to demonstrate whether these shared associations indicate shared biological mechanisms or simply reflect the epidemiological correlations among these traits.
In addition to platelet traits, there was substantial overlap of genetic associations with RBC and WBC traits examined by the BCX.18, 19 The shared genetic associations with the two other primary blood cell lineages further supports other studies proposing that mechanisms that govern platelet size and number also influence RBC and WBC traits.61 In BCX analyses, rs1050331 in the 3′ UTR of ZMIZ2 was associated with increased PLT, mean corpuscular hemoglobin (MCH), and mean corpuscular volume (MCV), as well as with decreased WBC count.18, 19 rs1050331 is also an eQTL for ZMIZ2 expression in whole blood (Table S21).62 There are known sex differences in cell counts, with females consistently having higher PLT and mixed results on MPV.63, 64 Similar to well-established PLT- and MPV-associated transcriptional regulator JMJD1C, ZMIZ2 directly interacts with AR to modulate AR-mediated transcription and influences mesodermal development, and thus genetic variation in ZMIZ2 could potentially contribute to hormonally mediate differences in PLT across genders.65, 66, 67 Also associated with increased PLT and decreased RBC indices was rs55707100 in MAP1A.18 Though typically examined in a neurological context, MAP1A is involved in microtubule assembly, a process important in blood cell development and function.68 Our observed association of MAP1A and its expression in platelets and RBCs suggests that the known role of MAP1A in developmental and cytoskeletal processes in neural tissues may extend to blood cells (Table S22). How these shared genetic factors specifically influence the development, maintenance, or clearance of multiple blood cell types remains to be determined.
Overlap with Non-Blood Cell Traits
Although the overlap with other blood cell traits may be intuitive, we also observed overlap with quantitative lipids traits. In cross-trait lookups, several known PLT/MPV loci confirmed in this study (e.g., JMJD1C, GCKR, and SH2B3) showed associations with lipids traits, and several known lipids loci showed association to PLT/MPV (e.g., FADS1, FADS2, APOH, and TMEM50A). Moreover, SH2B3, which is also expressed in human vascular endothelial cells where it modulates inflammation, has been associated with blood pressure and the risk of MI.69, 70, 71 Our study further suggests that a regulation of platelets could also contribute to potential implication of SH2B3 in the development of cardiovascular diseases. The associated SNVs in the FADS1/FADS2 locus (rs174546 and rs174583) are eQTLs for multiple lipid-related transcripts in blood-related tissues, including TMEM258, FADS1, FADS2, and LDLR (Table S21).62 Intriguingly, expression of TMEM258 has also been shown to be a transcriptional regulatory target of cardiovascular disease implicated CDKN2B-AS1 (MIM: 613149), a region marginally associated with PLT (discovery EA p = 1.00 × 10−6, replication EA p = 0.0577, combined EA p = 1.56 × 10−7) (Table S14).72, 73 Our genetic association results link the underlying genetic architecture of platelet and lipids traits as suggested by previous epidemiological, genetic, and animal studies.63, 74, 75, 76, 77 However, these observed shared genetic associations do not demonstrate whether these reflect direct genetic pleiotropy or indirect relationships. Several variants previously implicated in lipids (e.g., FADS1, FADS2, SH2B3, TMEM50A, and GCKR) have stronger associations with lipids traits relative to our platelet associations, suggesting that their primary effects are on lipids pathways (Table S20). Determining the directionality and causality among genetic variants, lipids, and platelets remains an important future step in dissecting which genetic variants may reveal new insights into platelet biology.
Conclusions
By performing a large meta-analysis of Exomechip association results, we identified rare, low-frequency, and common variants that influence PLT and MPV. Despite our ability to detect numerous associations with SNVs across a wide range of allele frequencies, the Exomechip interrogated a limited fraction of genomic variation. Sequencing-based studies across the genome in large sample sizes will be necessary to fully assess the contribution of variants across the allele frequency spectrum, particularly of rare variants in intergenic regions. Nonetheless, our results identify several intriguing genes and genetic mechanisms of platelet biology. Many of these associations overlapped with related blood cell and lipids traits, pointing to common mechanisms underlying their development and maintenance. Because blood cells share developmental lineages and several of our platelet-associated genes have known developmental or transcriptional regulatory functions, we hypothesize that the origins of these shared genetic associations are mainly in blood cell development in the bone marrow. How these genes function and interact in RBC, WBC, and platelet development will need to be tested in future experiments in both functional and human-based studies. Advances in these domains could provide key insights into genes that influence human blood disorders and reveal new mechanisms for the development of novel therapeutic applications.
Acknowledgments
We thank all participants and study coordinating centers. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute, the NIH, or the U.S. Department of Health and Human Services. The Framingham Heart Study (FHS) authors acknowledge that the computational work reported on in this paper was performed on the Shared Computing Cluster, which is administered by Boston University’s Research Computing Services. The MHI Biobank acknowledges the technical support of the Beaulieu-Saucier MHI Pharmacogenomic Center. We would like to thank Liling Warren for contributions to the genetic analysis of the SOLID-TIMI-52 and STABILITY datasets. The University Medicine Greifswald is a member of the Caché Campus program of the InterSystems GmbH. The SHIP and SHIP-TREND samples were genotyped at the Helmholtz Zentrum München. Estonian Genome Center, University of Tartu (EGCUT) would like to acknowledge Mr. V. Soo, Mr. S. Smith, and Dr. L. Milani. The Airwave Health Monitoring Study thanks Louisa Cavaliero who assisted in data collection and management as well as Peter McFarlane and the Glasgow CARE, Patricia Munroe at Queen Mary University of London, and Joanna Sarnecka and Ania Zawodniak at Northwick Park. FINCAVAS thanks the staff of the Department of Clinical Physiology for collecting the exercise test data. Young Finns Study (YFS) acknowledges the expert technical assistance in statistical analyses by Irina Lisinen.
Published: June 23, 2016
Footnotes
Supplemental Data include a note on eQTL analyses and additional funding information, 6 figures, and 23 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.05.005.
Web Resources
1000 Genomes, http://www.1000genomes.org
BCX ExomeChip association results, http://www.mhi-humangenetics.org/en/resources
CheckVCF, https://github.com/zhanxw/checkVCF
OMIM, http://www.omim.org/
RareMETALS, http://genome.sph.umich.edu/wiki/RareMETALS
RareMetalWorker, http://genome.sph.umich.edu/wiki/RAREMETALWORKER
Research Computing Services, http://www.bu.edu/tech/support/research/
Supplemental Data
References
- 1.Chu S.G., Becker R.C., Berger P.B., Bhatt D.L., Eikelboom J.W., Konkle B., Mohler E.R., Reilly M.P., Berger J.S. Mean platelet volume as a predictor of cardiovascular risk: a systematic review and meta-analysis. J. Thromb. Haemost. 2010;8:148–156. doi: 10.1111/j.1538-7836.2009.03584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutcliffe P., Connock M., Gurung T., Freeman K., Johnson S., Kandala N.B., Grove A., Gurung B., Morrow S., Clarke A. Aspirin for prophylactic use in the primary prevention of cardiovascular disease and cancer: a systematic review and overview of reviews. Health Technol. Assess. 2013;17:1–253. doi: 10.3310/hta17430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hennekens C.H., Dyken M.L., Fuster V. Aspirin as a therapeutic agent in cardiovascular disease: a statement for healthcare professionals from the American Heart Association. Circulation. 1997;96:2751–2753. doi: 10.1161/01.cir.96.8.2751. [DOI] [PubMed] [Google Scholar]
- 4.Schick U.M., Jain D., Hodonsky C.J., Morrison J.V., Davis J.P., Brown L., Sofer T., Conomos M.P., Schurmann C., McHugh C.P. Genome-wide association study of platelet count identifies ancestry-specific loci in Hispanic/Latino Americans. Am. J. Hum. Genet. 2016;98:229–242. doi: 10.1016/j.ajhg.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soranzo N., Rendon A., Gieger C., Jones C.I., Watkins N.A., Menzel S., Döring A., Stephens J., Prokisch H., Erber W. A novel variant on chromosome 7q22.3 associated with mean platelet volume, counts, and function. Blood. 2009;113:3831–3837. doi: 10.1182/blood-2008-10-184234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shameer K., Denny J.C., Ding K., Jouni H., Crosslin D.R., de Andrade M., Chute C.G., Peissig P., Pacheco J.A., Li R. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum. Genet. 2014;133:95–109. doi: 10.1007/s00439-013-1355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qayyum R., Snively B.M., Ziv E., Nalls M.A., Liu Y., Tang W., Yanek L.R., Lange L., Evans M.K., Ganesh S. A meta-analysis and genome-wide association study of platelet count and mean platelet volume in African Americans. PLoS Genet. 2012;8:e1002491. doi: 10.1371/journal.pgen.1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gieger C., Radhakrishnan A., Cvejic A., Tang W., Porcu E., Pistis G., Serbanovic-Canic J., Elling U., Goodall A.H., Labrune Y. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soranzo N., Spector T.D., Mangino M., Kühnel B., Rendon A., Teumer A., Willenborg C., Wright B., Chen L., Li M. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y.K., Oh J.H., Kim Y.J., Hwang M.Y., Moon S., Low S.K., Takahashi A., Matsuda K., Kubo M., Lee J., Kim B.J. Influence of genetic variants in EGF and other genes on hematological traits in Korean populations by a genome-wide approach. BioMed Res. Int. 2015;2015:914965. doi: 10.1155/2015/914965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh J.H., Kim Y.K., Moon S., Kim Y.J., Kim B.J. Genome-wide association study identifies candidate loci associated with platelet count in Koreans. Genomics Inform. 2014;12:225–230. doi: 10.5808/GI.2014.12.4.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., Glessner J.T., Zhang H., Hou C., Wei Z., Bradfield J.P., Mentch F.D., Guo Y., Kim C., Xia Q. GWAS of blood cell traits identifies novel associated loci and epistatic interactions in Caucasian and African-American children. Hum. Mol. Genet. 2013;22:1457–1464. doi: 10.1093/hmg/dds534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerrero J.A., Rivera J., Quiroga T., Martinez-Perez A., Antón A.I., Martínez C., Panes O., Vicente V., Mezzano D., Soria J.M., Corral J. Novel loci involved in platelet function and platelet count identified by a genome-wide study performed in children. Haematologica. 2011;96:1335–1343. doi: 10.3324/haematol.2011.042077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nürnberg S.T., Rendon A., Smethurst P.A., Paul D.S., Voss K., Thon J.N., Lloyd-Jones H., Sambrook J.G., Tijssen M.R., Italiano J.E., Jr., HaemGen Consortium A GWAS sequence variant for platelet volume marks an alternative DNM3 promoter in megakaryocytes near a MEIS1 binding site. Blood. 2012;120:4859–4868. doi: 10.1182/blood-2012-01-401893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson A.D. The genetics of common variation affecting platelet development, function and pharmaceutical targeting. J. Thromb. Haemost. 2011;9(Suppl 1):246–257. doi: 10.1111/j.1538-7836.2011.04359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auer P.L., Johnsen J.M., Johnson A.D., Logsdon B.A., Lange L.A., Nalls M.A., Zhang G., Franceschini N., Fox K., Lange E.M. Imputation of exome sequence variants into population- based samples and blood-cell-trait-associated loci in African Americans: NHLBI GO Exome Sequencing Project. Am. J. Hum. Genet. 2012;91:794–808. doi: 10.1016/j.ajhg.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auer P.L., Teumer A., Schick U., O’Shaughnessy A., Lo K.S., Chami N., Carlson C., de Denus S., Dubé M.P., Haessler J. Rare and low-frequency coding variants in CXCR2 and other genes are associated with hematological traits. Nat. Genet. 2014;46:629–634. doi: 10.1038/ng.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chami N., Chen M.-H., Slater A.J., Eicher J.D., Evangelou E., Tajuddin S.M., Love-Gregory L., Kacprowski T., Schick U.M., Nomura A. Exome genotyping identifies pleiotropic variants associated with red blood cell traits. Am. J. Hum. Genet. 2016;99:8–21. doi: 10.1016/j.ajhg.2016.05.007. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tajuddin S.M., Schick U.M., Eicher J.D., Chami N., Giri A., Brody J.A., Hill W.D., Kacprowski T., Li J., Lyytikäinen L.-P. Large-scale exome-wide association analysis identifies loci for white blood cell traits and pleiotropy with immune-mediated diseases. Am. J. Hum. Genet. 2016;99:22–39. doi: 10.1016/j.ajhg.2016.05.003. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grove M.L., Yu B., Cochran B.J., Haritunians T., Bis J.C., Taylor K.D., Hansen M., Borecki I.B., Cupples L.A., Fornage M. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS ONE. 2013;8:e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler T.W., Day F.R., Croteau-Chonka D.C., Wood A.R., Locke A.E., Mägi R., Ferreira T., Fall T., Graff M., Justice A.E., Genetic Investigation of Anthropometric Traits (GIANT) Consortium Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu D.J., Peloso G.M., Zhan X., Holmen O.L., Zawistowski M., Feng S., Nikpay M., Auer P.L., Goel A., Zhang H. Meta-analysis of gene-level tests for rare variant association. Nat. Genet. 2014;46:200–204. doi: 10.1038/ng.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price A.L., Kryukov G.V., de Bakker P.I., Purcell S.M., Staples J., Wei L.J., Sunyaev S.R. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson A.D., Yanek L.R., Chen M.H., Faraday N., Larson M.G., Tofler G., Lin S.J., Kraja A.T., Province M.A., Yang Q. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat. Genet. 2010;42:608–613. doi: 10.1038/ng.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N. Engl. J. Med. 2016;374:1898. doi: 10.1056/NEJMx160012. [DOI] [PubMed] [Google Scholar]
- 30.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S., Global Lipids Genetics Consortium Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., Gierman H.J., Levy D., Plump A., Dobrin R., Goring H.H., Curran J.E., Johnson M.P., Blangero J., Kim S.K. Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs. BMC Genomics. 2014;15:532. doi: 10.1186/1471-2164-15-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eicher J.D., Wakabayashi Y., Vitseva O., Esa N., Yang Y., Zhu J., Freedman J.E., McManus D.D., Johnson A.D. Characterization of the platelet transcriptome by RNA sequencing in patients with acute myocardial infarction. Platelets. 2016;27:230–239. doi: 10.3109/09537104.2015.1083543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fehrmann R.S., Jansen R.C., Veldink J.H., Westra H.J., Arends D., Bonder M.J., Fu J., Deelen P., Groen H.J., Smolonska A. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandomenico A., Monti S.M., Marasco D., Dathan N., Palumbo R., Saviano M., Ruvo M. IgE-binding properties and selectivity of peptide mimics of the FcvarepsilonRI binding site. Mol. Immunol. 2009;46:3300–3309. doi: 10.1016/j.molimm.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 35.Mackay G.A., Hulett M.D., Cook J.P., Trist H.M., Henry A.J., McDonnell J.M., Beavil A.J., Beavil R.L., Sutton B.J., Hogarth P.M., Gould H.J. Mutagenesis within human FcepsilonRIalpha differentially affects human and murine IgE binding. J. Immunol. 2002;168:1787–1795. doi: 10.4049/jimmunol.168.4.1787. [DOI] [PubMed] [Google Scholar]
- 36.Cook J.P., Henry A.J., McDonnell J.M., Owens R.J., Sutton B.J., Gould H.J. Identification of contact residues in the IgE binding site of human FcepsilonRIalpha. Biochemistry. 1997;36:15579–15588. doi: 10.1021/bi9713005. [DOI] [PubMed] [Google Scholar]
- 37.Garman S.C., Kinet J.P., Jardetzky T.S. The crystal structure of the human high-affinity IgE receptor (Fc epsilon RI alpha) Annu. Rev. Immunol. 1999;17:973–976. doi: 10.1146/annurev.immunol.17.1.973. [DOI] [PubMed] [Google Scholar]
- 38.Granada M., Wilk J.B., Tuzova M., Strachan D.P., Weidinger S., Albrecht E., Gieger C., Heinrich J., Himes B.E., Hunninghake G.M. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J. Allergy Clin. Immunol. 2012;129:840–845.e21. doi: 10.1016/j.jaci.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiner A.P., Lettre G., Nalls M.A., Ganesh S.K., Mathias R., Austin M.A., Dean E., Arepalli S., Britton A., Chen Z. Genome-wide association study of white blood cell count in 16,388 African Americans: the continental origins and genetic epidemiology network (COGENT) PLoS Genet. 2011;7:e1002108. doi: 10.1371/journal.pgen.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page C., Pitchford S. Platelets and allergic inflammation. Clin. Exp. Allergy. 2014;44:901–913. doi: 10.1111/cea.12322. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt V.A., Scudder L., Devoe C.E., Bernards A., Cupit L.D., Bahou W.F. IQGAP2 functions as a GTP-dependent effector protein in thrombin-induced platelet cytoskeletal reorganization. Blood. 2003;101:3021–3028. doi: 10.1182/blood-2002-09-2807. [DOI] [PubMed] [Google Scholar]
- 42.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nurden A.T., Pillois X., Fiore M., Heilig R., Nurden P. Glanzmann thrombasthenia-like syndromes associated with macrothrombocytopenias and mutations in the genes encoding the αIIbβ3 integrin. Semin. Thromb. Hemost. 2011;37:698–706. doi: 10.1055/s-0031-1291380. [DOI] [PubMed] [Google Scholar]
- 44.Obinata D., Takayama K., Urano T., Murata T., Ikeda K., Horie-Inoue K., Ouchi Y., Takahashi S., Inoue S. ARFGAP3, an androgen target gene, promotes prostate cancer cell proliferation and migration. Int. J. Cancer. 2012;130:2240–2248. doi: 10.1002/ijc.26224. [DOI] [PubMed] [Google Scholar]
- 45.Kartberg F., Asp L., Dejgaard S.Y., Smedh M., Fernandez-Rodriguez J., Nilsson T., Presley J.F. ARFGAP2 and ARFGAP3 are essential for COPI coat assembly on the Golgi membrane of living cells. J. Biol. Chem. 2010;285:36709–36720. doi: 10.1074/jbc.M110.180380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weimer C., Beck R., Eckert P., Reckmann I., Moelleken J., Brügger B., Wieland F. Differential roles of ArfGAP1, ArfGAP2, and ArfGAP3 in COPI trafficking. J. Cell Biol. 2008;183:725–735. doi: 10.1083/jcb.200806140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Begonja A.J., Pluthero F.G., Suphamungmee W., Giannini S., Christensen H., Leung R., Lo R.W., Nakamura F., Lehman W., Plomann M. FlnA binding to PACSIN2 F-BAR domain regulates membrane tubulation in megakaryocytes and platelets. Blood. 2015;126:80–88. doi: 10.1182/blood-2014-07-587600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li T., Shi Y., Wang P., Guachalla L.M., Sun B., Joerss T., Chen Y.S., Groth M., Krueger A., Platzer M. Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J. 2015;34:1630–1647. doi: 10.15252/embj.201489947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butler M., Morel A.S., Jordan W.J., Eren E., Hue S., Shrimpton R.E., Ritter M.A. Altered expression and endocytic function of CD205 in human dendritic cells, and detection of a CD205-DCL-1 fusion protein upon dendritic cell maturation. Immunology. 2007;120:362–371. doi: 10.1111/j.1365-2567.2006.02512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao L., Shi X., Chang H., Zhang Q., He Y. pH-dependent recognition of apoptotic and necrotic cells by the human dendritic cell receptor DEC205. Proc. Natl. Acad. Sci. USA. 2015;112:7237–7242. doi: 10.1073/pnas.1505924112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karpatkin S. Heterogeneity of human platelets. VI. Correlation of platelet function with platelet volume. Blood. 1978;51:307–316. [PubMed] [Google Scholar]
- 52.Catani L., Sollazzo D., Ricci F., Polverelli N., Palandri F., Baccarani M., Vianelli N., Lemoli R.M. The CD47 pathway is deregulated in human immune thrombocytopenia. Exp. Hematol. 2011;39:486–494. doi: 10.1016/j.exphem.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 53.Olsson M., Bruhns P., Frazier W.A., Ravetch J.V., Oldenborg P.A. Platelet homeostasis is regulated by platelet expression of CD47 under normal conditions and in passive immune thrombocytopenia. Blood. 2005;105:3577–3582. doi: 10.1182/blood-2004-08-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamao T., Noguchi T., Takeuchi O., Nishiyama U., Morita H., Hagiwara T., Akahori H., Kato T., Inagaki K., Okazawa H. Negative regulation of platelet clearance and of the macrophage phagocytic response by the transmembrane glycoprotein SHPS-1. J. Biol. Chem. 2002;277:39833–39839. doi: 10.1074/jbc.M203287200. [DOI] [PubMed] [Google Scholar]
- 55.Qayyum R., Becker L.C., Becker D.M., Faraday N., Yanek L.R., Leal S.M., Shaw C., Mathias R., Suktitipat B., Bray P.F. Genome-wide association study of platelet aggregation in African Americans. BMC Genet. 2015;16:58. doi: 10.1186/s12863-015-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lewis J.P., Ryan K., O’Connell J.R., Horenstein R.B., Damcott C.M., Gibson Q., Pollin T.I., Mitchell B.D., Beitelshees A.L., Pakzy R. Genetic variation in PEAR1 is associated with platelet aggregation and cardiovascular outcomes. Circ Cardiovasc Genet. 2013;6:184–192. doi: 10.1161/CIRCGENETICS.111.964627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eicher J.D., Xue L., Ben-Shlomo Y., Beswick A.D., Johnson A.D. Replication and hematological characterization of human platelet reactivity genetic associations in men from the Caerphilly Prospective Study (CaPS) J. Thromb. Thrombolysis. 2016;41:343–350. doi: 10.1007/s11239-015-1290-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stone J.C. Regulation and function of the RasGRP family of Ras activators in blood cells. Genes Cancer. 2011;2:320–334. doi: 10.1177/1947601911408082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Loo B., Martin J.F. A role for changes in platelet production in the cause of acute coronary syndromes. Arterioscler. Thromb. Vasc. Biol. 1999;19:672–679. doi: 10.1161/01.atv.19.3.672. [DOI] [PubMed] [Google Scholar]
- 60.Kauskot A., Vandenbriele C., Louwette S., Gijsbers R., Tousseyn T., Freson K., Verhamme P., Hoylaerts M.F. PEAR1 attenuates megakaryopoiesis via control of the PI3K/PTEN pathway. Blood. 2013;121:5208–5217. doi: 10.1182/blood-2012-10-462887. [DOI] [PubMed] [Google Scholar]
- 61.Bertin A., Mahaney M.C., Cox L.A., Rogers J., VandeBerg J.L., Brugnara C., Platt O.S. Quantitative trait loci for peripheral blood cell counts: a study in baboons. Mamm. Genome. 2007;18:361–372. doi: 10.1007/s00335-007-9022-8. [DOI] [PubMed] [Google Scholar]
- 62.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sloan A., Gona P., Johnson A.D. Cardiovascular correlates of platelet count and volume in the Framingham Heart Study. Ann. Epidemiol. 2015;25:492–498. doi: 10.1016/j.annepidem.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Panova-Noeva M., Schulz A., Hermanns M.I., Grossmann V., Pefani E., Spronk H.M., Laubert-Reh D., Binder H., Beutel M., Pfeiffer N. Sex-specific differences in genetic and nongenetic determinants of mean platelet volume: results from the Gutenberg Health Study. Blood. 2016;127:251–259. doi: 10.1182/blood-2015-07-660308. [DOI] [PubMed] [Google Scholar]
- 65.Daly M.E. Determinants of platelet count in humans. Haematologica. 2011;96:10–13. doi: 10.3324/haematol.2010.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno-Ayala R., Schnabel D., Salas-Vidal E., Lomelí H. PIAS-like protein Zimp7 is required for the restriction of the zebrafish organizer and mesoderm development. Dev. Biol. 2015;403:89–100. doi: 10.1016/j.ydbio.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 67.Peng Y., Lee J., Zhu C., Sun Z. A novel role for protein inhibitor of activated STAT (PIAS) proteins in modulating the activity of Zimp7, a novel PIAS-like protein, in androgen receptor-mediated transcription. J. Biol. Chem. 2010;285:11465–11475. doi: 10.1074/jbc.M109.079327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Y., Lee J.W., Ackerman S.L. Mutations in the microtubule-associated protein 1A (Map1a) gene cause Purkinje cell degeneration. J. Neurosci. 2015;35:4587–4598. doi: 10.1523/JNEUROSCI.2757-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ganesh S.K., Tragante V., Guo W., Guo Y., Lanktree M.B., Smith E.N., Johnson T., Castillo B.A., Barnard J., Baumert J., CARDIOGRAM, METASTROKE. LifeLines Cohort Study Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum. Mol. Genet. 2013;22:1663–1678. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Newton-Cheh C., Johnson T., Gateva V., Tobin M.D., Bochud M., Coin L., Najjar S.S., Zhao J.H., Heath S.C., Eyheramendy S., Wellcome Trust Case Control Consortium Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gudbjartsson D.F., Bjornsdottir U.S., Halapi E., Helgadottir A., Sulem P., Jonsdottir G.M., Thorleifsson G., Helgadottir H., Steinthorsdottir V., Stefansson H. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 72.Bochenek G., Häsler R., El Mokhtari N.E., König I.R., Loos B.G., Jepsen S., Rosenstiel P., Schreiber S., Schaefer A.S. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum. Mol. Genet. 2013;22:4516–4527. doi: 10.1093/hmg/ddt299. [DOI] [PubMed] [Google Scholar]
- 73.Deloukas P., Kanoni S., Willenborg C., Farrall M., Assimes T.L., Thompson J.R., Ingelsson E., Saleheen D., Erdmann J., Goldstein B.A., CARDIoGRAMplusC4D Consortium. DIAGRAM Consortium. CARDIOGENICS Consortium. MuTHER Consortium. Wellcome Trust Case Control Consortium Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gomes A.L., Carvalho T., Serpa J., Torre C., Dias S. Hypercholesterolemia promotes bone marrow cell mobilization by perturbing the SDF-1:CXCR4 axis. Blood. 2010;115:3886–3894. doi: 10.1182/blood-2009-08-240580. [DOI] [PubMed] [Google Scholar]
- 75.Su Y., Wang Z., Yang H., Cao L., Liu F., Bai X., Ruan C. Clinical and molecular genetic analysis of a family with sitosterolemia and co-existing erythrocyte and platelet abnormalities. Haematologica. 2006;91:1392–1395. [PubMed] [Google Scholar]
- 76.Wang Z., Cao L., Su Y., Wang G., Wang R., Yu Z., Bai X., Ruan C. Specific macrothrombocytopenia/hemolytic anemia associated with sitosterolemia. Am. J. Hematol. 2014;89:320–324. doi: 10.1002/ajh.23619. [DOI] [PubMed] [Google Scholar]
- 77.Murphy A.J., Bijl N., Yvan-Charvet L., Welch C.B., Bhagwat N., Reheman A., Wang Y., Shaw J.A., Levine R.L., Ni H. Cholesterol efflux in megakaryocyte progenitors suppresses platelet production and thrombocytosis. Nat. Med. 2013;19:586–594. doi: 10.1038/nm.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.