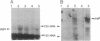Abstract
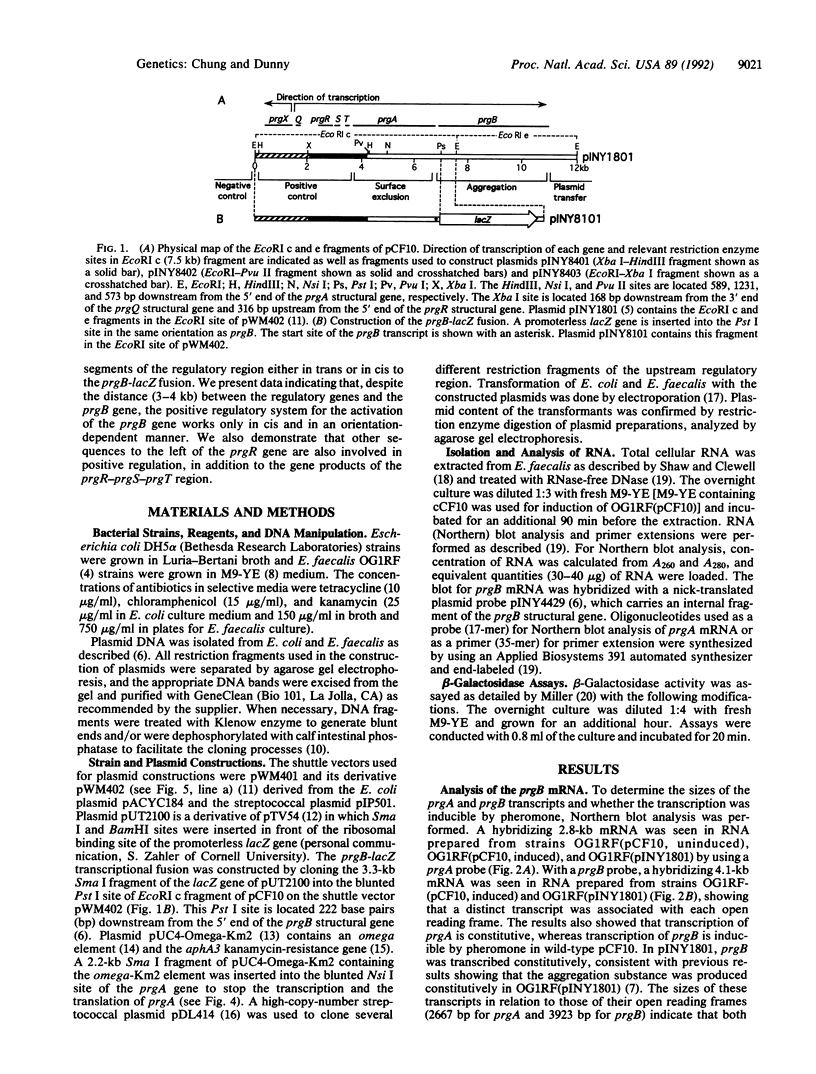
The prgB gene encodes the surface protein, Asc10, which mediates cell aggregation, resulting in high-frequency conjugative transfer of the pheromone-inducible tetracycline-resistance plasmid pCF10 in Enterococcus faecalis. Messenger RNA analysis by Northern blot hybridization and primer extension indicates that prgB transcription is pheromone-inducible and monocistronic. Previous transposon mutagenesis and sequencing analysis of a 12-kilobase (kb) region of pCF10 indicated that several genes including prgR and prgS are required to activate expression of prgB. The distance (3-4 kb) between these regulatory genes and prgB suggested that the activation might function in trans. To test this, a promoterless lacZ gene fusion to prgB was constructed and cloned without some or all of the regulatory genes. Several restriction fragments of the regulatory region were cloned in a higher copy-number plasmid, and numerous complementation studies were carried out in E. faecalis. Complementation in trans was not observed in any of these experiments. However, when the regulatory region and target genes were cloned in different sites of the same plasmid, separated by as much as 12 kb, activation of prgB was observed. Interestingly, this activation occurred only when the regions were cloned in the same relative orientation in which they exist on wild-type pCF10. These results suggest that one or more regulatory molecules may bind to an upstream cis-acting site and track along the DNA to reach a target site to activate prgB transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S. Multipartite genetic control elements: communication by DNA loop. Annu Rev Genet. 1989;23:227–250. doi: 10.1146/annurev.ge.23.120189.001303. [DOI] [PubMed] [Google Scholar]
- Berg D. E., Weiss A., Crossland L. Polarity of Tn5 insertion mutations in Escherichia coli. J Bacteriol. 1980 May;142(2):439–446. doi: 10.1128/jb.142.2.439-446.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P. J., Dunny G. M. Identification of regions of the Streptococcus faecalis plasmid pCF-10 that encode antibiotic resistance and pheromone response functions. Plasmid. 1986 May;15(3):230–241. doi: 10.1016/0147-619x(86)90041-7. [DOI] [PubMed] [Google Scholar]
- Christie P. J., Kao S. M., Adsit J. C., Dunny G. M. Cloning and expression of genes encoding pheromone-inducible antigens of Enterococcus (Streptococcus) faecalis. J Bacteriol. 1988 Nov;170(11):5161–5168. doi: 10.1128/jb.170.11.5161-5168.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Weaver K. E. Sex pheromones and plasmid transfer in Enterococcus faecalis. Plasmid. 1989 May;21(3):175–184. doi: 10.1016/0147-619x(89)90041-3. [DOI] [PubMed] [Google Scholar]
- Dunny G. M., Brown B. L., Clewell D. B. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G. M. Genetic functions and cell-cell interactions in the pheromone-inducible plasmid transfer system of Enterococcus faecalis. Mol Microbiol. 1990 May;4(5):689–696. doi: 10.1111/j.1365-2958.1990.tb00639.x. [DOI] [PubMed] [Google Scholar]
- Dunny G. M., Lee L. N., LeBlanc D. J. Improved electroporation and cloning vector system for gram-positive bacteria. Appl Environ Microbiol. 1991 Apr;57(4):1194–1201. doi: 10.1128/aem.57.4.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G. M., Zimmerman D. L., Tortorello M. L. Induction of surface exclusion (entry exclusion) by Streptococcus faecalis sex pheromones: use of monoclonal antibodies to identify an inducible surface antigen involved in the exclusion process. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8582–8586. doi: 10.1073/pnas.82.24.8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey J., Krisch H. M. Omega mutagenesis in gram-negative bacteria: a selectable interposon which is strongly polar in a wide range of bacterial species. Gene. 1985;36(1-2):143–150. doi: 10.1016/0378-1119(85)90078-2. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Geiduschek E. P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992 May 29;256(5061):1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- Kao S. M., Olmsted S. B., Viksnins A. S., Gallo J. C., Dunny G. M. Molecular and genetic analysis of a region of plasmid pCF10 containing positive control genes and structural genes encoding surface proteins involved in pheromone-inducible conjugation in Enterococcus faecalis. J Bacteriol. 1991 Dec;173(23):7650–7664. doi: 10.1128/jb.173.23.7650-7664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc D. J., Inamine J. M., Lee L. N. Broad geographical distribution of homologous erythromycin, kanamycin, and streptomycin resistance determinants among group D streptococci of human and animal origin. Antimicrob Agents Chemother. 1986 Apr;29(4):549–555. doi: 10.1128/aac.29.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. J., Moav B., Faras A. J., Guise K. S., Kapuscinski A. R., Hackett P. B. Functional analysis of elements affecting expression of the beta-actin gene of carp. Mol Cell Biol. 1990 Jul;10(7):3432–3440. doi: 10.1128/mcb.10.7.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M., Sugishita A., Furukawa K. Cloning and nucleotide sequences of histidase and regulatory genes in the Bacillus subtilis hut operon and positive regulation of the operon. J Bacteriol. 1988 Jul;170(7):3199–3205. doi: 10.1128/jb.170.7.3199-3205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted S. B., Kao S. M., van Putte L. J., Gallo J. C., Dunny G. M. Role of the pheromone-inducible surface protein Asc10 in mating aggregate formation and conjugal transfer of the Enterococcus faecalis plasmid pCF10. J Bacteriol. 1991 Dec;173(23):7665–7672. doi: 10.1128/jb.173.23.7665-7672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Casal J., Caparon M. G., Scott J. R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991 Apr;173(8):2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. H., Clewell D. B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985 Nov;164(2):782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3'5"-aminoglycoside phosphotransferase type III. Gene. 1983 Sep;23(3):331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Giaever G. N. Action at a distance along a DNA. Science. 1988 Apr 15;240(4850):300–304. doi: 10.1126/science.3281259. [DOI] [PubMed] [Google Scholar]
- Wirth R., An F. Y., Clewell D. B. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J Bacteriol. 1986 Mar;165(3):831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P., Zuber P., Perkins J. B., Sandman K., Igo M., Losick R. New ways to study developmental genes in spore-forming bacteria. Science. 1985 Apr 19;228(4697):285–291. doi: 10.1126/science.228.4697.285. [DOI] [PubMed] [Google Scholar]