Abstract
The current diagnosis of West Nile virus (WNV) infection is primarily based on serology, since molecular identification of WNV RNA is unreliable due to the short viremia and absence of detectable virus in cerebrospinal fluid (CSF). Recent studies have shown that WNV RNA can be detected in urine for a longer period and at higher concentrations than in plasma. In this study, we examined the presence of WNV RNA in serum, plasma, whole-blood, CSF, and urine samples obtained from patients diagnosed with acute WNV infection during an outbreak which occurred in Israel in 2015. Our results demonstrate that 33 of 38 WNV patients had detectable WNV RNA in whole blood at the time of diagnosis, a higher rate than in any of the other sample types tested. Overall, whole blood was superior to all other samples, with 86.8% sensitivity, 100% specificity, 100% positive predictive value, and 83.9% negative predictive value. Interestingly, WNV viral load in urine was higher than in whole blood, CSF, serum, and plasma despite the lower sensitivity than that of whole blood. This study establishes the utility of whole blood in the routine diagnosis of acute WNV infection and suggests that it may provide the highest sensitivity for WNV RNA detection in suspected cases.
INTRODUCTION
West Nile virus (WNV) is a mosquito-borne flavivirus which is the causative agent of West Nile fever (WNF) and West Nile neuroinvasive disease (WNND) in humans (1). Currently, laboratory diagnosis of WNF is based primarily on the identification of specific IgM and IgG antibodies in serum or cerebrospinal fluid (CSF) (2). However, due to high cross-reactivity with other flaviviruses and IgM persistence, neutralization or other serological tests as well as convalescent-phase samples are frequently required, and therefore, confirmation of WNV infection can be time-consuming and labor-intensive (2).
WNF can also be diagnosed by detection of WNV RNA in serum, plasma, or CSF (2). However, WNV RNA is rarely detected in these sample types due to the low level or absence of viremia at the time of symptom onset. Recently, WNV RNA was demonstrated to be detectable in urine at a higher load and for a longer period than in plasma from patients with acute WNV infection: 43.8% of urine samples taken from symptomatic WNV-infected patients tested positive for WNV RNA, suggesting that urine might be useful for confirmation of WNV infection (3).
In this study, we examined the utility of WNV RNA detection in serum, plasma, whole-blood (WB), CSF and urine samples for diagnosis of WNV infection using samples obtained from 105 patients with acute WNF during an outbreak which occurred in Israel in 2015.
MATERIALS AND METHODS
Patients.
Between 1 July and 31 December 2015, 1,141 serum and CSF samples obtained from 863 patients were sent to the Israeli national center for zoonotic viruses in the Central Virology Laboratory (CVL) in Sheba Medical Center for WNV diagnosis. In accordance with our West Nile disease (WND) definitions (see below), 105 patients were diagnosed with either probable or confirmed acute WND. The average and range of time between onset of symptoms and specimen collection from the WNV patients was 9 ± 7 days.
West Nile disease definition.
WND could be defined as WNF or WNND. A suspected case of WNND was defined as any instance of a person having at least one of the following neurological symptoms: viral encephalitis, viral meningitis, viral meningoencephalitis, or acute flaccid paralysis. A suspected case of WNF was defined as a fever of ≥38°C and the absence of other concomitant diseases that could account for the febrile illness. A case of WNND or WNF was defined as probable if the patient met the clinical criteria and laboratory tests demonstrated a positive WNV immunoglobulin M (IgM) in serum. A case was defined as confirmed if the patient met the clinical criteria and laboratory tests demonstrated at least one of the following 4 results: (i) isolation of WNV from serum, plasma, or cerebrospinal fluid (CSF); (ii) detection of WNV RNA by quantitative real-time PCR (qRT-PCR) in serum, plasma, or CSF; (iii) presence of specific IgM antibodies in the CSF; and (iv) detection of WNV IgM in the first sample with seroconversion to IgG in a convalescent-phase sample, no WNV IgM detection in the first sample with detection of WNV IgM in the convalescent-phase sample, or detection of WNV IgG in both samples with confirmation of at least a 4-fold increase in the convalescent-phase sample by a neutralization (NT) assay.
Laboratory investigation of suspected cases of WND.
RNA extracted from samples was tested for the presence of WNV RNA by qRT-PCR, described in detail below. Detection of IgM and IgG antibodies against WNV in serum and CSF samples was performed by enzyme-linked immunosorbent assay (ELISA; WNV IgM capture DxSelect ELISA and IgG DxSelect ELISA kits by Focus Diagnostics Inc., Cypress, CA). All probable cases were further tested by a serum neutralization test, as described elsewhere (4).
Detection of WNV RNA in biological specimens by RT-PCR.
For WNV RNA detection, total nucleic acids were purified from 200 μl (CSF, whole blood, serum, and plasma) or 1 ml (urine) of specimen by using the NucliSENS easyMAG system (bioMérieux, Marcy l'Etoile, France) according to the manufacturer's instructions, with minor modifications. Briefly, MS2 coliphage (∼10,000 copies/ml) was added to the lysis buffer to control for proper extraction and sample inhibition (5), and external lysis was performed to inactivate the virus as recommended by the manufacturer. This was followed by nucleic acid (NA) extraction using the easyMAG extractor. Extracted NA was eluted in 55 μl of elution buffer and stored at −70°C pending analysis. The ABI Prism 7500 sequence detection system (Applied Biosystems Inc., Foster City, CA) was used for the amplification and detection of the MS2 (5) and WNV RNA by qRT-PCR as previously described (6, 7). Briefly, for MS2 qRT-PCR, 5 μl of RNA was added to the AgPath Mastermix (Ambion, Applied Biosystems Inc.), which contained the published concentrations of primers and probes and 5-carboxy-X-rhodamine succinimidyl ester (ROX) as an internal reference dye, whereas 10 μl of RNA was added for the WNV qRT-PCR assay using a specific primer probe set for the envelope (ENV) protein and the 5′ untranslated region and the capsid protein described in references 6 and 7. qRT-PCR was performed under the following conditions: 30 min at 48°C, 10 min at 95°C, and 50 cycles of 15 s at 95°C and 1 min at 60°C. The limit of detection of our qRT-PCR method is 3 copies/reaction in whole-blood, urine, CSF, serum, and plasma specimens, with a PCR efficiency of 98.7%. WNV viral loads in the patient samples were determined in every run after comparing the positive threshold cycle (CT) value with a serial dilution of commercially available West Nile viral RNA standard (QCMD).
Ethics statement.
The Sheba Medical Center Ethical Review Board approved this study (2861-15-SMC).
RESULTS
Between July and December of 2015, the Israeli National Center for Zoonotic Viruses at the Central Virology Laboratory (CVL) received 143 serum and CSF samples from 105 patients who were diagnosed with WND. In accordance with our diagnostic protocol, confirmed WND was established in 88 patients and probable WND in 17 patients. To assess the utility of different sample types for WNV RNA detection, all 105 WND-diagnosed patients were requested to submit additional samples. Therefore, whole-blood, plasma, and urine samples were obtained simultaneously from each patient 2 to 7 days after the initial serum and CSF samples and, on average, 11 days after symptom onset. For all the requested samples, the CVL received 38 whole-blood, 35 plasma, and 48 urine samples, which, together with the 77 serum and 66 CSF samples initially received, were examined for the presence of WNV RNA by qRT-PCR analysis. Analysis of the results (Table 1) showed that WNV RNA was detected in 16.6% of CSF, 26% of serum, 20% of plasma, 86.8% of whole-blood, and 58.3% of urine samples. These data suggest that WNV RNA is present in whole blood in the majority of patients with acute WNF, significantly more than any other sample type tested.
TABLE 1.
Detection of WNV RNA in samples from patients with acute WNV infection
| Sample type | No. of samples tested | No. (%) of positive samples |
|---|---|---|
| Whole blood | 38 | 33 (86.8) |
| Serum | 77 | 20 (26) |
| CSF | 66 | 11 (16.6) |
| Plasma | 35 | 7 (20) |
| Urine | 48 | 28 (58.3) |
Importantly, WNV RNA was detected only in whole blood or urine obtained from patients with acute WNV infection. qRT-PCRs did not detect WNV RNA in whole blood (26 samples) or urine (47 samples) of patients without a serologically confirmed diagnosis of acute WNV infection.
We next compared the performance of WNV RNA detection in whole blood and urine, with the serological assays, which are considered the “gold standard” methods for WNV infection diagnosis. The performance of whole blood was superior to that of urine, with 86.8% sensitivity, 100% specificity, 100% positive predictive value (PPV), and 83.9% negative predictive value (NPV). Detection in urine showed 58.3% sensitivity, 100% specificity, 100% PPV, and 70.1% NPV (Table 2).
TABLE 2.
Performance of whole-blood and urine samples for WNV RNA detection by RT-PCR
| Sample type (no. of samples) | RT-PCR result | No. of samples with serology results of acute WNV infection |
|
|---|---|---|---|
| Positive | Negative | ||
| Whole blood (64) | Positive | 33 | 0 |
| Negative | 5 | 26 | |
| Urine (95) | Positive | 28 | 0 |
| Negative | 20 | 47 | |
Since 34 patients had both urine and whole-blood samples, we examined these patients in more detail. The results, which are presented in Table 3, show that the average time that had passed from onset of symptoms until urine and whole-blood sample collection was 11 days and that all 34 patients were confirmed for WND by serology, either by the presence of WNV IgM antibodies in CSF, IgG seroconversion, or NT assay (Table 3). WNV RNA was detected in 30 whole-blood samples but only in 19 urine samples from these 34 patients. Moreover, 12 patients who were WNV RNA positive by whole blood were negative by urine, whereas only 1 patient who was positive by urine was negative by whole blood. Interestingly, despite the detection of WNV RNA more frequently in whole blood than in urine, no significant difference in the duration of WNV RNA presence in these samples was found in the first 19 days after symptom onset. However, 2 patients whose samples were obtained 34 and 35 days after symptom onset contained WNV RNA only in whole blood and not in urine.
TABLE 3.
Serological and molecular test results performed in confirmed cases of West Nile virus infectiona
| No. | Serology result |
qRT-PCR result |
No. of days from symptom onset until urine and WB collection | Serology diagnosis (test) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CSF |
1st serum sample |
2nd serum sample |
WB (no. of copies/ml) | Urine (no. of copies/ml) | ||||||
| IgM | IgG | IgM | IgG | IgM | IgG | |||||
| 1 | Pos | Neg | Pos | Pos | ND | ND | Pos (5,877) | Pos (10,771) | 11 | Confirmed (IgM in CSF) |
| 2 | Pos | Neg | Pos | Neg | Pos | Neg | Pos (68) | Neg | 5 | Confirmed (IgM in CSF) |
| 3 | Pos | Neg | Pos | Pos | ND | ND | Pos (4) | Pos (17,907) | 5 | Confirmed (IgM in CSF) |
| 4 | Pos | Equ | Pos | Pos | Pos | Pos | Neg | Pos (17) | 19 | Confirmed (IgM in CSF) |
| 5 | Pos | Neg | Pos | Neg | ND | ND | Pos (3,367) | Neg | 12 | Confirmed (IgM in CSF) |
| 6 | Pos | Neg | Pos | Neg | Pos | Pos | Pos (4,448) | Pos (34) | 9 | Confirmed (IgM in CSF) |
| 7 | ND | ND | Pos | Pos | Pos | Pos | Neg | Neg | 12 | Confirmed (NT) |
| 8 | ND | ND | Pos | Pos | Neg | Pos | Neg | Neg | 7 | Confirmed (NT) |
| 9 | Pos | Neg | ND | ND | ND | ND | Pos (1) | Pos (551) | 13 | Confirmed (IgM in CSF) |
| 10 | Pos | Neg | Pos | Neg | ND | ND | Pos (137) | Neg | 8 | Confirmed (IgM in CSF) |
| 11 | Pos | Neg | Pos | Pos | ND | ND | Pos (137) | Pos (2,217) | 10 | Confirmed (IgM in CSF) |
| 12 | ND | ND | Pos | Neg | Pos | Neg | Pos (551) | Neg | 10 | Confirmed (NT) |
| 13 | ND | ND | Pos | Neg | Pos | Pos | Pos (3,609) | Neg | 34 | Confirmed (seroconversion) |
| 14 | Pos | Neg | Pos | Neg | ND | ND | Pos (551) | Pos (34) | 12 | Confirmed (IgM in CSF) |
| 15 | Pos | Neg | Pos | Neg | ND | ND | Pos (68) | Neg | 8 | Confirmed (IgM in CSF) |
| 16 | Pos | Neg | Pos | Neg | ND | ND | Pos (4) | Pos (137) | 17 | Confirmed (IgM in CSF) |
| 17 | Pos | Neg | ND | ND | ND | ND | Neg | Neg | 6 | Confirmed (IgM in CSF) |
| 18 | Pos | Neg | ND | ND | ND | ND | Pos (290,238) | Pos (9,438,916) | 5 | Confirmed (IgM in CSF) |
| 19 | Pos | Neg | Pos | Neg | ND | ND | Pos (551) | Pos (137) | 15 | Confirmed (IgM in CSF) |
| 20 | Pos | Neg | Pos | Neg | Pos | Pos | Pos (727) | Neg | 7 | Confirmed (IgM in CSF) |
| 21 | Pos | Neg | Pos | Pos | ND | ND | Pos (7,447) | Pos (590) | 4 | Confirmed (IgM in CSF) |
| 22 | Pos | Pos | Pos | Pos | ND | ND | Pos (551) | Neg | 11 | Confirmed (IgM in CSF) |
| 23 | Pos | Pos | Pos | Pos | ND | ND | Pos (137) | Pos (95,249) | 13 | Confirmed (IgM in CSF) |
| 24 | Pos | Neg | Pos | Neg | ND | ND | Pos (1,105) | Pos (582,356) | 3 | Confirmed (IgM in CSF) |
| 25 | Pos | Neg | Pos | Neg | Pos | Neg | Pos (45) | Neg | 6 | Confirmed (IgM in CSF) |
| 26 | Pos | Pos | Pos | Pos | ND | ND | Pos (6,524) | Neg | 6 | Confirmed (IgM in CSF) |
| 27 | Pos | Neg | Pos | Neg | ND | ND | Pos (52,332) | Pos (16,819) | 13 | Confirmed (IgM in CSF) |
| 28 | Pos | Neg | Pos | Neg | ND | ND | Pos (1,244) | Pos (68,662) | 11 | Confirmed (IgM in CSF) |
| 29 | Pos | Neg | Pos | Neg | ND | ND | Pos (472,561) | Pos (216,706,927) | 8 | Confirmed (IgM in CSF) |
| 30 | Pos | Neg | Pos | Neg | Pos | Pos | Pos (884) | Pos (30,400) | 10 | Confirmed (IgM in CSF) |
| 31 | Pos | Pos | ND | ND | ND | ND | Pos (138) | Neg | 35 | Confirmed (IgM in CSF) |
| 32 | Pos | Neg | ND | ND | ND | ND | Pos (16,702) | Neg | 6 | Confirmed (IgM in CSF) |
| 33 | Pos | Neg | Pos | Neg | ND | ND | Pos (1) | Pos (58) | 13 | Confirmed (IgM in CSF) |
| 34 | Pos | Pos | Pos | Pos | ND | ND | Pos (48) | Pos (1,381) | 19 | Confirmed (IgM in CSF) |
Pos, positive; Neg, negative; ND, not preformed; WB, whole blood.
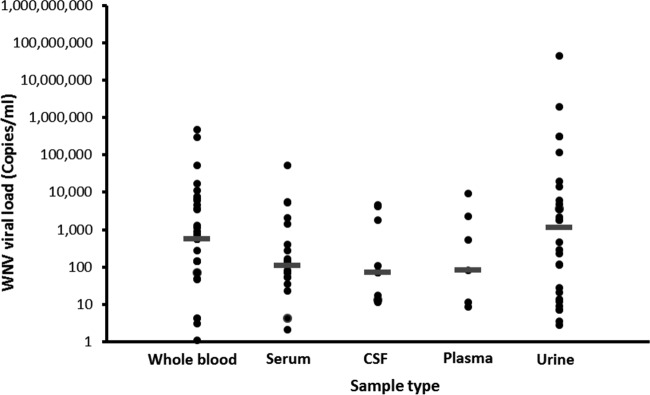
Regardless of the high percentage of WND patients with detectable WNV RNA in whole-blood samples, WNV viral load data (Fig. 1) demonstrated that the largest amount of WNV RNA was found in urine samples, with a median of 1,100 copies/ml. This is especially noteworthy since WNV viral load in whole blood was 2 times lower, with a median of 550 copies/ml (Fig. 1), while WNV viral loads in serum, plasma, and CSF were less than 100 copies/ml.
FIG 1.
WNV RNA viral load in samples from patients with acute WNV infection. Shown are WNV viral loads in serum (20 samples), plasma (7 samples), CSF (11 samples), whole blood (33 samples), and urine (28 samples), as determined by RT-PCR. All RT-PCR-negative samples were excluded from viral load analysis. Bars = median viral loads.
DISCUSSION
The introduction of RT-PCR assays ∼15 years ago has revolutionized laboratory diagnosis of many viral diseases (8). Instead of virus isolation in cell culture, which could take days, molecular detection of viral DNAs and RNAs can now be achieved in hours. For some viruses, however, such as WNV, it is much harder to confirm diagnosis since infection does not generate sufficient and long viremia (2). Here, we demonstrate that WNV RNA can be identified by RT-PCR in 86.8% of whole-blood samples during acute infection, thus highlighting the possible role of whole blood in WNV detection and WNF diagnosis.
The finding that WNV viral load in whole blood is greater than in plasma has been described previously for blood donors. Rios et al. (9) demonstrated that WNV bound to the red blood cell (RBC) component of blood represents a large proportion of the circulating virus, which may be higher than the proportion of circulating virus in plasma. Two later studies (10, 11) corroborated these data and revealed that WNV RNA concentrations in seropositive donations are 10-fold higher in whole blood than in plasma (10) and persist for a much longer period (11). Interestingly, the results from our study demonstrated that even though WNV viral load in whole blood samples was indeed higher than in serum or plasma, it was lower than in urine. Furthermore, since 86.8% of whole-blood samples but only 58.1% of urine samples tested positive for WNV RNA, our results suggest that higher viral load is not correlated with sensitivity of the clinical diagnosis. This suggests that WNV might not be retained in urine in all patients; however, when it is, it will exist in significant quantities.
Most interesting are the data obtained from WNV patients with both whole-blood and urine samples (Table 3). Since only 1 patient had detectable WNV RNA only in urine (and not in whole blood), whereas 12 patients had detectable WNV RNA only in whole blood (and not urine), our study indicates that in most cases testing WNV RNA in whole blood would be sufficient and additional sensitivity would not be achieved even if both whole-blood and urine samples are tested for the same patient.
As far as we know, this study represents the first comparison of whole-blood, serum, plasma, CSF, and urine samples for WNV RNA detection during acute infection. The results of this study have important implications for the routine diagnostics of WNV infection, as the detection of WNV RNA in whole blood by qRT-PCR was significantly more sensitive than testing of plasma, serum, CSF, or urine from patients with symptomatic WNV infection. Moreover, we would expect even higher sensitivity of detection once whole blood is obtained at the same time as serum and CSF samples. In fact, retrospectively, the results for whole blood would have allowed us to quickly confirm diagnosis of WNV infection in high proportions of patients for whom confirmation using other tests, such as neutralization, would take much longer. This study demonstrates that WNV RNA can be detected in whole blood with higher sensitivity and specificity than for any other sample type tested. Follow-up laboratory investigations should establish the utility of whole blood in the routine diagnosis of acute WNV infection.
ACKNOWLEDGMENTS
This work was supported by internal sources of the Central Virology Laboratory.
All authors declare that they have no competing interests or conflicts of interest.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Kramer LD, Styer LM, Ebel GD. 2008. A global perspective on the epidemiology of West Nile virus. Annu Rev Entomol 53:61–81. doi: 10.1146/annurev.ento.53.103106.093258. [DOI] [PubMed] [Google Scholar]
- 2.Sambri V, Capobianchi MR, Cavrini F, Charrel R, Donoso-Mantke O, Escadafal C, Franco L, Gaibani P, Gould EA, Niedrig M, Papa A, Pierro A, Rossini G, Sanchini A, Tenorio A, Varani S, Vazquez A, Vocale C, Zeller H. 2013. Diagnosis of West Nile virus human infections: overview and proposal of diagnostic protocols considering the results of external quality assessment studies. Viruses 5:2329–2348. doi: 10.3390/v5102329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barzon L, Pacenti M, Franchin E, Pagni S, Martello T, Cattai M, Cusinato R, Palu G. 2013. Excretion of West Nile virus in urine during acute infection. J Infect Dis 208:1086–1092. doi: 10.1093/infdis/jit290. [DOI] [PubMed] [Google Scholar]
- 4.Di Gennaro A, Lorusso A, Casaccia C, Conte A, Monaco F, Savini G. 2014. Serum neutralization assay can efficiently replace plaque reduction neutralization test for detection and quantitation of West Nile virus antibodies in human and animal serum samples. Clin Vaccine Immunol 21:1460–1462. doi: 10.1128/CVI.00426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shulman LM, Hindiyeh M, Muhsen K, Cohen D, Mendelson E, Sofer D. 2012. Evaluation of four different systems for extraction of RNA from stool suspensions using MS-2 coliphage as an exogenous control for RT-PCR inhibition. PLoS One 7:e39455. doi: 10.1371/journal.pone.0039455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage HM, Komar N, Panella NA, Allen BC, Volpe KE, Davis BS, Roehrig JT. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J Clin Microbiol 38:4066–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linke S, Ellerbrok H, Niedrig M, Nitsche A, Pauli G. 2007. Detection of West Nile virus lineages 1 and 2 by real-time PCR. J Virol Methods 146:355–358. doi: 10.1016/j.jviromet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 8.Leland DS, Ginocchio CC. 2007. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev 20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rios M, Daniel S, Chancey C, Hewlett IK, Stramer SL. 2007. West Nile virus adheres to human red blood cells in whole blood. Clin Infect Dis 45:181–186. doi: 10.1086/518850. [DOI] [PubMed] [Google Scholar]
- 10.Lai L, Lee TH, Tobler L, Wen L, Shi P, Alexander J, Ewing H, Busch M. 2012. Relative distribution of West Nile virus RNA in blood compartments: implications for blood donor nucleic acid amplification technology screening. Transfusion 52:447–454. doi: 10.1111/j.1537-2995.2011.03289.x. [DOI] [PubMed] [Google Scholar]
- 11.Lanteri MC, Lee TH, Wen L, Kaidarova Z, Bravo MD, Kiely NE, Kamel HT, Tobler LH, Norris PJ, Busch MP. 2014. West Nile virus nucleic acid persistence in whole blood months after clearance in plasma: implication for transfusion and transplantation safety. Transfusion 54:3232–3241. doi: 10.1111/trf.12764. [DOI] [PMC free article] [PubMed] [Google Scholar]