Abstract
Autosomal-dominant tubulo-interstitial kidney disease (ADTKD) encompasses a group of disorders characterized by renal tubular and interstitial abnormalities, leading to slow progressive loss of kidney function requiring dialysis and kidney transplantation. Mutations in UMOD, MUC1, and REN are responsible for many, but not all, cases of ADTKD. We report on two families with ADTKD and congenital anemia accompanied by either intrauterine growth retardation or neutropenia. Ultrasound and kidney biopsy revealed small dysplastic kidneys with cysts and tubular atrophy with secondary glomerular sclerosis, respectively. Exclusion of known ADTKD genes coupled with linkage analysis, whole-exome sequencing, and targeted re-sequencing identified heterozygous missense variants in SEC61A1—c.553A>G (p.Thr185Ala) and c.200T>G (p.Val67Gly)—both affecting functionally important and conserved residues in SEC61. Both transiently expressed SEC6A1A variants are delocalized to the Golgi, a finding confirmed in a renal biopsy from an affected individual. Suppression or CRISPR-mediated deletions of sec61al2 in zebrafish embryos induced convolution defects of the pronephric tubules but not the pronephric ducts, consistent with the tubular atrophy observed in the affected individuals. Human mRNA encoding either of the two pathogenic alleles failed to rescue this phenotype as opposed to a complete rescue by human wild-type mRNA. Taken together, these findings provide a mechanism by which mutations in SEC61A1 lead to an autosomal-dominant syndromic form of progressive chronic kidney disease. We highlight protein translocation defects across the endoplasmic reticulum membrane, the principal role of the SEC61 complex, as a contributory pathogenic mechanism for ADTKD.
Introduction
Autosomal-dominant tubulo-interstitial kidney disease (ADTKD) is a broad term that encompasses a group of largely monosystemic disorders that typically lead to progressive deterioration of kidney function and renal failure.1 To date, mutations in three genes, REN (MIM: 613092), UMOD (MIM: 603860), and MUC1 (MIM: 174000), have been implicated in ADTKD,2 accounting for the majority of affected individuals with ADTKD. Depending on the mutated gene involved, other phenotypes have also been observed. Individuals with REN mutations suffer from anemia, mild hyperkalaemia, and hyperuricemia due to a decreased renal urate excretion.3, 4, 5, 6, 7 Affected individuals with UMOD mutations often develop gout due to low urinary excretion of uric acid. Notably, clinical overlap also exists in individuals with mutations in TCF2 (MIM: 137920) encoding the transcription factor hepatocyte nuclear factor 1 (HNF1B), in whom hyperuricemia, gout, and cystic kidney disease have been observed.8 In contrast, families with MUC1 mutations have no other clinical manifestations.9
Here we describe two families with ADTKD and congenital anemia caused by missense mutations c.200T>G (p.Val67Gly) and c.553A>G (p.Thr185Ala) in SEC61A1 (GenBank: NM_013336.3), encoding the alpha subunit of the integral endoplasmatic reticular membrane translocon SEC61. Taking advantage of its recently reported tertiary structure, we demonstrate that p.Thr185Ala residue lies in the constriction ring of the SEC61 translocon pore and the p.Val67Gly residue is located in the plug domain. Since mutations in SEC61A1 have not been reported, to date, in ADTKD, we transiently expressed both mutant proteins in HEK293 cells. We found that both mutant and wild-type SEC61A1 proteins are localized in the endoplasmic reticulum; however, the mutant was delocalized partly to the Golgi. Likewise, SEC61A1 was abnormally expressed and localized in the Golgi also in a kidney biopsy from an affected individual. We also performed in vivo assays to determine the role of SEC61A1 in the development of the kidney. Knockdown or CRISPR-mediated deletions of the human SEC61A1 ortholog, sec61al2, in zebrafish perturbed the development of the pronephros leading to an absence of convolution of the pronephric tubules, confirming that the SEC61 complex and its translocon function is necessary for normal renal development. Further, in vivo complementation showed that both mutant mRNAs failed to rescue the renal phenotype compared to the observation of a complete rescue by the wild-type, suggesting that both variants are pathogenic mutations. Taken together, our data expand the genetic spectrum of tubulo-interstitial kidney disorders and highlight a hitherto unknown role for SEC61A1 in the formation of the minimal filtration unit of the kidney, the nephron.
Material and Methods
Clinical Evaluation
Family 1 was ascertained by A.R., J.V.D.W., and B.L.L. at the University Hospital Ghent. Family 2 was ascertained by A.J.B. at the Section on Nephrology, Wake Forest University School of Medicine. Medical histories were obtained as a part of the affected individual’s clinical workup. Investigations were approved by the Institutional Review Boards of the participating centers and were conducted according to the principles of the Declaration of Helsinki. Karyotyping, array comparative genomic hybridization, and fluorescent in situ hybridization for the velocardiofacial syndrome were performed by standard methods.
Genome-wide Linkage Analysis and Sequencing Analysis
DNA was extracted by standard methods. 12 family members (family 1) were genotyped using the Illumina Human CytoSNP12, containing approximately 300,000 SNPs, according to the manufacturer’s guidelines (Illumina). Assuming complete penetrance and dominant inheritance, we estimated power to detect linkage via FastSLINK.10, 11, 12 The maximum LOD score encountered in simulation replicates was 2.71. Multipoint parametric linkage analysis was carried out with Merlin13 using a subset of 10,521 SNPs, with an average inter-marker spacing of 250 kb.
Whole-exome sequencing was performed using the 50 Mb SureSelect Enrichment Kit (Agilent Technologies) for enrichment followed by next-generation sequencing on a SOLiD 4 (Life Technologies). The resulting data were analyzed using an in-house analysis pipeline for variant identification and interpretation (Radboud University Medical Center).14, 15, 16, 17, 18, 19
A custom gene panel was designed to target coding regions (6.88 Mb) of 3,616 genes associated with known inherited metabolic diseases, all known genes encoding mitochondrial proteins, all OMIM genes with known phenotype, and a set of genes causing nephrologic (including SEC61A1) and neurologic diseases. DNA enrichment was performed using the Nimblegen SeqCap EZ Choice Library kit (Roche) according to the manufacturer’s protocol. DNA sequencing was performed on the captured barcoded DNA library via an Illumina Hiseq 1500 system. The resulting FASTQ files were aligned to the human Genome Reference (hg19) via Novoalign (3.02.10). After genome alignment, conversion of SAM format to BAM and duplicate removal were performed using Picard Tools (1.129). The Genome Analysis Toolkit, GATK (3.3),20, 21, 22 was used for local realignment around indels, base recalibration, and variant recalibration and genotyping. Variant annotation was performed with SnpEff23 and GEMINI.24
The SEC61A1 and the NPHP3 (MIM: 267010, 604387) candidate causal variants were amplified using the primers listed in Table S1 and standard PCR conditions. PCR products were bi-directionally sequenced using the BigDye Terminator Cycle Sequencing kit (Applied Biosystems) and separated on an ABI 3130XL Genetic Analyzer (Applied Biosystems). Sequence numbering for SEC61A1 is based on Ensembl transcript ENST00000243253, with the A nucleotide of the start codon ATG indicated as position +1.
Immunohistochemistry of Kidney Biopsies
Paraformaldehyde-fixed kidney biopsies of family members 1-II:3 and 1-III:5 were investigated. Immunodetections of UMOD, REN, and MUC1 were performed as previously described.3, 9, 25 URAT-1 was detected with rabbit polyclonal SLC22A12 antibody (Sigma-Aldrich cat# HPA024575, RRID: AB_1858650) diluted 1:25 in 5% BSA in PBS. SEC61A1 was detected with rabbit recombinant monoclonal SEC61A antibody (EPR14379) (ab183046, Abcam) diluted 1:100 in 5% BSA in PBS. The sections were incubated with primary antibodies overnight at 4°C. Detection of bound primary antibody was achieved using Dako EnVision + TM Peroxidase Rabbit Kit (DAKO) with 3,3′-diaminobenzidine as substrate.
Transient Transfection of SEC61A1 in Human Embryonic Kidney 293 Cells
Wild-type SEC61A1 cDNA was synthesized by GenScript. A single C-terminal flag-tag sequence was appended to the originally synthesized wild-type cDNA using polymerase chain reaction (PCR) with specific oligonucleotide primers. Resulting PCR product was cloned into pCR3.1 vector (Invitrogen) and introduced into the Escherichia coli TOP 10′F strain (Invitrogen). The wild-type SEC61A1-flag clones (SEC61A1_FLAG/wt) were selected by sequencing. Mutant constructs c.553A>G (SEC61A1_FLAG /185A) and c.200T>G (SEC61A1_FLAG /67G) were prepared by site-directed mutagenesis. HEK293 cells were maintained in DMEM High Glucose medium supplemented with 10% (vol/vol) fetal calf serum (PAA), 100 U/mL penicillin G (Sigma), and 100 μg/mL streptomycin sulfate (PAA Laboratories GmbH). Transfections were carried out using Lipofectamine 2000TM (Invitrogen) with either 1.5 μg or 4 μg DNA for 1.5 × 105 or 8 × 105 cells, respectively. Expression of SEC61A1_FLAG proteins was assessed by standard western blot analysis with mouse monoclonal anti-FLAG antibody (F1804, Sigma-Aldrich). Endogenously expressed SEC61A1, SEC61B, and tubulin were detected with rabbit monoclonal Anti-SEC61A antibody (ab183046, Abcam), rabbit polyclonal anti-SEC61B antibody (ABIN2707013, Antibodies Online), and mouse monoclonal anti-acetylated tubulin antibody (Sigma-Aldrich cat# T7451; RRID: AB_609894), respectively. Goat anti-rabbit IgG (Thermo) and goat anti-mouse IgG (Sigma) were used as secondary antibodies. Chemiluminiscent signal for SEC61B and SEC61A1-FLAG detection was generated using Super Signal West Femto (Pierce). Super Signal West Pico (BioRad) was used for detection of SEC61A and acetylated tubulin. Corresponding band intensities were measured by GeneTools (Syngene) and normalized to acetylated tubulin. The measurements were performed at 36 hr after transfection in three biological replicates.
Intracellular Localization of SEC61A1 in Transiently Transfected HEK Cells and Kidney Biopsies
For immunofluorescence labeling, the transfected cells were grown on 1.8 cm2 glass 4-chamber slides (BD Falcon) for 24 hr. The cells were fixed with 100% ice cold methanol for 10 min in −20°C, washed, blocked with 5% BSA in PBS, and incubated in a humidified chamber at 4°C overnight with one of the following antibodies: the mouse monoclonal anti-FLAG M2 antibody (Sigma-Aldrich cat# F1804; RRID: AB_262044) for SEC61A1_FLAG detection, the rabbit polyclonal anti-PDI antibody (Cell Signaling cat# 3501; RRID: AB_2156433) for endoplasmic reticulum detection, the rabbit polyclonal anti-GM130 antibody (Sigma-Aldrich cat# G7295; RRID: AB_532244) for Golgi detection, and the rabbit polyclonal pan Cadherin antibody (Abcam cat# ab6529; RRID: AB_305545) for plasma membrane detection.
For intracellular localization in kidney biopsies, SEC61A1 was detected with rabbit recombinant monoclonal SEC61A antibody as described above. The Golgi was detected with mouse monoclonal anti Golgi 58K Protein/Formiminotransferase Cyclodeaminase (FTCD) antibody (Sigma-Aldrich cat# G2404; RRID: AB_477002). Endoplasmic reticulum was detected with the mouse monoclonal anti-PDI antibody (Enzo Life Sciences cat# ADI-SPA-891; RRID: AB_10615355).
For fluorescence detection, species-specific secondary antibodies Alexa Fluor 488 or 555 (Molecular Probes, Invitrogen) were used. Slides were mounted in the fluorescence mounting medium ProLong Gold Antifade Mountant with DAPI (Molecular Probes, Invitrogen) and analyzed by confocal microscopy. XYZ images were sampled according to Nyquist criterion using a Leica SP8X laser scanning confocal microscope, HC PL Apo objective (63×, N.A.1.40), 405 nm diode/50 mW DMOD Flexibl, and 488 and 555 laser lines in 470–670 nm 80 MHz pulse continuum WLL2. Images were restored using a classic maximum likelihood restoration algorithm in the Huygens Professional Software (SVI). The colocalization maps employing single pixel overlap coefficient values ranging from 0 to 1 were created in the Huygens Professional Software. The resulting overlap coefficient values are presented as the pseudo color whose scale is shown in the corresponding lookup tables (LUT).
Structural Impact of Identified Mutations
Mutations were mapped into the recently resolved 3D structure of Sec6126 determined in various states, namely, Sec61 complex translating hydrophilic peptide (PDB: 4CG5), translating peptide being inserted into membrane (PDB: 4CG6), and an idle protein complex (PDB: 4CG7). Structural models were visualized with Pymol Viewer (DeLano Scientific).
Knockdown, Rescue Experiments, Immunostaining, and Embryo Manipulations
Zebrafish (Danio rerio) embryos were raised and maintained as described.27 A splice-blocking morpholino (MO) against the fourth exon-intron junction of sec61al2: (5′-TGCTCATTCAGTAGTACACACCTCT-3′) was designed and obtained from Gene Tools. We injected 4 ng of MO and/or 100 pg RNA into wild-type zebrafish embryos at the 1- to 2-cell stage. Injected embryos were scored at 4 days post-fertilization (dpf) and classified into three groups—normal, V-shaped, and straight—based on the phenotype compared with an age-matched control group from the same clutch. For wild-type and mutant rescue experiments, site-directed mutagenesis was performed to introduce the variants into the mouse wild-type sec61a1 transcript (100% identity with the human transcript at the protein level). Wild-type and mutant full-length messages were Sanger sequenced and were cloned into the pCS2 vector and transcribed in vitro using the SP6 Message Machine kit (Ambion). All the experiments were repeated three times, and a chi-square test was used to determine significance. Suppression of endogenous message was shown by PCR amplification of cDNA reverse transcribed from extracted total mRNA (primers available upon request). Whole-mount immunostaining with the anti-Na+/K+ ATPase alpha-1 subunit antibody (α6F; DSHB) was performed for examining pronephric tubules and ducts. Embryos were fixed in Dent’s fixative (80% methanol, 20% dimethylsulphoxide [DMSO]) overnight. After rehydration with decreasing series of methanol in PBS, embryos were washed with PBS, permeabilized with 10 μg/mL proteinase K, and postfixed with 4% PFA. Embryos were then washed twice with IF buffer (0.1% Tween-20, 1% BSA in 1× PBS) for 10 min at room temperature. After incubation in blocking solution (10% FBS, 1% BSA in 1× PBS) for 1 hr at room temperature, embryos were incubated with the anti-α6F antibody (1:20) in blocking solution overnight at 4°C. After two washes in IF buffer for 10 min each, embryos were incubated in the secondary antibody solution, 1:1,000 Alexa Fluor rabbit anti-mouse IgG (Invitrogen), in blocking solution for 1 hr at room temperature.
Plasmids and Generation of sec61al2 Guide RNAs
The pT7-gRNA were generated by the W. Chen lab28 and obtained from Addgene (Addgene plasmid numbers 46759). Guide RNA (gRNA) was designed to target exon 4 of sec61al2 utilizing the CHOP CHOP design tool. Oligonucleotides were annealed and cloned into the pT7-gRNA vector digested by BsmBI: sec61al2-gRNA2, 5′-GTGATCATGGCATCCAACAG-3′ and 5′-CTGTTGGATGCCATGATCAC-3′. For making gRNA, template DNA was linearized with BamHI and gRNA was generated by in vitro transcription using MEGAshortscript T7 kit (Invitrogen). To test the efficiency of the gRNA, a mix of gRNA (100 ng) and purified CAS9 protein (PNABio, cat# CP01) was injected directly into 100 1-cell-stage embryos. We phenotyped three independent clutches. To observe directly genetic editing mediated by our gRNA2 s61al2-CRISPR/Cas9 system in injected embryos (founders, F0), genomic DNA was prepared from 30 randomly selected individuals and a short stretch of DNA (exon 4 of zebrafish sec61al2) flanking the target site was PCR amplified from the genomic DNA. A T7 endonuclease I (T7EI) assay was then performed as described28 followed by Sanger sequencing; the percentage of mosaicism was estimated at ∼50% in the F0 population.
Results
Clinical Investigations
A three-generation family (family 1; Figure 1A) presented with autosomal-dominant progressive chronic kidney disease associated with congenital anemia and intrauterine and postnatal growth retardation (Table 1). At term, birth weights ranged from 1,800 to 2,400 g (normal range 2,700–4,600 g). Variable findings on physical examination included cleft palate or bifid uvula (1-I:2 and 1-II:6), velopharyngeal insufficiency (1-II:6), pre-axial polydactyly (1-II:6), and mild cognitive impairment (1-II:1, 1-II:3, 1-II:5, and 1-II:6). Blood pressures were consistently in the normal range. Significant laboratory findings included normochromic, normocytic anemia that was responsive to erythropoietin, elevated serum creatinine levels, mild hyperuricemia, and the absence of proteinuria. Hematological and biochemical parameters for the three youngest affected individuals (1-III:4, 1-III:5, 1-III:7) are summarized in Table 1. Affected individuals 1-III:4 and 1-III:7 were lost to follow-up.
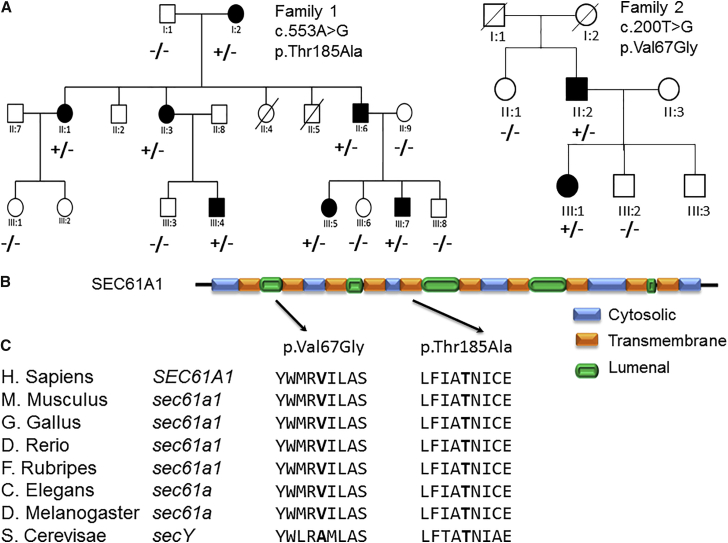
Figure 1.
Pedigrees, Schematic Protein Representation, and Conservation of SEC61A1
(A) Circles indicate women, squares indicate males; filled symbols indicate affected individuals; plus sign indicates presence of SEC61A1 variation; minus sign indicates absence of SEC61A1 variation.
(B) Position of the p.Val67Gly change in the plug region and the p.Thr185Ala change in the fifth transmembrane region of the translocase-transmembrane domain of SEC61A1.
(C) Conservation of the Val67 and Thr185 amino acids throughout evolution.
Table 1.
Clinical, Hematological, Biochemical, and Genetic Investigation
| Individual | 1-III:4 | 1-III:5 | 1-III:7 | 2-III:1 | 2-II:2 | Unit | Reference Value |
|---|---|---|---|---|---|---|---|
| Age at investigation | 11 | 8 | 4 | 27 | 61 | ||
| Hematology | |||||||
| Leukocytes | 8.45 | 8.4 | 7.89 | 3.4 | 3.5 | 103/μL | 4.5–13.5 |
| Hemoglobin | 10.3 | 10 | 9.1 | 9.5 | 12.3 | g/dL | 11.5–15.5 |
| Hematocrit | 32.5 | 29.6 | 27.9 | 28.7 | 36.9 | % | 35.0–45.0 |
| Biochemistry | |||||||
| Ureum | 39.2 | 36.9 | 42.9 | 47 | 42 | mg/dL | 13–43 |
| Creatinin | 1.20 | 1.17 | 0.9 | 1.65 | 2.6 | mg/dL | 0.53–0.79 |
| Uric acid | 9.0 | 6.9 | 5.6 | 7.8 | 5.8a | mg/dL | 3.4–7.0 |
| eGFR | 52.2 | ||||||
| Other characteristics | |||||||
| Birth weight | 2,220 | 2,400 | 1,800 | 3,430 | g | ||
| Birth length | 45 | 47 | 45 | cm | |||
| Onset nephropathy | 11 | 1 | 4 | 18 | year | ||
| Current weight | P3 | P3 | P50 | ||||
| Current length | P10 | <P3 | P25 | ||||
Abbreviations are as follows: eGFR, estimated glomerular filtration; P, percentile
Individual with a history of hyperuricemia; value obtained while on allopurinol.
The proband (1-III:5) presented at 12 years with height of 132 cm (< P3). Physical exam was otherwise unremarkable. Laboratory studies revealed a serum creatinine concentration of 1.46 mg/dL, blood urea nitrogen concentration of 93 mg/dL, and serum uric acid level of 6.7 mg/dL at last follow-up. The fractional excretion rates of sodium and uric acid were normal and the urinary potassium/(urinary potassium + sodium) ratio was low. Renin (5–14 pcg/mL) and aldosterone (5–14 ng/dL) were always in the low normal range. There was no hematuria or proteinuria. Ultrasound examinations revealed small, dysplastic kidneys without cysts.
At the age of 53 years, individual 1-II:1 has a serum creatinine concentration of 1.96 mg/dL, blood urea nitrogen level of 109.6 mg/dL, and estimated glomerular filtration rate (eGFR) of 27 mL/min/1.73 m2. She has a normal hemoglobin level (12.7 g/dL) under erythropoietin treatment. No proteinuria or hematuria was detected. Blood pressure was borderline 150/80 mmHg. She measured 147 cm for a weight of 63 kg. Affected individual 1-I:2 died at the age of 73 years while being on dialysis. She was admitted with acute sepsis (CRP 112 mg/L), lung edema, and kidney failure (creatinine 5.13 mg/dL). No gout or neutropenia was observed in family 1.
Kidney biopsies of family members 1-II:3 and 1-III:5 revealed multiple small foci of tubulo-interstitial lesions: clusters of atrophic tubules, either featured by thickened tubular basement membranes or those of endocrine type, occasionally protein casts in tubular lumina and discrete interstitial fibrosis (Figure 2A). Glomerular changes were dominated by cystic dilatation of Bowmann’s spaces with collapsed or rudimentary glomerular tufts and often filled with precipitate of plasma proteins (Figure 2B). Focal periglomerular fibrosis and focal glomerular sclerosis, arterial intimal thickening, and luminal narrowing of blood vessels were present. Cytogenetic analyses on affected individual 1-III:5, including both karyotyping and 22q11-fluorescent in situ hybridization, to exclude velocardiofacial syndrome, revealed normal results. In addition, whole-genome microarray analysis did not reveal any subchromosomal deletions or duplications.
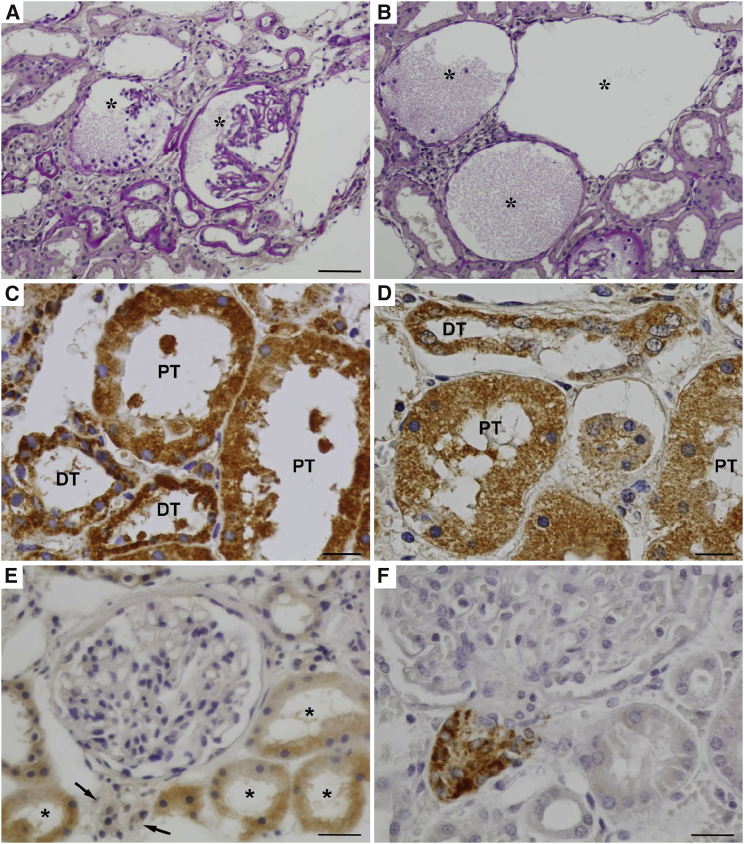
Figure 2.
Kidney Biopsy from Individual 1-II:3 with the p.Thr185Ala Substitution
(A) Clusters of atrophic tubules surrounding glomeruli with collapsed or rudimentary capillary tufts (marked by asterisks). A part of affected tubules shows thickening of tubular basement membranes; the remaining are endocrine-type atrophic tubules with narrowed lumina and simplified epithelium. PAS staining.
(B) Prominent cystic dilatation of Bowmann’s spaces (marked by asterisks), which are sometimes filled with finely granular proteinaceous material. PAS staining.
(C and D) SEC61A protein was detected immunohistochemically in proximal tubules (PT) and distal tubules (DT) in kidney tissue in both affected individual (C) and a control subject (D). Compare coarsely granular intracytoplasmic staining in the affected individual with finely granular and less intensive pattern in the control subject.
(E) Renin was undetectable immunohistochemically in juxtaglomerular apparatus (marked by arrows) in an affected individual but weak finely granular staining was found in the cytoplasm of renal tubules.
(F) Comparison with strong renin positivity in juxtaglomerular apparatus and negativity in tubular epithelium in a control subject.
Scale bars represent 100 μm in (A) and (B), 30 μm in (C) and (D), and 50 μm in (E) and (F).
Family 2 was referred for evaluation of ADTKD occurring in the father and daughter, but no involvement of other affected family members (Figure 1A). Both affected relatives suffered from congenital anemia and neutropenia, gout in the second decade of life, and chronic kidney disease. They had normal growth and intelligence.
The proband 2-III:1 presented at 2 weeks of age, when she was admitted for an abscess on the right buttock. The white blood count at that time was 14.3 × 109/L with 4% neutrophils, 92% lymphocytes, and 4% monocytes. The affected individual had persistent granulocyte stimulating factor-unresponsive neutropenia and recurrent admissions for cutaneous abscesses. Although the neutropenia persisted, infections were no longer present after age 12. The affected individual developed gout at age 18 years, with a serum urate of 7.2 mg/dL and serum creatinine of 1.1 mg/dL (eGFR 73 mL/min/1.73 m2). At age 28, she was diagnosed with hypotension (systolic blood pressure readings in the high 80s) and hyperkalemia (serum potassium 5.7 mEq/L). There was no proteinuria or hematuria. The plasma renin activity was 1.0 ng/mL/hr (normal range 0.5–5.9 ng/mL/hr), and the serum aldosterone level was 9.0 ng/dL (normal range 5–80 ng/dL). She was treated with fludrocortisone. Renal ultrasound showed the left kidney to be 9 cm and right kidney 9.4 cm with hyperechogenic parenchyma.
The father (2-II:2) also suffered from cutaneous abscesses requiring hospitalization until age 12 years. He demonstrated anemia and neutropenia. A bone marrow biopsy performed at age 12 showed a decrease in normoblasts and white blood cell precursors with normal megakaryocytes. Gout was diagnosed at age 21, and the affected individual presented for evaluation of chronic kidney disease at 51 years with a serum creatinine of 1.4 mg/dL (eGFR 58 mL/min/1.73 m2). Over 10 years the serum creatinine increased to 2.8 mg/dL (eGFR 23 mL/min/1.73 m2). Renal ultrasound revealed that the right kidney is 11.6 cm and the left 13.9 cm, with multiple bilateral simple cysts throughout the kidney. Urinary uromodulin was decreased in both individuals (Figure S1).
Molecular Genetic Analysis
Linkage Analysis and Whole-Exome Sequencing in Family 1
Based on the phenotypic similarities between the clinical presentation of family 1 and individuals with REN mutations (early-onset anemia, hyperuricemia, and progressive kidney failure), we first excluded REN in the proband (1-III:5) by Sanger sequencing. Subsequently, we collected DNA from 12 family members, including 7 affected individuals, and performed genome-wide multipoint parametric linkage analysis using SNP microarrays. Under a dominant model of inheritance, we identified a candidate region with a maximum LOD score of 2.7 on chromosome 3q14-25.1 (Figure S2). The region was confirmed by microsatellite marker analysis (between markers D3S3513 and D3S1299; results not shown). This 32-Mb candidate region (genomic positions: 120M–152M; NCBI build 37 [hg 19]) was gene rich with a total of 357 annotated genes. We next performed whole-exome sequencing on the proband and focused on rare (<1% minor allele frequency in the general population) and non-synonymous coding changes within the critical interval. We found two heterozygous nucleotide changes within the linked region on chromosome 3: c.1189C>T (p.Arg397Cys) in exon 7 of NPHP3 and c.553A>G (p.Thr185Ala) in exon 7 of SEC61A1. The former did not segregate with the disease in the family whereas the latter did (Figure 1A). The Thr185 position is conserved at the nucleotide (PhyloP score: 4.95) and amino acid levels (Figure 1C). The variant was predicted to be pathogenic by two prediction tools, SIFT29 and MutationTaster,30 although it was benign according to PolyPhen.31 In addition, the variant was absent from the 1000 Genomes Project database,32 the Exome Variant Server, the ExAC database, a series of 204 Belgian control chromosomes, and internal exome databases curated in Prague (∼600 exomes and 140 custom gene panels) and Nijmegen (∼500 exomes).
Screening for SEC61A1 Mutations in Individuals with a Similar Phenotype
The second SEC61A1 variant was identified in 1 out of 46 unrelated probands with an ADTKD-like phenotype using a custom gene panel sequencing developed at the Institute for Inherited Metabolic Disorders in Prague. The proband from family 2 carried a heterozygous missense variant c.200T>G (p.Val67Gly) in SEC61A1. The variant is also conserved at the nucleotide (PhyloP score: 6.32) and amino acid levels (Figure 1C). The variant was also predicted to be damaging by SIFT and PolyPhen and disease causing by MutationTaster. The p.Val67Gly change was reported once in the ExAC database (1/121,410 alleles) and was absent from all other databases mentioned above. Co-segregation of the p.Val67Gly change in the family was confirmed by Sanger sequencing (Figure 1A).
Prediction of the Structural and Functional Impact of the SEC61A1 Mutations
SEC61A1 encodes the protein transport protein Sec61 subunit alpha isoform 1 (SEC61α) that assembles with the protein transport protein Sec61 subunit beta (SEC61β) and the protein transport protein Sec61 subunit gamma (SEC61γ) to form the heterotrimeric protein-conducting channel SEC61 that is a part of the mammalian ER translocon. The Thr185 residue lies in the transmembrane helix 5 (TM5) that is located in the vicinity of translocated peptides.33 Residues Ile179 and Ile183 are involved in the formation of the constriction ring within a pore of the SEC61. As such, the mutation p.Thr185Ala is likely to disrupt the intrahelical hydrogen bond between the Thr185 side chain and the carbonyls of Leu181 and Phe182 and thus affects the structural integrity of the channel.34 Structural mapping of Val67 showed that this residue is located directly in the translocon pore and is part of a plug domain that seals and stabilizes the pore during the closed state (Figure 3). The increased flexibility of the glycine residue in this position may destabilize the pore structure and alter ion permeability of the channel, affecting translocation efficiency and/or inducing abnormal signal peptide orientation. A similar effect on the post-translational modification and sorting of secretory and transmembrane proteins has been described for the neighboring Arg66 residue in SEC61A1.35, 36 Systematic mutagenesis of the conserved and charged residues forming the constriction ring and plug domain in bacteria and yeast demonstrated that these alterations had no effect on translocon functionality and cell viability but affected translocation efficiency, integration of hydrophobic sequences into lipid bilayers, and stability of the translocon in the closed state.37, 38
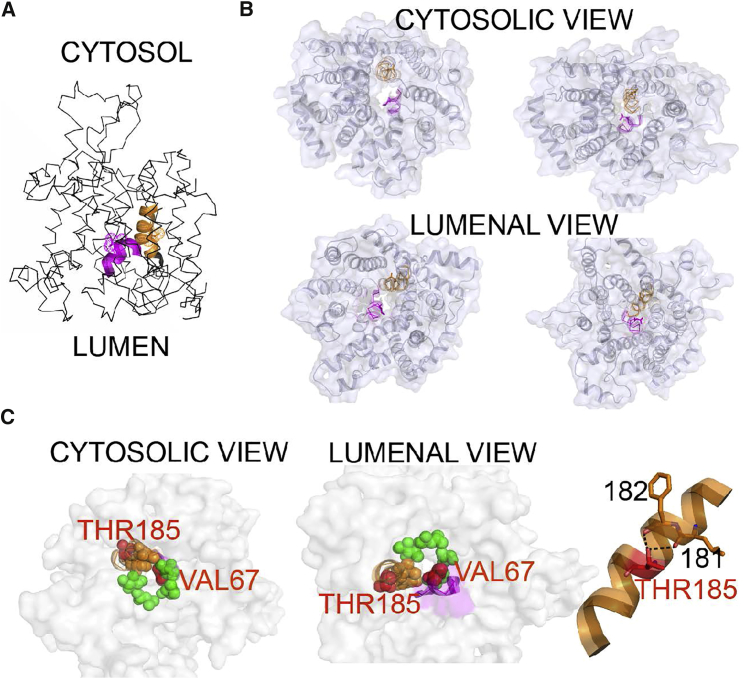
Figure 3.
Structural Topology of the Mutations in Sec61
The cryo-EM based models of translocons translating hydrophilic peptide with opened pore (PDB: 4CG5) and translating peptide inserted to membrane possessing a pore sealed by a plug domain (PDB: 4CG6) were used for illustration. Regions containing mutations are depicted with cartoon representation, namely plug domain (p.Val67Gly) in magenta and TM5 (p.Thr185Val) in orange. Mutated residues are highlighted as dots and/or sticks.
(A) Overall structure of Sec61 and location of the mutated residues.
(B) Orientation of the mutated residues in the Sec61 structure with opened translocation pore (left) and with pore sealed by a plug domain (right).
(C) Left: constriction ring formed by apolar residues represented by green (TM2, TM7, and TM10) and orange (positioned at TM5 near Thr185 residue) spheres; mutated residues are shown as red spheres. Right: detail of TM5 illustrating hydrogen bonding of hydroxyl Thr185 with carbonyls of Leu181 and Phe182.
Immunohistochemical Analysis of SEC61A1 in Kidney Biopsies
To assess the effect of identified mutations on the expression of SEC61A1, we performed immunohistochemical analysis of a kidney biopsy obtained from family member 1-II:3 with the p.Thr185Ala substitution. We observed a strong coarsely granular cytoplasmic staining in proximal and distal tubules and collecting ducts (Figure 2C). Upon comparison to the finely granular and less intensive intracellular staining observed in control subjects (Figure 2D), this was suggestive of abnormal intracellular localization and/or aggregation of SEC61A1.
Given that translocon malfunction may affect cotranslational translocation, post-translational processing, and cellular trafficking of secretory and transmembrane proteins, we also performed immunohistochemical analysis of renin, uromodulin, mucin-1, and urate transporter SLC22A12. In the affected kidney, we observed absence of renin staining in juxtaglomerular granular cells, which was accompanied by mild granular positivity of renin staining in the cytoplasm of tubular cells (Figure 2E). This observation was similar to that made in renal tissue from individuals with REN mutations.3 Expression patterns of the other proteins were comparable with controls (Figure S3). Atrophic cortical tubules were predominantly MUC1 and partially UMOD positive, suggesting their origin in distal tubules and collecting ducts in kidney biopsy.
Intracellular Localization of SEC61A1 Variants in Human Embryonic Kidney Cells and Kidney Biopsies
To determine their production, stability, and intracellular localization, we transiently expressed wild-type SEC61A1 (SEC61A1_FLAG/wt) and both mutant proteins (SEC61A1_FLAG /185A, SEC61A1_FLAG /67G) in HEK293 cells. Quantitative western blot analysis demonstrated that, compared to wild-type protein, production of the SEC61A1_FLAG/67G and SEC61A1_FLAG/185A is decreased significantly (Figures 4A and 4B). Overexpression of neither protein affected relative amounts of the endogeneously expressed SEC61A1 and SEC61B (Figure 4C). Immunofluorescence analysis and colocalization studies showed that wild-type protein was present in finely granular structures that were localized exclusively in the ER (Figure 4D). Both mutant proteins formed intracellular clumps localized in the ER (Figure 4D, panels I and R) and partly also in the Golgi (Figure 4D, panels I and O and in more detail in Figure S4).
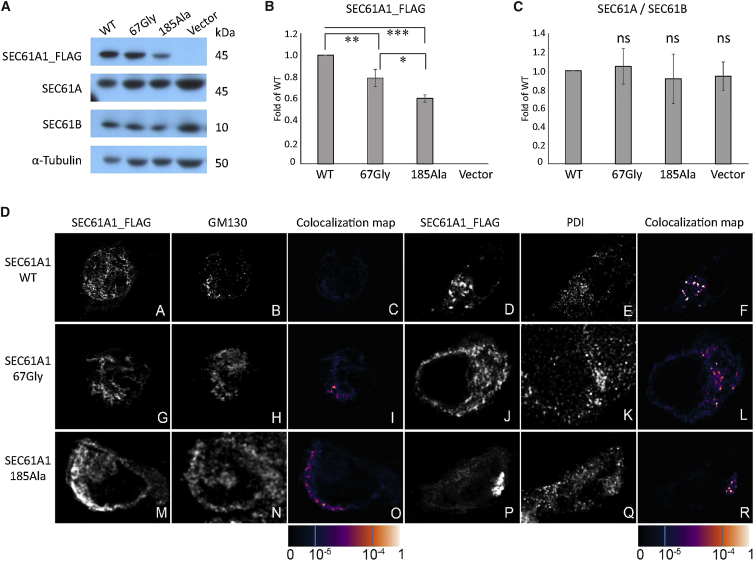
Figure 4.
Transient Expression and Intracellular Localization of SEC61A1_FLAG Variants in Human Embryonic Kidney 293 Cells
(A) Western blot detection of transiently expressed wild-type and mutated SEC61A1_FLAG proteins and endogenously expressed SEC61A, SEC61B, and tubulin at 36 hr post-transfection.
(B) Quantitative image analysis of wild-type and mutated SEC61A1_FLAG proteins demonstrating decreased amounts of mutated proteins compared to the wild-type. Results represent means of fold change ± SD of the relative signal intensities of mutated proteins to the wild-type protein from three biological replicates. Signal intensities of SEC61A1_FLAG proteins were normalized to that of α-tubulin. Statistical significance was assessed using Student’s t test; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(C) Quantitative image analysis of the endogenously expressed SEC61A and SEC61B demonstrating that overexpression of neither protein significantly affected relative amounts of the endogeneously expressed SEC61A1 and SEC61B transient expression of SEC61A1. Results represent means of fold change ± SD from three biological replicates of the relative signal intensities of SE61A over SEC61B in cells expressing individual mutated SEC61A1_FLAG protein and empty vector compare to that of cells expressing the wild-type protein. Signal intensities of SEC61A and SEC61B were normalized to that of α-tubulin. Statistical significance was assessed using Student’s t test.
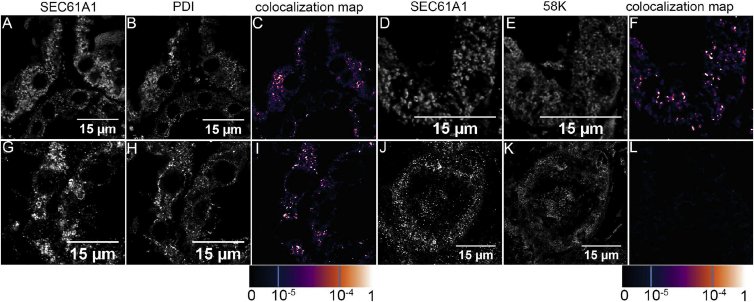
(D) Immunofluorescence analysis showing that the wild-type SEC61A1_FLAG is present in a finely granular (subpanel A) or coarsely granular (subpanel D) structures. Co-staining of wt-SEC61A1_FLAG with Golgi matrix protein GM130 (subpanel B) and with Protein disulphide isomerase (PDI) (subpanel E), a marker of endoplasmic reticulum (ER), demonstrating absence of the wt-SEC61A1_FLAG in the Golgi (subpanel C) but presence in the ER (subpanel F). p.Val67Gly (subpanels G and J) and p.Thr185Ala (subpanels M and P) variants of SEC61A1_FLAG are present in a form of intracellular clumps that are more pronounced in the latter. Co-staining with GM130 (subpanels H and N) and PDI (subpanels K and Q) demonstrating localization of both mutant proteins in the Golgi (subpanels I and O) as well as in ER (subpanels L and R). The degree of SEC6A1A_FLAG colocalization with selected markers is demonstrated by the fluorescent signal overlap coefficient values that range from 0 to 1. The resulting overlap coefficient values are presented as the pseudo color which scale is shown in corresponding lookup table.
To test the relevance of these findings in the setting of the tissue from the affected individual, we compared the intracellular localization of SEC61A1 in control and affected kidney obtained from family member 1-II:3 with p.Thr185Ala substitution. In the affected kidney, SEC61A1 is present in coarsely granular structures (Figures 5A and 5D). SEC61A1 was localized in the ER (Figure 5C) and a significant proportion of the protein was also localized in the Golgi (Figure 5F and in more detail in Figure S5). In contrast, in control kidney sections, SEC61A1 was present in finely granular structures (Figures 5G and 5J) and was located exclusively in the ER (Figure 5I).
Figure 5.
Intracellular Localization of SEC61A1 in Affected Kidney
(A and D) In affected kidney, SEC61A1 is present in coarsely granular structures.
(B, C, E, and F) Co-staining of SEC61A1 with Protein disulphide isomerase (PDI) (B), a marker of endoplasmic reticulum (ER), and with 58k Golgi-protein (E) demonstrate localization of the SEC61A1 in the ER (C) and in the Golgi (F).
(G and J) In control kidney, SEC6A1A is present in finely granular structures.
(H, I, K, and L) Co-staining with (H) PDI and (K) GM130 demonstrating localization of SEC61A1 in ER (I) but not in the Golgi (L). The degree of SEC6A1A colocalization with selected markers is demonstrated by the fluorescent signal overlap coefficient values that ranging from 0 to 1. The resulting overlap coefficient values are presented as the pseudo color which scale is shown in corresponding lookup table.
The presence of the mutant SEC1A1 proteins in the Golgi thus suggests that these proteins are recognized as abnormal and as such they are probably dislocated to the ER-Golgi intermediate compartment (ERGIC) and subjected to endoplasmic-reticulum-associated protein degradation (ERAD).39
Functional Testing of SEC61A1 Variants in Zebrafish Embryos
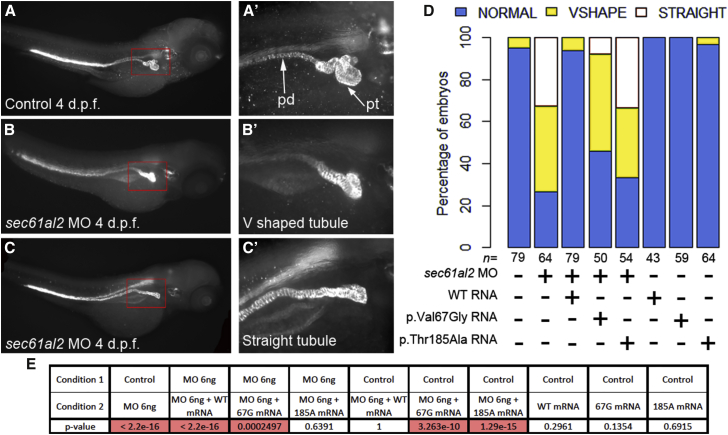
Our genetic data, the in silico analysis of the effect of the variants on the structure of SEC61A1, and the abnormal intracellular localization of the mutant proteins suggested that mutations in SEC61A1 is a strong candidate for the dominant familial nephropathy in both pedigrees. To our knowledge, mutations in SEC61A1 have not yet been associated with renal phenotypes in humans, nor has it been known to be necessary for the development of the kidney. Thus, we sought to determine the pathogenicity of the variants in vivo. Some of the pathologies observed in the affected individuals, such as fibrosis and gout, cannot be modeled reliably in early development. However, the structure of the nephron is conserved between human and zebrafish and one of the common features among affected individuals is tubular atrophy.40 Therefore, we asked whether SEC61A1 is involved in the development of the pronephros by suppressing the zebrafish ortholog of SEC61A1 and examining the complete system of paired pronephric tubules and ducts at 4 days post-fertilization (dpf).
Using reciprocal BLAST, we identified a single Danio rerio SEC61A1 ortholog (sec61al2 on chromosome 6; 96% amino acid identity). We designed a splice-blocking morpholino (sbMO), targeting the splice donor site of exon 4, which we injected into zebrafish eggs. Reverse-transcription polymerase chain reaction (RT-PCR) demonstrated that the sbMO affected the correct splicing of the sec61al2 transcript (i.e., skipping of exon 4; Figure S6). To test whether sec61a1 suppression affects the development of the pronephros, we stained the sbMO-injected embryos with an anti-Na+/K+ ATPase alpha-subunit monoclonal antibody (α6F), an enzyme expressed by the epithelial cells in the pronephric tubules and ducts.41 The embryos were scored and binned into three categories: normal convolution of the pronephric tubules, V-shaped (partial convolution of the tubules), or straight (absence of tubular convolution) (Figures 6A–6C). Masked scoring of embryos at 4 dpf (60 embryos on average per condition, repeated three times) showed an overall percentage of 70% of affected embryos that included 30% with an absence of convolution of the pronephric tubules and 40% with partial convolution (Figure 6D). By contrast, the pronephric ducts were extended along the trunk indistinguishably from control embryos (Figures 6A–6C). The tubular atrophy observed in the sec61al2 morphants was unlikely to be driven by overall developmental delay; morphants had a normal appearance with regard to their pigment cells and head size. In addition, there was no appreciable pathology in other internal organs, such as the heart or the swim bladder. Finally, their body length was indistinguishable from control embryos from the same clutch (Figures 6A–6C). The phenotype was also specific; the observed pathology could be rescued efficiently with co-injection of wild-type mouse full-length sec61a1 mRNA (100% identity between human and mouse at the protein level; p = 2.2 × 10−16 between morphants and co-injection with sbMO and wild-type message; Figures 6D and 6E). Next, to test whether the variants identified in the two pedigrees might be pathogenic, we co-injected sec61al2 sbMO with the mouse sec61a1 mRNA harboring the c.200T>G (p.Thr185Ala) or c.553A>G (p.Val67Gly) variant and we scored for presence or absence of convolution of the pronephric tubules. Masked scoring at 4 dpf showed that the mutant messages failed to rescue the tubular atrophy phenotype driven by the sbMO. Embryos co-injected with sbMO and c.553A>G (p.Thr185Ala) were indistinguishable from morphants, suggesting a complete loss of function of the c.553A>G (p.Thr185Ala) variant (p = 0.64, Figures 6D and 6E). The second variant, c.200T>G (p.Val67Gly), was found to be a partial loss-of-function allele: we observed a partial rescue of the phenotype in embryos co-injected with sbMO and (c.200T>G [p.Val67Gly]; Figures 6D and 6E). To validate our MO-derived data, we designed and tested guide RNAs to generate a sec61al2 CRISPR mutant. After validation of the guide RNA efficiency and evaluation of the percentage of mosaicism in the F0 founders (50%; Figure S7), we stained the pronephric tubules. We found that the F0 CRISPR mutants fully recapitulated the morphants; we observed convolution defects including V-shaped and straight tubules in 55% of the F0 founders injected with guide RNA and Cas9 (Figure S7). Taken together, our functional data suggest that SEC61A1 is necessary for the proper tubular organization of the nephron and that variants in SEC61A1 found in the two families are pathogenic.
Figure 6.
Suppression of SEC61A1 Leads to Pronephric Tubular Atrophy in Zebrafish
(A–C) Whole-mount immunostaining of 4 days post-fertilization (dpf) zebrafish larvae with anti-Na+/K+-ATPase alpha subunit monoclonal antibody (α6F) shows the overall anatomy of the pronephric ducts (pd) and pronephric tubules (pt), which become progressively convoluted in control larvae. Three levels of convolution were assessed: convoluted (normal) (A and A′), V-shaped (B and B′), and straight pronephric tubule (C and C′). The pronephric ducts are normal.
(D) Qualitative scoring of the tubular atrophy was performed in larvae batches injected with sec61al2 MO alone; MO and mutant (p.Thr185Ala) RNA or (p.Val67Gly) RNA; MO and wild-type (WT) capped-RNA; WT and mutant RNAs alone; and control.
(E) Results of Fisher’s exact test conducted between pairs of conditions. The significant p values (< 0.05) are highlighted in red.
Discussion
Here, we present evidence that mutations in SEC61A1 with complete or partial loss of function compared to wild-type can cause autosomal-dominant progressive tubulo-interstitial and glomerulocystic kidney disorder with anemia. Combining linkage analysis with WES in one family, followed by targeted testing and identification of a second family with a similar phenotype, indicated that missense mutations, predicted to affect the pore function of the SEC61 translocon complex, segregate with the disorder and are the likely drivers. Consistent with causality, one of the discovered alleles was absent from 121,410 control alleles, while the second was found only once. Notably, SEC61A1 is evolutionarily conserved and intolerant to deleterious variation in humans. There are no individuals with truncating or nonsense mutations in the ExAC database and most of the 94 identified alleles carrying missense variants represent changes that are structurally predicted to be neutral.
To interrogate further the functional candidacy of the discovered variants, we found that when expressed in HEK293 cells, the wild-type protein localized exclusively in the ER whereas both SEC6A1A variants are localized in the ER and abnormally in the Golgi. These in vitro data were then confirmed in kidney tissue from an affected individual. Taken together, this suggests that the mutant proteins probably do not interfere with translocons that are formed by the remaining pool of wild-type proteins, but rather they form translocons that are malfunctioning, which causes disease. We also utilized zebrafish embryo as an in vivo model. Consistent with our genetic and in vitro findings, suppression of sec61a1 recapitulates the tubular phenotype, while both discovered mutations impede the ability of the protein to function. Of note, the phenotype of the family bearing the functionally determined null allele (c.553A>G [p.Thr185Ala]) appears to have a more severe and more complex phenotype, although caution is obviously warranted in extrapolating genotype-phenotype correlations. The discovery of additional families with SEC61A1 mutations and the functional stratification of new alleles will be necessary to test this hypothesis. Likewise, we speculate that the SEC61A1 alleles probably contribute to some of the additional phenotypes (e.g., cognitive impairment, neutropenia) found in the two families, though further laboratory investigation will be required to confirm this.
In contrast to the phenotype observed in the Sec61a1Y344H/Y344H (GenBank: NM_016906.41) ENU-induced mouse homozygous for the mutant allele at codon 344 encoding a histidine-to-tyrosine substitution,42 we did not observe any glucose homeostasis or hepato-steatosis problems in our affected individuals. To the best of our knowledge, the renal tubular structures in homozygous and heterozygous mice have not been investigated. This discrepancy between phenotypic effects may be explained by the structural mapping of the mutations. Mutations described in our study affect directly the selectivity and permeability of the pore of the translocon channel. This implies that every translocon channel that contains the mutated SEC61A1 subunit is dysfunctional; the mutations are therefore acting as complete or partial loss of function. On the other hand, the recessive p.His344Tyr substitution is located at the luminal helix, forming a patch for the insertion of nascent peptides into membrane. Therefore, we can speculate that this mutation should not alter translocon pore functions per se. However, additional functional experiments both in vitro and in vivo are required to assess fully the pathogenicity of the p.His344Tyr substitution in the context of the renal development.
The SEC61 complex is a major component of the mammalian translocon, a complex that is needed to transport newly synthesized secretory proteins into the ER,43 integrate nascent proteins into membranes, and maintain ER ion permeability barrier.44 Our study of the structure of SEC61 translocon indicated that the two variants have the potential to disrupt the function of translocon pore by affecting the integrity of the plug domain or the constriction ring. We therefore speculate that altered structural properties of SEC61A1 may destabilize the translocon pore and lead to alterations in post-translational modifications, folding and sorting of various secretory and transmembrane proteins, and/or alterations in Ca2+ homeostasis. Such dysfunctions may induce ER stress or prevent the cell from responding appropriately to induced ER stress, with either ultimately leading to apoptosis. Chronic ER stress is the major pathogenic mechanism driving development and progression of ADTKD caused by REN3 and UMOD mutations.45 Of note, most identified REN mutations reduce the hydrophobicity of the signal peptide. Recent structural studies of Sec61 complexed with signal peptide indicates that aberrant interaction of the mutated signal peptides may destabilize the opened SEC61 translocon pore during co-translational translocation.46 This situation may therefore represent a common pathogenetic mechanism leading to similar clinical symptoms in affected individuals with REN and SEC61A1 mutations. ER stress has been postulated to play a mechanistic role in other disorders such as diabetes and open angle glaucoma.47, 48 Promising clinical trial results indicate that the chemical chaperone sodium phenylbutyrate (BPA) is efficient on these diseases and can alleviate the phenotypes in both mouse models and humans.49, 50 We therefore hypothesize that PBA may provide therapeutic benefits in individuals with SEC61A1 mutations and in other ER stress-related renal diseases.
In conclusion, we report two deleterious mutations in SEC61A1, a gene implicated in ADTKD. In light of previous studies,51, 52 our work provides additional evidence of the importance of the translocon in the normal development of the kidney.
Acknowledgments
B.L.L. is senior clinical investigator of the Fund for Scientific Research, Flanders (FWO, Belgium). This research was supported by funding from the Fund for Scientific Research, Flanders (FWO, Belgium) [G.0458.09; G.0221.12], the Fondation Leducq (MIBAVA 12CVD03), Starting grant (to B.L.L.) of the European Research Council (ERC- StG-2012-30972-BRAVE), and the University of Antwerp (Lanceringsproject). N.K. is a Distinguished Brumley Professor. This work was supported by grants P50DK096415 and P30DK096493 (to N.K.). S.K., A.P., K.H., and V.S. are funded by Charles University institutional programs PRVOUK-P24/LF1/3, UNCE 204011, and SVV2016/260148 and by the project LQ1604 NPU II from the Ministry of Education of the Czech Republic. This work was specifically supported by grants LH12015 from the Ministry of Education of the Czech Republic and NT13116-4/2012 from the Ministry of Education and Ministry of Health of the Czech Republic. A.P. was supported by GA UK No. 1402213. A. Hnízda was supported by the project LO1304 (program “NPU I”). A. Hoischen was supported by the Netherlands Organization for Health Research and Development (ZonMW 916-12-095). A.J.B. is supported by the US NIH grant 1R21DK106584-01.
We thank members of the Genomic Disorders Group Nijmegen, the Genomic facility in Motol University Hospital in Prague (OPPK.CZ.2.16/3.100/24022), and The National Center for Medical Genomics (LM2015091) for their technical support with the exome and gene panel sequencing. We thank Dr. David Tilstra from CentraCare Clinic, St. Cloud, who referred family 2 for evaluation and kindly provided medical records. We thank Corinne Antignac for critical revision of the manuscript.
Published: July 7, 2016
Footnotes
Supplemental Data include seven figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.05.028.
Web References
1000 Genomes, http://www.1000genomes.org
CHOPCHOP, https://chopchop.rc.fas.harvard.edu/
Ensembl Genome Browser, http://www.ensembl.org/index.html
ExAC Browser, http://exac.broadinstitute.org/
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Supplemental Data
References
- 1.Eckardt K.U., Alper S.L., Antignac C., Bleyer A.J., Chauveau D., Dahan K., Deltas C., Hosking A., Kmoch S., Rampoldi L. Autosomal dominant tubulointerstitial kidney disease: diagnosis, classification, and management--A KDIGO consensus report. Kidney Int. 2015;88:676–683. doi: 10.1038/ki.2015.28. [DOI] [PubMed] [Google Scholar]
- 2.Bleyer A.J., Kmoch S. Autosomal dominant tubulointerstitial kidney disease: of names and genes. Kidney Int. 2014;86:459–461. doi: 10.1038/ki.2014.125. [DOI] [PubMed] [Google Scholar]
- 3.Zivná M., Hůlková H., Matignon M., Hodanová K., Vylet’al P., Kalbácová M., Baresová V., Sikora J., Blazková H., Zivný J. Dominant renin gene mutations associated with early-onset hyperuricemia, anemia, and chronic kidney failure. Am. J. Hum. Genet. 2009;85:204–213. doi: 10.1016/j.ajhg.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bleyer A.J., Zivná M., Hulková H., Hodanová K., Vyletal P., Sikora J., Zivný J., Sovová J., Hart T.C., Adams J.N. Clinical and molecular characterization of a family with a dominant renin gene mutation and response to treatment with fludrocortisone. Clin. Nephrol. 2010;74:411–422. doi: 10.5414/cnp74411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck L.H., Jr. Childhood membranous nephropathy and dietary antigens. Am. J. Kidney Dis. 2012;59:174–176. doi: 10.1053/j.ajkd.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Stibůrková B., Majewski J., Hodanová K., Ondrová L., Jerábková M., Zikánová M., Vylet’al P., Sebesta I., Marinaki A., Simmonds A. Familial juvenile hyperuricaemic nephropathy (FJHN): linkage analysis in 15 families, physical and transcriptional characterisation of the FJHN critical region on chromosome 16p11.2 and the analysis of seven candidate genes. Eur. J. Hum. Genet. 2003;11:145–154. doi: 10.1038/sj.ejhg.5200937. [DOI] [PubMed] [Google Scholar]
- 7.Hodanová K., Majewski J., Kublová M., Vyletal P., Kalbácová M., Stibůrková B., Hůlková H., Chagnon Y.C., Lanouette C.M., Marinaki A. Mapping of a new candidate locus for uromodulin-associated kidney disease (UAKD) to chromosome 1q41. Kidney Int. 2005;68:1472–1482. doi: 10.1111/j.1523-1755.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- 8.Bingham C., Ellard S., van’t Hoff W.G., Simmonds H.A., Marinaki A.M., Badman M.K., Winocour P.H., Stride A., Lockwood C.R., Nicholls A.J. Atypical familial juvenile hyperuricemic nephropathy associated with a hepatocyte nuclear factor-1beta gene mutation. Kidney Int. 2003;63:1645–1651. doi: 10.1046/j.1523-1755.2003.00903.x. [DOI] [PubMed] [Google Scholar]
- 9.Bleyer A.J., Kmoch S., Antignac C., Robins V., Kidd K., Kelsoe J.R., Hladik G., Klemmer P., Knohl S.J., Scheinman S.J. Variable clinical presentation of an MUC1 mutation causing medullary cystic kidney disease type 1. Clin. J. Am. Soc. Nephrol. 2014;9:527–535. doi: 10.2215/CJN.06380613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ott J. Computer-simulation methods in human linkage analysis. Proc. Natl. Acad. Sci. USA. 1989;86:4175–4178. doi: 10.1073/pnas.86.11.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carothers A.D. Risk calculations under heterogeneity: comment on a letter by D. E. Weeks and J. Ott. Am. J. Hum. Genet. 1990;47:165–166. [PMC free article] [PubMed] [Google Scholar]
- 12.Cottingham R.W., Jr., Idury R.M., Schäffer A.A. Faster sequential genetic linkage computations. Am. J. Hum. Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 13.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 14.Hoischen A., Gilissen C., Arts P., Wieskamp N., van der Vliet W., Vermeer S., Steehouwer M., de Vries P., Meijer R., Seiqueros J. Massively parallel sequencing of ataxia genes after array-based enrichment. Hum. Mutat. 2010;31:494–499. doi: 10.1002/humu.21221. [DOI] [PubMed] [Google Scholar]
- 15.Gilissen C., Arts H.H., Hoischen A., Spruijt L., Mans D.A., Arts P., van Lier B., Steehouwer M., van Reeuwijk J., Kant S.G. Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am. J. Hum. Genet. 2010;87:418–423. doi: 10.1016/j.ajhg.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vissers L.E., de Ligt J., Gilissen C., Janssen I., Steehouwer M., de Vries P., van Lier B., Arts P., Wieskamp N., del Rosario M. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 17.Becker J., Semler O., Gilissen C., Li Y., Bolz H.J., Giunta C., Bergmann C., Rohrbach M., Koerber F., Zimmermann K. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2011;88:362–371. doi: 10.1016/j.ajhg.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vissers L.E., Fano V., Martinelli D., Campos-Xavier B., Barbuti D., Cho T.J., Dursun A., Kim O.H., Lee S.H., Timpani G. Whole-exome sequencing detects somatic mutations of IDH1 in metaphyseal chondromatosis with D-2-hydroxyglutaric aciduria (MC-HGA) Am. J. Med. Genet. A. 2011;155A:2609–2616. doi: 10.1002/ajmg.a.34325. [DOI] [PubMed] [Google Scholar]
- 19.Hoischen A., van Bon B.W., Rodríguez-Santiago B., Gilissen C., Vissers L.E., de Vries P., Janssen I., van Lier B., Hastings R., Smithson S.F. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat. Genet. 2011;43:729–731. doi: 10.1038/ng.868. [DOI] [PubMed] [Google Scholar]
- 20.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paila U., Chapman B.A., Kirchner R., Quinlan A.R. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput. Biol. 2013;9:e1003153. doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vylet’al P., Kublová M., Kalbácová M., Hodanová K., Baresová V., Stibůrková B., Sikora J., Hůlková H., Zivný J., Majewski J. Alterations of uromodulin biology: a common denominator of the genetically heterogeneous FJHN/MCKD syndrome. Kidney Int. 2006;70:1155–1169. doi: 10.1038/sj.ki.5001728. [DOI] [PubMed] [Google Scholar]
- 26.Gogala M., Becker T., Beatrix B., Armache J.P., Barrio-Garcia C., Berninghausen O., Beckmann R. Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature. 2014;506:107–110. doi: 10.1038/nature12950. [DOI] [PubMed] [Google Scholar]
- 27.Westerfield M. University of Oregon Press; 1995. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) p. 385. [Google Scholar]
- 28.Jao L.E., Wente S.R., Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 31.Sunyaev S., Ramensky V., Koch I., Lathe W., 3rd, Kondrashov A.S., Bork P. Prediction of deleterious human alleles. Hum. Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 32.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cannon K.S., Or E., Clemons W.M., Jr., Shibata Y., Rapoport T.A. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray T.M., Matthews B.W. Intrahelical hydrogen bonding of serine, threonine and cysteine residues within alpha-helices and its relevance to membrane-bound proteins. J. Mol. Biol. 1984;175:75–81. doi: 10.1016/0022-2836(84)90446-7. [DOI] [PubMed] [Google Scholar]
- 35.Goder V., Junne T., Spiess M. Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol. Biol. Cell. 2004;15:1470–1478. doi: 10.1091/mbc.E03-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Junne T., Schwede T., Goder V., Spiess M. The plug domain of yeast Sec61p is important for efficient protein translocation, but is not essential for cell viability. Mol. Biol. Cell. 2006;17:4063–4068. doi: 10.1091/mbc.E06-03-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Junne T., Schwede T., Goder V., Spiess M. Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J. Biol. Chem. 2007;282:33201–33209. doi: 10.1074/jbc.M707219200. [DOI] [PubMed] [Google Scholar]
- 38.Junne T., Kocik L., Spiess M. The hydrophobic core of the Sec61 translocon defines the hydrophobicity threshold for membrane integration. Mol. Biol. Cell. 2010;21:1662–1670. doi: 10.1091/mbc.E10-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamhi-Nesher S., Shenkman M., Tolchinsky S., Fromm S.V., Ehrlich R., Lederkremer G.Z. A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell. 2001;12:1711–1723. doi: 10.1091/mbc.12.6.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindstrand A., Davis E.E., Carvalho C.M., Pehlivan D., Willer J.R., Tsai I.C., Ramanathan S., Zuppan C., Sabo A., Muzny D. Recurrent CNVs and SNVs at the NPHP1 locus contribute pathogenic alleles to Bardet-Biedl syndrome. Am. J. Hum. Genet. 2014;94:745–754. doi: 10.1016/j.ajhg.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond I.A., Majumdar A., Hentschel H., Elger M., Solnica-Krezel L., Schier A.F., Neuhauss S.C., Stemple D.L., Zwartkruis F., Rangini Z. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development. 1998;125:4655–4667. doi: 10.1242/dev.125.23.4655. [DOI] [PubMed] [Google Scholar]
- 42.Lloyd D.J., Wheeler M.C., Gekakis N. A point mutation in Sec61alpha1 leads to diabetes and hepatosteatosis in mice. Diabetes. 2010;59:460–470. doi: 10.2337/db08-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haßdenteufel S., Klein M.C., Melnyk A., Zimmermann R. Protein transport into the human ER and related diseases, Sec61-channelopathies. Biochem. Cell Biol. 2014;92:499–509. doi: 10.1139/bcb-2014-0043. [DOI] [PubMed] [Google Scholar]
- 44.Lang S., Erdmann F., Jung M., Wagner R., Cavalie A., Zimmermann R. Sec61 complexes form ubiquitous ER Ca2+ leak channels. Channels (Austin) 2011;5:228–235. doi: 10.4161/chan.5.3.15314. [DOI] [PubMed] [Google Scholar]
- 45.Adam J., Bollée G., Fougeray S., Noël L.H., Antignac C., Knebelman B., Pallet N. Endoplasmic reticulum stress in UMOD-related kidney disease: a human pathologic study. Am. J. Kidney Dis. 2012;59:117–121. doi: 10.1053/j.ajkd.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 46.Voorhees R.M., Hegde R.S. Structure of the Sec61 channel opened by a signal sequence. Science. 2016;351:88–91. doi: 10.1126/science.aad4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hotamisligil G.S. Inflammation and endoplasmic reticulum stress in obesity and diabetes. Int. J. Obes. 2008;32(Suppl 7):S52–S54. doi: 10.1038/ijo.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joe M.K., Sohn S., Hur W., Moon Y., Choi Y.R., Kee C. Accumulation of mutant myocilins in ER leads to ER stress and potential cytotoxicity in human trabecular meshwork cells. Biochem. Biophys. Res. Commun. 2003;312:592–600. doi: 10.1016/j.bbrc.2003.10.162. [DOI] [PubMed] [Google Scholar]
- 49.Zode G.S., Kuehn M.H., Nishimura D.Y., Searby C.C., Mohan K., Grozdanic S.D., Bugge K., Anderson M.G., Clark A.F., Stone E.M., Sheffield V.C. Reduction of ER stress via a chemical chaperone prevents disease phenotypes in a mouse model of primary open angle glaucoma. J. Clin. Invest. 2011;121:3542–3553. doi: 10.1172/JCI58183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao C., Giacca A., Lewis G.F. Sodium phenylbutyrate, a drug with known capacity to reduce endoplasmic reticulum stress, partially alleviates lipid-induced insulin resistance and beta-cell dysfunction in humans. Diabetes. 2011;60:918–924. doi: 10.2337/db10-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davila S., Furu L., Gharavi A.G., Tian X., Onoe T., Qian Q., Li A., Cai Y., Kamath P.S., King B.F. Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat. Genet. 2004;36:575–577. doi: 10.1038/ng1357. [DOI] [PubMed] [Google Scholar]
- 52.Li D.H., Chan T., Satow R., Komazaki S., Hashizume K., Asashima M. The role of XTRAP-gamma in Xenopus pronephros development. Int. J. Dev. Biol. 2005;49:401–408. doi: 10.1387/ijdb.052005dl. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.