Abstract
A total of 1,056 native or Cary-Blair-preserved stool specimens were simultaneously tested by conventional stool culturing and by enteric bacterial panel (EBP) multiplex real-time PCR for Campylobacter jejuni, Campylobacter coli, Salmonella spp., and shigellosis disease-causing agents (Shigella spp. and enteroinvasive Escherichia coli [EIEC]). Overall, 143 (13.5%) specimens tested positive by PCR for the targets named above; 3 coinfections and 109 (10.4%) Campylobacter spp., 17 (1.6%) Salmonella spp., and 20 (1.9%) Shigella spp./EIEC infections were detected. The respective positive stool culture rates were 75 (7.1%), 14 (1.3%), and 7 (0.7%). The median threshold cycle (CT) values of culture-positive specimens were significantly lower than those of culture-negative ones (CT values, 24.3 versus 28.7; P < 0.001), indicating that the relative bacterial load per fecal specimen was significantly associated with the culture results. In Campylobacter infections, the respective median fecal calprotectin concentrations in PCR-negative/culture-negative (n = 40), PCR-positive/culture-negative (n = 14), and PCR-positive/culture-positive (n = 15) specimens were 134 mg/kg (interquartile range [IQR], 30 to 1,374 mg/kg), 1,913 mg/kg (IQR, 165 to 3,813 mg/kg), and 5,327 mg/kg (IQR, 1,836 to 18,213 mg/kg). Significant differences were observed among the three groups (P < 0.001), and a significant linear trend was identified (P < 0.001). Furthermore, the fecal calprotectin concentrations and CT values were found to be correlated (r = −0.658). Our results demonstrate that molecular screening of Campylobacter spp., Salmonella spp., and Shigella spp./EIEC using the BD Max EBP assay will result in timely diagnosis and improved sensitivity. The determination of inflammatory markers, such as calprotectin, in fecal specimens may aid in the interpretation of PCR results, particularly for enteric pathogens associated with mucosal damage and colonic inflammation.
INTRODUCTION
Acute bacterial gastroenteritis is a common global health problem. Infections with Salmonella spp., Campylobacter spp., and Shiga toxin-producing Escherichia coli (STEC) are spread to humans from animal reservoirs. In contrast, humans are the primary reservoir of Shigella spp. These four microorganisms are the most commonly reported enteric bacterial pathogens responsible for community-acquired diarrhea (1, 2). Conventional bacterial stool culturing is considered the traditional gold standard for diagnosis of the most common bacterial pathogens (3, 4), and current guidelines recommend one to two culture specimens for adult patients and one specimen for pediatric patients (5). Fresh stool specimens should be transported to the laboratory and processed within 2 h of collection (6, 7). If the specimen cannot be processed within 2 h, it should be placed in a transport medium to preserve the viability of bacterial pathogens, particularly Shigella and Campylobacter. Antibiotics administered at the time of specimen collection may compromise the outcome of stool culturing (8).
Several novel molecular assays for detecting enteric bacterial pathogens in stool specimens have been developed. These assays are very specific, and their sensitivities are superior to those of conventional methods (3, 7, 9, 10). Among these tests, the BD Max enteric bacterial panel (EBP) is an FDA-cleared, multiplex PCR assay designed for the detection of Salmonella spp., Shigella spp./enteroinvasive E. coli (EIEC), Campylobacter spp. (Campylobacter jejuni and Campylobacter coli), and Shiga-like toxin genes (stx1 and/or stx2) in preserved and unpreserved stool specimens using the BD Max system (BD Diagnostics, Allschwil, Switzerland). The BD Max system is a walkaway PCR instrument that can extract, amplify, and detect nucleic acids in batches of up to 24 samples within 3 h, requiring less than 2 min of hands-on time per sample (11).
In this study, we aimed to achieve the following: (i) to evaluate the performance of molecular testing compared with that of conventional culture methods in a privately owned clinical microbiology laboratory, currently serving approximately 2,200 hospital beds and several hundred physicians in private practice; (ii) to study the association between the relative bacterial load, as assessed using the threshold cycle (CT) values of pathogen-specific PCR targets, and the respective culture results; and (iii) to explore the association between bacterial load, as assessed using CT values, and the extent of intestinal inflammation, as assessed using stool calprotectin concentrations, in Campylobacter infections (C. jejuni and C. coli).
MATERIALS AND METHODS
Study design.
A total of 1,056 native or Cary-Blair-preserved stool specimens received between 4 July and 26 August 2013 (n = 262) and between 12 November and 12 March 2014 (n = 794), respectively, were included in this study. The specimens were simultaneously tested by conventional stool culturing and EBP multiplex real-time PCR for C. jejuni, C. coli, Salmonella spp., and shigellosis disease-causing agents (Shigella spp. and EIEC). STEC was excluded from the study because in Switzerland general stool bacteriology only includes culturing of Salmonella, Shigella, and Campylobacter. The study protocol was approved by the ethics committee of the Canton of Bern, Switzerland.
Enteric bacterial panel.
Ten microliters of each stool specimen was transferred with a loop to a sample buffer tube according to the manufacturer's instructions. Stool specimens were tested within 48 h of receipt and were stored at 2 to 8°C prior to testing. The fully automated process took less than 3 h.
Culture.
Cary-Blair-preserved and native fecal specimens were processed on the day of arrival with a BD Kiestra Total Laboratory Automation System (Becton-Dickinson). The following media were inoculated: selenite broth, Hektoen enteric (HE) agar, xylose-lysine-deoxycholate (XLD) agar, and Campylosel agar (all from bioMérieux Suisse S.A., Geneva, Switzerland). Campylosel agar was incubated in a microaerobic atmosphere (5% O2, 10% CO2, and 85% N2) at 42°C for 48 h, and results were read after 24 and 48 h. HE and XLD agar media were incubated aerobically for 48 h, and results were read after 24 and 48 h. Selenite broth was subcultured after 15 h of aerobic incubation onto HE and XLD agar. Potential pathogens were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS; Bruker Daltonics GmbH, Bremen, Germany) or Vitek-2 (bioMérieux Suisse S.A., Geneva, Switzerland). Salmonella and Shigella isolates were confirmed by serological testing (Siemens Healthcare Diagnostics Products GmbH, Marburg, Germany; Sifin Diagnostics GmbH, Berlin, Germany).
BD Max EBP calibration curves.
Privileged access to the CT values generated by the BD Max system was obtained from the manufacturer for the duration of the study to establish a correlation between the BD Max CT values and CFU counts. Serial 10-fold dilutions (1.5 × 108 cells ml−1 to 1.5 × 101 cells ml−1) in 0.85% sodium chloride solution of C. jejuni ATCC 33291, Salmonella enterica subsp. enterica serovar Enteritidis ATCC 13076, and Shigella flexneri DSM 4782 were plated on the appropriate agar media, and colonies were counted after 24 h (Salmonella and Shigella) or 48 h (Campylobacter) of incubation. In parallel, 10 μl from each dilution step was tested in triplicate using the EBP. The CT values and CFU counts per milliliter were compared.
Calprotectin.
Calprotectin levels were determined in 69 (6.5%) stool specimens by a Ridascreen assay (R-Biopharm, Darmstadt, Germany) to assess the intestinal inflammatory response induced by Campylobacter, a classical agent of inflammatory diarrhea. Specimens were subdivided into three groups according to the respective Campylobacter culture and PCR results as follows: group 1 (n = 40), culture negative/PCR negative; group 2 (n = 14), culture negative/PCR positive; and group 3 (n = 15), culture positive/PCR positive.
Statistical analysis.
A statistical software program (MedCalc, version 15.6; MedCalc, Ostend, Belgium) was used to conduct statistical analysis. To determine whether CT values are related to bacterial counts, univariate linear regression analysis, with the CT values as dependent variables and the CFU counts of the different pathogens as independent variables, was performed. A Mann-Whitney test was used to assess differences in the CT values between two groups, and a Kruskal-Wallis test was used to assess differences in the calprotectin levels among three groups. The trends in the calprotectin concentration were analyzed using a Jonckheere-Terpstra trend test. P values of <0.05 were considered statistically significant. Correlations between the bacterial load, as determined according to the CT value, and the calprotectin level in campylobacteriosis were determined using a Spearman rank correlation test.
RESULTS
A total of 1,056 stool specimens were included in the study. Of them, 913 (86.5%) were negative for Campylobacter spp., Salmonella spp., and Shigella spp./EIEC according to the EBP assay, and 143 (13.5%) were positive for Campylobacter spp., Salmonella spp., and Shigella spp./EIEC, including three coinfections, Campylobacter spp. along with Salmonella spp. (n = 1) or Shigella spp./EIEC (n = 2). In detail, molecular-based screening yielded 109 (10.4%) specimens positive for Campylobacter spp., 17 (1.6%) specimens positive for Salmonella spp., and 20 (1.9%) specimens positive for Shigella spp./EIEC.
In comparison, the overall positive stool culture rate for C. jejuni/C. coli, Salmonella spp., and Shigella spp. was 8.9%, i.e., 7.1% (n = 75) C. jejuni/C. coli, 1.3% (n = 14) Salmonella spp., and 0.7% (n = 7) Shigella spp. None of the culture-positive cases were missed by the EBP assay, and the assay's negative predictive value for Campylobacter spp., Salmonella spp., and Shigella spp. was as high as 100% (95% confidence interval [CI], 99 to 100%).
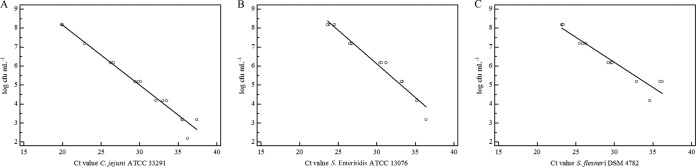
Privileged access to the CT values generated by the BD Max system was obtained from the manufacturer for the duration of the study. The CT values were strongly correlated with the CFU counts for the type strains of C. jejuni (R2 = 0.9809), S. enterica subsp. enterica serovar Enteritidis (R2 = 0.9654), and S. flexneri (R2 = 0.9205) (Fig. 1).
FIG 1.
Correlation between BD Max enteric bacterial panel CT values and CFU counts for Campylobacter jejuni ATCC 33291 (A), Salmonella enterica subsp. enterica serovar Enteritidis ATCC 13076 (B), and Shigella flexneri DSM 4782 (C).
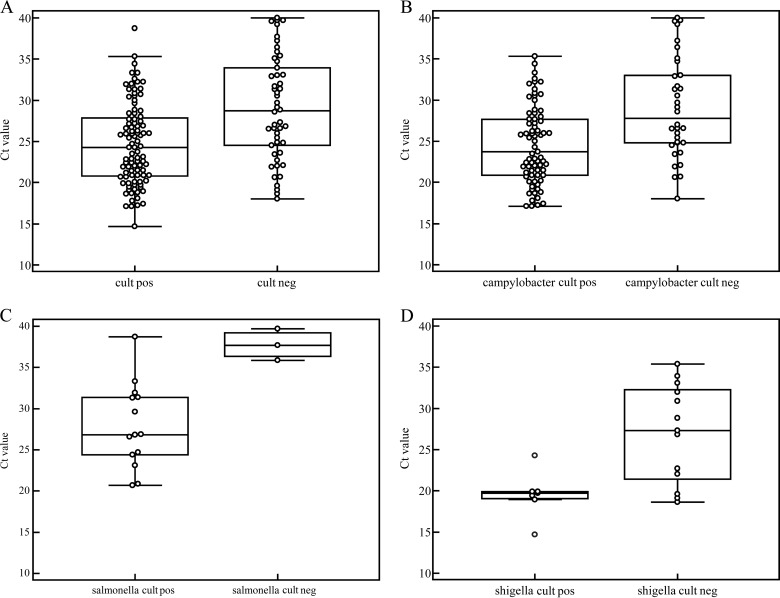
To assess the effect of the relative bacterial load per specimen on the culture results, we compared the median CT values of concordant (group 3) and discordant (group 2) specimens (Fig. 2). The median CT values of the concordant specimens were significantly lower than those of the discordant ones (CT values, 24.3 versus 28.7; P < 0.001). The pathogen-specific CT values of the concordant and discordant specimens, respectively, were 23.7 and 27.8 (P < 0.001) for C. jejuni/C. coli, 26.9 and 37.7 (P = 0.02) for Salmonella spp., and 19.7 and 27.3 (P = 0.02) for Shigella spp.
FIG 2.
Comparison of all CT values of concordant (culture positive/PCR positive; group 3) and discordant (culture negative/PCR positive; group 2) specimens for C. jejuni/C. coli, Salmonella spp., and Shigella spp (A), C. jejuni/C. coli only (B), Salmonella spp. only (C), and Shigella spp. only (D). The median CT values of the concordant specimens were lower than those of the discordant specimens; this difference was statistically significant (C. jejuni/C. coli, P < 0.001; Salmonella spp., P = 0.02; and Shigella spp., P = 0.02).
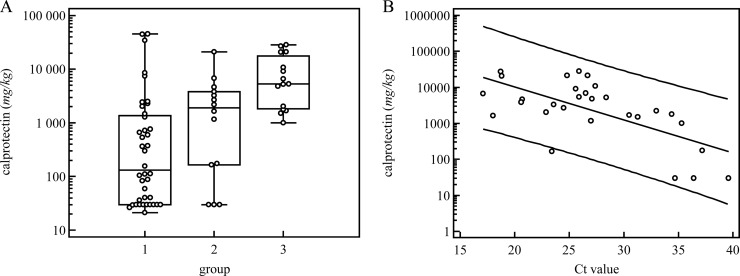
To verify the discordant-positive Campylobacter PCR results, we determined the concentrations of fecal calprotectin, a surrogate marker of intestinal inflammation, in the concordant-negative (group 1), discordant-positive (group 2), and concordant-positive (group 3) stool specimens (Fig. 3). The median calprotectin concentrations in the concordant-negative, discordant-positive, and concordant-positive specimens were 134 mg/kg (interquartile range [IQR], 30 to 1,374), 1,913 mg/kg (IQR, 165 to 3,813), and 5,327 mg/kg (IQR, 1,836 to 18,213), respectively. Significant differences were observed among the three groups (P < 0.001), and a significant linear trend was detected (P < 0.001) (Fig. 3A). Furthermore, the calprotectin concentrations and CT values were found to be correlated (r = −0.658) (Fig. 3B).
FIG 3.
(A) Calprotectin concentrations were compared in stool specimens that were concordant negative (culture negative/PCR negative; group 1), discordant positive (culture negative/PCR positive; group 2), and concordant positive (culture positive/PCR positive; group 3) for Campylobacter jejuni/C. coli. The respective median calprotectin concentrations were 134 mg/kg (group 1), 1,913 mg/kg (group 2), and 5,327 mg/kg (group 3); the concentration was significantly higher in group 3 than in groups 1 and 2 (P < 0.05), revealing a significant trend across the three groups (P < 0.001). (B) Calprotectin concentrations and CT values were correlated, with a Spearman r of −0.658.
DISCUSSION
This study evaluated the feasibility and performance of a molecular-based screening method for detecting C. jejuni/C. coli, Salmonella spp., and Shigella spp. compared with conventional stool culturing in a diagnostic laboratory. Furthermore, the association between the relative bacterial load and the respective culture results was examined, as well as the association between the bacterial load and the extent of intestinal inflammation in Campylobacter infections. The BD Max EBP assay was run on a fully automated BD Max system with minimal hands-on time and a short turnaround time.
In a previous study, C. jejuni/C. coli were the most frequently detected pathogens, similar to the results of our study (4). We observed an overall prevalence of 13.5% for C. jejuni/C. coli, Salmonella spp., and Shigella spp./EIEC by molecular testing. Earlier studies based on conventional stool cultures reported a prevalence of approximately 6% for C. jejuni/C. coli, Salmonella spp., and Shigella spp. (8, 9). In our investigation, the prevalence of Campylobacter spp. alone was 10.4%, that of Salmonella spp. was 1.6%, and that of Shigella spp./EIEC was 1.9%, as determined by PCR. Buchan et al. also identified an increased prevalence of gastroenteritis pathogens using a competitor molecular-based assay with the same targets as the EBP (9). Of note, in our study, we detected a prevalence of 1.9% Shigella spp./EIEC by PCR but only a prevalence of only 0.7% by culturing. The PCR target gene ipaH of Shigella is shared by EIEC. Therefore, it is unclear whether the discrepant culture results were due to the higher than expected prevalence of EIEC or to the rather poor sensitivity for Shigella cultures, as has been reported by several authors (7, 9, 12–14). Difficulties with culturing Shigella were also illustrated in a case in our study. S. flexneri was cultured from one out of three stool specimens submitted, despite indications of relatively high bacterial loads in all three specimens (CT values of 19.9, 18.6, and 22.0, respectively).
Inappropriate specimen storage and transportation and previous antibiotic treatment are among the known factors that compromise culture results by affecting the viability of bacterial pathogens. Jansen et al. (1) have demonstrated that culture-based methods miss a substantial proportion of Campylobacter infections. Although current guidelines restrict antibiotic therapy to patients at risk of severe disease, in Switzerland, more than 60% of laboratory-confirmed Campylobacter cases are treated with antibiotics (15).
In addition, we detected an association between the relative bacterial loads and the culture results, further corroborating the conclusion that PCR is more sensitive than culturing. This effect was the least marked for Salmonella, likely due to the standard broth enrichment step used to increase Salmonella culture sensitivity. In contrast, Shigella was rarely cultured at CT values exceeding 24.3. Campylobacter was missed by culturing at both at high and low CT values. This finding is suggestive of issues of reduced pathogen viability, which would directly affect culture results but would not affect results obtained using molecular-based methods (12).
As a pathogen, Campylobacter causes mucosal damage and inflammation of the colon. To quantify the inflammatory responses in the colons of patients with suspected Campylobacter-associated diarrhea, we measured the fecal calprotectin concentration (16, 17). Fecal calprotectin (18, 19), a protein dimer of the neutrophil cytosol, is a surrogate marker for distinguishing organic inflammatory diseases of the gastrointestinal tract, such as Crohn's disease and ulcerative colitis, from functional diseases, such as irritable bowel syndrome. A calprotectin level of >100 μg/g stool is indicative of active organic disease, whereas a level of <50 μg/g stool is not indicative of inflammation in the gastrointestinal tract. Levels of between 50 and 100 μg/g stool may indicate the presence of a variety of mild organic diseases, such as inflammation caused by nonsteroidal anti-inflammatory drugs (NSAIDs) and mild diverticulitis.
We found that the Campylobacter bacterial load, as assessed by the CT value, and the respective intestinal inflammatory response, as determined by the fecal calprotectin concentration, were correlated (Fig. 3). Patients without Campylobacter infection (group 1) possessed a broad range of fecal calprotectin concentrations, suggesting that pathogens other than Campylobacter or noninfectious inflammatory bowel diseases, such as Crohn's disease or ulcerative colitis, might be responsible for the elevated calprotectin levels.
In the patients with Campylobacter infections (groups 2 and 3), we found a positive correlation between the bacterial load and the extent of the intestinal inflammatory response, as confirmed using the Jonckheere-Terpstra proportion test, which revealed significant trends among all groups (Fig. 3). However, in the absence of complementary clinical and epidemiological data that may assist in assessing confounding factors, cautious interpretation of the laboratory data is advisable.
In conclusion, our study demonstrated that the molecular-based screening of Campylobacter spp., Salmonella spp., and Shigella spp. using the BD Max EBP assay resulted in timely and sensitive detection of these major bacterial enteric pathogens. This assay is easy to perform; therefore, it is not restricted to molecular laboratories. Still, conventional stool culturing of PCR-positive specimens will remain crucial for antibiotic susceptibility testing and epidemiological purposes, such as serotype determination and subtyping in outbreaks (4).
ACKNOWLEDGMENTS
We thank the Microbiology Department of labormedizinisches zentrum Dr Risch for providing us with clinical samples and Dorothea Hillmann for performing calprotectin analysis. We thank the laboratory technicians from the Microbiology Department of labormedizinisches zentrum Dr Risch for their assistance.
We declare that we have no conflicts of interest.
N.W., T.B, L.R., and M.R. conceived and designed the approach of the study. N.W. and S.T. were analysts. L.R. and N.W. were study statisticians. N.W., T.B., and L.R. wrote the paper.
This study was supported by the labormedizinisches zentrum Dr Risch.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Jansen A, Stark K, Kunkel J, Schreier E, Ignatius R, Liesenfeld O, Werber D, Göbel UB, Zeitz M, Schneider T. 2008. Aetiology of community-acquired, acute gastroenteritis in hospitalised adults: a prospective cohort study. BMC Infect Dis 8:143. doi: 10.1186/1471-2334-8-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrington SM, Buchan BW, Doern C, Fader R, Ferraro MJ, Pillai DR, Rychert J, Doyle L, Lainesse A, Karchmer T, Mortensen JE. 2015. Multicenter evaluation of the BD Max enteric bacterial panel PCR assay for rapid detection of Salmonella spp., Shigella spp., Campylobacter spp. (C. jejuni and C. coli), and Shiga toxin 1 and 2 genes. J Clin Microbiol 53:1639–1647. doi: 10.1128/JCM.03480-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerrant RL, Van Gilder T, Steiner TS, Thielman NM, Slutsker L, Tauxe RV, Hennessy T, Griffin PM, DuPont H, Bradley Sack R, Tarr P, Neill M, Nachamkin I, Reller LB, Osterholm MT, Bennish ML, Pickering LK. 2001. Practice guidelines for the management of infectious diarrhea. Clin Infect Dis 32:331–351. doi: 10.1086/318514. [DOI] [PubMed] [Google Scholar]
- 5.Baron EJ, Miller JM, Weinstein MP, Richter SS, Gilligan PH, Thomson RB, Bourbeau P, Carroll KC, Kehl SC, Dunne WM, Robinson-Dunn B, Schwartzman JD, Chapin KC, Snyder JW, Forbes BA, Patel R, Rosenblatt JE, Pritt BS. 2013. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM). Clin Infect Dis 57:e22–e121. doi: 10.1093/cid/cit278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humphries RM, Linscott AJ. 2015. Laboratory diagnosis of bacterial gastroenteritis. Clin Microbiol Rev 28:3–31. doi: 10.1128/CMR.00073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutta P, Pramanik KC, Chatterjee A, Roy S, Dutta S, Rajendran K, Bhattacharya SK. 2001. Sensitivity and performance characteristics of a direct PCR with stool samples in comparison to conventional techniques for diagnosis of Shigella and enteroinvasive Escherichia coli infection in children with acute diarrhoea in Calcutta, India. J Med Microbiol 50:667–674. doi: 10.1099/0022-1317-50-8-667. [DOI] [PubMed] [Google Scholar]
- 8.Rohner P, Pittet D, Pepey B, Nije-Kinge T, Auckenthaler R. 1997. Etiological agents of infectious diarrhea: implications for requests for microbial culture. J Clin Microbiol 35:1427–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchan BW, Olson WJ, Pezewski M, Marcon MJ, Novicki T, Uphoff TS, Chandramohan L, Revell P, Ledeboer NA. 2013. Clinical evaluation of a real-time PCR assay for identification of Salmonella, Shigella, Campylobacter (Campylobacter jejuni and C. coli), and Shiga toxin-producing Escherichia coli isolates in stool specimens. J Clin Microbiol 51:4001–4007. doi: 10.1128/JCM.02056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuurman T, de Boer RF, van Zanten E, van Slochteren KR, Scheper HR, Dijk-Alberts BG, Möller AV, Kooistra-Smid AM. 2007. Feasibility of a molecular screening method for detection of Salmonella enterica and Campylobacter jejuni in a routine community-based clinical microbiology laboratory. J Clin Microbiol 45:3692–3700. doi: 10.1128/JCM.00896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stellrecht KA, Espino AA, Maceira VP, Nattanmai SM, Butt SA, Wroblewski D, Hannett GE, Musser KA, Espino A, Maceira VP, Nattanmai SM, Butt SA, Wroblewski D, Hannett GE, Musser KA. 2014. Premarket evaluations of the IMDx C. difficile for Abbott m2000 assay and the BD Max Cdiff assay. J Clin Microbiol 52:1423–1428. doi: 10.1128/JCM.03293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Kabir F, Manneh J, Lertsethtakarn P, Begum S, Gratz J, Becker SM, Operario DJ, Taniuchi M, Janaki L, Platts-Mills JA, Haverstick DM, Kabir M, Sobuz SU, Nakjarung K, Sakpaisal P, Silapong S, Bodhidatta L, Qureshi S, Kalam A, Saidi Q, Swai N, Mujaga B, Maro A, Kwambana B, Dione M, Antonio M, Kibiki G, Mason CJ, Haque R, Iqbal N, Zaidi AKM, Houpt ER. 2014. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis 14:716–724. doi: 10.1016/S1473-3099(14)70808-4. [DOI] [PubMed] [Google Scholar]
- 13.Houng HS, Sethabutr O, Echeverria P. 1997. A simple polymerase chain reaction technique to detect and differentiate Shigella and enteroinvasive Escherichia coli in human feces. Diagn Microbiol Infect Dis 28:19–25. doi: 10.1016/S0732-8893(97)89154-7. [DOI] [PubMed] [Google Scholar]
- 14.Anderson NW, Buchan BW, Ledeboer NA. 2014. Comparison of the BD Max enteric bacterial panel to routine culture methods for detection of Campylobacter, enterohemorrhagic Escherichia coli (O157), Salmonella, and Shigella isolates in preserved stool specimens. J Clin Microbiol 52:1222–1224. doi: 10.1128/JCM.03099-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bless PJ, Schmutz C, Suter K, Jost M, Hattendorf J, Mäusezahl-Feuz M, Mäusezahl D. 2014. A tradition and an epidemic: determinants of the campylobacteriosis winter peak in Switzerland. Eur J Epidemiol 29:527–537. doi: 10.1007/s10654-014-9917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Summerton CB, Longlands MG, Wiener K, Shreeve DR. 2002. Faecal calprotectin: a marker of inflammation throughout the intestinal tract. Eur J Gastroenterol Hepatol 14:841–845. doi: 10.1097/00042737-200208000-00005. [DOI] [PubMed] [Google Scholar]
- 17.Fagerberg UL, Lööf L, Lindholm J, Hansson LO, Finkel Y. 2007. Fecal calprotectin: a quantitative marker of colonic inflammation in children with inflammatory bowel disease. J Pediatr Gastroenterol Nutr 45:414–420. doi: 10.1097/MPG.0b013e31810e75a9. [DOI] [PubMed] [Google Scholar]
- 18.Burri E, Beglinger C. 2014. The use of fecal calprotectin as a biomarker in gastrointestinal disease. Expert Rev Gastroenterol Hepatol 8:197–210. doi: 10.1586/17474124.2014.869476. [DOI] [PubMed] [Google Scholar]
- 19.Manz M, Burri E, Rothen C, Tchanguizi N, Niederberger C, Rossi L, Beglinger C, Lehmann FS. 2012. Value of fecal calprotectin in the evaluation of patients with abdominal discomfort: an observational study. BMC Gastroenterol 12:5. doi: 10.1186/1471-230X-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]