Abstract
The tRNA splicing endonuclease is a highly evolutionarily conserved protein complex, involved in the cleavage of intron-containing tRNAs. In human it consists of the catalytic subunits TSEN2 and TSEN34, as well as the non-catalytic TSEN54 and TSEN15. Recessive mutations in the corresponding genes of the first three are known to cause pontocerebellar hypoplasia (PCH) types 2A-C, 4, and 5. Here, we report three homozygous TSEN15 variants that cause a milder version of PCH2. The affected individuals showed progressive microcephaly, delayed developmental milestones, intellectual disability, and, in two out of four cases, epilepsy. None, however, displayed the central visual failure seen in PCH case subjects where other subunits of the TSEN are mutated, and only one was affected by the extensive motor defects that are typical in other forms of PCH2. The three amino acid substitutions impacted the protein level of TSEN15 and the stoichiometry of the interacting subunits in different ways, but all resulted in an almost complete loss of in vitro tRNA cleavage activity. Taken together, our results demonstrate that mutations in any known subunit of the TSEN complex can cause PCH and progressive microcephaly, emphasizing the importance of its function during brain development.
Main Text
The production of functional tRNAs is highly regulated and critical for every tissue and cell.1, 2 Despite this ubiquitous requirement, mutations affecting various aspects of this process have been shown to cause distinct disorders.2 For instance, mutations in various aminoacyl-tRNA synthetases, which catalyze the loading of amino acids onto tRNAs, cause, among other disorders, Usher syndrome (MIM: 614504),3 hereditary spastic paraplegia,4 and pontocerebellar hypoplasia (PCH [MIM: 611523]).2, 5 Contrasting this heterogeneity, the latter phenotype is also associated with other steps of tRNA biogenesis and represents a case of connected biological processes that converge on a common phenotype.
Together with Karaca et al. we described in two independent studies that a single homozygous mutation in CLP1 (MIM: 608757), which encodes an RNA kinase, causes brain atrophy and peripheral neurodegeneration (PCH10 [MIM: 615803]).6, 7 The associated amino acid substitution, p.Arg104His, abrogated interaction with the tRNA-splicing endonuclease (TSEN) complex; this in turn caused a reduction in pre-tRNA cleavage and an accumulation of unspliced tRNA and linear fragments.6, 7 Likewise, the TSEN proteins themselves have been implicated in hindbrain malformations.8, 9, 10, 11, 12 Budde and colleagues described a large clinical cohort of 58 individuals suffering from different classes of pontocerebellar hypoplasia.8 The majority carried the same homozygous mutation in TSEN54 (MIM: 608755), probably due to a common founder event. Additionally, one person harbored a homozygous mutation in TSEN2 (MIM: 608753) and one in TSEN34 (MIM: 608754). This genetic evidence later prompted the subdivision of PCH2 into PCH2A (caused by TSEN54 mutations [MIM: 277470]), PCH2B (TSEN2 [MIM: 612389]), and PCH2C (TSEN34 [MIM: 612390]) (see GeneReviews in Web Resources).
The human tRNA splicing endonuclease consists of a heterotetramer complex formed by the four TSEN proteins, which were identified by homology with their yeast counterparts.13, 14 TSEN2 and TSEN34 are the catalytic subunits, which form a compound active site, whereas TSEN54 and TSEN15 are structural proteins.15 Although the other three components have been implicated in human disease for almost a decade, we only recently reported a recessive TSEN15 (MIM: 608756) candidate variant in global developmental delay.16 Here we describe two additional independent families with mutations in TSEN15 and redefine the clinical phenotypes as pontocerebellar hypoplasia with progressive microcephaly. We further demonstrate that all three mutations affect the integrity and function of the tRNA splicing endonuclease complex, thereby providing evidence for the involvement of the reported variants in the pathogenicity of disease.
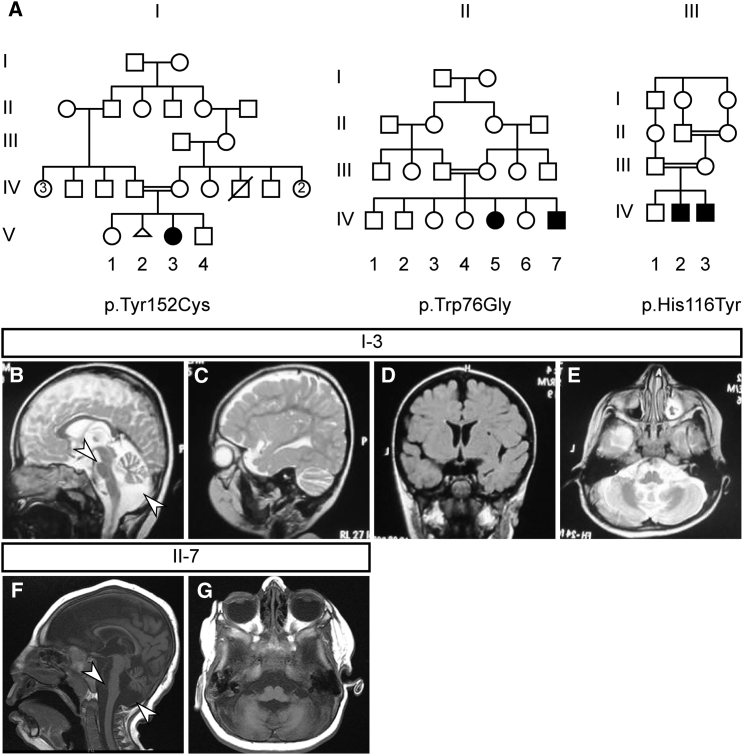
The affected individual from family I (I-V-3) was born to consanguineous parents of Pakistani origin (Figure 1A and Table 1). She was born via normal spontaneous vaginal delivery (NSVD) at full term without complications. She has one older and one younger sibling who were both healthy. Weight, length, and head circumference at birth were unremarkable, although the latter was slightly below average (−1.4 SD). The affected girl was diagnosed with microcephaly (−3.4 SD) and intellectual disability at 1.5 years of age. She was irritable at presentation and had already experienced two episodes of seizures, the first occurring at 11 months of age. She did not show any spasticity, jitteriness, or hypertonia, but displayed hypotonia and weak tendon reflexes. There was no central visual impairment, but there was intermittent strabismus. At 2.5 years, she had poor language skills and was restricted to non-specific sounds other than “mama” and “baba” (mom and dad). Her hand function was impaired without hand preference, she was non-ambulatory, and she was able to sit only with support. Brain MRI (Figures 1B–1E) revealed hypoplasia of the pons and cerebellum (Figure 1B) and an increase in extra-axial CSF spaces (Figures 1C and 1D), indicative of bilateral atrophy of the fronto-parietal cortex. Cortical architecture, white matter, and corpus callosum appeared normal.
Figure 1.
Characterization of Three Families with Progressive Microcephaly and Intellectual Disability
(A) Family tree of the three families (I–III) presented in this study. Consanguinity (double bar) of families suggests a recessive inheritance of the disease. Note that one individual in family II has been described previously.16 The procedures followed for recruitment and data collection were in accordance with the ethical standards of the responsible committee on human experimentation at the respective participating institute and proper informed consent was obtained.
(B–E) Panels show MRIs for the affected girl in family I (I-V-3). Shown are sagittal (B-C; T2 weighted), coronal (D; FLAIR), and axial (E; T2 weighted) images. Arrowheads indicate pontocerebellar hypoplasia (B).
(F and G) Panels show T1 weighted MRIs for the affected boy in family II (II-IV-7). Shown are sagittal (F) and axial (G) images. Arrowhead indicates pontocerebellar hypoplasia (F).
Table 1.
Clinical Information
|
Family ID |
I |
II |
III |
III |
|---|---|---|---|---|
| Mutation | p.Tyr152Cys | p.Trp76Gly | p.His116Tyr | p.His116Tyr |
| Evaluation | ||||
| Gender | female | male | male | male |
| Ethnic origin | Pakistani | Saudi | Syrian | Syrian |
| Pregnancy duration | full term | full term | full term | full term |
| Polyhydramnios | no | no | no | no |
| Weight at birth | 3.3 kg | 3.0 kg | ND | 2.3 kg |
| Length at birth | 50 cm | ND | normal (verbal) | normal (verbal) |
| HC at birth | 34 cm (−1.4 SD) | 31 cm (−2.5 SD) | normal (verbal) | normal (verbal) |
| Diagnosis age | 1.5 years | at birth | 18 months | 2 years |
| HC at latest examination | 1.5 years: 42 cm (−3.4 SD) | 16 months: 35 cm (−9.7 SD) | 5.5 years: 46 cm (−4.4 SD) | 12.5 years: 49.5 cm (−3.0 SD) |
| Intellectual disability | moderate | severe | mild/moderate | mild/moderate |
| Irritability | yes | ND | no | yes |
| Seizures | ||||
| Seizures | yes | yes | no | no |
| Seizure onset | 11 months | 8 months | – | – |
| Motor Findings | ||||
| Spasticity | no | peripheral | no | no |
| Jitteriness/clonus | no | yes | no | no |
| Hypertonia | no | yes (peripheral) | no | no |
| Hypotonia | yes | yes (axial) | no | no |
| Deep tendon reflexes | weak | increased (+3) | ND | ND |
| Visual Findings | ||||
| Central visual impairment | no | no | ND | ND |
| Primary optic atrophy | no | no | ND | ND |
| Nystagmus | no | no | no | no |
| Strabismus | intermittent | no | ND | ND |
| Fixation and following | yes | no | yes | yes |
| Developmental Milestones | ||||
| Gross motor | delayed | delayed | delayed | normal |
| Fine motor | delayed | absent | ND | ND |
| Language | delayed | absent | delayed | delayed |
| Cognitive | delayed | delayed | delayed | delayed |
| Social | absent | delayed | normal | normal |
| Diagnostic Tests | ||||
| EEG | yes | yes | ND | ND |
| MRI Findings | ||||
| Cerebellum | hypoplasia | hypoplasia | ND | ND |
| Pons | hypoplasia | hypoplasia | ND | ND |
| Cerebral cortex | atrophy | no structural abnormalities | ND | ND |
| Ventricles | increased extra-axial CSF spaces | increased intra- and extra-axial CSF spaces | ND | ND |
| White matter | normal | normal | ND | ND |
| Corpus callosum | normal | thin | ND | ND |
Abbreviations are as follows: ND, no data; verbal, information provided by the parents; SD, standard deviations; CSF, cerebrospinal fluid. Note that no detailed clinical information was available for the affected girl in family II.
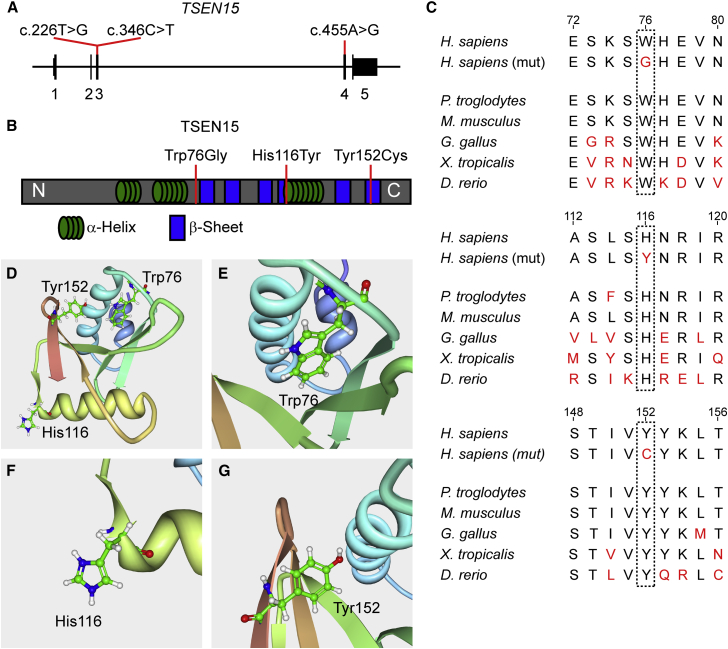
The affected individual and both parents were subjected to whole-exome sequencing (WES) for genetic analysis. Employing our internal pipeline for prioritization of potentially disease-causing mutations, we excluded all but two homozygous single-nucleotide variants (SNVs), one of which did not segregate with the phenotype.17 The remaining candidate, located in TSEN15 (GenBank: NM_052965.3; c.455A>G), was unique both in our unrelated cohort of more than 5,000 sequenced individuals in the GME Variome and in the ExAC database and resulted in a p.Tyr152Cys amino acid substitution (Figures 2A, 2B, and S3B). This variant was amid a stretch of extensive homozygosity (HomozygosityMapper, Figure S1) and predicted to be disease causing by the online program MutationTaster.19, 20, 21 Furthermore, the altered residue was highly conserved in vertebrates (Figure 2C).
Figure 2.
The Three Families Carry TSEN15 Missense Mutations that Affect Conserved Amino Acid Residues
(A) Schematic of TSEN15 depicting the coding sequence spanning 5 exons and the 5′ and 3′ UTR in exons 1 and 5, respectively. Red lines indicate the position of the three identified mutations and their coordinates within the cDNA (GenBank: NM_052965.3).
(B) Schematic of the TSEN15 protein depicting the location of α helices (green) and β sheets (blue) using information from the previously generated NMR structure from RCSB protein data bank.18 N and C indicate the N and C termini of the protein, respectively. Red lines indicate the position and the coordinates of the three amino acid substitutions resulting from the described mutations (GenBank: NP_443197.1).
(C) All three amino acids affected by the mutations are conserved in vertebrates.
(D) Ribbon model of TSEN15, as solved by NMR.18 Depicted are the three affected residues: Trp76, His116, and Tyr152.
(E) Magnification of Trp76, which is located in the core of the folded protein and might be critical for folding and internal hydrophobic interactions.
(F) Magnification of His116, which is located on the surface of the protein.
(G) Magnification of Tyr152, which is located in the last β sheet C-terminal to a critical interaction loop with TSEN2.
We previously reported an individual with microcephaly and global developmental delay harboring a candidate mutation in TSEN15.16 The clinical presentation and variant found in family I prompted us to revisit this affected boy (family II; individual II-IV-7). He was born to consanguineous Saudi parents and exhibited progressive microcephaly (−9.7 SD), and his MRI at 16 months showed hypoplasia of the cerebellum and pons (Figures 1A, 1F, and 1G and Table 1). He suffered from severe epilepsy since the age of 8 months. At his last examination at 16 months, he was not able to roll or sit. He showed no language development and did not visually fixate or follow. He further presented with appendicular hypertonia and axial hypotonia, and his deep tendon reflexes were increased with positive Babinski sign. The older sister presented with similar clinical symptoms and the parents previously had three spontaneous first-term abortions. As reported, the affected individual carried a homozygous missense mutation resulting in a p.Trp76Gly substitution in this highly conserved residue (Figures 2A–2C).16
The striking similarities of these two case subjects, which both showed PCH2-like phenotypes including progressive microcephaly, supported the pathogenicity of these mutations. To expand our clinical study, we re-analyzed our cohort of individuals with neurodevelopmental disorders, which have undergone WES, for other TSEN15 mutations.
Indeed, we discovered a third case in a consanguineous family of Syrian origin (family III) with one healthy older and two affected younger sons (III-IV-2 and III-IV-3), both of whom were diagnosed with intellectual disability and progressive microcephaly (Figure 1A and Table 1). They were born via NSVD after a full-term pregnancy with unremarkable length and head circumference, but at last examination (5.5 and 12.5 years) their head size was far below average (−4.4 SD and −3.0 SD). Epileptic seizures, spasticity, jitteriness, and hyper- or hypotonia were not reported. Nystagmus was absent and both displayed visual fixation and following. At the age of 12.5 years, the older affected boy (III-IV-2) responded to simple questions and orders. His pronunciation, however, was unclear and he used incorrect grammar. His motor development appeared unaffected: he crawled at the age of 5–7 months and walked at 13 months. Individual III-IV-3 showed similar speech development as his older brother. At the age of 5.5 years he could crawl but did not walk. Due to the ongoing conflict in Syria, we were not able to reach the family or the original clinicians and could not confirm a PCH phenotype by medical imaging.
The discovered homozygous TSEN15 SNV (GenBank: NM_052965.3; c.346C>T) resulted in a p.His116Tyr substitution (Figures 2A, 2B, and S3A). The mutations segregated with the phenotype and whole-genome SNP homozygosity mapping of all three siblings further supported the pathogenicity of this variant (HomozygosityMapper and Cyto Scan HD as described previously22) (Figure S2). Only three candidate regions were detected (hg19: chr1: 165,644,864–173,103,272; chr1: 179,756,638–185,885,630; chr4: 82,381,626–85,082,293), one of which encompassed the TSEN15 locus. Furthermore, this mutation was predicted to be disease causing by MutationTaster, and His116 is highly conserved among vertebrates (Figure 2C).
The three affected residues, although converging phenotypically, are structurally unrelated (Figure 2D; protein work bench using the RCSB protein data bank).18 Trp76 is located in the hydrophobic core of the protein and is critical for folding and stability (Figure 2E). His116, on the other hand, is positioned on the surface (Figure 2F) in an area that is putatively involved in TSEN34 interaction and has been shown by NMR to associate with a second TSEN15 molecule.14, 18 It is disputed, however, whether the latter has relevance for the endonuclease function. Tyr152 is an absolutely conserved residue located toward the carboxyl terminus, forming a critical β sheet via a hydrogen bond with Asp69, pulling loop 7 toward the carboxyl terminus of α helix 2 (Figure 2G).18 The archaeal equivalent of this loop is critical for the dimerization and configuration of the enzyme active sites.14, 23
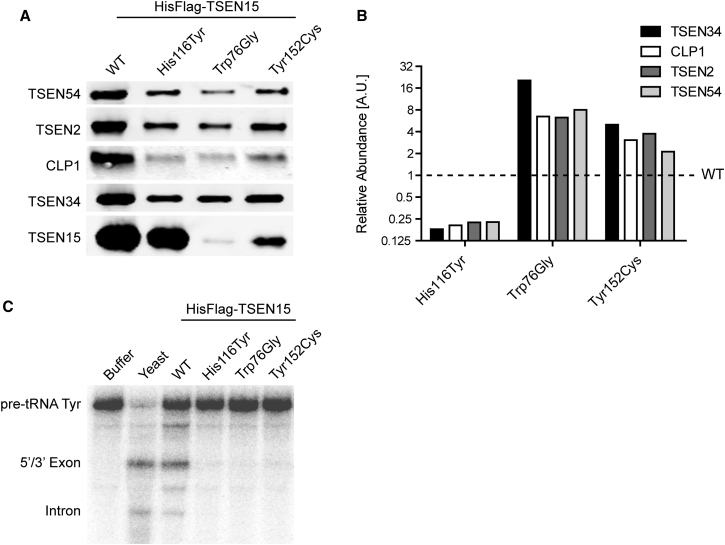
In order to understand the impact of these mutations on protein function, we generated SHY5Y cell lines stably expressing N-terminally HisFlag-tagged wild-type or mutant TSEN15. After cell lysis and tandem affinity purification using anti-Flag and Ni-NTA, we performed western blot analysis of the endonuclease complex.6 When probed with specific antibodies for TSEN54, TSEN2, CLP1, TSEN34, and the tagged TSEN15, the mutants showed distinct behaviors and impacted two key features of the expression of the TSEN complex (Figures 3A and 3B). First, whereas the p.His116Tyr substitution had no effect on the level of expression of TSEN15, the p.Trp76Gly and p.Tyr152Cys variants exhibited greatly reduced abundance, the former affecting protein levels more severely (Figure 3A). Second, all three mutants altered the stoichiometry of the heterotetrameric TSEN complex and its associations with the CLP1 protein (Figure 3B). The p.His116Tyr alteration resulted in a stark decrease of the TSEN partners and CLP1 relative to TSEN15 levels; in contrast, the other two substitutions resulted in a relatively increased association. We confirmed these results using a western blot variant (Peggy Sue, ProteinSimple) and a cellular overexpression system (Figure S4).
Figure 3.
The Three Described Amino Acid Substitutions Affect Complex Assembly and tRNA Cleavage
(A) Western blot analysis of purified samples obtained from stable cell lines expressing wild-type (WT) or mutant HisFlag-tagged TSEN15. All lysates were subjected to tandem affinity purification (Ni-NTA and Flag M2, Sigma, cat# F1804, RRID: AB_262044) and probed with antibodies detecting TSEN54 (Abcam, ab178696, 0.86 μg/mL), TSEN2 (Proteintech, cat# 13103-2-AP, RRID: AB_2272213; 4 μg/mL), CLP1 (HEAB antibody, Abcam, ab172683, 0.4 μg/mL), TSEN34 (Abcam, ab68868, 1 μg/mL), and TSEN15 (Flag M2, Sigma, cat# F1804, RRID: AB_262044; 0.2 μg/mL). Whereas the p.His116Tyr mutant TSEN15 can be recovered at levels comparable to WT, p.Trp76Gly and p.Tyr152Cys result in lower protein levels and affect protein folding, stability, or both.
(B) Quantification of TSEN54, TSEN2, CLP1, and TSEN34 levels, normalized to the respective mutant TSEN15 and wild-type (equals 1). 1:1:1:1 stoichiometry of the complex, as well as relative levels of CLP1, are severely affected by the substitutions. Whereas the p.His116Tyr substitution results in destabilization of the purified complex and relative reduction of its components and CLP1 protein, the other two mutants show a relative increase of the other proteins.
(C) tRNA cleavage assay using 32P-labeled tyrosine pre-tRNA (pre-tRNA Tyr) as substrate. Shown is an RNA gel of reactions incubated with buffer, purified yeast complex (Yeast), the purified WT complex, and the mutants. Yeast, as well as WT complex, catalyze a cleavage reaction that results in the production of a 5′ and a 3′ exon, as well as the intron. All three mutant complexes show near complete loss of cleavage activity.
Previously, it has been shown that mutation of CLP1 results in a decrease in the association of the entire TSEN complex, leading to impaired tRNA cleavage due to the lack of TSEN complex.6, 7 To determine the functional implications of the altered stoichiometry of the purified TSEN complexes, we employed an in vitro tRNA cleavage assay. Wild-type and mutant complexes were tested for enzymatic activity using 32P-labeled intron-containing tyrosine tRNA as substrate.6 The readout of this assay is the cleavage of the pre-tRNA into 5′ and 3′ exon “halves” and the intron after a 2 hr incubation with purified TSEN complexes. The wild-type complex, as well as a purified yeast tRNA endonuclease control, catalyzed this reaction as expected and produced the predicted RNA fragments (Figure 3C). Purified complexes from all three mutants, however, showed an almost complete loss of the enzymatic function and absence of cleaved substrate. These results highlight the importance of proper heterotetrameric assembly of the TSEN complex to achieve recognition and cleavage of tRNA substrates.
Our data suggest a converging molecular phenotype caused by mutations that affect the TSEN15 protein in different ways. The p.His116Tyr substitution, consistent with the residue’s location, did not affect the production and stability of the protein. Nevertheless, the mutant severely decreased assembly of stable endonuclease complex, most likely due to reduced interaction with its binding partner, the active site containing TSEN34. The other two residues are situated within the protein (Figures 2D, 2E, and 2G) and their alteration probably affects proper folding, stability, or both. Based on the location of Trp76 in the very center of the protein and the critical hydrogen bond provided by Tyr152, we suggest that the variants probably impair protein folding and result in degradation of the misfolded product. The more pronounced impact of the p.Trp76Gly substitution on TSEN15 stability seen here may underlie the congenital and overall more severe presentation of microcephaly in family II.
In all cases, however, the substitution resulted in changes of the complex’s stoichiometry. TSEN2 and TSEN34 could be detected in all three purifications at lower levels; nevertheless, the in vitro cleavage assay showed an almost complete dearth of cleaved tRNA. This finding reiterates the importance of the correct assembly of the tRNA splicing endonuclease subunits for its enzymatic activity.13, 14, 15
The conspicuous absence of homozygous null alleles for the other three members of the TSEN complex suggested early lethality upon complete loss of their function.8, 9, 10, 12 This hypothesis was supported by compound heterozygous individuals, carrying one nonsense and one missense mutation. These were classified as the more severe PCH4 (MIM: 225753) and PCH5 (MIM: 610204) or PCH2 with aspects of PCH4.8, 10, 11 These findings imply an essential function of the TSEN complex in humans, similar to what has been found in S. cerevisiae.24
The in vitro assay employed here showed compromised enzymatic activity of complexes purified by TSEN15 mutants. However, the phenotype of the identified affected individuals was milder than those seen in most other PCH2 case subjects. We propose two possibilities for this discrepancy. First, it is conceivable that the three TSEN15 mutants may retain tRNA splicing function in the cellular context. All three enzyme variants alter the subunit stoichiometry of the TSEN enzyme but do not completely abolish the interactions between the TSEN subunits. Therefore, there may remain sufficient TSEN activity within affected cells to allow for a basal production of tRNA from spliced precursors. Thus, the lack of in vitro cleavage activity might be a reflection of the slower reaction kinetics. Second, other members of the complex may have functions beyond the catalysis of tRNA cleavage which contribute to the more severe phenotype. Mutation of TSEN15 may only impair tRNA splicing, which can be tolerated through development, but ultimately leads to the described phenotypes. As cells from affected members of these families become available, it will be interesting to determine the levels of tRNA derived from intron-containing tRNA genes.
Humans express approximately 600 tRNAs, 34 of which undergo splicing (GtRNAdb). Of these, two isotypes are exclusively represented by the spliced population: tRNACys ACA and tRNAIle UAU. Although wobble base-pairing is possible for the Cys-UGU codon, shifting the decoding of all UGU to the tRNACys UGC isotype might still influence the translation kinetics of certain proteins and ultimately their folding. The recent observation that synonymous mutations can result in non-optimal codons and affect protein structure and function lends weight to this contention.1, 25 The AUA codon, on the other hand, is exclusively decoded by its corresponding tRNAIle UAU (GtRNAdb). Furthermore, in S. cerevisiae, the modification of the U34/U36 in tRNAIle UAU to pseudouridine requires the presence of the intron, highlighting the critical role splicing plays in the production of this tRNA isotype.26 Although it is difficult to pinpoint how alterations in the kinetics of translation will impact a given protein, it has recently been demonstrated that alterations of a brain-specific tRNA can cause neurodegeneration caused by ribosome stalling.27
All TSENs show constant expression in the cerebellum during and after development, which is comparable to other brain areas, such as the dorsolateral prefrontal cortex (BrainSpan Atlas). This suggests that pons and cerebellum are generally more sensitive to aberration in tRNA biology, an observation that is supported by previous work.5, 6, 7, 28, 29 Ultimately, the elucidation of the deleterious effects of these mutations will require mouse models for the individual members of the TSEN complex. These will help to determine the origin of the PCH2 phenotypes and will shed light on the hindbrain’s sensitivity to tRNA processing defects.
In this study we report pathogenic mutations in TSEN15 that cause pontocerebellar hypoplasia. Although we could not confirm the pontine and cerebellar phenotype for family III due to the lack of medical imaging, the similarity in clinical presentation and the molecular evidence suggest that the same structural abnormality underlies the phenotype. Thus, we propose to classify this phenotype as pontocerebellar hypoplasia type 2 (PCH2) and, according to the convention that uses genetic evidence as determinant, to refine it as PCH2F (see GeneReviews in Web Resources). This additional subclass shares several features with other cases of PCH2: hypoplasia of the hindbrain, progressive microcephaly, and often epilepsy. In contrast to, for instance, PCH2A, however, affected individuals did not suffer from central visual impairment and the overall clinical presentation was less severe.
The implication of the last remaining member of the tRNA splicing endonuclease complex in pontocerebellar hypoplasia underlines the importance of tRNA splicing in brain development and neurological disease. Moreover, the significant postnatal increase in clinical severity suggests a potential treatment window for drugs or genetic therapies that increase the function of this complex.
Acknowledgments
We are indebted to the families for their participation in this study. This work was supported by the US NIH (grants P01HD070494, R01NS098004, and R01NS083823; J.G.G.), the Simons Foundation for Autism Research (grant no. 275275; J.G.G.), the Howard Hughes Medical Institute (J.G.G.), the KACST grant 13-BIO1113-20 (F.S.A.), and the Saudi Human Genome Project (F.S.A.). M.W.B. is supported by an EMBO Long-Term Fellowship (ALTF 174-2015), which is co-funded by the Marie Curie Actions of the European Commission (LTFCOFUND2013, GA-2013-609409). R.A.J. was supported by the Deutsche Forschungsgemeinschaft (DFG; AB393/1-2 and AB393/2-2). We thank Guoliang Chai and Anide Johansson for helpful discussions.
Published: July 7, 2016; corrected online: August 30, 2016
Footnotes
Supplemental Data include four figures and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.05.023.
Accession Numbers
The accession number for the WES data set that includes individual I-V-3 and her parents is dbGAP: phs000288.v1.p1.
Web Resources
BrainSpan – Atlas of the Developing Human Brain, http://www.brainspan.org/
ExAC Browser, http://exac.broadinstitute.org/
GeneReviews, Namavar, Y., Eggens, V.R.C., Barth, P.G., and Baas, F. (1993). TSEN54-related pontocerebellar hypoplasia, http://www.ncbi.nlm.nih.gov/books/NBK9673/
GME Variome, http://igm.ucsd.edu/gme
GtRNAdb, http://gtrnadb.ucsc.edu
HomozygosityMapper software, http://www.homozygositymapper.org/
MutationTaster, http://www.mutationtaster.org/
OMIM, http://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Supplemental Data
References
- 1.Kirchner S., Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015;16:98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 2.Abbott J.A., Francklyn C.S., Robey-Bond S.M. Transfer RNA and human disease. Front. Genet. 2014;5:158. doi: 10.3389/fgene.2014.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puffenberger E.G., Jinks R.N., Sougnez C., Cibulskis K., Willert R.A., Achilly N.P., Cassidy R.P., Fiorentini C.J., Heiken K.F., Lawrence J.J. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS ONE. 2012;7:e28936. doi: 10.1371/journal.pone.0028936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novarino G., Fenstermaker A.G., Zaki M.S., Hofree M., Silhavy J.L., Heiberg A.D., Abdellateef M., Rosti B., Scott E., Mansour L. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343:506–511. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edvardson S., Shaag A., Kolesnikova O., Gomori J.M., Tarassov I., Einbinder T., Saada A., Elpeleg O. Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am. J. Hum. Genet. 2007;81:857–862. doi: 10.1086/521227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaffer A.E., Eggens V.R., Caglayan A.O., Reuter M.S., Scott E., Coufal N.G., Silhavy J.L., Xue Y., Kayserili H., Yasuno K. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell. 2014;157:651–663. doi: 10.1016/j.cell.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karaca E., Weitzer S., Pehlivan D., Shiraishi H., Gogakos T., Hanada T., Jhangiani S.N., Wiszniewski W., Withers M., Campbell I.M., Baylor Hopkins Center for Mendelian Genomics Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell. 2014;157:636–650. doi: 10.1016/j.cell.2014.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budde B.S., Namavar Y., Barth P.G., Poll-The B.T., Nürnberg G., Becker C., van Ruissen F., Weterman M.A., Fluiter K., te Beek E.T. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat. Genet. 2008;40:1113–1118. doi: 10.1038/ng.204. [DOI] [PubMed] [Google Scholar]
- 9.Maraş-Genç H., Uyur-Yalçın E., Rosti R.O., Gleeson J.G., Kara B. TSEN54 gene-related pontocerebellar hypoplasia type 2 presenting with exaggerated startle response: report of two cases in a family. Turk. J. Pediatr. 2015;57:286–289. [PMC free article] [PubMed] [Google Scholar]
- 10.Bierhals T., Korenke G.C., Uyanik G., Kutsche K. Pontocerebellar hypoplasia type 2 and TSEN2: review of the literature and two novel mutations. Eur. J. Med. Genet. 2013;56:325–330. doi: 10.1016/j.ejmg.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Namavar Y., Chitayat D., Barth P.G., van Ruissen F., de Wissel M.B., Poll-The B.T., Silver R., Baas F. TSEN54 mutations cause pontocerebellar hypoplasia type 5. Eur. J. Hum. Genet. 2011;19:724–726. doi: 10.1038/ejhg.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cassandrini D., Biancheri R., Tessa A., Di Rocco M., Di Capua M., Bruno C., Denora P.S., Sartori S., Rossi A., Nozza P. Pontocerebellar hypoplasia: clinical, pathologic, and genetic studies. Neurology. 2010;75:1459–1464. doi: 10.1212/WNL.0b013e3181f88173. [DOI] [PubMed] [Google Scholar]
- 13.Paushkin S.V., Patel M., Furia B.S., Peltz S.W., Trotta C.R. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell. 2004;117:311–321. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- 14.Abelson J., Trotta C.R., Li H. tRNA splicing. J. Biol. Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 15.Trotta C.R., Paushkin S.V., Patel M., Li H., Peltz S.W. Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature. 2006;441:375–377. doi: 10.1038/nature04741. [DOI] [PubMed] [Google Scholar]
- 16.Alazami A.M., Patel N., Shamseldin H.E., Anazi S., Al-Dosari M.S., Alzahrani F., Hijazi H., Alshammari M., Aldahmesh M.A., Salih M.A. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10:148–161. doi: 10.1016/j.celrep.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 17.Dixon-Salazar T.J., Silhavy J.L., Udpa N., Schroth J., Bielas S., Schaffer A.E., Olvera J., Bafna V., Zaki M.S., Abdel-Salam G.H. Exome sequencing can improve diagnosis and alter patient management. Sci. Transl. Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song J., Markley J.L. Three-dimensional structure determined for a subunit of human tRNA splicing endonuclease (Sen15) reveals a novel dimeric fold. J. Mol. Biol. 2007;366:155–164. doi: 10.1016/j.jmb.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 20.Seelow D., Schuelke M. HomozygosityMapper2012--bridging the gap between homozygosity mapping and deep sequencing. Nucleic Acids Res. 2012;40:W516–W520. doi: 10.1093/nar/gks487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seelow D., Schuelke M., Hildebrandt F., Nürnberg P. HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abou Jamra R., Wohlfart S., Zweier M., Uebe S., Priebe L., Ekici A., Giesebrecht S., Abboud A., Al Khateeb M.A., Fakher M. Homozygosity mapping in 64 Syrian consanguineous families with non-specific intellectual disability reveals 11 novel loci and high heterogeneity. Eur. J. Hum. Genet. 2011;19:1161–1166. doi: 10.1038/ejhg.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Trotta C.R., Abelson J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science. 1998;280:279–284. doi: 10.1126/science.280.5361.279. [DOI] [PubMed] [Google Scholar]
- 24.Trotta C.R., Miao F., Arn E.A., Stevens S.W., Ho C.K., Rauhut R., Abelson J.N. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell. 1997;89:849–858. doi: 10.1016/s0092-8674(00)80270-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhou M., Guo J., Cha J., Chae M., Chen S., Barral J.M., Sachs M.S., Liu Y. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature. 2013;495:111–115. doi: 10.1038/nature11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szweykowska-Kulinska Z., Senger B., Keith G., Fasiolo F., Grosjean H. Intron-dependent formation of pseudouridines in the anticodon of Saccharomyces cerevisiae minor tRNA(Ile) EMBO J. 1994;13:4636–4644. doi: 10.1002/j.1460-2075.1994.tb06786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishimura R., Nagy G., Dotu I., Zhou H., Yang X.L., Schimmel P., Senju S., Nishimura Y., Chuang J.H., Ackerman S.L. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaheen R., Han L., Faqeih E., Ewida N., Alobeid E., Phizicky E.M., Alkuraya F.S. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 2016;135:707–713. doi: 10.1007/s00439-016-1665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Ling J., Barcia G., Jing L., Wu J., Barry B.J., Mochida G.H., Hill R.S., Weimer J.M., Stein Q. Mutations in QARS, encoding glutaminyl-tRNA synthetase, cause progressive microcephaly, cerebral-cerebellar atrophy, and intractable seizures. Am. J. Hum. Genet. 2014;94:547–558. doi: 10.1016/j.ajhg.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.