Abstract
The sequence kernel association test (SKAT) is probably the most popular statistical test used in rare-variant association studies. Its null distribution involves unknown parameters that need to be estimated. The current estimation method has a valid type I error rate, but the power is compromised given that all subjects are used for estimation. I have developed an estimation method that uses only control subjects. Named SKAT+, this method uses the same test statistic as SKAT but differs in the way the null distribution is estimated. Extensive simulation studies and applications to data from the Genetic Analysis Workshop 17 and the Ocular Hypertension Treatment Study demonstrated that SKAT+ has superior power over SKAT while maintaining control over the type I error rate. This method is applicable to extensions of SKAT in the literature.
Keywords: SKAT, sequence data, rare variant, association test, power
Introduction
Since the proliferation of DNA microarray technology about a decade ago, genome-wide association studies (GWASs) have successfully discovered many genetic variants associated with numerous human diseases and traits.1 However, these identified variants explain only a small fraction of the overall heritability for most complex traits.2, 3, 4, 5 Because DNA microarray technology targets only common SNPs, it is possible that the “missing heritability” is due to variants that are rare. The rapid development of next-generation sequencing technology provides a great opportunity for studying rare variants.
Because a single SNP can provide only limited power, it is important to simultaneously leverage multiple SNPs in a gene or a region in rare-variant association studies. One approach is to test for the cumulative effect of rare variants.6, 7, 8, 9, 10 Such burden tests are most powerful when the effects of all variants are in the same direction. When some variants are protective but others are deleterious, this approach is no longer optimal.
An alternative approach is to test for the cumulative quadratic effect of rare variants. Examples include the C-alpha test11 and the sequence kernel association test (SKAT).12 SKAT is a score-based variance-component test and is computationally efficient. It includes the C-alpha test as a special case. It has a solid theoretical foundation.13, 14 It can handle both dichotomous traits and continuous traits and is able to control for covariates. SKAT has been generalized in many ways for the purposes of incorporating burden tests,15 conducting meta-analysis,16 analyzing extreme continuous traits17 and survival outcomes,18, 19, 20 performing family-based association tests,21 and studying gene-gene and gene-environmental interactions.22, 23
One advantage of SKAT is that the form of its limiting distribution is known. It is a linear combination of a certain number of independent and identically distributed random variables, each of which follows a chi-square distribution with 1 degree of freedom (df). Although these combination coefficients are unknown, once they are estimated, one is able to compute the theoretical p value right away24, 25 without using computation-intensive techniques such as permutation. The focus of this report concerns ways to estimate these coefficients.
The current practice used by SKAT is to use all subjects in estimating these coefficients. However, as I will show, although the type I error rate is maintained, the power of SKAT is compromised in such a practice. Recognizing this fact, I propose a general approach to estimating these coefficients by using only control subjects. I demonstrate how this approach can be applied to dichotomous traits with and without covariates, as well as continuous traits. Its performance is assessed by extensive simulation studies. I further illustrate the utility of this method by applying it to data from the Genetic Analysis Workshop 17 (GAW17)26 and the Ocular Hypertension Treatment Study (OHTS).27
Material and Methods
Let m be the number of SNPs in a candidate region or a pathway. The genotype score of subject i at SNP j is denoted by gij, which takes value 0, 1, or 2, the number of copies of the minor allele. The genotypes of all subjects can be summarized by an n × m matrix G = (gij), where n is the number of subjects.
Case-Control Data with No Covariates
Let n1 be the number of case subjects and n0 be the number of control subjects. The total number of subjects is n = n1 + n0. Phenotypes are represented by an n × 1 vector y, whose elements are equal to 1 for case subjects and 0 for control subjects. Define as the proportion of case subjects. The most popular version of the SKAT statistic is the one with linear kernel12 and has the following form:
When none of the m SNPs is associated with the case-control status (i.e., the null hypothesis), the distribution of SKAT can be approximated by that of , where are eigenvalues of the n × n matrix , which has , an n × n matrix, and as independent 1-df chi-square-distributed random variables.12 Here, I is an n × n identity matrix, and 1 is an n × 1 vector of values of 1. The rank of matrix is min {m, n}. Because the non-zero eigenvalues are the same as the non-zero eigenvalues from GtPG, the distribution of is completely determined by the eigenvalues of GtPG. Note that the dimension of GtPG, which is m × m, is smaller than that of in the usual case of m < n. It is computationally more convenient to use GtPG.
Define the sample variance matrix of G as S. We have
That is, . The matrix S is an estimate of the underlying variance matrix of m SNPs when the null hypothesis of no association is true. When none of the SNPs is associated with the phenotype (the null hypothesis), it makes sense to use all subjects to compute S for the null distribution because none of the subjects carries a variant, and all of them are expected to share a common variance of genotype scores. However, when some SNPs are associated with the phenotype, using all subjects is no longer appropriate because case and control subjects are expected to have different mean SNP scores at these SNPs. Their variance matrices of the genotype scores are expected to be different as well. In this situation, a reasonable estimation of the null variance matrix of the m SNPs is the sample variance matrix of the genotypes scores for the control subjects.
I propose the following method for approximating the null distribution of SKAT. Let S0 be the sample variance matrix of the genotype scores on the m SNPs among the n0 control subjects:
where G0 is an n0 × m matrix of genotype scores of control subjects, I0 is an n0 × n0 identity matrix, and 10 is an n0 × 1 vector of values of 1. Let be the eigenvalues of matrix . The null distribution of the SKAT statistic is approximated by provided that n0 > m. I call this method SKAT+.
Apparently, when none of the SNPs is associated with the phenotype, S0 and S converge to the same variance matrix as n0 and n get large. That is, using either S0 or S will give a valid type I error rate. However, when the null hypothesis is not true, S0 and S no longer converge to the same variance matrix. The trace of the limiting matrix of S is larger than that of S0. Using S instead of S0 tends to get a distribution with larger variance and a larger p value and hence a less powerful testing procedure.
Case-Control Data with Covariates
Let xi be a p × 1 vector of the values of p covariates for subject i. To remove their confounding effect, the following logistic regression is fit first:
Let be the estimated probability of yi = 1:
where and are estimates of and , respectively. Let . The SKAT12 statistic is now defined by
When the sample size is large enough, the distribution of SKAT can be approximated by that of , where are eigenvalues of or, equivalently, those of . Here,
which has , an n × (p + 1) matrix, and
Define as
Σ is the variance matrix of the residuals of the projection of on the linear space spanned by , where . Following the same idea as in the previous subsection, we substitute Σ with its counterpart computed with subjects whose value of is low. One decision we have to make is the threshold for selection. For instance, we can use those subjects whose value is below the median or third quartile of . Another choice is to use control subjects only. This is equivalent to using 0 as the threshold given that all control subjects satisfy .
Let n0 be the number of such chosen subjects. Let G0, V0, and denote the sub-matrices of G, V, and , respectively, corresponding to the selected subjects. Let P0 and Σ0 be defined as
and
respectively. Let be the eigenvalues of matrix (n − p − 1)Σ0. Because GtPG can be expressed as , the null distribution of the SKAT statistic is approximated by when n0 > m. This is because when the null is true, the residuals are independent of the covariate-corrected genotypes. Hence, Σ0 and Σ converge to the same matrix as and n get large.
When there are no covariates, we have and , where . This method reduces to the method proposed in the previous subsection.
Continuous Traits
First, we fit the following null linear regression that has covariates only (that is, no genotype scores are used):
where . Let denote the predicted value of and . The estimates of , , and are denoted by , , and , respectively. The SKAT statistic with linear kernel is defined as12
Its asymptotic distribution is , where are the eigenvalues of GtPG, in which and .
We write the matrix GtPG as , where
denotes the variance matrix of the genotype scores after the effects of the covariates are removed. Assuming that higher phenotypic value is positively associated with disease severity, we can choose subjects whose residual is low. Denote the matrices of the genotypes and the covariates of these selected subjects as G0 and X0, respectively. Define . Furthermore, denote their covariate-corrected genotype-score variance matrix as S0. The asymptotic distribution of SKAT is then estimated by , where are the eigenvalues of . S0 and S converge to the same matrix under the null given that the residuals are independent of the genotypes, and selection based on doesn’t bias S0 as an estimate of the underlying genotype covariance matrix.
A Resampling Procedure
To obtain a resampling p value, we can use the following procedure. Given the n0 selected subjects and their genotype-score matrix G0, we randomly select n0 phenotype residuals (or for continuous traits). Let denote the vector of selected phenotype residuals. A resampling version of the SKAT statistic is computed as
Repeat this process the desired number of times. The resampling p value is computed as the proportion of the resampled SKAT statistic that is equal to or larger than the observed SKAT statistic.
Results
Simulation Studies
I simulated dichotomous traits and continuous traits under various configurations in order to evaluate the performance of the proposed SKAT+ method. I calculated both the p value based on the asymptotic distribution and the resampling p value based on 1,000 resamples. I compared SKAT+ with SKAT, SKAT-O,28 and aSPU,29 a recent competitor to SKAT. The aSPU method depends on the best p value of a number of sums of powered score statistics at each SNP. For the SKAT method, I used the R package SKAT (version 1.1.2). Small-sample adjustment was turned off, and no weights were applied to SNPs, although such tricks can be used in SKAT+ as well. For the aSPU method, I downloaded (on November 30, 2015) the permutation-based aSPU R code aSPUperm.R from Dr. Wei Pan’s website. This aSPU R code works only for dichotomous traits, although in theory it works for continuous traits as well. The number of permutations for aSPU is fixed at 1,000. Under each configuration, the number of simulation replications for all methods is fixed at 1,000. Rejection rates at significance levels 0.05 and 0.01 are reported unless otherwise noted.
For the assessment of type I error rate and power for dichotomous traits, I used an R code used previously30 and many other publications coauthored by Dr. Wei Pan. This R code (simRareSNP.R) was downloaded (on November 30, 2015) from Dr. Wei Pan’s website. Specifically, there are eight disease-susceptibility variants whose odds ratios for each additional copy of the minor allele are pre-specified. For instance, an odds ratio equal to 1 for each of these variants implies that none of them is associated with the simulated trait. So, this is a setup used for the study of type I error rate. The disease probability can be expressed via the following logistic-regression model:
where corresponds to a background disease probability of 0.05, and are the odds ratios for the eight disease-susceptibility variants, respectively. In addition, a varying number of neutral rare variants whose minor allele frequencies (MAFs) were randomly chosen from a uniform distribution on interval (0.001, 0.01) were simulated. All variants were in linkage equilibrium. A total of 500 case and 500 control subjects were simulated. A similar simulation setup has also been used elsewhere.12, 31 Only control subjects were used for the SKAT+ method. Simulated type I error rates with odds ratios equal to 1 for each of the eight disease-susceptibility variants are reported in the first half of Table 1. They are under control for all four statistics except that the resampling p values for SKAT+ are slightly liberal in some situations.
Table 1.
Simulated Type I Error Rate for Dichotomous Traits over 1,000 Replicates
| Method |
α = 0.05 |
α = 0.01 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
No. of Neutral Rare Variants |
No. of Neutral Rare Variants |
|||||||||
| 0 | 4 | 8 | 16 | 32 | 0 | 4 | 8 | 16 | 32 | |
| Without Covariates | ||||||||||
| SKAT+ | 0.046 | 0.040 | 0.047 | 0.043 | 0.040 | 0.007 | 0.005 | 0.013 | 0.005 | 0.006 |
| SKAT+ (rs) | 0.063 | 0.046 | 0.059 | 0.062 | 0.049 | 0.017 | 0.012 | 0.017 | 0.014 | 0.013 |
| SKAT | 0.050 | 0.036 | 0.049 | 0.049 | 0.040 | 0.007 | 0.006 | 0.011 | 0.004 | 0.006 |
| SKAT-O | 0.041 | 0.041 | 0.052 | 0.052 | 0.045 | 0.009 | 0.008 | 0.010 | 0.004 | 0.005 |
| aSPU | 0.037 | 0.037 | 0.047 | 0.048 | 0.041 | 0.005 | 0.007 | 0.009 | 0.005 | 0.005 |
| With Covariates | ||||||||||
| SKAT+ | 0.050 | 0.042 | 0.038 | 0.051 | 0.039 | 0.004 | 0.011 | 0.002 | 0.012 | 0.004 |
| SKAT+ (rs) | 0.062 | 0.047 | 0.049 | 0.066 | 0.050 | 0.012 | 0.016 | 0.010 | 0.017 | 0.008 |
| SKAT | 0.042 | 0.039 | 0.044 | 0.049 | 0.045 | 0.006 | 0.008 | 0.003 | 0.013 | 0.008 |
| SKAT-O | 0.055 | 0.046 | 0.054 | 0.057 | 0.051 | 0.006 | 0.013 | 0.005 | 0.012 | 0.008 |
| aSPU | 0.045 | 0.046 | 0.049 | 0.053 | 0.051 | 0.005 | 0.011 | 0.007 | 0.016 | 0.011 |
SKAT+ (rs) represents the SKAT+ method based on resampling p values.
I also considered the case where there are two covariates, X1 and X2. The disease probability is determined by
where X1 follows a standard normal distribution, and X2 follows a binary distribution taking value 0 or 1 with probability 0.5. This way of adding covariates has been used elsewhere.12 To accommodate covariates X1 and X2, I modified Dr. Wei Pan’s R code accordingly. Subjects whose values were not higher than the third quartile of were selected for the SKAT+ method. Simulated type I error rates are reported in the second half of Table 1. Again, they are under control except that the resampling p values for SKAT+ are slightly liberal in some situations.
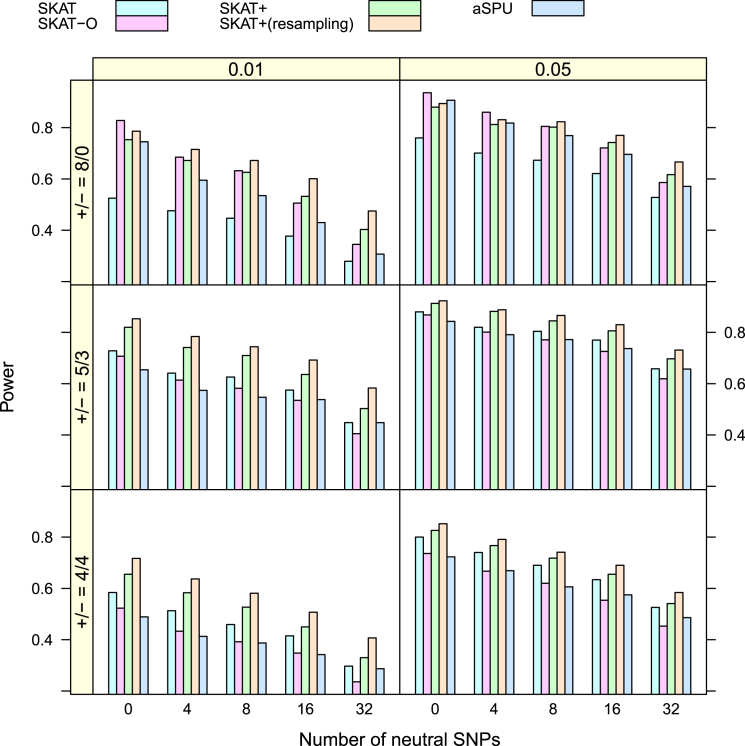
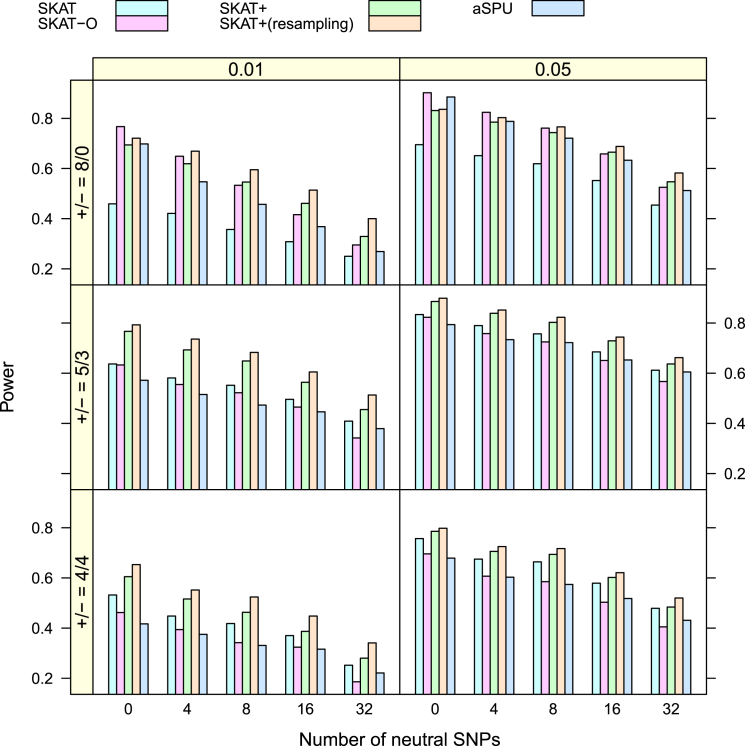
For the power analysis, I considered three scenarios in terms of the odds ratios of the eight disease-causing SNPs. They were (1) ORs = (2, 2, 2, 2, 2, 2, 2, 2), (2) ORs = (3, 3, 2, 2, 2, 1/2, 1/2, 1/2), and (3) ORs = (3, 1/3, 2, 2, 2, 1/2, 1/2, 1/2). These are typical scenarios used in the literature.29, 30 Scenario 1 mimics the situation where all causal variants work in the same direction, whereas the other two allow their directions to be different with different deleterious/protective ratios. The power is presented in Figure 1 and Figure 2 for situations without and with covariates, respectively. The SKAT+ method performed apparently better—in some cases much better—than SKAT across the board. It also performed better than SKAT-O and aSPU in almost all cases even when ORs = (2, 2, 2, 2, 2, 2, 2, 2), a case in favor of SKAT-O and aSPU because this is a situation for which they are optimized.28, 30
Figure 1.
Simulated Power of Dichotomous Traits over 1,000 Replicates
There are no covariates. Only control subjects were selected for SKAT+.
Figure 2.
Simulated Power of Dichotomous Traits over 1,000 Replicates
There are two covariates. The third quartile of was used for selecting subjects for SKAT+.
Continuous traits were simulated according to the following model:12
| (Equation 1) |
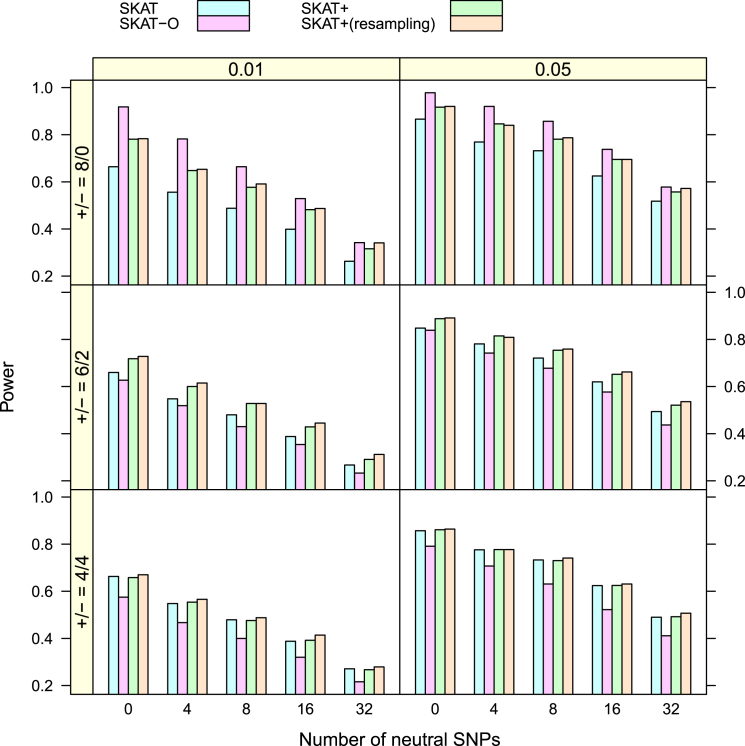
where X1 and X2 were generated in the same way as in the case of dichotomous traits, and g1, …, g8 were genotype scores at eight causal rare variants. The MAFs of these causal variants were selected uniformly from the interval (0.001, 0.01). For the study of type I error rate, were set at 0. For the study of power, the magnitude of was equal to , and the signs of these β values were in one of the following three configurations: (1) signs = (+, +, +, +, +, +, +, +), (2) signs = (+, +, +, +, +, +, −, −), or (3) signs = (+, +, +, +, −, −, −, −). Additional neutral SNPs were simulated with MAFs uniformly chosen from the interval (0.001, 0.01). Sample size was 1,000, and subjects whose values were below the third quartile of the residuals were selected for SKAT+. That is, 75% were selected. The type I error rates are presented in Table 2. All are in line with the nominal levels. The simulated power is presented in Figure 3. SKAT+ performed better than SKAT when the ratio of positive β values to negative β values was 8:0 (configuration 1) or 6:2 (configuration 2). When this ratio was 4:4 (configuration 3), the performance of SKAT+ was almost identical to that of SKAT. This is because at this ratio, the simulated trait according to model 1 is symmetric with respect to 0. After the effect of covariates is removed, the sample covariance matrix of the selected subjects is expected to be the same as that of the whole sample. Compared with SKAT-O, SKAT+ performed worse only in configuration 1, a situation optimized for SKAT-O. In other situations, SKAT-O performed even worse than SKAT.
Table 2.
Simulated Type I Error Rate for Continuous Traits over 1,000 Replicates
| Method |
α = 0.05 |
α = 0.01 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
No. of Neutral Rare Variants |
No. of Neutral Rare Variants |
|||||||||
| 0 | 4 | 8 | 16 | 32 | 0 | 4 | 8 | 16 | 32 | |
| SKAT+ | 0.054 | 0.055 | 0.052 | 0.042 | 0.048 | 0.009 | 0.010 | 0.007 | 0.011 | 0.006 |
| SKAT+ (rs) | 0.051 | 0.058 | 0.051 | 0.040 | 0.056 | 0.011 | 0.011 | 0.008 | 0.013 | 0.010 |
| SKAT | 0.048 | 0.048 | 0.045 | 0.036 | 0.043 | 0.008 | 0.006 | 0.005 | 0.009 | 0.006 |
| SKAT-O | 0.039 | 0.049 | 0.037 | 0.038 | 0.043 | 0.008 | 0.008 | 0.005 | 0.009 | 0.006 |
Subjects whose values were among the lowest 75% were selected for the SKAT+ method. SKAT+ (rs) represents the SKAT+ method based on resampling p values. The aSPU method was not computed because the R code from its authors doesn’t handle continuous traits.
Figure 3.
Simulated Power of Continuous Traits over 1,000 Replicates
There are two covariates. The third quartile of was used for selecting subjects for SKAT+.
Application to GAW17 Data
GAW17 provided 200 phenotype datasets26 simulated with mini-exome genotype data on 697 subjects selected from the 1000 Genomes Project. There were 24,487 SNPs in 3,205 genes. Four phenotypes were simulated on the basis of the genotypes of these subjects. Three of them (denoted by Q1, Q2, and Q4) were quantitative, and one was dichotomous such that the affection status was determined by Q1, Q2, and Q4 and a latent liability through a liability threshold model. Q1 was influenced by 39 SNPs in 9 genes. Q2 was influenced by 72 SNPs in 13 genes. Q4 was not influenced by any of the genotyped SNPs. The latent liability was influenced by 51 SNPs in 15 genes. As a result, the dichotomous trait was influenced by 162 genotyped SNPs in 36 genes (Q1 and the latent liability shared one common gene). Age, gender, and smoking status were confounders to Q1, Q4, and the latent liability, but not to Q2. I compared the proportion of times SKAT+, SKAT, and SKAT-O were significant at level 0.05 at each causal gene across 200 simulated datasets. I focused on the continuous traits Q1, Q2, and Q4. The following linear null model was considered:
Note that covariates were controlled for although they were known to have no effect on Q2. Q4 was used here for confirming that the type I error rate of SKAT+ was under control. The effects of the genotyped causal SNPs were all in the same direction of increasing the continuous traits. Only SNPs whose MAFs were no larger than 1% were used. Genes with only one genotyped SNP were excluded. Because the majority of the MAFs at the causal genes were extremely rare at a magnitude of 1 × 10−4, the 95th percentile of the residuals was used for selecting subjects for SKAT+. The proportion of rejecting the null hypothesis at level 0.05 out of the 200 simulated replicates is presented in Table 3. Given that the power was not high for most causal genes even at level 5% for all methods, only those genes at which the maximum power of the three statistics was greater than 10% are presented. It is clear that SKAT+ has higher power than SKAT. Its power is less than SKAT-O in most situations. This is very possibly because the direction of the causal variants in these genes is simulated to be the same, a situation SKAT-O is optimized for. The results for trait Q4 suggest that the type I error rates for all three methods are under control, especially given that there were only 200 replicates.
Table 3.
Proportion of Significant Test Results at Level 0.05 out of the 200 Simulated GAW17 Datasets
| Gene |
Q1 |
Q2 |
Q4 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| SKAT+ | SKAT | SKAT-O | SKAT+ | SKAT | SKAT-O | SKAT+ | SKAT | SKAT-O | |
| FLT1 (MIM: 165070) | 1.000 | 1.000 | 1.000 | – | – | – | 0.045 | 0.045 | 0.035 |
| FLT4 (MIM: 136352) | 0.500 | 0.430 | 0.570 | – | – | – | 0.055 | 0.065 | 0.045 |
| HIF1A (MIM: 603348) | 0.110 | 0.100 | 0.195 | – | – | – | 0.070 | 0.060 | 0.065 |
| KDR (MIM: 191306) | 0.995 | 0.995 | 1.000 | – | – | – | 0.075 | 0.065 | 0.075 |
| BCHE (MIM: 177400) | – | – | – | 0.400 | 0.340 | 0.410 | 0.050 | 0.040 | 0.055 |
| PDGFD (MIM: 609673) | – | – | – | 0.710 | 0.680 | 0.715 | 0.035 | 0.025 | 0.035 |
| RARB (MIM: 180220) | – | – | – | 0.440 | 0.420 | 0.345 | 0.065 | 0.055 | 0.050 |
| SIRT1 (MIM: 604479) | – | – | – | 0.625 | 0.600 | 0.640 | 0.080 | 0.055 | 0.075 |
| SREBF1 (MIM: 184756) | – | – | – | 0.425 | 0.375 | 0.670 | 0.065 | 0.065 | 0.075 |
| VLDLR (MIM: 192977) | – | – | – | 0.185 | 0.175 | 0.180 | 0.035 | 0.030 | 0.040 |
| VNN1 (MIM: 603570) | – | – | – | 0.195 | 0.185 | 0.150 | 0.075 | 0.065 | 0.085 |
| VWF (MIM: 613160) | – | – | – | 0.335 | 0.300 | 0.300 | 0.025 | 0.030 | 0.035 |
Application to OHTS
Primary open-angle glaucoma (POAG [MIM: 137760]) is a leading cause of irreversible blindness. Although a genetic basis has been established for a substantial fraction of POAG, no risk alleles of major effect have been identified.32 The etiology of POAG is likely to be complex. Because POAG is assessed through quantitative measures such as central corneal thickness (CCT), intraocular pressure, and cup-to-disc ratio, one promising research direction is to map genes underlying these quantitative measures. Indeed, large-scale GWASs have identified genes that affect CCT.33, 34, 35 Using data from the OHTS,27 I applied the methods SKAT+, SKAT, and SKAT-O to a gene-based GWAS of CCT.
OHTS is a multi-center, randomized clinical trial sponsored by the National Eye Institute. Its goal is to investigate the efficacy of medical treatment in delaying or preventing the onset of POAG in individuals with elevated intraocular pressure. A total of 1,636 individuals between 40 and 80 years old were enrolled, and 1,077 of them were genotyped in a subsequent study. Data for this genetic study are available from dbGaP (study accession number dbGaP: phs000240.v1.p1). Both genotype data and baseline phenotype data are available for 1,057 subjects. The vast majority of these subjects are non-Hispanic white (752) and black (249).
There were 1,051,295 genotyped SNPs and 30,562 autosomal genes. The HGNC gene symbols were obtained with the R/Bioconductor package biomaRt (version 2.26.1). As in Lee et al.,28 genes that contained fewer than three SNPs were excluded from further consideration. This reduced the number of genes to 23,778.
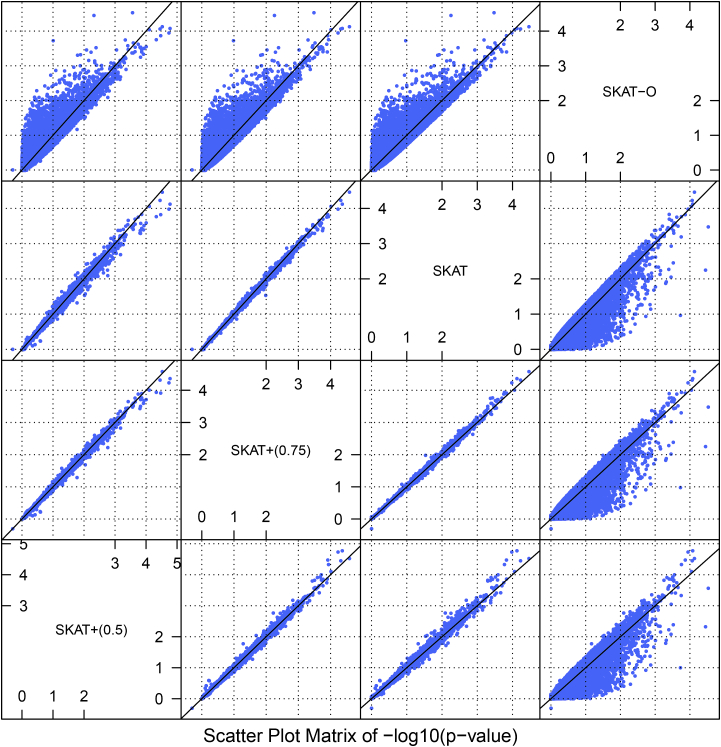
Because a thinner CCT increases the risk of POAG,32 subjects with higher CCT measurements were used as control subjects in calculations of the null distribution for SKAT+. In particular, two inclusion criteria were used: the first and second quartiles. CCT measurements from both eyes were averaged and used as the response, and age and gender were used as covariates. The analysis was done on non-Hispanic white samples only, given that this group is much larger than other ethnic groups. A scatterplot matrix of the genome-wide gene-based p values from SKAT+ (with two different inclusion criteria), SKAT, and SKAT-O is shown in Figure 4. SKAT-O behaved rather differently from the others. The other three behaved pretty similarly to each other, but they did differ, especially SKAT+ with 50% of the subjects, which is not surprising. Most importantly, there doesn’t seem to exist a systematic bias in favor of SKAT+. Those genes at which at least one of SKAT+, SKAT, and SKAT-O had a p value less than 0.0001 are listed in Table 4. These genes warrant further investigation. Unfortunately, they do not overlap ZNF469 (MIM: 612078), COL5A1 (MIM: 120215), COL8A2 (MIM: 120252), AKAP13 (MIM: 604686), or AVGR8, genes for which association with CCT has been reported previously.33, 34, 35 I also conducted a simulation study to confirm that the type I error rate of SKAT+ is under control at a much higher level. For this purpose, genes TRERF1 (MIM: 610322) and IQUB in Table 4 were arbitrarily selected. A continuous trait following a standard normal distribution and a dichotomous trait with disease probability 0.3 were each generated with 5 × 106 replicates. For the continuous trait, 75% of the subjects were used as control subjects. For the dichotomous trait, all simulated control subjects were used for the SKAT+ method. The expected proportion of control subjects is equal to 1 − 0.3 = 0.7. The type I error rates are presented in Table 5. They are clearly under control.
Figure 4.
Scatterplots of Genome-wide Gene-Based p Values for the OHTS
SKAT+ (0.5) refers to the SKAT+ method using 50% of the subjects for its p value calculation. SKAT+ (0.75) uses 75% of the subjects.
Table 4.
A Summary of Gene-Based Association p Values with Data from the OHTS
| Chromosome | Gene | SKAT+ (0.5) | SKAT+ (0.75) | SKAT | SKAT-O |
|---|---|---|---|---|---|
| 2 | HAGLROS | 3.525 × 10−5 | 4.821 × 10−5 | 5.918 × 10−5 | 8.619 × 10−5 |
| 6 | EXOC2 (MIM: 615329) | 2.725 × 10−4 | 3.285 × 10−4 | 3.327 × 10−4 | 2.951 × 10−5 |
| TRERF1 (MIM: 610322) | 2.976 × 10−5 | 2.586 × 10−5 | 3.427 × 10−5 | 7.390 × 10−5 | |
| 7 | NDUFA5 (MIM: 601677) | 1.822 × 10−5 | 5.848 × 10−5 | 1.042 × 10−4 | 1.129 × 10−4 |
| IQUB | 1.680 × 10−5 | 4.322 × 10−5 | 7.473 × 10−5 | 8.301 × 10−5 | |
| 15 | MTMR10 (MIM: 208500) | 4.767 × 10−3 | 5.528 × 10−3 | 5.592 × 10−3 | 3.536 × 10−5 |
| SNAP23 (MIM: 602534) | 3.355 × 10−5 | 1.217 × 10−4 | 1.486 × 10−4 | 1.359 × 10−4 | |
| HAUS2 (MIM: 613429) | 9.859 × 10−5 | 1.930 × 10−4 | 2.581 × 10−4 | 2.784 × 10−4 | |
| MYL12BP1 | 4.672 × 10−5 | 1.155 × 10−4 | 1.693 × 10−4 | 1.748 × 10−4 | |
| 16 | TXNDC11 | 7.867 × 10−5 | 9.746 × 10−5 | 8.708 × 10−5 | 9.013 × 10−5 |
Genes were selected if any statistic had a p value less than 0.0001 in the non-Hispanic white-only sample. SKAT+ (0.5) refers to SKAT+ method using 50% of the subjects for its p value calculation. SKAT+ (0.75) uses 75% of the subjects.
Table 5.
Simulated Type I Error Rates of SKAT+ for Two Top Genes in Table 4 over 5 × 106 Replicates
| Trait Type | Gene |
Nominal Level |
||
|---|---|---|---|---|
| 10−4 | 10−5 | 2.5 × 10−6 | ||
| Continuous | TRERF1 | 8.1800 × 10−5 | 2.8000 × 10−6 | 1.2000 × 10−6 |
| IQUB | 1.0120 × 10−4 | 7.6000 × 10−6 | 2.0000 × 10−6 | |
| Dichotomous | TRERF1 | 9.7600 × 10−5 | 4.6000 × 10−6 | 3.0000 × 10−6 |
| IQUB | 1.1128 × 10−4 | 9.6000 × 10−6 | 2.0000 × 10−6 | |
Only non-Hispanic white subjects were used. The continuous trait followed a standard normal distribution, and the dichotomous trait had a disease probability equal to 0.3. 75% of subjects were used as controls subjects.
Discussion
I have proposed the SKAT+ method for gene-based association testing. This method uses the same test statistic as SKAT but estimates the null distribution differently. By using a properly selected subset of subjects, this estimation method leads to a more powerful testing procedure. The selection is based on the residuals of the phenotype after the effect of covariates has been removed. The null distribution depends only on the second moments of the phenotype and the genotypes controlling for the effect of covariates. Selection based on phenotype residuals does not affect the validity of the test but has an effect on the power.
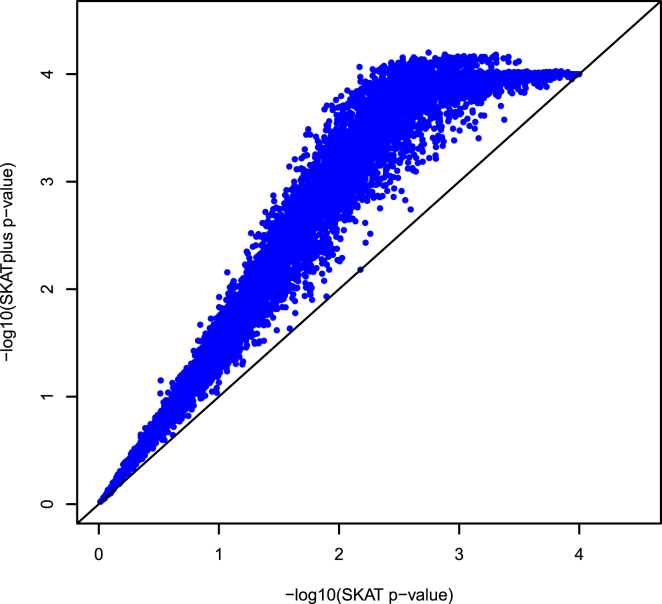
The current estimation method has a valid type I error rate, but the power is compromised because the estimated distribution does not correspond to the desired one when the null hypothesis is not true. It is contaminated by the distribution of SKAT under the alternative hypothesis. Because of this, the proposed SKAT+ method is almost surely destined to be more powerful than SKAT as sample size increases. No other SKAT competitors can make such a statement given that they perform better in only certain situations. To demonstrate this point, I simulated 10,000 datasets from the covariate-free dichotomous-trait model (Simulation Studies) used in the simulation studies with ORs = (2, 2, 2, 2, 2, 2, 2, 2) and no neutral SNPs. The −log10-transformed 10,000 p values for SKAT+ were plotted against SKAT p values transformed in the same way (Figure 5). The vast majority of the 10,000 p values from SKAT+ were much more significant than those from SKAT, and almost none of them were apparently worse.
Figure 5.
The p Values from SKAT+ versus the p Values from SKAT over 10,000 Replicates
The vast majority of the 10,000 p values from SKAT+ are much more significant than those from SKAT, given that the points are almost all above the 45° line. The simulation model is 1, with ORs = (2, 2, 2, 2, 2, 2, 2, 2) and no neutral SNPs.
Selection of subjects for the SKAT+ method is critical for power improvement. For case-control studies without covariates, the selection seems to be obvious. When there are covariates or the trait is continuous, the selection is less so. For the simulation studies, I selected subjects whose trait values were not higher than the third quartile after the effect of covariates had been removed. I also tried other thresholds, such as the median and the 80% quantile. The results are not shown, but they were consistent with the intuition that SKAT+ becomes more similar to SKAT as more subjects are selected. Generally, a lower threshold leads to fewer subjects and less accuracy in p value computation. The larger the sample size, the lower the threshold one can afford while still having enough selected subjects for estimating the null distribution with certain accuracy. SKAT+ contains SKAT as a special case. If small sample size is a concern, one is recommended to use SKAT instead of SKAT+. Furthermore, one can use the recently proposed small-sample method.36 A previous method28 for small-sample situations “can overcorrect and lead to inflated type I error.”36
A related issue in selecting control subjects is the allele frequencies of the variants. The rarer they are, the more sensitive the sample variance matrix of genotype scores is to the selected control subjects. Hence, a larger number of control subjects should be used. For an MAF of 1%, there are on average two copies of the minor allele per 100 subjects. So, 500 subjects would have ten copies of the minor allele. If the MAFs are rarer, having a more stable sample variance matrix would require more subjects for the control group.
Note that when there are no covariates, it is possible to estimate the unknown null-distribution parameters by using data from a reference database, such as the 1000 Genomes Project or the International HapMap Project. One can compute the variance matrix for the SNPs used in the study and use it in place of the matrix S0. In this way, there is no need to select subjects for the SKAT+ method, but one needs to use an appropriate matching data sample. When there are covariates, generally it is not possible to use a reference database this way.
Also note that the basic principle behind SKAT+ is applicable to extensions of SKAT, as mentioned in the Introduction. It can also be generalized to phenotype with a distribution in the exponential family if the diagonal matrix V is replaced with a diagonal matrix of proper estimates of .14
An implementation of the SKAT+ method is provided in the R package iGasso.
Acknowledgments
This work is dedicated to the memory of the late Dr. Leon F. Burmeister. Leon was a nice gentleman, a great colleague, and a wise mentor. I thank Leon for his long-time support and inspiration. Preparation of Genetic Analysis Workshop 17 (GAW17) data was partly supported by NIH grant R01 MH059490 and used data from the 1000 Genomes Project. The GAW17 was supported by NIH grant R01 GM031575 from the National Institute of General Medical Sciences. I also thank the handling editor and three anonymous reviewers for their useful comments.
Published: June 9, 2016
Web Resources
1000 Genomes Project, http://www.1000genomes.org/
Dr. Wei Pan’s website, http://www.biostat.umn.edu/∼weip/prog
International HapMap Project, https://hapmap.ncbi.nlm.nih.gov/
OMIM, http://www.omim.org
R package iGasso, http://cran.r-project.org/web/packages/iGasso/index.html
References
- 1.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuk O., Hechter E., Sunyaev S.R., Lander E.S. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. USA. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S.H., Wray N.R., Goddard M.E., Visscher P.M. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichler E.E., Flint J., Gibson G., Kong A., Leal S.M., Moore J.H., Nadeau J.H. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgenthaler S., Thilly W.G. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST) Mutat. Res. 2007;615:28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Madsen B.E., Browning S.R. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price A.L., Kryukov G.V., de Bakker P.I., Purcell S.M., Staples J., Wei L.-J., Sunyaev S.R. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li B., Leal S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris A.P., Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet. Epidemiol. 2010;34:188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neale B.M., Rivas M.A., Voight B.F., Altshuler D., Devlin B., Orho-Melander M., Kathiresan S., Purcell S.M., Roeder K., Daly M.J. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu D., Lin X., Ghosh D. Semiparametric regression of multidimensional genetic pathway data: least-squares kernel machines and linear mixed models. Biometrics. 2007;63:1079–1088. doi: 10.1111/j.1541-0420.2007.00799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu D., Ghosh D., Lin X. Estimation and testing for the effect of a genetic pathway on a disease outcome using logistic kernel machine regression via logistic mixed models. BMC Bioinformatics. 2008;9:292. doi: 10.1186/1471-2105-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S., Emond M.J., Bamshad M.J., Barnes K.C., Rieder M.J., Nickerson D.A., Christiani D.C., Wurfel M.M., Lin X., NHLBI GO Exome Sequencing Project—ESP Lung Project Team Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S., Teslovich T.M., Boehnke M., Lin X. General framework for meta-analysis of rare variants in sequencing association studies. Am. J. Hum. Genet. 2013;93:42–53. doi: 10.1016/j.ajhg.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnett I.J., Lee S., Lin X. Detecting rare variant effects using extreme phenotype sampling in sequencing association studies. Genet. Epidemiol. 2013;37:142–151. doi: 10.1002/gepi.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin X., Cai T., Wu M.C., Zhou Q., Liu G., Christiani D.C., Lin X. Kernel machine SNP-set analysis for censored survival outcomes in genome-wide association studies. Genet. Epidemiol. 2011;35:620–631. doi: 10.1002/gepi.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai T., Tonini G., Lin X. Kernel machine approach to testing the significance of multiple genetic markers for risk prediction. Biometrics. 2011;67:975–986. doi: 10.1111/j.1541-0420.2010.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H., Meigs J.B., Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genet. Epidemiol. 2013;37:196–204. doi: 10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Lee S., Zhu X., Redline S., Lin X. GEE-based SNP set association test for continuous and discrete traits in family-based association studies. Genet. Epidemiol. 2013;37:778–786. doi: 10.1002/gepi.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X., Lee S., Christiani D.C., Lin X. Test for interactions between a genetic marker set and environment in generalized linear models. Biostatistics. 2013;14:667–681. doi: 10.1093/biostatistics/kxt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin X., Lee S., Wu M.C., Wang C., Chen H., Li Z., Lin X. Test for rare variants by environment interactions in sequencing association studies. Biometrics. 2016;72:156–164. doi: 10.1111/biom.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies R. Algorithm AS 155: The distribution of a linear combination of χ2 random variables. J. R. Stat. Soc. Ser. C Appl. Stat. 1980;29:323–333. [Google Scholar]
- 25.Imhof J. Computing the distribution of quadratic forms in normal variables. Biometrika. 1961;48:419–426. [Google Scholar]
- 26.Almasy L., Dyer T.D., Peralta J.M., Kent J.W., Jr., Charlesworth J.C., Curran J.E., Blangero J. Genetic Analysis Workshop 17 mini-exome simulation. BMC Proc. 2011;5(Suppl 9):S2. doi: 10.1186/1753-6561-5-S9-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon M.O., Kass M.A. The Ocular Hypertension Treatment Study: design and baseline description of the participants. Arch. Ophthalmol. 1999;117:573–583. doi: 10.1001/archopht.117.5.573. [DOI] [PubMed] [Google Scholar]
- 28.Lee S., Wu M.C., Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13:762–775. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan W., Kim J., Zhang Y., Shen X., Wei P. A powerful and adaptive association test for rare variants. Genetics. 2014;197:1081–1095. doi: 10.1534/genetics.114.165035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basu S., Pan W. Comparison of statistical tests for disease association with rare variants. Genet. Epidemiol. 2011;35:606–619. doi: 10.1002/gepi.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang T., Elston R.C. Improved power by use of a weighted score test for linkage disequilibrium mapping. Am. J. Hum. Genet. 2007;80:353–360. doi: 10.1086/511312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fingert J.H. Primary open-angle glaucoma genes. Eye (Lond.) 2011;25:587–595. doi: 10.1038/eye.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y., Dimasi D.P., Hysi P.G., Hewitt A.W., Burdon K.P., Toh T., Ruddle J.B., Li Y.J., Mitchell P., Healey P.R. Common genetic variants near the Brittle Cornea Syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. PLoS Genet. 2010;6:e1000947. doi: 10.1371/journal.pgen.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitart V., Bencić G., Hayward C., Skunca Herman J., Huffman J., Campbell S., Bućan K., Navarro P., Gunjaca G., Marin J. New loci associated with central cornea thickness include COL5A1, AKAP13 and AVGR8. Hum. Mol. Genet. 2010;19:4304–4311. doi: 10.1093/hmg/ddq349. [DOI] [PubMed] [Google Scholar]
- 35.Vithana E.N., Aung T., Khor C.C., Cornes B.K., Tay W.-T., Sim X., Lavanya R., Wu R., Zheng Y., Hibberd M.L. Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum. Mol. Genet. 2011;20:649–658. doi: 10.1093/hmg/ddq511. [DOI] [PubMed] [Google Scholar]
- 36.Chen J., Chen W., Zhao N., Wu M.C., Schaid D.J. Small sample kernel association tests for human genetic and microbiome association studies. Genet. Epidemiol. 2016;40:5–19. doi: 10.1002/gepi.21934. [DOI] [PMC free article] [PubMed] [Google Scholar]