Abstract
This study was conducted to explore the effects of interferon tau (IFNT) on the intestinal microbiota and expression of interleukin 17 (IL-17) in the intestine of mice. IFNT supplementation increased microbial diversity in the jejunum and ileum but decreased microbial diversity in the feces. IFNT supplementation influenced the composition of the intestinal microbiota as follows: (1) decreasing the percentage of Firmicutes and increasing Bacteroidetes in the jejunum and ileum; (2) enhancing the percentage of Firmicutes but decreasing Bacteroidetes in the colon and feces; (3) decreasing Lactobacillus in the jejunum and ileum; (4) increasing the percentage of Blautia, Bacteroides, Alloprevotella, and Lactobacillus in the colon; and (5) increasing the percentage of Lactobacillus, Bacteroides, and Allobaculum, while decreasing Blautia in the feces. Also, IFNT supplementation decreased the expression of IL-17 in the intestines of normal mice and of an intestinal pathogen infected mice. In conclusion, IFNT supplementation modulates the intestinal microbiota and intestinal IL-17 expression, indicating the applicability of IFNT to treat the intestinal diseases involving IL-17 expression and microbiota.
1. Introduction
Interferon tau (IFNT) is produced by trophectoderm cells of conceptuses of ruminant species and is the maternal recognition of the pregnancy signal. Besides its critical roles in implantation and establishment of pregnancy in ruminants [1, 2], it has a plethora of physiological functions in various cell types such as macrophages, lymphocytes, and epithelial cells in humans and mice [3–5]. It is a type I interferon (IFN), which includes IFN alpha (IFNA), IFN beta (IFNB), IFN delta (IFND), and IFN omega (IFNW). After binding to a common receptor, IFNA receptor 1 (IFNAR1), and IFNAR2, type I IFNs affect the production of inflammatory cytokines such as interleukin- (IL-) 1β and tumor necrosis factor α (TNF-α) [6, 7]. Thus, type I IFNs have widely recognized roles in inflammatory diseases, such as experimental allergic encephalomyelitis, multiple sclerosis, and spontaneous autoimmune diabetes [5, 8–10]. Notably, unlike other members of type I IFN family, IFNT has few adverse effects and low cytotoxicity even at high dosages [11, 12], suggesting its therapeutic potential as an alternative to other type I IFNs due to its anti-inflammatory effects. Recent compelling findings about the anti-inflammatory effects of IFNT include lower NLRP3 (nucleotide-binding oligomerization domain-like receptor, pyrin domain-containing 3) inflammasome-driven IL-1β secretion by human macrophages [4], mitigation of obesity-associated systemic tissue inflammation in mice [5], and promotion of Th2 biased immune response in mice [9].
The influence of IFNT on intestinal microbiota is unknown. The intestinal microbiota provides important benefits for the development of immune responses; however, the disturbances in the intestinal microbiota are associated with numerous chronic inflammatory diseases [13, 14]. Also, the effect of IFNT on expression of IL-17 in the intestine is not known. The potential effect of IFNT on expression of IL-17 is important as IL-17 promotes local chemokine production to recruit monocytes and neutrophils to sites of inflammation that leads to development and pathogenesis of various autoimmune diseases, including rheumatoid arthritis, psoriasis vulgaris, multiple sclerosis, and inflammatory bowel diseases [15, 16]. In this study, the intestinal microbiota and expression of IL-17 in the intestine were explored after two weeks of IFNT supplementation in a mouse model. The hypothesis is that IFNT supplementation alters intestinal microbiota and intestinal innate immunity in mouse model.
2. Materials and Methods
2.1. Bacterial Strains
This study used the Escherichia coli F4-producing strain W25K (hereafter referred as ETEC; O149:K91, K88ac; LT, STb, EAST), which was originally isolated from a piglet with diarrhea [17].
2.2. IFNT Supplementation for Mice
This study was conducted according to the guidelines of the Laboratory Animal Ethical Commission of the Chinese Academy of Sciences. ICR (Institute for Cancer Research) mice (six weeks of age) were purchased from SLAC Laboratory Animal Central (Changsha, China). The mice were housed individually in a pathogen-free animal vivarium (temperature, 25°C; relative humidity, 53%; 12 h dark/12 h light) and had free access to a standard rodent diet [18] and drinking water. After three days of accommodation, mice were assigned randomly into two groups (IFNT and control; n = 10/group). Mice in the control group were fed the basal diet [18] and normal water, while mice in IFNT group were fed the basal diet and water containing recombinant IFNT (40 μg/L) for two weeks. The effective supplemental dosage of IFNT was established in previous study [5, 19]. At the end of the two weeks of experimental period, mice were sacrificed to collect contents of the lumens of the jejunum, ileum, and colon, as well as feces. The tissues including jejunum, ileum, and colon were also collected. Feed and water intake and body weight gain were monitored throughout the experiment. Samples were collected and stored at −80°C until processed.
2.3. ETEC Infection of Mice
After three days of accommodation to the conditions of the vivarium, ICR mice were assigned randomly into two groups (ETEC and IFNT+ETEC; n = 10/group). Mice in IFNT+ETEC group were fed the basal diet and recombinant IFNT-supplemented water (40 μg/L) for two weeks, while mice in ETEC group were fed the basal diet and had normal water. After two weeks of feeding, mice in both groups were inoculated with 108 CFUs of ETEC W25K by oral gavage. At 6 hours after infection, all active mice were sacrificed to collect the jejunum, and the samples were stored at −80°C until processed.
2.4. 16S rDNA Sequencing with Illumina MiSeq Sequencing
DNA was extracted from the luminal contents of the jejunum, ileum and colon, and feces using the Qiagen QIAamp DNA Stool Mini Kit according to the protocol for isolation of DNA. Equal amounts of DNA from six different mice were pooled to generate one common sample for each type of sample (i.e., control versus IFNT, intestinal source, and feces). The V4-V5 region of the bacterial 16S ribosomal RNA gene was amplified by PCR using primers 515F 5′-barcode-GTGCCAGCMGCCGCGG-3′ and 907R 5′-CCGTCAATTCMTTTRAGTTT-3′, where barcode is an eight-base sequence unique to each sample. Illumina MiSeq sequencing and general data analyses were performed by a commercial company (Biotree, Shanghai, China). Miseq PE Libraries, Miseq Sequencing, and further analyses were based on previous work [20].
2.5. RT-PCR
Total RNA was isolated from liquid nitrogen frozen and ground jejunum, ileum, and colon using TRIZOL regent (Invitrogen, USA) and then treated with DNase I (Invitrogen, USA) according to the manufacturer's instructions. Synthesis of the first strand (cDNA) was performed using oligo (dT) 20 and Superscript II reverse transcriptase (Invitrogen, USA). Primers were selected according to previous references [18, 21]. β-actin was used as an internal control to normalize expression of target gene transcripts. The RT-PCR experiment was conducted according to previous studies [18, 21].
2.6. Statistical Analyses
Data shown are the means ± the standard error of the mean (SEM). All statistical analyses for data were performed using SPSS 16.0 software (Chicago, IL, USA). Data were analyzed for the two treatment groups using Student's t-test. Differences of P < 0.05 are considered significant.
3. Results
3.1. IFNT Treatment Increases Feed Intake
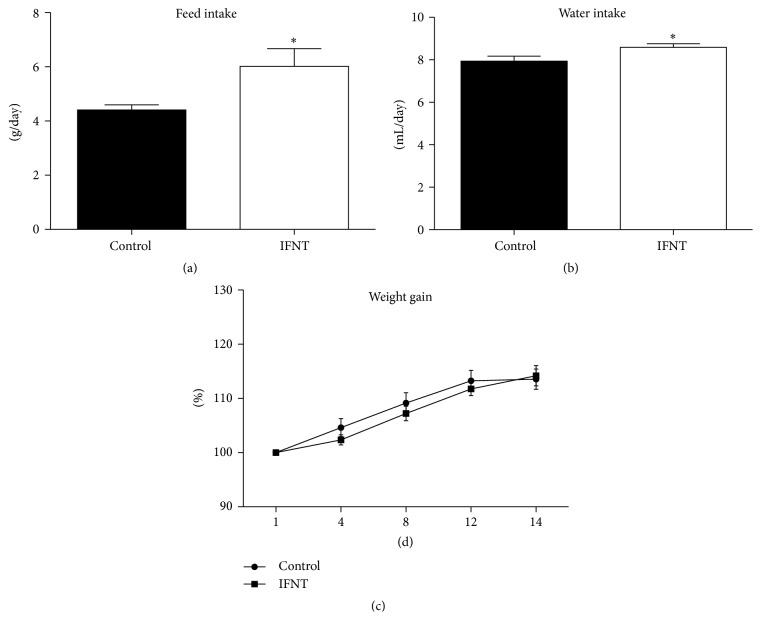
To investigate the effect of IFNT supplementation on mouse growth performance, feed intake, water intake, and body weight were monitored in IFNT-supplemented mice and control mice. With two weeks of IFNT supplementation, the averages for feed intake and water intake for IFNT-supplemented mice were significant (P < 0.05) higher than for control mice (Figures 1(a) and 1(b)). However, IFNT supplementation had no significant effect on body weight of mice (Figure 1(c)).
Figure 1.
IFNT supplementation has little effect on mouse body weight. (a) Average feed intake in the control and IFNT-supplemented mice (n = 10). (b) Average water intake for control and IFNT-supplemented mice (n = 10). (c) Relative body weight gains for control and IFNT-supplemented mice (n = 10). Control mice were fed the basal diet and normal water, while mice in IFNT group were fed the basal diet and IFNT-supplemented water for two weeks. The asterisk (∗) indicates a statistically significant difference between two treatment groups (P < 0.05). Data were analyzed using Student's t-test.
3.2. Changes in Bacterial Diversity of the Intestinal Microbiota Associated with IFNT Supplementation
To explore the influence of IFNT supplementation on the intestinal microbiota, we analyzed the intestinal microbiota at end of two weeks of IFNT supplementation with 16S rDNA sequencing (Table 1). For microbiota in the jejunum, both Shannon and Simpson indices demonstrated that the diversity of microbiota in mice with IFNT supplementation was higher than the control mice, while the richness indices (Ace and Chao) suggested that the community richness in IFNT-supplemented and control mice was similar (Table 1). For the microbiota in the ileum, the diversity of microbiota (Shannon and Simpson) and richness indices (Ace) for mice with IFNT supplementation were higher than that of control mice (Table 1). For the microbiota in the colon, the diversity of microbiota (Shannon) and richness indices (Ace and Chao) were similar for IFNT-supplemented and control mice (Table 1). For the fecal microbiota, microbial diversity (Shannon and Simpson) in mice with IFNT supplementation was lower than for control mice, while the community richness (Ace and Chao) for IFNT-supplemented mice was similar to that for control mice (Table 1). Collectively, IFNT supplementation increases the diversity of microbiota in small intestine, while decreasing the diversity of microbiota in the feces.
Table 1.
Comparison of phylotype coverage and diversity estimation of the 16S rDNA gene libraries at 97% similarity from the pyrosequencing analysis.
| Group | Number of readings | Number of OTU | Coverage | Richness estimator | Diversity index | ||
|---|---|---|---|---|---|---|---|
| Ace (95% CI) | Chao (95% CI) | Shannon (95% CI) | Simpson (95% CI) | ||||
| Jejunum | |||||||
| Control | 12323 | 51 | 99.85% | 78 (62–120) | 70 (57–108) | 1.06 (1.04–1.09) | 0.52 (0.51–0.53) |
| IFNT | 15653 | 64 | 99.95% | 70 (66–83) | 67 (65–78) | 1.32 (1.30–1.35) | 0.44 (0.43–0.45) |
|
| |||||||
| Ileum | |||||||
| Control | 12442 | 37 | 99.89% | 52 (42–84) | 57 (42–110) | 0.73 (0.70–0.75) | 0.70 (0.69–0.71) |
| IFNT | 12264 | 66 | 99.79% | 124 (99–167) | 174 (101–400) | 1.06 (1.03–1.08) | 0.56 (0.55–0.56) |
|
| |||||||
| Colon | |||||||
| Control | 10327 | 298 | 99.58% | 323 (311–343) | 322 (309–349) | 4.32 (4.29–4.35) | 0.032 (0.030–0.034) |
| IFNT | 11276 | 288 | 99.68% | 306 (297–324) | 313 (299–345) | 4.32 (4.29–4.34) | 0.027 (0.026–0.028) |
|
| |||||||
| Feces | |||||||
| Control | 12613 | 314 | 99.69% | 336 (326–356) | 339 (325–369) | 4.39 (4.36–4.41) | 0.026 (0.025–0.027) |
| IFNT | 12493 | 312 | 99.58% | 345 (331–371) | 363 (337–414) | 4.16 (4.13–4.19) | 0.046 (0.044–0.049) |
3.3. IFNT-Associated Alterations in Intestinal Microbiota
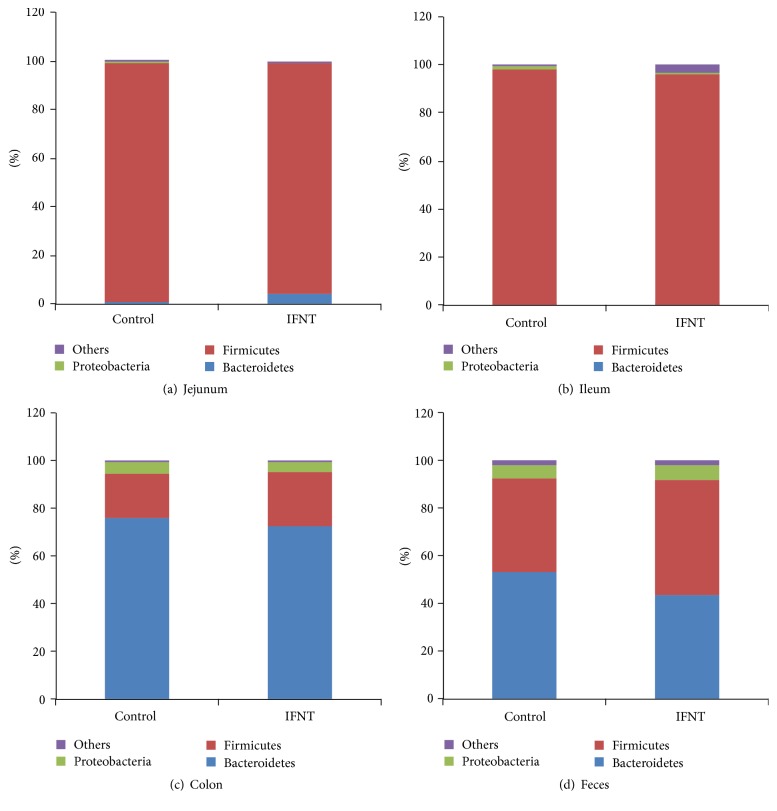
The taxonomy of the intestinal microbiota was assessed using a taxon-dependent analysis and the RDP classifier. Seven phyla, including one candidate division (TM7), were found in the microbiota of the jejunum for all samples, including six phyla in the control mice and seven phyla in mice with IFNT supplementation. Eight phyla were found in the microbiota of the ileum of all samples, including six phyla in control mice and seven phyla in mice with IFNT supplementation. Ten phyla were found in the microbiota of the colon of all samples, including ten phyla in control mice and eight phyla in mice with IFNT supplementation. Ten phyla were found in the microbiota of the feces of all samples, including nine phyla in control mice and ten phyla in mice with IFNT supplementation.
For the jejunum, the two most abundant phyla in IFNT-supplemented mice, accounting for approximately 99% of all assigned sequence readings, were Firmicutes (94.5%) and Bacteroidetes (4.4%) (Figure 2(a)). In control mice, most abundant phyla were Firmicutes (97.6%) and Bacteroidetes (1.2%) (Figure 2(a)). For the ileum, the three most abundant phyla in IFNT-supplemented mice were Firmicutes (95.3%), Candidate_division_TM7 (2.3%), and Proteobacteria (1.1%) (Figure 2(b)), while in control mice, they were Firmicutes (97.7%), Proteobacteria (1.5%), and Bacteroidetes (0.5%) (Figure 2(b)). For the microbiota in the colon, the three most abundant phyla in IFNT-supplemented mice were Bacteroidetes (72.2%), Firmicutes (23.0%), and Proteobacteria (4.1%) (Figure 2(c)), while they were Bacteroidetes (75.8%), Firmicutes (18.8%), and Proteobacteria (4.3%) in control mice (Figure 2(c)). For feces, the three most abundant phyla in IFNT-supplemented mice were Bacteroidetes (43.6%), Firmicutes (48.1%), and Proteobacteria (5.9%) (Figure 2(d)), while Bacteroidetes (53.2%), Firmicutes (39.2%), and Proteobacteria (5.3%) were most abundant for control mice (Figure 2(d)).
Figure 2.
Composition of the intestinal microbiota at the phylum level after IFNT supplementation. (a) The microbial composition in the jejunum. (b) The microbial composition in the ileum. (c) The microbial composition in the colon. (d) The microbial composition in the feces.
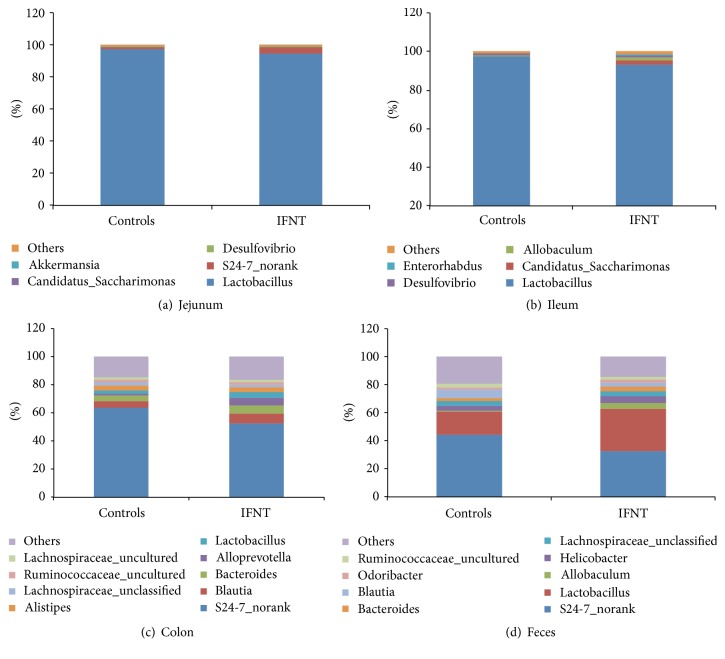
For the microbiota of the jejunum, the two most abundant genera in IFNT-supplemented mice, accounting for approximately 99% of all assigned sequence readings, were Lactobacillus (94.3%) and S24-7_norank (4.9%) (Figure 3(a)). In control mice, they were Lactobacillus (97.3%) and S24-7_norank (1.2%) (Figure 3(a)). For the ileum, the five most abundant genera in IFNT-supplemented mice were Lactobacillus (93.3%), Candidatus-Saccharimonas (2.3%), Allobaculum (1.2%), Desulfovibrio (1.1%), and Enterorhabdus (0.6%), while they were Lactobacillus (97.3%), Candidatus-Saccharimonas (0.3%), Allobaculum (0.1%), Desulfovibrio (1.5%), and Enterorhabdus (0.07%) in control mice (Figure 3(b)). For the microbiota in the colon, IFNT supplementation increased the percentages of Blautia (7.0% versus 5.1%), Bacteroides (6.4% versus 3.7%), Alloprevotella (5.2% versus 1.3%), and Lactobacillus (4.0% versus 2.5%), compared with control mice (Figure 3(c)). For the fecal microbiota, IFNT supplementation increased the percentages of Lactobacillus (30.0% versus 16.7%), Bacteroides (3.3% versus 1.8%), and Allobaculum (4.5% versus 0.4%), while decreasing the Blautia (2.7% versus 6.5%) compared with control mice (Figure 3(d)).
Figure 3.
The composition of the intestinal microbiota at the genus level after IFNT supplementation. (a) The microbial composition in the jejunum. (b) The microbial composition in the ileum. (c) The microbial composition in the colon. (d) The microbial composition in the feces.
Collectively, IFNT supplementation affects the composition of intestinal microbiota in mice, especially those for the colon and feces.
3.4. IFNT Inhibits Expression IL-17 in the Intestine
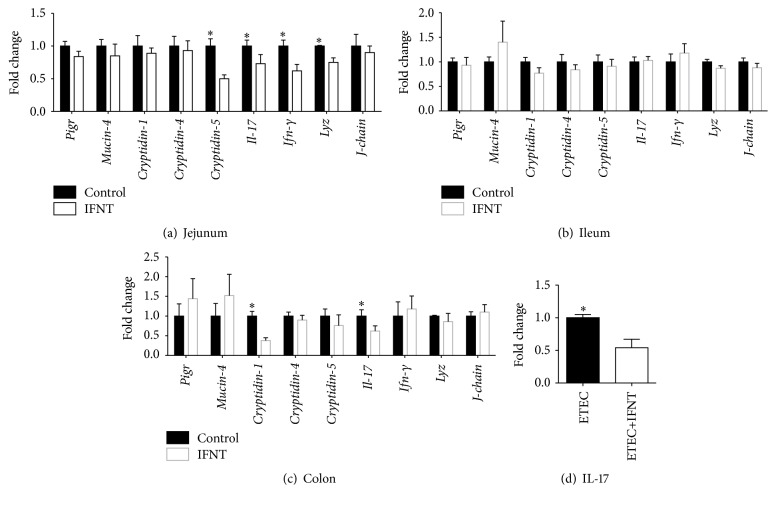
The effect of IFNT supplementation on activation of intestinal innate immunity in mice was further explored, focusing on the expression of polymeric immunoglobulin receptor (Pigr), Mucin-4, Cryptidin-1, Cryptidin-4, Cryptidin-5, Il-17, interferon gamma (Ifn-γ), lysozyme (Lyz), and J-chain in the jejunum, ileum, and colon [18, 21]. In the jejunum, IFNT supplementation significantly decreased the expression of Cryptidin-5, Il-17, Ifn-γ, and Lyz, while it had little effect on the expression of the other transcripts (Figure 4(a)). IFNT supplementation had no significant effect on the expression of those transcripts in the ileum of mice (Figure 4(b)). In the colon, IFNT supplementation significantly lowered the expression of Cryptidin-1 and Il-17 but had little effect on the expression of the other transcripts (Figure 4(c)). As IFNT supplementation decreased the expression of Il-17 in the jejunum and colon, we further validated the effect of IFNT to decrease expression of IL-17 in ETEC infected mouse model. We found that ETEC infection promotes the Il-17 expression in the mouse jejunum at 6 hours after infection (W. Ren and Y. Yin, unpublished results). After two weeks of IFNT supplementation, expression of Il-17 in the jejunum was significantly lower in IFNT-supplemented mice, compared to that of nonsupplemented mice during ETEC infection (Figure 4(d)). Thus, IFNT supplementation reduces the expression of the inflammatory cytokine, IL-17, in the intestine of mice.
Figure 4.
IFNT supplementation decreases expression of IL-17. (a) Expression of innate immune factors (Pigr, Mucin-4, Cryptidin-1, Cryptidin-4, Cryptidin-5, Il-17, Ifn-γ, Lyz, and J-chain) in the jejunum of mice (n = 10). (b) Expression of innate immune factors in the ileum of mice (n = 10). (c) Expression of innate immune factors in the colon of mice (n = 10). (d) IFNT decreases expression of IL-17 in the jejunum of mice following ETEC infection (n = 10). Data were analyzed using Student's t-test. An asterisk (∗) indicates a statistically significant difference between treatment groups (P < 0.05).
4. Discussion
In this study, although two weeks of IFNT supplementation increases the mouse feed and water intake but has little effect on body weight of mice. Results of a previous study revealed that IFNT supplementation (8 μg/kg BW/day) reduces body weight beginning at 3 weeks after IFNT supplementation in Zucker Diabetic Fatty rats, while lower dose of IFNT supplementation (4 μg/kg BW/day) has no significant effect on body weight during 8 weeks of IFNT treatment [19], indicating that the effect of IFNT on body weight depends on dosage and duration of IFNT treatment. However, in a mouse model with high-fat or low-fat diet, 12 weeks of IFNT treatment does not significantly affect body weight [5]. However, IFNT supplementation has little effect on feed intake and water intake in those investigations [4, 5].
In the present study, IFNT supplementation increases the microbial diversity in the jejunum and ileum, while decreasing the microbial diversity in the feces of mice. The gut microbiota affects numerous biological functions [22, 23] and is linked to the pathogenesis of various diseases, such as obesity [24], cancer [25], and liver cirrhosis [26]. The influence of the gut microbiome on host physiological functions and the pathogenesis of disease in hosts may result from the activities of the microbiome and its metabolic products [22]. It is widely accepted that body weight is associated with the composition of intestinal microbiome and its metabolic capacity [27]. An increase in the relative proportion of Firmicutes is linked to obesity as Firmicutes ferments plant polysaccharides to produce short-chain fatty acids (SCFA), which provides additional energy for the host [28]. In phyla, IFNT supplementation decreases the percentage of Firmicutes, while increasing the Bacteroidetes in the jejunum and ileum. However, IFNT supplementation increases the percentage of Firmicutes, while decreasing the Bacteroidetes in the colon and feces. Thus, IFNT supplementation may regulate body weight and metabolism through effects on the intestinal microbiota. At the genus level, IFNT supplementation decreases the Lactobacillus in the jejunum and ileum but increases the percentage of Lactobacillus and Bacteroides in the colon and feces. Lactobacillus has critical roles in the intestine to combat gastrointestinal bacterial pathogens and rotaviruses through competitive metabolic interactions and the production of antimicrobial molecules [29]. Bacteroides are known for their capacity to metabolize a wide variety of oligosaccharides from the intestinal luminal, such as xylan, starch, and host-derived glycans [30]. Thus, results of the present study suggest that IFNT supplementation affects those functions of the intestinal microbiome in mice.
IFNT supplementation inhibits intestinal expression of IL-17, which suggests that IFNT reduces intestinal inflammation. IL-17 is produced by inducible Th17 (iTh17) cells and natural Th17 (nTh17) cells and regarded as an intestinal proinflammatory cytokine [31]. IL-17 can activate nuclear factor κB (NF-κB) transcription factors, extracellular signal-regulated protein kinase (ERK1 and ERK2), c-Jun N-terminal kinases (JNK-1 and JNK-2), and mitogen-activated protein kinases (p38 MAPKs) pathways, leading to upregulation of expression of inflammatory cytokines, such as IL-6 and IL-1 [32]. Recent investigations have revealed that mammalian target of rapamycin (mTOR) is a critical signaling pathway for Th17 responses and IL-17 expression [33–37]. The mTOR signaling regulates IL-17 expression through hypoxia-inducible factor 1 α (HIF-1α) and ribosomal protein S6 kinase (S6K: S6K1 and S6K2) [37–40]. mTOR signaling activates HIF-1α, which promotes IL-17 expression by activating RORγt (a key transcriptional regulator of Th17 cells) and mediating degradation of Foxp3 (a key transcriptional regulator of Treg cells) [40]. S6K1 promotes the expression of early growth response protein 2 (EGR2), which then inhibits growth factor independent 1 transcription repressor (GFI1), which can negatively regulate expression of IL-17 without affecting Rorc expression [37, 38]. S6K2 (the nuclear-localized counterpart of S6K1) binds to RORγt to promote nuclear translocation of RORγt, which can complex with HIF-α and p300 in the nucleus to promote expression of IL-17 [37–39]. Thus, the underlying mechanism by which IFNT supplementation reduces intestinal IL-17 expression is of interest. The effect of IFNT supplementation to decrease expression of IL-17 in the intestine indicates a potential therapeutic application of IFNT to mitigate intestinal inflammatory diseases associated with expression of IL-17.
In conclusion, IFNT supplementation affects the diversity and composition of the intestinal microbiota and decreases expression of IL-17 in mice. The findings from this study are significant in understanding the physiological and immunological functions of IFNT in treatment of inflammatory diseases.
Acknowledgments
This study was supported by National Natural Science Foundation of China (nos. 31330075, 31372326, and 31272463), National Key Basic Research Program of China (2013CB127302), and the National Science and Technology Ministry (2014BAD08B11).
Abbreviations
- EGR2:
Early growth response protein 2
- ERK:
Extracellular signal-regulated protein kinase
- ETEC:
Enterotoxigenic Escherichia coli
- FoxP3:
Forkhead box P3
- GFI1:
Growth factor independent 1 transcription repressor
- HIF-1α:
Hypoxia-inducible factor 1α
- mTOR:
Mammalian target of rapamycin
- RORγt:
Retinoic acid receptor-related orphan receptor gamma t
- S6K:
Ribosomal protein S6 kinase.
Competing Interests
The authors have no competing interests.
Authors' Contributions
Wenkai Ren and Bie Tan designed the experiment; Wenkai Ren and Shuai Chen conducted the experiment; Wenkai Ren, Liwen Zhang, and Gang Liu analyzed the data; Tarique Hussain, Xiao Hao, Jie Yin, and Jielin Duan helped in the experiment; Wenkai Ren wrote the paper; Guoyao Wu, Fuller W. Bazer, and Yulong Yin revised the paper.
References
- 1.Brooks K., Spencer T. E. Biological roles of interferon tau (IFNT) and type I IFN receptors in elongation of the ovine conceptus. Biology of Reproduction. 2015;92, article 47 doi: 10.1095/biolreprod.114.124156. [DOI] [PubMed] [Google Scholar]
- 2.Bazer F. W., Ying W., Wang X., et al. The many faces of interferon tau. Amino Acids. 2015;47(3):449–460. doi: 10.1007/s00726-014-1905-x. [DOI] [PubMed] [Google Scholar]
- 3.Alexenko A. P., Ealy A. D., Roberts R. M. The cross-species antiviral activities of different IFN-tau subtypes on bovine, murine, and human cells: contradictory evidence for therapeutic potential. Journal of Interferon & Cytokine Research. 1999;19(12):1335–1341. doi: 10.1089/107999099312795. [DOI] [PubMed] [Google Scholar]
- 4.Hara K., Shirasuna K., Usui F., et al. Interferon-tau attenuates uptake of nanoparticles and secretion of interleukin-1β in macrophages. PLoS ONE. 2014;9(12) doi: 10.1371/journal.pone.0113974.e113974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ying W., Kanameni S., Chang C.-A., et al. Interferon tau alleviates obesity-induced adipose tissue inflammation and insulin resistance by regulating macrophage polarization. PLoS ONE. 2014;9(6) doi: 10.1371/journal.pone.0098835.e98835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.González-Navajas J. M., Lee J., David M., Raz E. Immunomodulatory functions of type i interferons. Nature Reviews Immunology. 2012;12(2):125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boscá L., Bodelón O. G., Hortelano S., Casellas A., Bosch F. Anti-inflammatory action of type I interferons deduced from mice expressing interferon β . Gene Therapy. 2000;7(10):817–825. doi: 10.1038/sj.gt.3301179. [DOI] [PubMed] [Google Scholar]
- 8.Gough D. J., Messina N. L., Clarke C. J. P., Johnstone R. W., Levy D. E. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36(2):166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soos J. M., Stüve O., Youssef S., et al. Cutting edge: oral type I IFN-τ promotes a Th2 bias and enhances suppression of autoimmune encephalomyelitis by oral glatiramer acetate. The Journal of Immunology. 2002;169(5):2231–2235. doi: 10.4049/jimmunol.169.5.2231. [DOI] [PubMed] [Google Scholar]
- 10.Inoue M., Shinohara M. L. The role of interferon-β in the treatment of multiple sclerosis and experimental autoimmune encephalomyelitis—in the perspective of inflammasomes. Immunology. 2013;139(1):11–18. doi: 10.1111/imm.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pontzer C. H., Bazer F. W., Johnson H. M. Antiproliferative activity of a pregnancy recognition hormone, ovine trophoblast protein-1. Cancer Research. 1991;51(19):5304–5307. [PubMed] [Google Scholar]
- 12.Soos J. M., Subramaniam P. S., Hobeika A. C., Schiffenbauer J., Johnson H. M. The IFN pregnancy recognition hormone IFN-τ blocks both development and superantigen reactivation of experimental allergic encephalomyelitis without associated toxicity. Journal of Immunology. 1995;155(5):2747–2753. [PubMed] [Google Scholar]
- 13.Chassaing B., Koren O., Goodrich J. K., et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519(7541):92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West C. E., Renz H., Jenmalm M. C., et al. The gut microbiota and inflammatory noncommunicable diseases: associations and potentials for gut microbiota therapies. Journal of Allergy and Clinical Immunology. 2015;135(1):3–14. doi: 10.1016/j.jaci.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Ouyang W., Kolls J. K., Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28(4):454–467. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koga T., Hedrich C. M., Mizui M., et al. CaMK4-dependent activation of AKT/mTOR and CREM-α underlies autoimmunity-associated Th17 imbalance. Journal of Clinical Investigation. 2014;124(5):2234–2245. doi: 10.1172/JCI73411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren W., Liu G., Yin J., et al. Draft genome sequence of enterotoxigenic Escherichia coli strain W25K. Genome Announcements. 2014;2(3) doi: 10.1128/genomea.00593-14.e00593-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren W., Chen S., Yin J., et al. Dietary arginine supplementation of mice alters the microbial population and activates intestinal innate immunity. Journal of Nutrition. 2014;144(6):988–995. doi: 10.3945/jn.114.192120. [DOI] [PubMed] [Google Scholar]
- 19.Tekwe C. D., Lei J., Yao K., et al. Oral administration of interferon tau enhances oxidation of energy substrates and reduces adiposity in Zucker diabetic fatty rats. BioFactors. 2013;39(5):552–563. doi: 10.1002/biof.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson J., Shokralla S., Porter T. M., et al. Simultaneous assessment of the macrobiome and microbiome in a bulk sample of tropical arthropods through DNA metasystematics. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(22):8007–8012. doi: 10.1073/pnas.1406468111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren W., Duan J., Yin J., et al. Dietary l-glutamine supplementation modulates microbial community and activates innate immunity in the mouse intestine. Amino Acids. 2014;46(10):2403–2413. doi: 10.1007/s00726-014-1793-0. [DOI] [PubMed] [Google Scholar]
- 22.Lee W.-J., Hase K. Gut microbiota-generated metabolites in animal health and disease. Nature Chemical Biology. 2014;10(6):416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian S., Huq S., Yatsunenko T., et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510(7505):417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anhê F. F., Roy D., Pilon G., et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut. 2015;64(6):872–883. doi: 10.1136/gutjnl-2014-307142. [DOI] [PubMed] [Google Scholar]
- 25.Louis P., Hold G. L., Flint H. J. The gut microbiota, bacterial metabolites and colorectal cancer. Nature Reviews Microbiology. 2014;12(10):661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 26.Qin N., Yang F., Li A., et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513(7516):59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 27.Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 28.Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 29.Liévin-Le Moal V., Servin A. L. Anti-infective activities of Lactobacillus strains in the human intestinal microbiota: from probiotics to gastrointestinal anti- infectious biotherapeutic agents. Clinical Microbiology Reviews. 2014;27(2):167–199. doi: 10.1128/cmr.00080-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcobal A., Sonnenburg J. L. Human milk oligosaccharide consumption by intestinal microbiota. Clinical Microbiology and Infection. 2012;18(supplement 4):12–15. doi: 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shabgah A. G., Fattahi E., Shahneh F. Z. Interleukin-17 in human inflammatory diseases. Postepy Dermatologii i Alergologii. 2014;31(4):256–261. doi: 10.5114/pdia.2014.40954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang S. H., Dong C. Signaling of interleukin-17 family cytokines in immunity and inflammation. Cellular Signalling. 2011;23(7):1069–1075. doi: 10.1016/j.cellsig.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gulen M. F., Kang Z., Bulek K., et al. The receptor SIGIRR suppresses Th17 cell proliferation via inhibition of the interleukin-1 receptor pathway and mTOR kinase activation. Immunity. 2010;32(1):54–66. doi: 10.1016/j.immuni.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J. S., Sklarz T., Banks L. B., et al. Natural and inducible TH17 cells are regulated differently by Akt and mTOR pathways. Nature Immunology. 2013;14(6):611–618. doi: 10.1038/ni.2607. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Park Y., Jin H.-S., Lopez J., et al. TSC1 regulates the balance between effector and regulatory T cells. The Journal of Clinical Investigation. 2013;123(12):5165–5178. doi: 10.1172/jci69751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagai S., Kurebayashi Y., Koyasu S. Role of PI3K/Akt and mTOR complexes in Th17 cell differentiation. Annals of the New York Academy of Sciences. 2013;1280(1):30–34. doi: 10.1111/nyas.12059. [DOI] [PubMed] [Google Scholar]
- 37.Ren W., Yin J., Duan J., et al. mTORC1 signaling and IL-17 expression: defining pathways and possible therapeutic targets. European Journal of Immunology. 2016;46(2):291–299. doi: 10.1002/eji.201545886. [DOI] [PubMed] [Google Scholar]
- 38.Kurebayashi Y., Nagai S., Ikejiri A., et al. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORγ . Cell Reports. 2012;1(4):360–373. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Shi L. Z., Wang R., Huang G., et al. HIF1α-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. The Journal of Experimental Medicine. 2011;208(7):1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dang E. V., Barbi J., Yang H.-Y., et al. Control of TH17/Treg balance by hypoxia-inducible factor 1. Cell. 2011;146(5):772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]