Abstract
Objective
The objective of the study was to systematically review the literature for studies reporting gene expression analyses (GEA) of the biological processes involved in early human peri-implant bone healing.
Methods
Electronic databases (MEDLINE, EMBASE) were searched in duplicate. Controlled and uncontrolled studies reporting GEA of human peri-implant tissues - including ≥5 patients and ≥2 time points - during the first 4 weeks of healing were eligible for inclusion. Methodological quality and risk of bias were also assessed.
Results
Four exploratory studies were included in reporting GEA of either tissues attached to SLA or SLActive implants after 4 to 14 days or cells attached to TiOBlast or Osseospeed implants after 3 to 7 days. A total of 111 implants from 43 patients were analyzed using validated array methods; however, considerable heterogeneity and risk of bias were detected. A consistent overall pattern of gene expression was observed; genes representing an immuno-inflammatory response were overexpressed at days 3 to 4, followed by genes representing osteogenic processes at day 7. Genes representing bone remodeling, angiogenesis, and neurogenesis were expressed concomitantly with osteogenesis. Several regulators of these processes, such as cytokines, growth factors, transcription factors, and signaling pathways, were identified. Implant surface properties seemed to influence the healing processes at various stages via differential gene expression.
Conclusion
Limited evidence from gene expression studies in humans indicates that osteogenic processes commence within the first post-operative week and they appear influenced at various stages by implant surface properties.
Keywords: Dental implant, Osseointegration, Gene expression, Molecular assessment
Review
Introduction
Osseointegrated oral implants are an integral part of modern reconstructive dentistry and are associated with favorable long-term therapeutic outcomes [1]. Osseointegration was originally defined as the direct contact between vital bone and a load-bearing implant observed at the light microscopic, i.e., histological, level [2]. Morphogenesis of implant osseointegration has been assessed in several preclinical in vivo and clinical histological studies [3-6], providing the basis for understanding the biological process.
The biological events during the early phase of osseointegration are directly influenced by the osseous microenvironment (i.e., cells, signaling molecules, and matrix) into which the implant is placed and have many similarities with general wound healing mechanisms [7]. Implant surgery induces trauma, resulting in bleeding and fibrin clot formation and an inflammatory reaction that dominate the events of the first post-operative week. The deposition of vital new bone on the implant surface by osteoblasts (osteogenesis), a fundamental requirement for osseointegration, occurs via secretion of a complex extracellular matrix (ECM) of proteins, which subsequently undergoes mineralization to form bone [8,9]. Primary (woven) bone lined by osteoblasts can indeed be observed on the implant surface already after 1 week [3,5]. In parallel, removal of the created bone debris and remodeling of necrotized bone (due to the pressure exerted by the implant) is underway. Replacement of woven bone by organized and mechanically superior lamellar bone can be observed from the second to fourth week (depending on the species) and progressively increases until woven bone is almost entirely replaced (8 to 12 weeks). These events, including the nutrition of the newly formed tissue, are sustained through concomitantly occurring angiogenesis, i.e., formation of new blood vessels from existing ones [10,11]. Thus, osseointegration is a dynamic process whereby bone formation and remodeling occur in parallel around the implant [4,6].
Morphogenesis of osseointegration and assessment of the degree of bone-to-implant contact is usually performed by means of histological evaluation [12], while the underlying molecular processes may be more precisely evaluated at genetic level [13,14]. Data from gene expression analyses of fracture healing provide the basis for understanding these processes [15]. These studies have identified the cells, signals, and interactions governing the key processes of bone regeneration. Bone-forming osteoblasts are primarily derived from marrow-resident multipotent progenitor cells (mesenchymal stem cells (MSCs)), which are recruited to the regeneration site. This process of MSC recruitment and differentiation along the osteogenic lineage is termed as osteoinduction and is controlled primarily by various pro/anti-inflammatory cytokines (CKs) and by growth factors (GFs) secreted by inflammatory cells and/or osteoblasts or by GF resident within the extracellular matrix (e.g., bone morphogenetic proteins (BMPs)) in response to injury [16-18]. Moreover, CKs and GFs act as signaling molecules via specific signaling pathways and guide the process of cell differentiation in the proper temporal sequence [19,20]. Intermediaries in this process are various bone-specific transcription factors (TFs), which act as ‘molecular switches’ during cell differentiation and are targets of CKs and GFs [21]. TFs facilitate bone-specific gene transcription and ultimately gene expression by which MSCs undergo differentiation and acquire the osteoblastic phenotype [22]. While GFs regulate mainly osteoinduction and osteogenesis, pro-inflammatory CKs regulate the antagonist process of bone resorption by inducing the differentiation of hematopoietic stem cells (HSCs) into osteoclasts and macrophages [23], contributing to the dynamic nature of bone regeneration and remodeling.
Recent in vitro [24] and preclinical in vivo [25] studies have focused on the early molecular biological responses to various titanium implant surfaces. Understanding these early responses is essential for efforts aiming to accelerate and enhance the process of osseointegration [26]. Upregulation or downregulation of specific genes in peri-implant tissues identified by analyses of genetic material (DNA, RNA) reflects the nature and timing of the various healing processes, which in turn could be potential ‘molecular targets’ for enhancing osseointegration [27,28]. The aim of the present study was to systematically review the available literature on gene expression analyses of the biological processes involved in early human peri-implant bone healing.
Methods
Study design
A study protocol for a systematic qualitative literature review was developed based on recommended methods [29]. The focused question was ‘what biological processes are reflected by gene expression analyses in peri-implant tissues of humans during the early stages (up to 4 weeks) of healing?’
Inclusion and exclusion criteria
All studies, controlled (using different implants) or uncontrolled, reporting gene expression analyses of peri-implant tissues harvested from ≥5 human patients at ≥2 time points during the first 4 weeks of healing, were eligible for inclusion. Studies reporting the use of either ‘experimental’ (micro) or standard implants with clear description of implant surface properties, placed in the maxilla or mandible and retrieved at a later time point, were eligible for inclusion. Studies reporting (1) analyses of peri-implant mucosa or sulcular fluid or peri-implant tissues of failing or infected implants (peri-implantitis), (2) only histological or immunohistochemical analyses without gene expression of harvested tissues, and (3) in vitro and preclinical in vivo studies were excluded. Primary outcome of interest was the biological process (or processes) reflected by gene expression at a particular time point of peri-implant tissue healing.
Search strategy
Electronic databases of MEDLINE (via PubMed) and EMBASE were searched by one author (SS) for relevant English-language literature up to and including June 2014. The search strategy used for MEDLINE was ((((("gene expression" OR transcriptome OR transcriptional OR molecular OR microarray))) AND ((osseointegration OR healing OR "peri implant"))) AND implants) AND ((human OR humans OR patients OR subjects)). Unpublished literature was searched via the Google and Google Scholar search engines. Additionally, the bibliographies of all relevant studies and review articles were searched.
Study selection
Titles and abstracts of the search identified studies were screened by two authors (SS and VS) based on the inclusion criteria, and full texts of all eligible studies were obtained. Differences in assessment of eligibility were resolved by discussion with the third author (AS). Full texts were independently reviewed by both reviewers, and final inclusion was based on the aforementioned inclusion criteria.
Data extraction
Both reviewers independently extracted data from the full texts of included articles using specially designed forms. Data on author(s), study design, implant type/surface, any additional procedures performed, number of patients (in each group), presence of a control group, procedure and time of implant retrieval, methods of gene expression analysis, and main results, were extracted. Descriptive summaries of the studies were entered into tables, and a qualitative synthesis of evidence was planned. Any disagreement between the reviewers regarding data extraction was resolved by discussion.
Assessment of methodology and risk of bias
Assessment of the methodological validity of the included studies was performed using criteria adapted from previous reports [30,31]. Aspects of study design, genotyping methods, and data analyses were considered using nine criteria (Table 1).
Table 1.
Assessment of the genotyping methodology in the included studies
| Methodology | Ivanovski et al. [ 34 ] | Donos et al. [ 35 ] | Bryington et al. [ 36 ] | Thalji et al. [ 37 ] |
|---|---|---|---|---|
| Tissue harvesting | Tissue attached to implant carefully removed with a curette, preexisting hard tissue discarded | Tissue attached to implant carefully removed with a curette and homogenized | Implants removed by reverse threading and homogenized; cell lysates isolated | Implants removed by reverse threading and homogenized; cell lysates isolated |
| Sample preparation | Total RNA isolation, purification, quantity/quality analysis and biotin-labeling | Total RNA isolation, purification, quantity/quality analysis and biotin-labeling | Total RNA isolation, quantification | Total RNA isolation, purification, quantity/quality analysis and biotin-labeling |
| Array technique | Microarray hybridization (Human WG-6 V3) | Microarray hybridization (Human WG-6 V3) | RT-PCR (custom RT-PCR array for osteogenesis genes; human inflammatory cytokines and receptors PCR array) | Microarray hybridization (Affymetrix Human Gene 1.1 ST) |
| Scanning, data preparation | Bead Station 500/Bead Studio v3 software, raw probe expression values extracted | Bead Station 500/ Bead Studio v3 software, raw probe expression values extracted | RT2 SYBR Green qPCR Master Mix/7500 Real-Time PCR system | Affymetrix Gene Chip Scanner |
| Processing | Noisy data discarded | Noisy data discarded | Normalization of osteogenesis and cytokine array | Unclear |
| Clustering | GO categories (DAVID tool) | GO categories (DAVID tool) | Osteogenesis genes; cytokine-related genes | GO categories (Gene Spring) |
| Statistical analysis | Gene Spring software | Gene Spring software | RT2 Profiler software | Gene Spring software |
| Comparisons | Pair-wise comparisons between three time points (4 vs. 7 days, 7 vs. 14 days, and 4 vs. 14 days) | Pair-wise comparisons at each time point (4, 7, and 14 days) between SLA and SLActive surfaces | T-test to evaluate differences between each implant surface per time point | Two-way ANOVAs to determine differences between implant surface type and time points; pair-wise comparisons of each implant surface independently at different time points (day 7 vs. day 3) |
GO, gene ontology; DAVID, Database for Annotation, Visualization and Integrated Discovery.
The risk of bias in the included studies was assessed using an adaptation of published guidelines for reporting systematic reviews of periodontal genetic association studies [32]. Mainly, aspects of study design and methodological validity were assessed using 15 criteria and scored as ‘yes,’ ‘no,’ or ‘unclear’ based on the information provided in the study manuscript (Table 2). Moreover, published guidance [33] regarding the qualitative and quantitative syntheses of results from genetic association studies was consulted, and heterogeneity across the included studies was assessed to explore the possibility of a meta-analysis.
Table 2.
Assessment of risk of bias and heterogeneity within and across the included studies
| Category | Ivanovski et al. [ 34 ] | Donos et al. 2011 [ 35 ] | Bryington et al. [ 36 ] | Thalji et al. [ 37 ] |
|---|---|---|---|---|
| Study design | ||||
| Comparison | None (only SLActive) | SLA vs. SLActive | TiOBlast vs. Osseospeed | TiOBlast vs. Osseospeed |
| Setting | University | University | University | University |
| Population, inclusion criteria | 9 healthy volunteers with no mandibular third molars, no contraindications for oral surgery; age 21 to 48, median 29 years | 9 healthy volunteers with no mandibular third molars; age 21 to 48, median 29 years | 6 women, 4 men; implant patients, systemically healthy (no HTN, diabetes, CVD); age 25 to 58, mean 36.2 years | 9 women, 2 men; implant patients, systemically healthy; age 47 to 69, mean 60.2 years |
| Exclusion criteria | Smokers | Smokers | Smokers, pregnancy, periodontal/periapical disease, subjects taking bisphosphonates, hormone replacement therapy, corticosteroids | Smokers, uncontrolled diabetes, history of head/neck radiotherapy, taking corticosteroids, bisphosphonates |
| Comparability of groups | Unclear | Unclear | Unclear | Unclear |
| Potential confounders, e.g., post-op medication | Unclear | Unclear | Unclear | Unclear |
| Power calculation | No | No | No | No |
| Statistical correction | For multiple sampling | For multiple sampling | Unclear | For multiple sampling |
| Methods | ||||
| Tissue analyzed | Peri-implant tissue | Peri-implant tissue | Implant-adherent cells | Implant-adherent cells |
| Genetic material analyzed | Total RNA | Total RNA | Total RNA | Total RNA |
| Success rate | Unclear | 16/18 samples (88.8%) | 7/10 subject samples (70%) | Unclear |
| Genotyping method | Microarray | Microarray | RT-PCR | Whole-genome microarray |
| Genotype counts | Yes | Yes | Yes | Yes |
| Blinding | Unclear | Unclear | Yes | Unclear |
| Reproducibility, validated genotyping accuracy | No | No | No | No |
All studies were judged to be at a high risk of bias with substantial heterogeneity across studies.
Results and discussion
The included studies basically report on commercially available implants from two major manufacturers and involve comparisons of different implant surface technologies in regard with topography and/or chemistry modifications within each implant system. Various analyses were performed in the included studies; however, an attempt has been made to synthesize the various findings and discuss them herein irrespective of the specific implant systems, based on the assumption that basic biological mechanisms of peri-implant bone wound healing are largely implant system independent.
Search results and study characteristics
Of the 242 search identified studies, only four studies were finally included in the review, all focusing on the impact of implant surface on early human peri-implant bone healing (Figure 1; Table 3). Genetic analyses of total RNA isolated from either newly formed peri-implant bone harvested by trephination [34,35] or from cells adherent to implants retrieved by reverse threading [36,37] were performed. In total, 111 implants from 43 patients were analyzed. All four studies reported the use of commercially existing implant surfaces, i.e., either a chemically modified, hydrophilic, sand-blasted, acid-etched surface (SLActive®, Institute Straumann AG, Basel, Switzerland); or a hydrophilic (SLActive®) versus a hydrophobic unmodified SLA® (Institute Straumann AG, Basel, Switzerland) surface; or a micro-topographic titanium-oxide grit-blasted surface (TiOBlast®, AstraTech, Molndal, Sweden) versus a chemically modified nano-topographic grit-blasted surface (Osseospeed®, AstraTech, Molndal, Sweden). Implant retrieval times were at 3 or 4 days and 7 days in all studies and additionally at 14 days in two studies [34,35].
Figure 1.
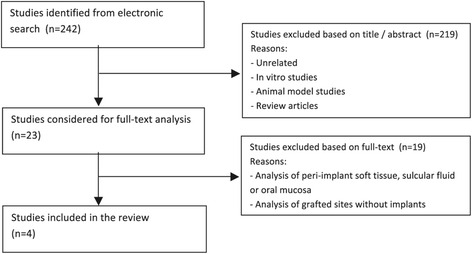
Flowchart for study selection ( n = number of studies).
Table 3.
Summary of findings from the included studies ( n = 4)
| Study | Ivanovski et al. [ 34 ] | Donos et al. [ 35 ] | Bryington et al. [ 36 ] | Thalji et al. [ 37 ] |
|---|---|---|---|---|
| Design | 9 patients; 9 implants placed | 18 patients; 18 implants placed | 10 patients; 60 implants placed | 11 patients; 44 implants placed |
| Total RNA extracted from peri-implant tissue (trephine) | 16 samples analyzed | 42 samples analyzed | Total RNA extracted from implant adherent cells (reverse thread) | |
| Total RNA extracted from peri-implant tissue (trephine) | Total RNA extracted from implant adherent cells (reverse thread) | |||
| Surface | SLActive | SLA vs. SLActive | TiOBlast vs. Osseospeed | TiOBlast vs. Osseospeed |
| GE day 3/4 | Upregulated | Upregulated on SLA | Upregulated on both surfaces | No significant differences between surfaces at any time point (P > 0.05) |
| CKs (TNF-a, IL-6, IL-2) | Neurogenesis | Osteogenesis (Runx2, Osx, BMP6, OPN) | Results presented as GE at day 7 vs. day 3 for each surface | |
| Immune-inflammatory cells (LC, MP) | Collagen organization | Inflammatory CKs (IL-1A,B, TNF) | ||
| Inflammatory NF-kB p/w | Upregulated on SLActive | MP activity | ||
| Ras protein p/w | Upregulated on Osseospeed | |||
| Collagen organization | Chemotaxis (CCL18, CXCL10, CXCL14) | |||
| CK response | Anti-inflammatory CKs (TOLLIP, IL9, IL22) | |||
| GE day 7 | Upregulated | Upregulated on both surfaces | Upregulated on both surfaces | Upregulated on both surfaces |
| MSC genes (HOX, Sp3) | Inflammatory CKs (IL1, IL2, IL6, TNFS) | Osteogenesis (Runx2, Osx*, OCN*, OPN, BMP6, BSP) | ECM (Coll, GPs, PGs) | |
| GF (TGF-B receptor) | Neurogenesis | [* Osseo > TiOB; P < 0.05] | Collagen organization (PLODs, LOX, PCOLCE) | |
| VEGF sig. (vs. day 14) | Upregulated on SLActive | Angiogenesis/VEGF sig. (ANXA, EPAS1) | ||
| Wnt p/w | Neurogenesis (BDNF, NTF3) | Ossification | ||
| Downregulated | ECM (OPN) | Remodeling (MMPs, TIMPs) | ||
| Inflammatory NF-kB p/w (vs. day 4) | BMP p/w (BMP4, BMP2K) | Osteoclastic (CTSK, ACP5) | ||
| MAPK sig. | Chemotaxis (CKs, MP activity) | |||
| Mineralization | Anti-inflammatory CKs (CCL22, CCL18) | |||
| Focal adhesion (integrins) | Downregulated on both surfaces | |||
| Angiogenesis (VEGF sig., P13-Akt p/w) | Inflammatory CKs (IL1A, IL1B) | |||
| Downregulated on SLActive | ||||
| Inflammatory cells (LC) | ||||
| GE day 14 | Upregulated | Upregulated on both surfaces | - | |
| ECM (Coll, OC, ON, ALP) | BMP p/w (BMP4, BMP2K) | |||
| TFs (Osx, Dlx5, Twist1, Smad6) | Downregulated on both surfaces | |||
| Remodeling (MMP, CTSK) | Inflammatory cells (LC) | |||
| GFs (BMP, GDF) | ||||
| Angiogenesis (VEGF sig.) | ||||
| Neurogenesis | ||||
| TGF-b/BMP, Notch p/w | ||||
| Ras protein p/w | ||||
| Wnt-receptor genes | ||||
| Notch genes (up/down) | ||||
| Downregulated | ||||
| Inflammatory response (vs. day 7) |
GE, gene expression; CKs, cytokines; p/w, pathway; MSC, mesenchymal stem cells; GF, growth factors; sig., signaling; ECM, extracellular matrix; TFs, transcription factors; MP, macrophage; LC, lymphocytes; GPs, glycoproteins; PGs, proteoglycans.
Assessment of methodology and risk of bias
All studies used validated methods for gene expression analysis; genetic data was analyzed using microarray (three studies) or real-time PCR (RT-PCR) (one study) methods (Table 1). Total RNA was isolated from lysates of either trephined peri-implant tissues or implant-adherent cells, and subjected to microarray processing or RT-PCR. Although moderate-to-good agreement has been reported between the two methods, validation of DNA microarray results by the more sensitive PCR array is generally recommended [38]. None of the microarray studies identified have validated their results using RT-PCR. Genotyping data (gene lists) were imported and analyzed using computer software and further condensed into functionally and biologically relevant categories. Nevertheless, differential gene expression in relation to a particular cell type or region of tissue analyzed was not performed [35]. Gene ‘upregulation’ was reported when genes were expressed at a higher level on one implant surface in comparison to another; in context, differentiation between gene expression and over-expression may be difficult to define. Statistical methods were used to compare differences in gene expression between different time points and/or implant surfaces (P < 0.05 significance level), while correcting for possible errors, i.e., false gene discovery rate due to multiple sampling [39]. There was considerable heterogeneity across the included studies in terms of study design, population, implant surface technology, genotyping methods, and data analyses (Table 2). Therefore, no meta-analysis of association between gene expression and implant surface properties was relevant.
Thus, high risk of bias should be considered when interpreting the results, due to the above methodological limitations and the overall limited information (four studies) available.
Biological processes identified through gene expression in peri-implant tissues
Conventional implant surgery involves osteotomy preparation and insertion of the implant into the alveolar bone. The immediate local effects of this procedure, functionally relevant to subsequent healing processes, are (1) bone trauma, (2) formation of bone debris, (3) hemostasis and clot formation, and (4) hypoxia. These effects involve the release of specific CKs and GFs within the local environment [7], resulting in recruitment of two primary cell types to the site, inflammatory cells and progenitor cells (MSCs and HSCs) [19], which in turn regulate the subsequent healing processes. A summary of differentially regulated genes relating to the involved biological processes is presented in Table 4, while Figure 2 represents an evidence-based illustrative model summarizing these processes.
Table 4.
Summary of biological processes and associated genes reported in the included studies
| Process | Upregulated genes |
|---|---|
| Category (gene code) | |
| Inflammation/immune response | |
| Pro-inflammatory cytokines | Tumor necrosis factor (TNF-a, TNFSF9) |
| Interleukin (IL-6, IL-2, IL-1 F9, IL-23A, IL-6ST) | |
| Interferon (IFNA2) | |
| Nuclear factor-kB (I-kB kinase/NF-kB) | |
| Anti-inflammatory cytokines | Interleukin (IL-22, IL-9) |
| Toll interacting protein (TOLLIP) | |
| Cells | Lymphocyte, macrophage negative proliferation (BTLA, LST1) |
| Macrophage scavenger receptor (MSR1) | |
| Chemotaxis | Chemokines (CCR8, CCL18, CCL22, CXCL10, CXCL14) |
| Osteoinduction/osteogenesis | |
| Growth factors (GF)/signaling pathways | Insulin-like GF (IGF1) |
| Transforming GF (TGF-b, TGF-b receptor 1, 2 and 3, TGF-a) | |
| Platelet-derived GF (PDGF receptor) | |
| Bone morphogenetic proteins (BMP4, BMP6, BMP receptor 1A, BMP2-K) | |
| Growth and differentiation factor (GDF10) | |
| Wnt frizzled receptor (FZD3, FZD8, FRZB) | |
| Notch (NOTCH2) | |
| Ras-protein signal transduction (RAP1B, RAP1A, RASGRP4) | |
| Mitogen activated protein kinase (MAP3K7IP2, MAPK9, MAP2K3, MAP3K2) | |
| Transcription factors | ‘Master switches’ [RUNX2, SP7 (OSX)] |
| Homeobox (DLX1, DLX5, HOXD12, MSX1, HOXA5, HOXB1, HOXB6, HOXC6) | |
| SP [SP1, SP3, SP7 (Osx)] | |
| Twist (TWIST 1-receptor) | |
| ECM deposition/organization | Collagen (Col1A1, Col12A1, Col6A3, Col3A1, Col6A1, Col11A1, Col11A2, Col13A1, Col5A2) |
| Non-collagen proteins [BGLAP (OC), SPARC (ON), SPP1 (OP), BSP, IBSP, POSTN, ECM1] | |
| Small leucine-rich proteoglycans (SLRP) (DCN, BGN, LUM) | |
| Heat-shock protein 47 (HSP47) | |
| Alkaline phosphatase (ALPL) | |
| Cadherin (CDH11) | |
| Integrin (ITGB4, ITGB5) | |
| Laminin (LAMA2, LAMA3) | |
| Pro-collagen lysyl-hydroxylase (PLOD1, PLOD2, PLOD3) | |
| Pro-collagen C-endopeptidase enhancer (PCOLCE) | |
| Lysyl-oxidase (LOX) | |
| Osteoclast activity/remodeling | Cathepsin K (CTSK, CTSK-receptor) |
| Tartarate-resistant acid phosphatase (TRAP/ACP5) | |
| Matrix metallopeptidase (MMP2, MMP12, MMP9, MMP7, MMP13) | |
| Tissue inhibitor metallopeptidase (TIMP2, TIMP3) | |
| Angiogenesis | Vascular endothelial GF-signaling (EPAS1, ANXA2, EGR1-binding protein) |
| Phosphatidyl-inositol 3-kinase (PI3K)-Akt signaling | |
| Neurogenesis | Brain-derived neurotrophic factor (BDNF) |
| Neurotrophin 3 (NTF3) | |
| NK2 homeobox 2(NKX2-2) | |
| Tubby-like protein 3 (TULP3) | |
Figure 2.
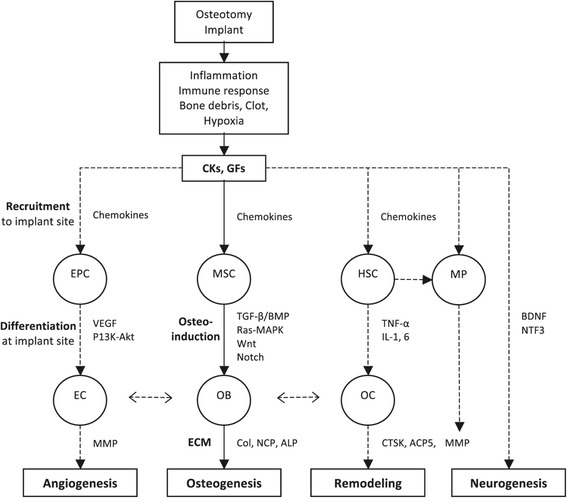
Summary of biological processes identified via gene expression during early peri-implant bone healing. CKs, cytokines; GFs, growth factors; EPC, endothelial progenitor cells; EC, endothelial cells; MSC, mesenchymal stem cells; OB, osteoblasts; ECM, extracellular matrix; HSC, haematopoietic stem cells; MP, macrophages; OC, osteoclasts.
Inflammation
All studies reported a significant upregulation of genes associated with inflammation during the first time point of observation (day 3 or 4) regardless of the implant surface. Specifically, upregulation regarded pro-inflammatory cytokines of the interleukin (IL), tumor necrosis factor (TNF), and interferon (IFN) families, as well as genes associated with proliferation of lymphocytes and macrophages (MPs). Previous in vitro [40,41] and animal [42] studies have reported the significance of MPs at the bone-implant interface and identified favorable MP activity in relation to modified rough surfaces as demonstrated by in vitro gene expression that was associated with increased in vivo bone formation. Also, the nuclear factor-kB (NF-kB) inflammatory pathway was upregulated at day 4 [34], while macrophage activity and chemokines of the CCL and CXL families in the peri-implant tissues continued to remain prominent at day 7.
However, this inflammatory response was generally downregulated at later time points (day 7 or 14). For example, in one study, genes associated with pro-inflammatory cytokines (IL-1B, IL-1A, IL-1R2) and chemokines (CCL22, CCL18) were downregulated and upregulated, respectively, on day 7, at both implant surface technologies examined (Osseospeed and TiOBlast) [37]. Moreover, the anti-inflammatory response seemed to be modulated by surface properties. In one study, genes related to anti-inflammatory cytokines such as IL-9, IL-22, toll-like receptor inhibitor protein (TOLLIP), and several key chemokines (CCL18, CXCL10, CXCL18) were significantly upregulated on Osseospeed surfaces but not TiOBlast, at day 3 [36]. In another study, genes associated with inflammatory cell proliferation were significantly downregulated earlier on SLActive surfaces compared to the SLA, i.e., at day 7 instead of day 14 [35]. Therefore, the initial inflammatory response seems to be important for the recruitment of cells that govern subsequent healing processes and is regulated by a natural biological immune response which may be further modified by implant surface properties.
Osteogenic differentiation
Cells along the osteogenic differentiation pathway may be artificially categorized as (1) undifferentiated MSCs, (2) osteo-chondro-progenitor cells, (3) pre-osteoblasts, and (4) osteoblasts; although in reality, a developmental continuum without distinct boundaries may exist [43]. While pre-differentiated osteoblasts in the marrow compartment only play a minor role in bone wound healing, a more prominent role is that of undifferentiated MSCs which are recruited to the regeneration site where they differentiate into osteoblasts [16]. The recruitment and differentiation of MSCs is regulated by CKs and GFs [17,19]. The GFs most commonly implicated in bone wound healing are BMPs, members of the TGF-β family, PDGF, and IGF-1 [19,20]. Moreover, the bone debris created during implant surgery, the peri-implant blood clot (i.e., platelets) and the differentiating MSCs themselves further contribute to release of GFs at the site [44,45].
All studies reported some evidence of osteogenic differentiation at an early time point (day 3 or 4) via expression of genes associated with key growth factors (bone morphogenetic proteins (BMP4, BMP6, BMP2-kinase), growth and differentiation factor-10 (GDF10), transforming growth factors (TGF-α, TGF-β), platelet-derived growth factor (PDGF), and insulin-like growth factor-1 (IGF1)), transcription factors (Runx2, Osx, Dlx3, Dlx5, Msx1, HOX genes, Sp1, Sp3), and/or osteogenic signaling pathways (TGF-β/BMP signaling, Wnt-receptors, Ras-protein/mitogen-activated protein kinase (Ras/MAPK) signal transduction). In all studies, these genes were further upregulated at day 7. Upregulation of osteogenic factors seemed regulated by implant surface. The key transcription factor osterix (Osx) was upregulated on the Osseospeed surface, but not TiOBlast at day 7 [36], while tissues adjacent to SLActive surfaces demonstrated comparatively greater BMP and Ras/MAPK expression compared to SLA surfaces at day 7 [35]. Previous in vivo animal studies have reported correlations between upregulated osteogenic gene expression in peri-implant tissues and enhanced histological and biomechanical measures of osseointegration during early (1- to 4-week) healing times [27,46]; nevertheless, it is unclear whether upregulation and/or overexpression of genes at a specific time point directly correlates to increased protein production in vivo.
The key signaling pathways, via which GFs guide osteogenic cell differentiation, are the TGF-β/BMP- and Wnt-mediated pathways [19,47]. While the BMP pathway ensures differentiation of MSCs into osteo-chondro-progenitors (OCPs), the Wnt pathway is essential for subsequent osteoblastic commitment, i.e., Wnt acts ‘downstream’ of BMP to ensure that OCPs differentiate into osteoblasts and not chondroblasts [47]. Genes associated with both TGF-β/BMP and Wnt pathway (Wnt receptors) were upregulated at day 7 [34,35] and day 14 [34] on SLA and SLActive surfaces, suggesting the occurrence of osteogenic differentiation at these time points.
GF-regulated signaling pathways exert their effects on differentiating cells via activation of TFs. The TFs Runx2 and Osx are considered as ‘master switches’ and absolute requirements for osteoblast differentiation [21] - while Runx2 is essential for MSC differentiation, Osx acting ‘downstream’ of Runx2 controls osteoblastic fate determination [48,49]. An upregulation of these genes was observed in relation to the TiOBlast, Osseospeed, and SLActive surfaces in the present review. However, at day 7, expression of Osx was significantly greater on Osseospeed than TiOBlast surfaces. This finding is consistent with previous animal [50,51] and human studies [52] where superior in vivo osseointegration (i.e., larger amount of bone-to-implant contact occurring earlier) of Osseospeed versus TiOBlast implants was reported. Thus, it appears that implant surface topography and/or chemistry influence peri-implant bone healing in humans both at the signaling pathway and transcription factor level.
ECM production
Deposition of new bone on the implant surface involves the secretion of a complex ECM (scaffold) of proteins by osteoblasts, which subsequently undergoes mineralization [9]. Expression of ECM proteins is a reliable indicator of early osteogenic activity [19] and was identified in all four studies at days 7 and 14. All studies reported some evidence of ECM production and/or organization at days 7 and 14. Upregulated genes associated with ECM deposition included various collagens (Col 1 to 11), non-collagen proteins (osteopontin (OPN), osteonectin (ON), osteocalcin (OCN), bone sialoprotein (IBSP), periostin (POSTN), and ECM protein-1), alkaline phosphatase (ALP), and bone-specific adhesion proteins (integrins (ITGB4, ITGB5), laminins (LAMA2, LAMA3), and cadherins (CDH11)). Osteocalcin, the most bone-specific ECM protein and a late marker of osteogenic differentiation [19], was significantly upregulated on Osseospeed (versus TiOBlast) surfaces at day 7 [36]. Osteopontin, an ECM protein essential for mineralization [53], was significantly upregulated on SLActive comparing to SLA surfaces at day 7 [35]. The possibility that implant surface features enhance osteogenic differentiation of MSCs via upregulation of specific genes (e.g., SLActive versus SLA in regard with BMP and Wnt signaling) has been demonstrated in vitro [54].
Furthermore, genes associated with collagen fibril formation/organization (heat-shock protein-47 (HSP-47), pro-collagen C-endopeptidase enhancer (PCOLCE), small leucine-rich proteoglycans (SLRP)) and post-translational modification (pro-collagen lysyl-hydroxylases (PLOD1, PLOD2, PLOD3) and lysyl-oxidase (LOX)) were upregulated on Osseospeed and TiOBlast surfaces [37]. Collagen comprises approximately 90% of the ECM and collagen fibrillogenesis and organization directly determine the biomechanical properties of bone [55,56]. Genes associated with collagen fibril formation, maturation, and post-translational modification expressed by osteoblasts [57,58] were upregulated on TiOBlast and Osseospeed implants, representing early ECM organization at the bone-implant interface. These modifications determine the pattern of collagen cross-linking which in turn influences tissue organization, mineralization, and ultimately mechanical bone strength [56], and in the case of osseointegration, the integrity of the bone-implant interface [37].
Osteoclastic activity and remodeling
While GFs regulate osteogenesis, pro-inflammatory CKs (e.g., IL-1, IL-6, TNF-α) simultaneously regulate the antagonist process of bone resorption via osteoclasts [23]. Moreover, osteoblasts themselves stimulate osteoclastogenesis via macrophage colony stimulating factor (M-CSF) and receptor activator of NF-kB ligand (RANKL) genes but also closely regulate this process via osteoprotegerin (OPG), an inhibitor of RANKL [59].
Two studies reported expression of genes associated with osteoclastic activity and ECM degradation (cathepsin-K (CTSK), tartarate-resistant acid phosphatase (ACP5), and/or matrix metalloproteinases (MMPs)), on Osseospeed and TiOBlast surfaces at day 7 [37], and SLActive surfaces at day 14. However, upregulation of MMP inhibitors (TIMP-2, -3) was also reported on TiOBlast and Osseospeed surfaces suggesting a control of the resorption process. Although no studies reported differential RANKL/OPG expression, a previous in vitro study [60] reported significant downregulation of osteoclastogenic genes on SLActive surfaces. Collectively, these data reaffirm the dynamic nature of bone formation and resorption at the implant-bone interface, even in early healing stages, and suggest the possibility for implant surface technology modulation of bone remodeling.
Angiogenesis
Angiogenesis is closely related to osteogenesis and occurs simultaneously during bone regeneration [11]. Physiological oxygen tensions in bone are about 12.5% O2 but fall to 1% O2 in regeneration sites due to disruption of the local vasculature as a result of injury and/or surgery [61,62]. A key event that stimulates angiogenesis (and osteogenesis) at regeneration sites is hypoxia, via the hypoxia inducible (transcription) factor-1 (HIF-1) that regulates expression of angiogenic genes [63]. The key cells involved in angiogenesis are macrophages, which in response to hypoxia and inflammation release chemotactic and angiogenic growth factors (e.g., VEGF) [40,64], and endothelial progenitor cells (EPCs) which differentiate into endothelial cell lining blood vessels [65]. VEGF is the single most important regulator of EPC differentiation and vessel formation [66]. Moreover, a role for VEGF in osteogenic differentiation has also been suggested mainly via interaction with the BMP signaling pathway [67].
In the present review, a significant simultaneous upregulation of several angiogenesis-related genes was identified at day 7 in all included studies. Pro-angiogenic factors (ANXA2, EPAS-1) were upregulated at TiOBlast and Osseospeed surfaces at day 7 [37]. Genes associated with VEGF and P13K-AKT signaling pathways were upregulated at SLActive (but not SLA) surfaces on day 7 and continued to be upregulated on day 14 [35]. The P13K-AKT pathway is reported to be important for endothelial cell survival, migration, and vessel formation, in addition to aiding VEGF-mediated angiogenesis [68]. Previous in vitro studies have reported the pro-angiogenic effects of SLActive surfaces by promoting VEGF expression in EPCs and osteoblasts [65,69], while enhanced histological osseointegration of SLActive implants has been directly correlated with increased angiogenesis in a dog model [70,71]. Thus, implant surface technology appears to have the possibility to also influence angiogenesis at early stages of wound healing.
Neurogenesis
Bone innervation includes both myelinated and unmyelinated nerve fibers located in the periosteum, bone cortex, Haversian systems, Volkmann’s canals, and the marrow spaces [72]. An interesting finding in the present review was the significant upregulation of genes associated with neurogenesis, more than any other biological process, on SLActive and SLA surfaces at all time points [34,35]. Specific processes represented were axon formation, growth and differentiation, and the neural signaling pathway. This is consistent with previous in vivo reports of murine fracture healing [73] and calvarial defect regeneration in relation to SLA surfaces [74,75]. Key neurotrophic factors (brain-derived neurotrophic factor (BDNF) and neurotrophin 3 (NTF3)), essential for neuronal survival and differentiation during development [76], were significantly upregulated on SLActive versus SLA surfaces at day 7 suggesting an effect of surface modulation. The P13K-AKT pathway, upregulated on SLActive surfaces (in relation to angiogenesis), has also been implicated in neuronal survival and subsequent neural development [77,78] and could have contributed to upregulation of neurogenic genes at these surfaces. Indeed, previous histologic reports have described changes in bone innervation after implant placement (and loading) and the presence of nerve fibers within the peri-implant bone, in animals and humans [79-81].
It can be hypothesized that peri-implant neurogenesis is one of the underlying mechanisms governing the phenomenon of osseoperception, defined as the tactile sensibility of osseointegrated implants to occlusal forces induced via activation of nerve endings and/or receptors in the peri-implant environment [82,83]. Moreover, recent evidence suggests that implant surface properties may influence the degree of osseoperception in humans [84], which can be correlated with the genomic evidence for implant surface modulation of neurogenesis during osseointegration.
Finally, the present review findings are consistent with a recent gene expression study of healing extraction sockets in humans [85]. This study reported an initial upregulation of pro-inflammatory cytokines (IL-1, IL-6) at day 1, but by day 7, genes suggestive of immune response (IL-10), osteogenesis (TGF, BMP4, BMP7, OCN and ALP), and angiogenesis (VEGF) were upregulated, continuing until day 14, suggesting that the basic biological processes governing alveolar wound healing and osseointegration are the same.
Conclusions
Based on limited evidence of gene expression data from four studies involving 43 patients, the following remarks can be made:
Early peri-implant healing (2 weeks) involves a sequence of biological events which are similar to those observed in other bone wound healing scenarios (fractures, extraction-sockets).
Osseointegration depends on osteogenesis at the implant interface, but other simultaneously occurring processes such as inflammation, bone resorption, angiogenesis and neurogenesis also play an important role, as evidenced by consistent and concomitant gene expression.
Several genes associated with key regulators of biological processes, such as cells, cytokines, growth factors, transcription factors, signaling pathways, and secretory products, were shown to be differentially regulated during peri-implant healing in a manner that was largely consistent - in terms of nature and timing - with previous in vitro and preclinical in vivo histological studies of osseointegration.
Implant surface technology can influence osseointegration, at every step of the early wound healing process, i.e., anti-inflammatory response, progenitor cell recruitment, osteoinduction, growth factor/transcription factor expression, signaling pathway regulation, and extracellular matrix production. However, the relevance of those observations is questionable; no distinct differences have been demonstrated in terms of histological outcomes at later time points or short- and long-term clinical performance among the various implant surface technologies discussed herein.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SS conceived and carried out the study and drafted the manuscript. VS and AS participated in carrying out the study and drafting the manuscript. All authors read and approved the final manuscript.
Contributor Information
Siddharth Shanbhag, Email: sidshanbhag@gmail.com.
Vivek Shanbhag, Email: dr.vshanbhag@gmail.com.
Andreas Stavropoulos, Email: andreas.stavropoulos@mah.se.
References
- 1.Setzer FC, Kim S. Comparison of long-term survival of implants and endodontically treated teeth. J Dent Res. 2014;93(1):19–26. doi: 10.1177/0022034513504782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrektsson T, Brånemark PI, Hansson HA, Lindström J. Osseointegrated titanium implants. Requirements for ensuring a long-lasting, direct bone-to-implant anchorage in man. Acta Orthop Scand. 1981;52(2):155–70. doi: 10.3109/17453678108991776. [DOI] [PubMed] [Google Scholar]
- 3.Berglundh T, Abrahamsson I, Lang NP, Lindhe J. De novo alveolar bone formation adjacent to endosseous implants. Clin Oral Implants Res. 2003;14(3):251–62. doi: 10.1034/j.1600-0501.2003.00972.x. [DOI] [PubMed] [Google Scholar]
- 4.Abrahamsson I, Berglundh T, Linder E, Lang NP, Lindhe J. Early bone formation adjacent to rough and turned endosseous implant surfaces. An experimental study in the dog. Clin Oral Implants Res. 2004;15(4):381–92. doi: 10.1111/j.1600-0501.2004.01082.x. [DOI] [PubMed] [Google Scholar]
- 5.Lang NP, Salvi GE, Huynh-Ba G, Ivanovski S, Donos N, Bosshardt DD. Early osseointegration to hydrophilic and hydrophobic implant surfaces in humans. Clin Oral Implants Res. 2011;22(4):349–56. doi: 10.1111/j.1600-0501.2011.02172.x. [DOI] [PubMed] [Google Scholar]
- 6.Bosshardt DD, Salvi GE, Huynh-Ba G, Ivanovski S, Donos N, Lang NP. The role of bone debris in early healing adjacent to hydrophilic and hydrophobic implant surfaces in man. Clin Oral Implants Res. 2011;22(4):357–64. doi: 10.1111/j.1600-0501.2010.02107.x. [DOI] [PubMed] [Google Scholar]
- 7.Terheyden H, Lang NP, Bierbaum S, Stadlinger B. Osseointegration - communication of cells. Clin Oral Implants Res. 2012;23(10):1127–35. doi: 10.1111/j.1600-0501.2011.02327.x. [DOI] [PubMed] [Google Scholar]
- 8.Marco F, Milena F, Gianluca G, Vittoria O. Peri-implant osteogenesis in health and osteoporosis. Micron. 2005;36(7–8):630–44. doi: 10.1016/j.micron.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Sela J, Gross UM, Kohavi D, Shani J, Dean DD, Boyan BD, et al. Primary mineralization at the surfaces of implants. Crit Rev Oral Biol Med. 2000;11(4):423–36. doi: 10.1177/10454411000110040301. [DOI] [PubMed] [Google Scholar]
- 10.Raghavendra S, Wood MC, Taylor TD. Early wound healing around endosseous implants: a review of the literature. Int J Oral Maxillofac Implants. 2005;20(3):425–31. [PubMed] [Google Scholar]
- 11.Kanczler JM, Oreffo RO. Osteogenesis and angiogenesis: the potential for engineering bone. Eur Cell Mater. 2008;15:100–14. doi: 10.22203/ecm.v015a08. [DOI] [PubMed] [Google Scholar]
- 12.Sağirkaya E, Kucukekenci AS, Karasoy D, Akça K, Eckert SE, Çehreli MC. Comparative assessments, meta-analysis, and recommended guidelines for reporting studies on histomorphometric bone-implant contact in humans. Int J Oral Maxillofac Implants. 2013;28(5):1243–53. doi: 10.11607/jomi.3166. [DOI] [PubMed] [Google Scholar]
- 13.Kojima N, Ozawa S, Miyata Y, Hasegawa H, Tanaka Y, Ogawa T. High-throughput gene expression analysis in bone healing around titanium implants by DNA microarray. Clin Oral Implants Res. 2008;19(2):173–81. doi: 10.1111/j.1600-0501.2007.01432.x. [DOI] [PubMed] [Google Scholar]
- 14.Lin Z, Rios HF, Volk SL, Sugai JV, Jin Q, Giannobile WV. Gene expression dynamics during bone healing and osseointegration. J Periodontol. 2011;82(7):1007–17. doi: 10.1902/jop.2010.100577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ai-Aql ZS, Alagl AS, Graves DT, Gerstenfeld LC, Einhorn TA. Molecular mechanisms controlling bone formation during fracture healing and distraction osteogenesis. J Dent Res. 2008;87(2):107–18. doi: 10.1177/154405910808700215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albrektsson T, Johansson C. Osteoinduction, osteoconduction and osseointegration. Eur Spine J. 2001;10(Suppl 2):S96–101. doi: 10.1007/s005860100282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miron RJ, Zhang YF. Osteoinduction: a review of old concepts with new standards. J Dent Res. 2012;91(8):736–44. doi: 10.1177/0022034511435260. [DOI] [PubMed] [Google Scholar]
- 18.Ito H. Chemokines in mesenchymal stem cell therapy for bone repair: a novel concept of recruiting mesenchymal stem cells and the possible cell sources. Mod Rheumatol. 2011;21(2):113–21. doi: 10.3109/s10165-010-0357-8. [DOI] [PubMed] [Google Scholar]
- 19.Hughes FJ, Turner W, Belibasakis G, Martuscelli G. Effects of growth factors and cytokines on osteoblast differentiation. Periodontol 2000. 2006;41:48–72. doi: 10.1111/j.1600-0757.2006.00161.x. [DOI] [PubMed] [Google Scholar]
- 20.Scheller EL, Krebsbach PH. The use of soluble signals to harness the power of the bone microenvironment for implant therapeutics. Int J Oral Maxillofac Implants. 2011;26(Suppl):70–9. [PMC free article] [PubMed] [Google Scholar]
- 21.Marie PJ. Transcription factors controlling osteoblastogenesis. Arch Biochem Biophys. 2008;473(2):98–105. doi: 10.1016/j.abb.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 22.Fakhry M, Hamade E, Badran B, Buchet R, Magne D. Molecular mechanisms of mesenchymal stem cell differentiation towards osteoblasts. World J Stem Cells. 2013;5(4):136–48. doi: 10.4252/wjsc.v5.i4.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minkin C, Marinho VC. Role of the osteoclast at the bone-implant interface. Adv Dent Res. 1999;13:49–56. doi: 10.1177/08959374990130011401. [DOI] [PubMed] [Google Scholar]
- 24.Thalji G, Cooper L. Molecular assessment of osseointegration in vitro: a review of the current literature. Oral & Craniofacial Tissue Engineering. 2012;2(3):221–49. [Google Scholar]
- 25.Thalji G, Cooper LF. Molecular assessment of osseointegration in vivo: a review of the current literature. Int J Oral Maxillofac Implants. 2013;28(6):e521–34. doi: 10.11607/jomi.te33. [DOI] [PubMed] [Google Scholar]
- 26.Cooper LF. Biologic determinants of bone formation for osseointegration: clues for future clinical improvements. J Prosthet Dent. 1998;80(4):439–49. doi: 10.1016/S0022-3913(98)70009-5. [DOI] [PubMed] [Google Scholar]
- 27.Omar OM, Lennerås ME, Suska F, Emanuelsson L, Hall JM, Palmquist A, et al. The correlation between gene expression of proinflammatory markers and bone formation during osseointegration with titanium implants. Biomaterials. 2011;32(2):374–86. doi: 10.1016/j.biomaterials.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Nishimura I. Genetic networks in osseointegration. J Dent Res. 2013;92(12 Suppl):109S–18. doi: 10.1177/0022034513504928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins JPT, Green S, editors. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 [updated March 2011]. The Cochrane Collaboration, 2011. Available from www.cochrane-handbook.org; 2011.
- 30.Tarca AL, Romero R, Draghici S. Analysis of microarray experiments of gene expression profiling. Am J Obstet Gynecol. 2006;195(2):373–88. doi: 10.1016/j.ajog.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slonim DK, Yanai I. Getting started in gene expression microarray analysis. PLoS Comput Biol. 2009;5(10):e1000543. doi: 10.1371/journal.pcbi.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nibali L. Suggested guidelines for systematic reviews of periodontal genetic association studies. J Clin Periodontol. 2013;40(8):753–6. doi: 10.1111/jcpe.12128. [DOI] [PubMed] [Google Scholar]
- 33.Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE statement. PLoS Med. 2009;6(2):e22. doi: 10.1371/journal.pmed.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanovski S, Hamlet S, Salvi GE, Huynh-Ba G, Bosshardt DD, Lang NP, et al. Transcriptional profiling of osseointegration in humans. Clin Oral Implants Res. 2011;22(4):373–81. doi: 10.1111/j.1600-0501.2010.02112.x. [DOI] [PubMed] [Google Scholar]
- 35.Donos N, Hamlet S, Lang NP, Salvi GE, Huynh-Ba G, Bosshardt DD, et al. Gene expression profile of osseointegration of a hydrophilic compared with a hydrophobic microrough implant surface. Clin Oral Implants Res. 2011;22(4):365–72. doi: 10.1111/j.1600-0501.2010.02113.x. [DOI] [PubMed] [Google Scholar]
- 36.Bryington M, Mendonça G, Nares S, Cooper LF. Osteoblastic and cytokine gene expression of implant-adherent cells in humans. Clin Oral Implants Res. 2012; doi:10.1111/clr.12054. [DOI] [PubMed]
- 37.Thalji GN, Nares S, Cooper LF. Early molecular assessment of osseointegration in humans. Clin Oral Implants Res. 2013; doi:10.1111/clr.12266. [DOI] [PubMed]
- 38.Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–93. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1–2):279–84. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 40.Alfarsi MA, Hamlet SM, Ivanovski S. Titanium surface hydrophilicity modulates the human macrophage inflammatory cytokine response. J Biomed Mater Res A. 2013; doi:10.1002/jbm.a.34666. [DOI] [PubMed]
- 41.Pajarinen J, Kouri VP, Jämsen E, Li TF, Mandelin J, Konttinen YT. The response of macrophages to titanium particles is determined by macrophage polarization. Acta Biomater. 2013;9(11):9229–40. doi: 10.1016/j.actbio.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 42.Chehroudi B, Ghrebi S, Murakami H, Waterfield JD, Owen G, Brunette DM. Bone formation on rough, but not polished, subcutaneously implanted Ti surfaces is preceded by macrophage accumulation. J Biomed Mater Res A. 2010;93(2):724–37. doi: 10.1002/jbm.a.32587. [DOI] [PubMed] [Google Scholar]
- 43.Heng BC, Cao T, Stanton LW, Robson P, Olsen B. Strategies for directing the differentiation of stem cells into the osteogenic lineage in vitro. J Bone Miner Res. 2004;19(9):1379–94. doi: 10.1359/JBMR.040714. [DOI] [PubMed] [Google Scholar]
- 44.Dhore CR, Snel SJ, Jacques SV, Naert IE, Walboomers XF, Jansen JA. In vitro osteogenic potential of bone debris resulting from placement of titanium screw-type implants. Clin Oral Implants Res. 2008;19(6):606–11. doi: 10.1111/j.1600-0501.2007.01519.x. [DOI] [PubMed] [Google Scholar]
- 45.Kim DH, Yoo KH, Choi KS, Choi J, Choi SY, Yang SE, et al. Gene expression profile of cytokine and growth factor during differentiation of bone marrow-derived mesenchymal stem cell. Cytokine. 2005;31(2):119–26. doi: 10.1016/j.cyto.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Omar O, Svensson S, Zoric N, Lenneras M, Suska F, Wigren S, et al. In vivo gene expression in response to anodically oxidized versus machined titanium implants. J Biomed Mater Res A. 2010;92(4):1552–66. doi: 10.1002/jbm.a.32475. [DOI] [PubMed] [Google Scholar]
- 47.Marcellini S, Henriquez JP, Bertin A. Control of osteogenesis by the canonical Wnt and BMP pathways in vivo: cooperation and antagonism between the canonical Wnt and BMP pathways as cells differentiate from osteochondroprogenitors to osteoblasts and osteocytes. Bioessays. 2012;34(11):953–62. doi: 10.1002/bies.201200061. [DOI] [PubMed] [Google Scholar]
- 48.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108(1):17–29. doi: 10.1016/S0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 49.Komori T. Regulation of osteoblast differentiation by Runx2. Adv Exp Med Biol. 2010;658:43–9. doi: 10.1007/978-1-4419-1050-9_5. [DOI] [PubMed] [Google Scholar]
- 50.Guo J, Padilla RJ, Ambrose W, De Kok IJ, Cooper LF. The effect of hydrofluoric acid treatment of TiO2 grit blasted titanium implants on adherent osteoblast gene expression in vitro and in vivo. Biomaterials. 2007;28(36):5418–25. doi: 10.1016/j.biomaterials.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 51.Mendonça G, Mendonça DB, Simões LG, Araújo AL, Leite ER, Duarte WR, et al. Nanostructured alumina-coated implant surface: effect on osteoblast-related gene expression and bone-to-implant contact in vivo. Int J Oral Maxillofac Implants. 2009;24(2):205–15. [PubMed] [Google Scholar]
- 52.Rocci M, Rocci A, Martignoni M, Albrektsson T, Barlattani A, Gargari M. Comparing the TiOblast and Osseospeed surfaces. Histomorphometric and histological analysis in humans. Oral Implantol (Rome) 2008;1(1):34–42. [PMC free article] [PubMed] [Google Scholar]
- 53.Qin C, Baba O, Butler WT. Post-translational modifications of sibling proteins and their roles in osteogenesis and dentinogenesis. Crit Rev Oral Biol Med. 2004;15(3):126–36. doi: 10.1177/154411130401500302. [DOI] [PubMed] [Google Scholar]
- 54.Vlacic-Zischke J, Hamlet SM, Friis T, Tonetti MS, Ivanovski S. The influence of surface microroughness and hydrophilicity of titanium on the up-regulation of TGFβ/BMP signalling in osteoblasts. Biomaterials. 2011;32(3):665–71. doi: 10.1016/j.biomaterials.2010.09.025. [DOI] [PubMed] [Google Scholar]
- 55.Viguet-Carrin S, Garnero P, Delmas PD. The role of collagen in bone strength. Osteoporos Int. 2006;17(3):319–36. doi: 10.1007/s00198-005-2035-9. [DOI] [PubMed] [Google Scholar]
- 56.Saito M, Marumo K. Collagen cross-links as a determinant of bone quality: a possible explanation for bone fragility in aging, osteoporosis, and diabetes mellitus. Osteoporos Int. 2010;21(2):195–214. doi: 10.1007/s00198-009-1066-z. [DOI] [PubMed] [Google Scholar]
- 57.Pornprasertsuk S, Duarte WR, Mochida Y, Yamauchi M. Lysyl hydroxylase-2b directs collagen cross-linking pathways in MC3T3-E1 cells. J Bone Miner Res. 2004;19(8):1349–55. doi: 10.1359/JBMR.040323. [DOI] [PubMed] [Google Scholar]
- 58.Kaku M, Mochida Y, Atsawasuwan P, Parisuthiman D, Yamauchi M. Post-translational modifications of collagen upon BMP-induced osteoblast differentiation. Biochem Biophys Res Commun. 2007;359(3):463–8. doi: 10.1016/j.bbrc.2007.05.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyce BF. Advances in the regulation of osteoclasts and osteoclast functions. J Dent Res. 2013;92(10):860–7. doi: 10.1177/0022034513500306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mamalis AA, Markopoulou C, Vrotsos I, Koutsilirieris M. Chemical modification of an implant surface increases osteogenesis and simultaneously reduces osteoclastogenesis: an in vitro study. Clin Oral Implants Res. 2011;22(6):619–26. doi: 10.1111/j.1600-0501.2010.02027.x. [DOI] [PubMed] [Google Scholar]
- 61.Heppenstall RB, Grislis G, Hunt TK. Tissue gas tensions and oxygen consumption in healing bone defects. Clin Orthop Relat Res. 1975;106:357–65. doi: 10.1097/00003086-197501000-00048. [DOI] [PubMed] [Google Scholar]
- 62.Potier E, Ferreira E, Andriamanalijaona R, Pujol JP, Oudina K, Logeart-Avramoglou D, et al. Hypoxia affects mesenchymal stromal cell osteogenic differentiation and angiogenic factor expression. Bone. 2007;40(4):1078–87. doi: 10.1016/j.bone.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 63.Mamalis AA, Cochran DL. The role of hypoxia in the regulation of osteogenesis and angiogenesis coupling in intraoral regenerative procedures: a review of the literature. Int J Periodontics Restorative Dent. 2013. doi:10.11607/prd.0868. [DOI] [PubMed]
- 64.Nucera S, Biziato D, De Palma M. The interplay between macrophages and angiogenesis in development, tissue injury and regeneration. Int J Dev Biol. 2011;55(4–5):495–503. doi: 10.1387/ijdb.103227sn. [DOI] [PubMed] [Google Scholar]
- 65.Ziebart T, Schnell A, Walter C, Kämmerer PW, Pabst A, Lehmann KM, et al. Interactions between endothelial progenitor cells (EPC) and titanium implant surfaces. Clin Oral Investig. 2013;17(1):301–9. doi: 10.1007/s00784-012-0691-7. [DOI] [PubMed] [Google Scholar]
- 66.Clarkin CE, Gerstenfeld LC. VEGF and bone cell signalling: an essential vessel for communication? Cell Biochem Funct. 2013;31(1):1–11. doi: 10.1002/cbf.2911. [DOI] [PubMed] [Google Scholar]
- 67.Zhang F, Qiu T, Wu X, Wan C, Shi W, Wang Y, et al. Sustained BMP signaling in osteoblasts stimulates bone formation by promoting angiogenesis and osteoblast differentiation. J Bone Miner Res. 2009;24(7):1224–33. doi: 10.1359/jbmr.090204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shiojima I, Walsh K. Role of Akt signaling in vascular homeostasis and angiogenesis. Circ Res. 2002;90(12):1243–50. doi: 10.1161/01.RES.0000022200.71892.9F. [DOI] [PubMed] [Google Scholar]
- 69.Raines AL, Olivares-Navarrete R, Wieland M, Cochran DL, Schwartz Z, Boyan BD. Regulation of angiogenesis during osseointegration by titanium surface microstructure and energy. Biomaterials. 2010;31(18):4909–17. doi: 10.1016/j.biomaterials.2010.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwarz F, Ferrari D, Herten M, Mihatovic I, Wieland M, Sager M, et al. Effects of surface hydrophilicity and microtopography on early stages of soft and hard tissue integration at non-submerged titanium implants: an immunohistochemical study in dogs. J Periodontol. 2007;78(11):2171–84. doi: 10.1902/jop.2007.070157. [DOI] [PubMed] [Google Scholar]
- 71.Schwarz F, Herten M, Sager M, Wieland M, Dard M, Becker J. Histological and immunohistochemical analysis of initial and early osseous integration at chemically modified and conventional SLA titanium implants: preliminary results of a pilot study in dogs. Clin Oral Implants Res. 2007;18(4):481–8. doi: 10.1111/j.1600-0501.2007.01341.x. [DOI] [PubMed] [Google Scholar]
- 72.Lambrichts I. Histological and ultrastructural aspects of bone innervation. In: Jacobs R, editor. Osseoperception. Leuven, Belgium: UZ Leuven; 1998. pp. 13–20. [Google Scholar]
- 73.Bais M, McLean J, Sebastiani P, Young M, Wigner N, Smith T, et al. Transcriptional analysis of fracture healing and the induction of embryonic stem cell-related genes. PLoS One. 2009;4(5):e5393. doi: 10.1371/journal.pone.0005393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ivanovski S, Hamlet S, Retzepi M, Wall I, Donos N. Transcriptional profiling of “guided bone regeneration” in a critical-size calvarial defect. Clin Oral Implants Res. 2011;22(4):382–9. doi: 10.1111/j.1600-0501.2010.02104.x. [DOI] [PubMed] [Google Scholar]
- 75.Donos N, Retzepi M, Wall I, Hamlet S, Ivanovski S. In vivo gene expression profile of guided bone regeneration associated with a microrough titanium surface. Clin Oral Implants Res. 2011;22(4):390–8. doi: 10.1111/j.1600-0501.2010.02105.x. [DOI] [PubMed] [Google Scholar]
- 76.Skaper SD. The neurotrophin family of neurotrophic factors: an overview. Methods Mol Biol. 2012;846:1–12. doi: 10.1007/978-1-61779-536-7_1. [DOI] [PubMed] [Google Scholar]
- 77.Vaillant AR, Mazzoni I, Tudan C, Boudreau M, Kaplan DR, Miller FD. Depolarization and neurotrophins converge on the phosphatidylinositol 3-kinase-Akt pathway to synergistically regulate neuronal survival. J Cell Biol. 1999;146(5):955–66. doi: 10.1083/jcb.146.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilkins A, Kemp K, Ginty M, Hares K, Mallam E, Scolding N. Human bone marrow-derived mesenchymal stem cells secrete brain-derived neurotrophic factor which promotes neuronal survival in vitro. Stem Cell Res. 2009;3(1):63–70. doi: 10.1016/j.scr.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 79.Buma P, Elmans L, Oestreicher AB. Changes in innervation of long bones after insertion of an implant: immunocytochemical study in goats with antibodies to calcitonin gene-related peptide and B-50/GAP-43. J Orthop Res. 1995;13(4):570–7. doi: 10.1002/jor.1100130412. [DOI] [PubMed] [Google Scholar]
- 80.Corpas Dos Santos L, Lambrichts I, Quirynen M, Collaert B, Politis C, Vrielinck L, et al. Peri-implant bone innervation: histological findings in humans. Eur J Oral Implantol. 2014;7(3):283–92. [PubMed] [Google Scholar]
- 81.Huang Y, Jacobs R, Van Dessel J, Bornstein MM, Lambrichts I, Politis C. A systematic review on the innervation of peri-implant tissues with special emphasis on the influence of implant placement and loading protocols. Clin Oral Implants Res. 2014; doi:10.1111/clr.12344. [DOI] [PubMed]
- 82.Abarca M, Van Steenberghe D, Malevez C, Jacobs R. The neurophysiology of osseointegrated oral implants. A clinically underestimated aspect. J Oral Rehabil. 2006;33(3):161–9. doi: 10.1111/j.1365-2842.2005.01556.x. [DOI] [PubMed] [Google Scholar]
- 83.Klineberg I, Calford MB, Dreher B, Henry P, Macefield V, Miles T, et al. A consensus statement on osseoperception. Clin Exp Pharmacol Physiol. 2005;32(1–2):145–6. doi: 10.1111/j.1440-1681.2005.04144.x. [DOI] [PubMed] [Google Scholar]
- 84.Enkling N, Utz KH, Bayer S, Stern RM. Osseoperception: active tactile sensibility of osseointegrated dental implants. Int J Oral Maxillofac Implants. 2010;25(6):1159–67. [PubMed] [Google Scholar]
- 85.Canuto RA, Pol R, Martinasso G, Muzio G, Gallesio G, Mozzati M. Hydroxyapatite paste Ostim, without elevation of full-thickness flaps, improves alveolar healing stimulating BMP- and VEGF-mediated signal pathways: an experimental study in humans. Clin Oral Implants Res. 2013;24 Suppl A100:42–8. doi:10.1111/j.1600-0501.2011.02363.x. [DOI] [PubMed]


