Abstract
Crisponi syndrome (CS)/cold-induced sweating syndrome type 1 (CISS1) is a very rare autosomal-recessive disorder characterized by a complex phenotype with high neonatal lethality, associated with the following main clinical features: hyperthermia and feeding difficulties in the neonatal period, scoliosis, and paradoxical sweating induced by cold since early childhood. CS/CISS1 can be caused by mutations in cytokine receptor-like factor 1 (CRLF1). However, the physiopathological role of CRLF1 is still poorly understood. A subset of CS/CISS1 cases remain yet genetically unexplained after CRLF1 sequencing. In five of them, exome sequencing and targeted Sanger sequencing identified four homozygous disease-causing mutations in kelch-like family member 7 (KLHL7), affecting the Kelch domains of the protein. KLHL7 encodes a BTB-Kelch-related protein involved in the ubiquitination of target proteins for proteasome-mediated degradation. Mono-allelic substitutions in other domains of KLHL7 have been reported in three families affected by a late-onset form of autosomal-dominant retinitis pigmentosa. Retinitis pigmentosa was also present in two surviving children reported here carrying bi-allelic KLHL7 mutations. KLHL7 mutations are thus associated with a more severe phenotype in recessive than in dominant cases. Although these data further support the pathogenic role of KLHL7 mutations in a CS/CISS1-like phenotype, they do not explain all their clinical manifestations and highlight the high phenotypic heterogeneity associated with mutations in KLHL7.
Main Text
Crisponi syndrome (CS)/cold-induced sweating syndrome type 1 (CISS1 [MIM: 272430]) is a rare autosomal-recessive disorder, known to be caused by mutations in CRLF1, encoding for the cytokine receptor-like factor-1 (CRLF1 [MIM: 604237]).1, 2 CS/CISS1 usually manifests in the neonatal period, when individuals present with hyperthermia and abnormal paroxysmal contractions of the facial and oropharyngeal muscles, as well as feeding and respiratory difficulties, often requiring the use of nasogastric feeding. Physical dysmorphism including a large face, broad nose, and camptodactyly have been described in most of CS/CISS1-affected individuals. Hyperthermia and acute respiratory crises are frequently associated with death within the first months of life. In the surviving CS/CISS1 case subjects, although feeding difficulties and hyperthermia often resolve after infancy, the development of scoliosis and sometimes psychomotor retardation occur, along with paradoxical sweating at cold ambient temperatures with onset at 3 years of age. Recently, a new family of “CNTF receptor-related disorders” was termed, comprising CS/CISS1, cold-induced sweating syndrome type 2 (CISS2 [MIM: 610313]) due to mutations in cardiotrophin-like cytokine factor 1 (CLCF1 [MIM: 607672]), and Stüve-Wiedemann syndrome (SWS [MIM: 601559]) due to mutations in leukemia inhibitory factor receptor (LIFR [MIM: 151443]), all showing overlapping clinical features.1 CRLF1 and CLCF1 proteins are involved in the ciliary neurotrophic factor (CNTF)-receptor pathway, important for embryonic development and maintenance of the nervous system.3 This pathway supports the differentiation and survival of a wide range of neural cell types during development and in adulthood. The stable heterodimeric complex of CRLF1 and CLCF1 forms a ligand for CNTFR, which, along with gp130 and LIFR, comprise the CNTF-receptor complex. Binding of CRLF1/CLCF1 to CNTFR leads to dimerization of gp130/LIFR, which in turn induces downstream signaling events, including activation of the JAK1/STAT3 pathway.4
To date, 35 distinct CRLF1 mutations have been found overall either as homozygous or compound heterozygous sequence changes in 56 CS/CISS1 case subjects.5 Although the identification of mutations in CRLF1 provides a definite diagnosis in subjects with suspected diagnosis of CS/CISS1 syndrome, a subset of suspected cases remains yet genetically unexplained after sequencing of CRLF1. Among the 64 case subjects referred to our center with a suspected clinical diagnosis of CS/CISS1 between 2007 and 2014, molecular genetic tests failed to detect mutations in CRLF1 in 25 case subjects with the method employed. In the present study, we ranked these case subjects based on their phenotype similarity to CS/CISS1. We focused on the infantile phenotype and in particular on the four main typical CS criteria: hyperthermia in the first months of life, feeding difficulties, contraction of oropharyngeal muscles, and camptodactyly. This allowed us to assign the 25 suspected CS/CISS1 case subjects to the following ranks: (1) very likely: 9 individuals (2 sibs), who fulfilled all of the four main criteria; (2) questionable: 14 individuals who fulfilled two or three of them; or (3) unlikely: 2 individuals who fulfilled only one of main typical CS criteria. Subjects assigned to rank 1 and their parents, where available, were selected for exome sequencing. Individuals within the rank 1 group were further prioritized for the exome analysis based on consanguinity. This allowed us to identify five individuals (CS_144, CS_258, CS_259, CS_260, and CS_267) from four families (all of Turkish origin) as the top ranked candidates on whom we performed the exome analysis. All participants (probands and parents) provided written informed consent to participate in this study according to a protocol reviewed and approved by the institutional review board at the Münster University Hospital Ethical Committee in Germany.
Genomic DNA was extracted from whole blood samples and sheared by sonication or tagmentation, and sequencing libraries were prepared using the TruSeq DNA Library Preparation Kit and Truseq Exome Enrichment kit or Nextera Rapid Capture Expanded Exome Kit (Illumina) according to manufacturer instructions. Libraries were loaded into Paired Ends v3 flow cells on an Illumina cBot followed by indexed paired-end sequencing (101+7+101 bp) on a HiSeq 2000 using SBS Kit v3 chemistry (Illumina). Success of tagmentation and quality of post-amplification libraries were assessed using DNA-1000 chips on the BioAnalyzer 2100 (Agilent) and Qubit fluorimetric quantitation using Qubit dsDNA BR Assay Kits (Invitrogen).
This approach achieved a 80× average coverage over the 62 Mb of genomic regions sequenced, with more than 95% regions covered. The observed transition to transversion rate (Ti/Tv) was 2.2, according to what is expected for expanded exome capture kit. Data analysis has been performed using an analysis pipeline implemented in Orione.6 In brief, paired-end sequence reads were aligned to the human genome (hg19) with the Burrows-Wheeler Aligner (BWA v.0.7.5a)7 using default settings. Initial mappings were processed using the GATK framework (v.2.8.1)8 according to their Best Practices recommendations.9 Variant sites were identified using GATK Unified Genotyper module. Variants were classified as known or novel based on dbSNP146 and annotated using GATK Variant Annotator, SnpSift/SnpEff,10 and KGGSeq.11 Annotations included positions in UCSC, RefGene, GENCODE, and ENSEMBL transcripts, OMIM and ClinVar annotations, potential false positive signals,12 allele frequency in dbSNP, ESP6500 (release SI-V2), 1000 Genomes Project (release 05/2013), and ExAC, functional predictions for the amino-acid changes according to different models (SIFT, PolyPhen2, LRT, MutationTaster, MutationAssessor, and FATHMM) retrieved from dbNSFP v.2.9 (database of human nonsynonymous SNPs and their functional predictions).13 Regions of homozygosity were identified by H3M2.14 Variants were filtered according to variant and genotype quality, recessive pattern of inheritance, gene feature (frameshift, non frameshift, stop loss/gain, splicing, missense) and MAF < 1% in reference databases (dbSNP138, dbSNP141, 1000 Genomes, ESP6500, ExAC, and an in-house database of 300 sequenced individuals). The numbers of variants remaining after filtering are provided in Table S1.
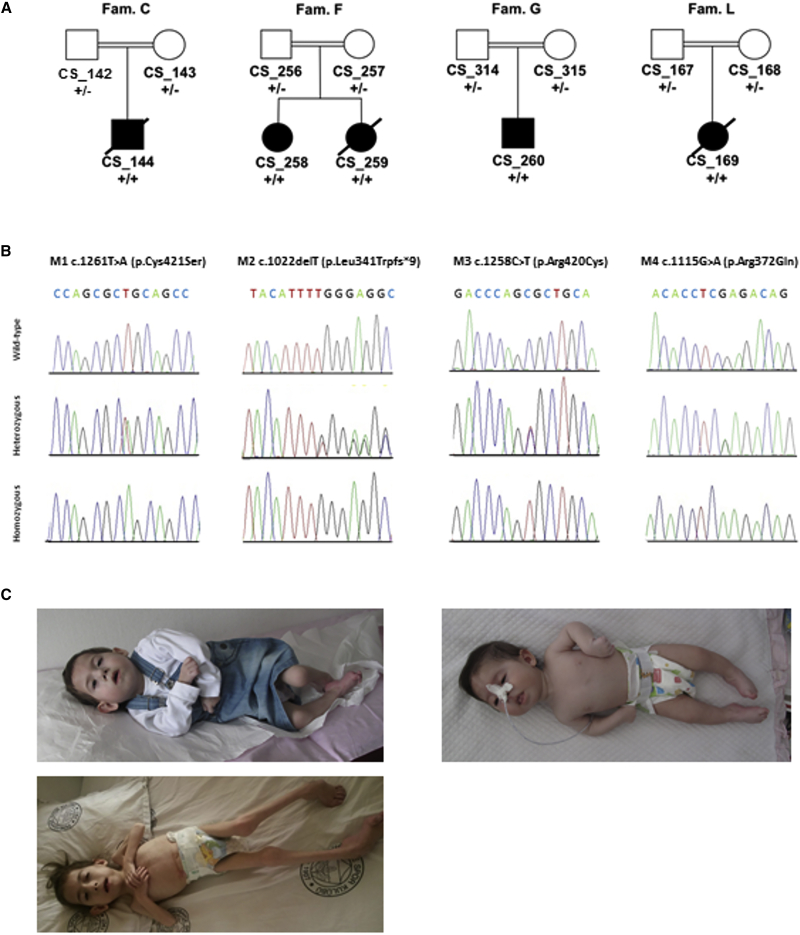
Before starting the analysis of the exome data, we re-checked for pathogenic variants in CRLF1 as well as in CLCF1 and LIFR, all belonging to the family of the “CNTF receptor-related disorders.” We also looked for variants in CNTFR. All these genes were efficiently covered by probes except for CRLF1 exon 1. We also looked at possible CNVs in these loci. For this analysis we employed the CoNIFER pipeline15 using as input the same BAM files of the SNVs pipeline. The samples, and 67 control exomes, were analyzed simultaneously (SVD [singular value decomposition] = 2) and scanned particularly to find CNVs. The CNV calls were annotated with the pathogenicity info from ClinGen database through a custom script. The results of these analyses allowed us to exclude their involvement in the disorder. Successively, extending the analysis to the whole-exome data, we identified a common gene in three out of the four families, kelch-like family member 7 (KLHL7 [MIM: 611119]; GenBank: NM_001031710.2), harboring the following private and potential deleterious mutations, all in homozygous state: M1 (c.1261T>A [p.Cys421Ser]) in family C, M2 (c.1022delT [p.Leu341Trpfs∗9]) in family F, and M3 (c.1258C>T [p.Arg420Cys]) in family G (Figure 1). All variants were confirmed by Sanger sequencing in all the family members and were absent in the Geno2MP database containing data from ∼5,000 individuals, including both persons affected with a Mendelian condition and unaffected individuals who are relatives of persons with a Mendelian condition. The variant M3 (c.1258C>T [p.Arg420Cys]) in family G was present as rs780705654 in dbSNP146 and in ExAC with 2 heterozygotes (1 East Asian, 1 European non-Finnish) on 121,411 total alleles (C 0.99998355; T 0.00001647). To further investigate the contribution of KLHL7 to the total genetic load of CS/CISS1-like phenotype, we analyzed the 20 remaining samples of the entire cohort of case subjects with no mutations detected in CRLF1 (rank 1, 2, and 3). We analyzed all the coding sequence and intron-exon junctions of KLHL7 with Big Dye Terminator Ready Reaction Mix according to the manufacturer’s (Thermo Fisher Scientific) instructions. All the primer pairs used for Sanger sequencing were the same reported in Friedman et al.16 We identified one additional KLHL7 missense variant, M4 (c.1115G>A [p.Arg372Gln]) in the affected individual from family L, belonging to the rank 2 group. All families carrying mutations in KLHL7 were of Turkish origin and consanguineous (Table 1). All the parents were found to be heterozygous carriers (Figure 1B). Clinical features of the two sisters CS_258 and CS_259 from family F are depicted in Figure 1C. Considering that the CS/CISS1 cohort consists of 64 case subjects clustered in 60 families, the fraction of cases caused by KLHL7 mutations is around 6.7%.
Figure 1.
Rank 1 Families Selected for Exome Sequencing and Sanger Sequencing of the Four KLHL7 Mutations Found
(A) Family pedigrees of individuals with KLHL7 mutations. Symbols and colors are defined as follows: square, male; circle, female; white, unaffected; dot, unaffected carrier; black, affected.
(B) Sanger sequencing of four KLHL7 mutations found in the four Turkish consanguineous families. Mutation status of KLHL7 is indicated beneath symbols for each subject: +/−, heterozygous carriers; +/+, homozygous for KLHL7 mutation. Panels shows wild-type, heterozygous, and homozygous status for M1 (c.1261T>A [p.Cys421Ser]), M2 (c.1022delT [p.Leu341Trpfs∗9]), M3 (c.1258C>T [p.Arg420Cys]), and M4 (c.1115G>A [p.Arg372Gln]), respectively.
(C) Clinical features of CS_258 (top left at the age of 3 years and 10 months and bottom left at the age of 7 years old) and CS_259 (right at the age of 5 months) from family F. Written informed consent for publication of their clinical images was obtained from their parent.
Table 1.
Clinical and Mutational Findings in Five Individuals with a CS/CISS1-like Phenotype Associated with Bi-allelic Mutations in KLHL7
| Family | C | F | F | G | L |
|---|---|---|---|---|---|
| Individual code | CS_144 | CS_258 | CS_259 | CS_260 | CS_169 |
| Sex | F | F | F | M | M |
| Age at diagnosis | 7 months | 4 years | 6 months | 11 months | 5.5 months |
| Ethnic origin | Turkey | Turkey | Turkey | Turkey | Turkey |
| Age at death | 21 months | 24 months | 7 months | ||
| Consanguinity | yes | yes | yes | yes | yes |
| Hyperthermia | + | + | ? | + | − |
| Contraction of oropharyngeal muscles | + | + | + | + | + |
| Feeding difficulties | + | + | + | + | + |
| Camptodactyly | + | + | + | + | + |
| Retinitis pigmentosa | − (12 mo) | + (6 years) | NA | + (4 years) | NA |
| Cyanosis | − | − | + | − | − |
| Swallowing difficulties | + | + | + | + | + |
| Full cheeks | − | + | + | + | + |
| Foot anomalies | ? | + | − | NA | + |
| Highly arched palate | + | + | + | + | |
| Depressed nasal bridge | + | + | + | + | + |
| Cold-induced sweating | − | + | − | − | − |
| Scoliosis | − | − | − | NA | − |
| Joint contractures | + | + | + | NA | − |
| Psychomotor retardation | NA | − | − | NA | + |
| KLHL7 mutation | exon9: c.1261T>A (p.Cys421Ser) | exon8: c.1022delT (p.Leu341Trpfs∗9) | exon8: c.1022delT (p.Leu341Trpfs∗9) | exon9: c.1258C>T (p.Arg420Cys) | exon8: c.1115G>A (p.Arg372Gln) |
Abbreviation is as follows: NA, not available.
Mutations in KLHL7 have recently been reported to cause an autosomal-dominant form of retinitis pigmentosa (adRP) (RP42 [MIM: 612943]), preferentially affecting the rod photoreceptors.16 Through linkage analysis and subsequent mutation screening, three disease-causing substitutions have been identified: c.449G>A (p.Ser150Asn), c.457G>A (p.Ala153Thr), and c.458C>T (p.Ala153Val) (Figure 2A). A detailed phenotypic characterization with long-term follow up of 11 individuals from a large family with the c.458C>T (p.Ala153Val) mutation revealed a phenotype with a varying degree of retinal photoreceptor dysfunction, with a late onset in some members and slow progression.17 A consecutive study reported that the visual phenotype is similar among individuals with these mutations. Late onset (beyond 50 years of age), slow progression, strong retention of foveal function, and bilateral concentric constriction of the visual fields with far periphery sparing may guide mutation screening in adRP.18
Figure 2.
KLHL7 Protein Structure and In Silico Analysis of the Mutants Found in CS/CISS1 Phenotype
(A) Schematic representation of the KLHL7 protein. Substitutions reported here and in Friedman et al.16 are indicated. The corresponding positions are referred to the KLHL7 splice isoform 1.
(B) Protein alignment of the KLHL7 KELCH domain. All substitutions are marked in red with an asterisk. Position indicated as follows: asterisk (∗), identical; colon (:), strongly conserved; period (.), conserved; stabilized loops underlined. Sequence data used from Uniprot: human, Q8IXQ5; bovine (Bos taurus), Q0VCQ5; frog (Xenopus tropicalis), F6UPT8; bird (Gallus gallus), Q5ZI33; fly (Musca domestica), T1PLA9; worm (Caenorhabditis remanei), E3MGH6; plant (Oryza sativa), Q84S70; KEAP1: human Kelch-like ECH-associated protein 1, Q14145.
(C) Structure of human KLHL7 Kelch domain (PDB: 3II7) in schematic representation with substitutions p.Arg372Gln and p.Arg420Cys involved in salt bridges (stabilized loops in dark green); p.Cys421Ser with partial exposition to the solvent.
KLHL7 encodes a 586-aa BTB-Kelch-related protein, containing one BTB and BACK and one Kelch domain (also known as Kelch repeat β-propeller domain), the latter consisting of six Kelch motifs (UniProtKB: Q8IXQ5; Figure 2A). KLHL7 has been recently identified as a novel substrate recognition subunit of a CUL3-based ubiquitin ligase complex. In particular KLHL7 forms a dimer, assembles with CUL3 through its BTB and BACK domains, and exerts E3 activity, poly-ubiquitinating target proteins for proteasome-mediated degradation.19
Failure to ubiquitinate target proteins could result in accumulation of the substrate, leading to long-term cellular toxicity within the photoreceptors.16 KLHL7 is widely expressed in human tissues, including rod photoreceptors, but its physiological function is not well understood.16, 20
All the three substitutions found associated to adRP are clustered in the BACK domain, which is required, along with the BTB domain, for CUL3 binding (Figures 2A and S1A). Two substitutions, p.Ala153Val and p.Ala153Thr, do not affect KLHL7 dimerization but attenuate its interaction and co-localization with CUL3, in a dominant-negative manner with reduced E3 ligase activity.19 In contrast, the four variants here found associated with the phenotype under study are located close to or within the Kelch domain, which should not be required for CUL3 binding according to Kigoshi et al. (Figure 2A).19
To identify a potential disease-causing effect of these variants, we investigated their pathogenicity score, species conservation, and predicted impact on the protein structure (Table S2 and Figures 2B and 2C). An inspection of the corresponding positions within the protein structure shows that the p.Leu341Trpfs∗9 substitution results in a premature termination codon and an almost complete loss of the Kelch domain from the protein.
The p.Arg420Cys substitution is situated within a connecting loop between Kelch motifs III and IV. The Arg420 is not exposed to the protein surface. Instead, it forms a salt bridge with Glu452, the carboxyl group of which is within hydrogen bridge distance from the arginine side chain. Those non-exposed internal salt bridges contribute substantially to the stability of the adjacent fold and in this case probably to one of the loop positions that contribute to the specificity of substrate recognition. Loss of this stabilization may result in diminishing or loss of substrate binding or even in lowered or altered specificity due to higher plasticity of this loop. The Arg420 position (together with the Glu452 position) is highly conserved within KLHL7 or KLHL7-like proteins from different organisms, even those very distinct from vertebrates (where KLHL7 sequences are surprisingly conserved as a whole, e.g., 99% between the mammalians human and cow and 96% between humans and frog; Figure 2B). Also the corresponding positions in other human KELCH proteins (e.g., KEAP1) are conserved, thus indicating an important function of this salt bridge for proper maintenance of structure and thus most probably of substrate recognition.
The p.Cys421Ser substitution is adjacent to the p.Arg420Cys substitution but in this case, this position is not conserved within the animal kingdom and for the fly, there is even a serine at this position (Figure 2B). This is not very surprising because Ser and Cys behave physically very similarly concerning polarity (with Cys a little less polar than Ser), residue volume, and hydrogen bridge formation potency. In vertebrate KLHL7, this Cys is not involved in a disulfide bridge but forms a hydrogen bridge with Ser395 C = O, which would also be possible for Ser at this position. This Cys421 is partially exposed to the solvent within the binding region of the Kelch domain and could thus influence the substrate binding to some extent.
The situation with p.Arg372Gln is quite similar to p.Arg420Cys: again Arg372 forms an internal electrostatic interaction (“salt bridge”) in hydrogen bridge distance with Glu401, thus stabilizing a loop between motifs II and III at the Kelch domain surface (Leu366 to Ala376), which connects two β strands of the beta-propeller structure (Figure 2C), which may be involved in protein ligand (“substrate”) recognition. Again, this salt bridge is conserved over a broad range of organisms even at overall low sequence similarity, as it is the case with the fly, plant, or worm sequence, pointing to a strong consequence for local structural situation with a loss of this interaction by exchanging the positively charged arginine by the uncharged glutamine. The fact that all four variants are located within the Kelch domain of KLHL7 supports a role for this domain in the pathogenesis of the CS/CISS1-like phenotype.
Previous immunocytochemical analyses in HeLa cells transfected with plasmids encoding KLHL7:WT (wild-type) and the three KLHL7 mutants associated to adRP showed that p.Ser150Asn has subcellular localization similar to KLHL7:WT in perinuclear punctate structures and co-localizes with CUL3, whereas p.Ala153Val and p.Ala153Thr were more peripheral with no formation of such punctate structures and no prominent co-localization with CUL3.19 These data have been further confirmed by coIP studies, showing that whereas p.Ser150Asn could interact with CUL3, p.Ala153Val and p.Ala153Thr could not.19 The CUL3-KLHL7 punctate structures could be inclusion bodies containing target proteins ubiquitinated and degraded by proteasome.19
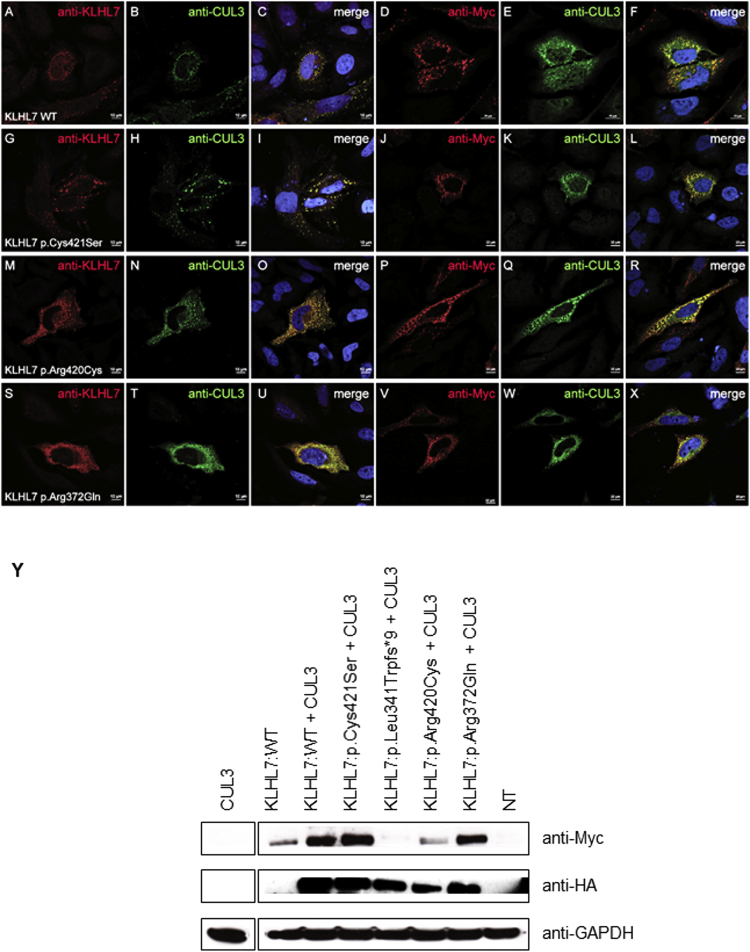
We also investigated the expression and the subcellular localization of the three substitutions found in CS/CISS1-like phenotype and their co-localization with CUL3 (Figure 3). Human DDK-Myc-KLHL7:WT clone (#RC208038 Origene) was mutagenized using the QuickChange Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer’s protocol. Immunocytochemical analyses in HeLa cells transfected with plasmids encoding KLHL7:WT and the KLHL7 mutants associated to a CS/CISS1-like phenotype, alone or in combination with CUL3, followed by confocal microscopy analysis, showed that KLHL7:WT is localized in punctate structures in the cytoplasm around the nucleus as reported also in Kigoshi et al. (Figures 3A–3F)19 and that KLHL7 mutants have similar subcellular localization as KLHL7:WT (Figures 3G–3X). Furthermore, all KLHL7:WT and mutants co-localize with CUL3 (Figures 3C, 3F, 3I, 3L, 3O, 3R, 3U, and 3X). KLHL7 (c.1022delT [p.Leu341Trpfs∗9]) protein is absent, as seen by western blotting, most likely resulting in nonsense-mediated mRNA decay with a loss of function as a putative disease mechanism (Figure 3Y).
Figure 3.
Subcellular Localization of the KLHL7 Mutants Found in CS/CISS1 Phenotype and Their Co-localization with CUL3
(A–X) HeLa cells cultured on coverslips were transfected with plasmids encoding either KLHL7:WT (A–F), KLHL7:p.Cys421Ser (G–L), KLHL7:p.Arg420Cys (M–R), KLHL7:p.Arg372Gln (S–X), and HA-CUL3 (kindly donated by Prof. Chiba). The cells were then fixed and stained with either anti-KLHL7 (Abnova Corp., cat# H00055975-B01P, RRID: AB_1576675; dilution 1:200) or anti-Myc (Abcam cat# ab18185, RRID: AB_444307; dilution 1:1,000) and anti-CUL3 antibodies (Bethyl cat# A301-109A, RRID: AB_873023; dilution 1:250). The nucleus was stained with DAPI. Confocal microscopy analysis shows KLHL7:WT and mutants in red and CUL3 in green. Scale bars represent 10 μm.
(Y) HeLa cells were transfected with plasmids encoding either KLHL7:WT, KLHL7:p.Cys421Ser, KLHL7:p.Leu341Trpfs∗9, KLHL7:p.Arg420Cys, KLHL7:p.Arg372Gln, and HA-CUL3, and the cell lysates were immunoblotted with anti-Myc, anti-HA, and anti-GAPDH. NT = not transfected.
KLHL7 belongs to the Kelch superfamily, and several proteins belonging to this superfamily act as mediators between E3 ubiquitin ligases and ubiquitinated substrates. It was hypothesized that substitutions in KLHL7 could affect homeostasis by altering ubiquitination of target substrates such as photoreceptor-specific proteins, resulting in inappropriate accumulation of the substrates targeted for proteasomal degradation that could lead to cellular toxicity within the highly metabolically active photoreceptors.16
Considering that mono-allelic mutations are responsible for adRP prompted us to review the retinal phenotype in our five probands. Unfortunately, three individuals (CS_144, CS_259, CS_169) died in their first years of life. In CS_144, the eye examination performed when the proband was 12 months old revealed normal results.
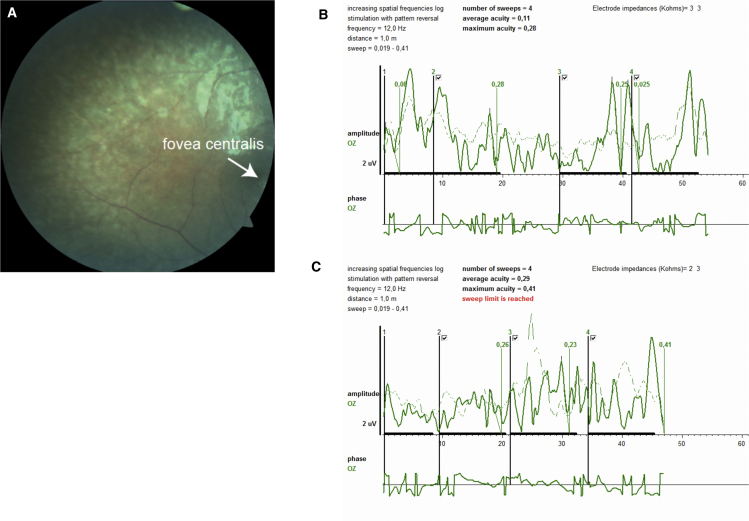
In CS_258, sister of CS_259 (both presenting with a CS/CISS1-like phenotype), the eye examination (done at the age of 6 years) revealed normal cornea, lens, and anterior chamber with bilateral subclinical chorioretinal atrophy and bilateral pale cupping optic discs (Figure 4A). Sweep visual evoked potentials (sVEP) were performed for estimation of visual acuity. VEP represents the response of the visual cortex to visual stimuli, and sVEP has been used as an important technique to measure visual acuity in nonverbal children and malingering individuals. It evaluates visual acuity in a short recording time by obtaining amplitudes in four separate phases and gives an estimated visual acuity with a value corresponding to equivalent Snellen visual acuity. Potentials for mean and maximum visual acuity in both eyes were obtained from sVEP recordings (Figures 4B and 4C). Maximal visual acuity and mean visual acuity for the left eye was consistent with 0.28 and 0.11, respectively (Figure 4B). Maximum visual acuity and mean visual acuity for the right eye was consistent with 0.41 and 0.29, respectively (Figure 4C). Electroretinography was attempted to be recorded from the proband, but it could not be obtained due to her noncooperation. The eye movements were normal and there was bilateral ptosis of the eyelids. Parents’ eye examination (done at 30 years old for the mother and at 34 for the father) revealed no signs of retinal anomaly.
Figure 4.
Pigmentary Retinopathy and Loss of Visual Acuity in CS_258
(A) Fundus photography taken by digital Fundus Camera (Carl Zeiss Meditec, VISUCAM 500) showed retinal pigmentary changes, and an abnormal appearance of the macula with attenuated arteriolar vessels (the arrow indicates the fovea centralis).
(B and C) Left (B) and right (C) eye electrophysiological examination done to assess visual function of the proband: sVEP (sweep visual evoked potential) was performed 1 m from the optoelectronic stimulator (Metrovision-Vision Monitor). Potential for mean and maximum visual acuity in both eyes was obtained from sVEP recordings. A VEP produced by a pattern stimulus that is altered at a high temporal frequency rate between 5 and 15 Hz was obtained. Recorded amplitudes were consistent with an equivalent Snellen visual acuity of 0.11 (mean) and 0.28 (maximal) for the left eye and 0.29 (mean) and 0.41 (maximal) for the right eye. The sVEP shows maximal amplitude values in four phases corresponding to Snellen equivalent. Maximal visual acuity and mean visual acuity for the left eye was consistent with 0.28 and 0.11, respectively (B). Maximum visual acuity and mean visual acuity for the right eye was consistent with 0.41 and 0.29, respectively (C).
In CS_260, the result of eye examination (performed at the age of 4 years) was also consistent with pigmented retinopathy/retinal dystrophy (data not shown). Unfortunately, we could not test all the parents for the retinal phenotype because they live in remote villages far away from the reference hospital. Interestingly, there are no retinal problems ascribed so far in individuals with CS/CISS1 or SWS phenotypes associated with CRLF1 or LIFR mutations, respectively. The only known ocular abnormalities in CS/CISS1 case subjects are non-specific “erosive keratitis” and reduced lacrimation with keratitis.1 Absent corneal reflex, decreased blink reflexes, and corneal opacities are recognized features also of SWS with poor visual outcome.21, 22 Very recently, a case of vitreoretinal pathology has been reported in SWS. It has been hypothesized that LIFR mutations may cause intrinsic weakness of the neurosensory retina, predisposing it to injury.23
Current evidence establishes CNTF/LIF as potent growth factor agents that can affect retinal neuronal differentiation and survival. The CNTF/LIF signaling pathway regulates developmental programmed cell death and differentiation of rod precursor cells in the mouse retina in vivo24 by the JAK/STAT pathway.25, 26, 27
A critical evaluation of the retinal phenotype in our two surviving probands revealed early-onset retinitis pigmentosa (before 6 years of age). KLHL7 mutations could thus show greater severity in recessive than in dominant cases. Although these data further support the pathogenic role of bi-allelic KLHL7 mutations in a CS/CISS1-like phenotype, they do not explain all their clinical manifestations and highlight the high phenotypic heterogeneity associated to mutations in KLHL7. Based on interaction studies reported by Kigoshi et al.,19 where KLHL7:WT and the mutant lacking the Kelch domain are able to bind to CUL3, and on our localization data (co-localization of all mutants with CUL3), we hypothesize that the loss-of-function mutants identified here in KLHL7 interfere with recognition and binding to the substrate(s). Thus, bi-allelic mutations in KLHL7 may lead to complete failure in the ubiquitination of the target substrate(s), resulting in the severe outcome of a Crisponi/CISS1-like phenotype associated with early-onset retinitis pigmentosa (Figure S1). Unfortunately, the substrate(s) bound to KLHL7 are so far unknown, and our proposed hypothesis is one among several. The disruption of the structural bridges in the Kelch domain could also affect the catalytic E3 activity or the stability of the protein. Therefore, future studies determining the target substrates of KLHL7 will be of great importance in better understanding the pathophysiologic role of KLHL7. Uncovering whether the different proteins such as KLHL7, CRLF1, CLCF1, and LIFR converge functionally to associate with disorders of overlapping phenotype can have important implications in the search for novel disease-associated genes and pharmacological targets.
Acknowledgments
We are grateful to all family members for their participation to the study and to their treating physicians. In particular we thank the following physicians who supplied DNA material and clinical information from individuals with CS/CISS1-like phenotype: Margot Van Allen, Jaime Campos Castello, Francesco Muntoni, Goran Anneren, Fernando Santos, Mohnish Suri, Davide Firinu, Peter Toth-Heyn, Rosario Casalone, Giuseppina Timpani, Maria Teresa Sinelli, Charu Deshpande, Robert Robinson, Adnan Manzur, Arnaud Isapof, Nelida Montano, J. Van Harssel, Livia Garavelli, Paolo Villani, Cigdem Seher Kasapkara, and Irina Snoeck. We thank Prof. Francesco Cucca, director of IRGB-CNR, for his continuous encouragement and support to the study. We are thankful to the CRS4 HPC group for IT support. We thank Prof. Tomoki Chiba, University of Tsukuba, Japan, for kindly providing the HA-CUL3 plasmid used in this study. In particular, we would like to acknowledge the Associazione Sindrome di Crisponi e Malattie Rare for its constant support, encouragement, and active participation both with us and the involved families. This work was supported by TELETHON exploratory grant GEP13093 to L.C., by a grant from the Associazione Sindrome di Crisponi e Malattie Rare, Italy, to L.C., and by a grant from Innovative Medical Research, Münster, Germany, to I.B. and F.R. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Published: July 7, 2016
Footnotes
Supplemental Data include one figure and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.05.026.
Accession Numbers
The four pathogenic variants reported here have been deposited in ClinVar under the following accession numbers: c.1261T>A (p.Cys421Ser), SCV000268116; c.1022delT (p.Leu341Trpfs∗9), SCV000268118; c.1258C>T (p.Arg420Cys), SCV000268117; and c.1115G>A (p.Arg372Gln), SCV000268119.
Web Resources
1000 Genomes, http://www.1000genomes.org
ClinGen, https://www.clinicalgenome.org/
dbNSFP v.2.0, https://sites.google.com/site/jpopgen/dbNSFP
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
The Human Protein Atlas, http://www.proteinatlas.org/
UniProt, http://www.uniprot.org/
Supplemental Data
References
- 1.Crisponi L., Crisponi G., Meloni A., Toliat M.R., Nurnberg G., Usala G., Uda M., Masala M., Hohne W., Becker C. Crisponi syndrome is caused by mutations in the CRLF1 gene and is allelic to cold-induced sweating syndrome type 1. Am. J. Hum. Genet. 2007;80:971–981. doi: 10.1086/516843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dagoneau N., Bellais S., Blanchet P., Sarda P., Al-Gazali L.I., Di Rocco M., Huber C., Djouadi F., Le Goff C., Munnich A., Cormier-Daire V. Mutations in cytokine receptor-like factor 1 (CRLF1) account for both Crisponi and cold-induced sweating syndromes. Am. J. Hum. Genet. 2007;80:966–970. doi: 10.1086/513608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forger N.G., Prevette D., deLapeyrière O., de Bovis B., Wang S., Bartlett P., Oppenheim R.W. Cardiotrophin-like cytokine/cytokine-like factor 1 is an essential trophic factor for lumbar and facial motoneurons in vivo. J. Neurosci. 2003;23:8854–8858. doi: 10.1523/JNEUROSCI.23-26-08854.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heinrich P.C., Behrmann I., Haan S., Hermanns H.M., Müller-Newen G., Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piras R., Chiappe F., Torraca I.L., Buers I., Usala G., Angius A., Akin M.A., Basel-Vanagaite L., Benedicenti F., Chiodin E. Expanding the mutational spectrum of CRLF1 in Crisponi/CISS1 syndrome. Hum. Mutat. 2014;35:424–433. doi: 10.1002/humu.22522. [DOI] [PubMed] [Google Scholar]
- 6.Cuccuru G., Orsini M., Pinna A., Sbardellati A., Soranzo N., Travaglione A., Uva P., Zanetti G., Fotia G. Orione, a web-based framework for NGS analysis in microbiology. Bioinformatics. 2014;30:1928–1929. doi: 10.1093/bioinformatics/btu135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. John Wiley & Sons, Inc.; Hoboken, NJ: 2002. Current Protocols in Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cingolani P., Patel V.M., Coon M., Nguyen T., Land S.J., Ruden D.M., Lu X. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front. Genet. 2012;3:35. doi: 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M.-X., Gui H.-S., Kwan J.S.H., Bao S.-Y., Sham P.C. A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res. 2012;40:e53. doi: 10.1093/nar/gkr1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuentes Fajardo K.V., Adams D., Mason C.E., Sincan M., Tifft C., Toro C., Boerkoel C.F., Gahl W., Markello T., NISC Comparative Sequencing Program Detecting false-positive signals in exome sequencing. Hum. Mutat. 2012;33:609–613. doi: 10.1002/humu.22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X., Jian X., Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magi A., Tattini L., Palombo F., Benelli M., Gialluisi A., Giusti B., Abbate R., Seri M., Gensini G.F., Romeo G., Pippucci T. H3M2: detection of runs of homozygosity from whole-exome sequencing data. Bioinformatics. 2014;30:2852–2859. doi: 10.1093/bioinformatics/btu401. [DOI] [PubMed] [Google Scholar]
- 15.Krumm N., Sudmant P.H., Ko A., O’Roak B.J., Malig M., Coe B.P., Quinlan A.R., Nickerson D.A., Eichler E.E., NHLBI Exome Sequencing Project Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman J.S., Ray J.W., Waseem N., Johnson K., Brooks M.J., Hugosson T., Breuer D., Branham K.E., Krauth D.S., Bowne S.J. Mutations in a BTB-Kelch protein, KLHL7, cause autosomal-dominant retinitis pigmentosa. Am. J. Hum. Genet. 2009;84:792–800. doi: 10.1016/j.ajhg.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugosson T., Friedman J.S., Ponjavic V., Abrahamson M., Swaroop A., Andréasson S. Phenotype associated with mutation in the recently identified autosomal dominant retinitis pigmentosa KLHL7 gene. Arch. Ophthalmol. 2010;128:772–778. doi: 10.1001/archophthalmol.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen Y., Locke K.G., Hood D.C., Birch D.G. Rod photoreceptor temporal properties in retinitis pigmentosa. Exp. Eye Res. 2011;92:202–208. doi: 10.1016/j.exer.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kigoshi Y., Tsuruta F., Chiba T. Ubiquitin ligase activity of Cul3-KLHL7 protein is attenuated by autosomal dominant retinitis pigmentosa causative mutation. J. Biol. Chem. 2011;286:33613–33621. doi: 10.1074/jbc.M111.245126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bredholt G., Storstein A., Haugen M., Krossnes B.K., Husebye E., Knappskog P., Vedeler C.A. Detection of autoantibodies to the BTB-kelch protein KLHL7 in cancer sera. Scand. J. Immunol. 2006;64:325–335. doi: 10.1111/j.1365-3083.2006.01821.x. [DOI] [PubMed] [Google Scholar]
- 21.Di Rocco M., Stella G., Bruno C., Doria Lamba L., Bado M., Superti-Furga A. Long-term survival in Stuve-Wiedemann syndrome: a neuro-myo-skeletal disorder with manifestations of dysautonomia. Am. J. Med. Genet. A. 2003;118A:362–368. doi: 10.1002/ajmg.a.10242. [DOI] [PubMed] [Google Scholar]
- 22.Injarie A.M., Narang A., Idrees Z., Saggar A.K., Nischal K.K. Ocular treatment of children with Stuve-Wiedemann syndrome. Cornea. 2012;31:269–272. doi: 10.1097/ICO.0b013e3182182089. [DOI] [PubMed] [Google Scholar]
- 23.Palejwala N.V., Stempel A.J., Stout J.T. Bilateral giant retinal tears in a pediatric patient with leukemia inhibitory factor receptor deficiency (Stuve-Wiedemann syndrome) Retin. Cases Brief Rep. 2015;9:245–247. doi: 10.1097/ICB.0000000000000150. [DOI] [PubMed] [Google Scholar]
- 24.Elliott J., Cayouette M., Gravel C. The CNTF/LIF signaling pathway regulates developmental programmed cell death and differentiation of rod precursor cells in the mouse retina in vivo. Dev. Biol. 2006;300:583–598. doi: 10.1016/j.ydbio.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Ozawa S., Ishikawa K., Ito Y., Nishihara H., Yamakoshi T., Hatta Y., Terasaki H. Differences in macular morphology between polypoidal choroidal vasculopathy and exudative age-related macular degeneration detected by optical coherence tomography. Retina. 2009;29:793–802. doi: 10.1097/IAE.0b013e3181a3b7d9. [DOI] [PubMed] [Google Scholar]
- 26.Rhee K.D., Goureau O., Chen S., Yang X.J. Cytokine-induced activation of signal transducer and activator of transcription in photoreceptor precursors regulates rod differentiation in the developing mouse retina. J. Neurosci. 2004;24:9779–9788. doi: 10.1523/JNEUROSCI.1785-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S.S.-M., Wei J., Qin H., Zhang L., Xie B., Hui P., Deisseroth A., Barnstable C.J., Fu X.-Y. STAT3-mediated signaling in the determination of rod photoreceptor cell fate in mouse retina. Invest. Ophthalmol. Vis. Sci. 2004;45:2407–2412. doi: 10.1167/iovs.04-0003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.