Abstract
The epigenetic clock, defined as the DNA methylome age (DNAmAge), is a candidate biomarker of ageing. In this study, we aimed to characterize the behaviour of this marker during the human lifespan in more detail using two follow-up cohorts (the Young Finns study, calendar age i.e. cAge range at baseline 15–24 years, 25-year-follow-up, N = 183; The Vitality 90+ study, cAge range at baseline 19–90 years, 4-year-follow-up, N = 48). We also aimed to assess the relationship between DNAmAge estimate and the blood cell distributions, as both of these measures are known to change as a function of age. The subjects’ DNAmAges were determined using Horvath’s calculator of epigenetic cAge. The estimate of the DNA methylome age acceleration (Δ-cAge-DNAmAge) demonstrated remarkable stability in both cohorts: the individual rank orders of the DNAmAges remained largely unchanged during the follow-ups. The blood cell distributions also demonstrated significant intra-individual correlation between the baseline and follow-up time points. Interestingly, the immunosenescence-associated features (CD8+CD28− and CD4+CD28− cell proportions and the CD4/CD8 cell ratio) were tightly associated with the estimate of the DNA methylome age. In summary, our data demonstrate that the general level of Δ-cAge-DNAmAge is fixed before adulthood and appears to be quite stationary thereafter, even in the oldest-old ages. Moreover, the blood DNAmAge estimate seems to be tightly associated with ageing-associated shifts in blood cell composition, especially with those that are the hallmarks of immunosenescence. Overall, these observations contribute to the understanding of the longitudinal aspects of the DNAmAge estimate.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-016-9927-9) contains supplementary material, which is available to authorized users.
Keywords: DNA methylation, DNAmAge, Epigenetic clock, Follow-up, Immunosenescence
Background
In the field of biogerontology, one important objective has been to develop a universal biomarker of ageing that describes an individual’s biological age as opposed to calendar age (cAge). For example, one promising biomarker has been telomere length, which is conveniently obtainable. However, the correlation between telomere length and cAge age is only moderate (Benetos et al. 2001), and the relevance of telomere length as an unambiguous biomarker of ageing is questionable (Mather et al. 2011). Recently, based on large-scale epigenome-wide association studies, the DNA methylation levels at certain CpG sites (Garagnani et al. 2012; Horvath 2013; Hannum et al. 2013; Steegenga et al. 2014; Kananen et al. 2016) have been shown to be strongly associated with cAge in multiple tissues; i.e. methylation at these sites shows clocklike behaviour. These findings have been further refined to produce DNA methylome age (DNAmAge) calculation algorithms. These algorithms provide surprisingly accurate age predictions, with correlation coefficients between cAge and DNAmAge as high as 0.9 and median absolute deviations (MADs, the median of the differences between DNAmAge and cAge) of age predictions as low as 3 years (Bocklandt et al. 2011; Hannum et al. 2013; Horvath 2013; Weidner et al. 2014; Florath et al. 2014). Horvath (2013)) generated a multi-tissue age predictor algorithm (available at https://dnamage.genetics.ucla.edu/) based on elastic net regression analysis using 21,000 probes from the HumanMethylation450 and HumanMethylation27 BeadChips. This predictor algorithm was trained with 8000 samples of various tissue types and cAge ranges in 82 Illumina DNA methylation array data sets. On the grounds of the training results, the ‘epigenetic clock’, i.e. the regression model, was constructed and the model consists of 353 CpG sites whose methylation level best explains the variation in cAge. Using this algorithm, the difference between cAge and DNAmAge estimate (Δ-cAge-DNAmAge) might be considered as a quantitative measure of a person’s biological ageing.
Horvath’s epigenetic clock ticking rate has been shown to be highest during organismal growth (Horvath 2013). However, observations in non-proliferative and immortalized tissues have indicated that DNAmAge is different from mitotic age and that DNAmAge might rather be a measure of the magnitude of epigenetic maintenance (Horvath 2013; Horvath and Levine 2015), although the available evidence is limited. In addition, analysis of twins provided some evidence that the heritability of DNAmAge is 100 % at the time of birth but decreases to 39 % by late adulthood (Horvath 2013). Therefore, non-genetic factors appear to influence the epigenetic clock ticking rate, but it remains unclear when and how these inter-individual differences in the epigenetic clock arise and how stabile these differences during distinct stages of the human lifespan are.
In recent studies, accelerated biological ageing, as indicated by DNAmAge estimate, has been demonstrated regarding several ageing-associated phenotypes (physical and mental fitness (Marioni et al. 2015b), mortality (Marioni et al. 2015a) or phenotypes with ageing-resembling features (HIV1 (Horvath and Levine 2015) and Down syndrome (Horvath et al. 2015a)). In addition, obesity induces an increase in DNAmAge in hepatocytes (Horvath et al. 2014). Nevertheless, some of these results are conflicting; e.g. military personnel suffering from more severe post-traumatic stress symptoms have been demonstrated to have a lower DNAmAges (Boks et al. 2015). However, because of the limited number of longitudinal studies on DNAmAge, the behaviour and utility of the DNAmAge as a biomarker of ageing are currently unknown. As different blood cell types display distinct DNA methylation profiles (Houseman et al. 2012; Jaffe and Irizarry 2014) and the immune cell distribution changes as a function of age (Weiskopf et al. 2009; Pawelec et al. 2010), the relationship between alterations in methylation patterns and blood cell distributions is specifically relevant to ageing and age-related phenomena.
The immune cell repertoire in individuals aged 70 years and older demonstrates a notable decline; the classic manifestation of immunosenescence is the inability to generate appropriate responses to vaccines and infections (Weiskopf et al. 2009). The phenomena underlying immunosenescence include clonal expansion of CD4+CD28− and CD8+CD28− cells, which lack the costimulatory surface receptor CD28, a key factor in T cell activation. CD28− cells exhibit shortened telomeres and produce excessive levels of proinflammatory cytokines. Furthermore, the lifespan of CD28− cells is prolonged and these cells may survive for years in the periphery because they undergo apoptosis less frequently than e.g. CD28+ cells. (Weiskopf et al. 2009; Arnold et al. 2011) Although CD28− cells constitute a minor proportion of the total leukocyte pool, they are key players in immune system ageing. Thus, the CD28− cells should be considered when examining the implications of ageing on DNA methylation in leukocytes.
Horvath’s DNAmAge has been reported to be relatively independent from variation in the composition of the major blood cell types, but these considerations apply only to middle-aged men and to samples from individuals with a mean cAge between 66 and 79 years (Horvath 2013; Marioni et al. 2015a). Furthermore, few population-based cross-sectional studies have examined genome-wide methylation patterns accounting for blood cell type distributions, yet typically only the major blood cell types have been analysed (Lam et al. 2012; Zilbauer et al. 2013; Jaffe and Irizarry 2014; Marttila et al. 2015). Thus, longitudinal studies including more detailed cell type characterizations and covering wider age ranges of individuals of both sexes are needed. Hence, using two Finnish follow-up cohorts, The Vitality 90+ study (V90) (age of nonagenarians at baseline of 90 years and age range of young controls of 19–34 years, 4-year follow-up, N = 48) and The Young Finns study (YFS) (age range at baseline of 15–24 years, 25-year follow-up, N = 183), we aimed to elucidate the longitudinal characteristics of the DNAmAge estimate. Specifically, we addressed the individual trajectories of the DNA methylome age acceleration estimate throughout adulthood in both sexes and examined how the DNAmAge estimate parallels with the age-related fluctuation of blood cell composition. With specific relevance to ageing, our data included also the oldest-old individuals and we assessed the distributions of the specific immunosenescence-representing parameters: the CD4+/CD8+ cell ratio and CD4+CD28− and CD8+CD28− lymphocytes.
Results
DNA methylome age in individuals in the YFS
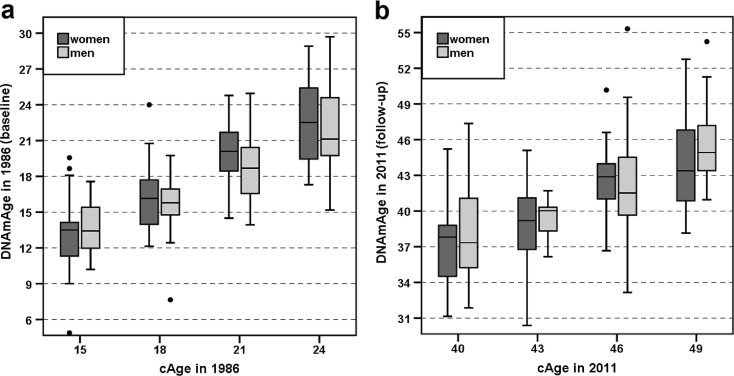
Horvath’s DNAmAge was determined for each individual using a multi-tissue age predictor (Horvath 2013). First, the resulting DNAmAges of the YFS participants were evaluated separately in relation to cAge and gender using samples collected in years 1986 and 2011. As shown in Fig. 1a, b and Supplementary Table 1 (Additional File 2), the gender-wise medians of DNAmAge were less than the corresponding cAge at both time points. The median ± MAD DNAmAges of the subjects in the YFS_1986 cohort and the corresponding values of the same subjects in the YFS_2011 cohort are shown in Supplementary Table 1 (Additional File 2). Spearman’s rank sum correlation coefficients between cAge and DNAmAge were 0.785 (P = 8.10 × 10−25) among women and 0.702 (P = 1.25 × 10−11) among men in the YFS_1986 cohort. The corresponding values in the YFS_2011 cohort were 0.635 (P = 4.24 × 10−14) among women and 0.591 (P = 7.36 × 10−8) among men.
Fig. 1.
a, b The DNAmAge values in the YFS samples at baseline and at follow-up after 25 years are visualized as boxplots. The YFS_1986 sample (N = 183) is shown in panel a, and the YFS_2011 sample (N = 183) is shown in panel b. The subjects were categorized by cAge and gender. The boxes for women and men are coloured dark and light grey, respectively. The differences in the median DNAmAge values between men and women at each cAge group were not prominent (Mann-Whitney U test, P > 0.05). More detailed information can be found in Additional Files 1 and 2. (Colour figure online)
Table 1.
Summary of DNAmAges and PBMC counts for the V90 sample in 2010 and 2014
| Sample collection year | Group | Number | cAge | DNAmAge of PBMCs | CD3+ of live-gated cells (%) | CD4+ of CD3+ (%) | CD8+ of CD3+ (%) | CD4+/CD8+ ratio | CD28− of CD4+ (%) | CD28− of CD8+ (%) | CD19 of live-gated cells (%) | CD14+ of live-gated cells (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2010 | 19–28 years, women | 14 | 22.5 (3.0) | 22.3 (2.5) | 71.6 (3.3) | 59.0 (4.5) | 30.1 (6.0) | 2.1 (0.5) | 0.2 (0.1) | 14.7 (6.8) | na | 2.0 (1.11) |
| 20–29 years, men | 7 | 24.0 (4.5) | 22.5 (4.4) | 67.0 (3.0) | 60.2 (2.7) | 31.8 (4.7) | 1.9 (0.5) | 0.3 (0.4) | 18.1 (13.3) | na | 2.4 (0.44) | |
| 90 years, women | 84 | 90.0 (0) | 81.7 (7.3) | 60.5 (9.6) | 65.6 (15.6) | 27.1 (13.0) | 2.4 (1.7) | 9.3 (9.7) | 65.3 (17.5) | na | 8.5 (4.52) | |
| 90 years, men | 31 | 90.0 (0) | 87.2 (5.3) | 54.3 (10.2) | 57.2 (12.5) | 28.7 (13.0) | 2.0 (1.1) | 9.2 (8.5) | 69.0 (16.0) | na | 7.5 (4.30) | |
| 2014 | 19–34 years, women | 27 | 26.0 (4.4) | 29.4 (6.8) | 67.0 (7.0) | 58.9 (5.6) | 30.2 (6.8) | 1.9 (0.5) | 2.3 (2.4) | 30.0 (10.8) | 5.8 (2.5) | 4.4 (4.6) |
| 22–32 years, men | 13 | 28.0 (4.4) | 35.1 (9.2) | 59.0 (6.2) | 55.1 (4.2) | 32.8 (4.7) | 1.6 (0.3) | 1.4 (1.9) | 39.2 (17.0) | 8.0 (4.4) | 8.5 (9.9) | |
| 94 years, women | 27 | 94 (0) | 89.6 (7.2) | 58.8 (11.7) | 49.1 (18.2) | 35.6 (19.1) | 1.3 (1.1) | 14.3 (12.2) | 74.4 (16.5) | 2.8 (2.3) | 8.4 (8.7) | |
| 94 years, men | 14 | 94 (0) | 91.5 (11.8) | 62.5 (8.9) | 58.3 (19.3) | 28.1 (14.6) | 2.1 (1.5) | 10.0 (11.6) | 75.0 (16.1) | 2.2 (0.7) | 10.8 (11.6) |
The values were categorized by cAge group and gender. The table shows the median (median absolute deviation) values. Data not available are denoted as ‘na’
The changes in DNAmAge from 1986 to 2011 (i.e. how many years the DNA methylome of a person had aged in 25 years: DNAmAge in 2011 − DNAmAge in 1986) were 23.23 ± 2.74 years among women (N = 113), 23.92 ± 3.10 years among men (N = 70) and 23.26 ± 2.97 among all individuals (N = 183). This change corresponds to 3.7 years of methylome ageing in 4 calendar years. The changes in DNAmAge did not significantly differ between men and women (Mann-Whitney U test, P = 0.076).
DNA methylome ages in individuals in the V90
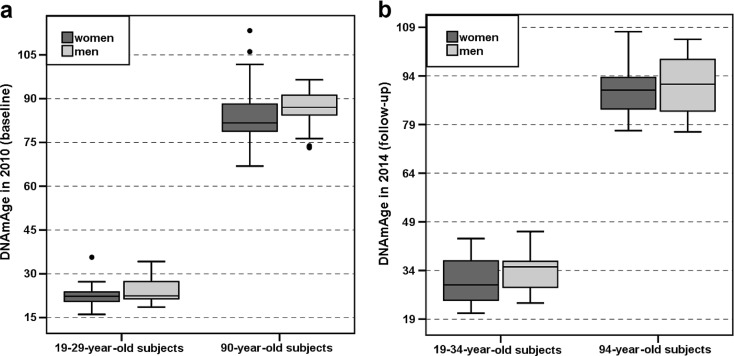
Similarly to the analysis using the YFS data, the resulting DNAmAges of the V90 participants were first evaluated separately in relation to cAge and gender using samples collected in years 2010 and 2014. Spearman’s rank sum correlation coefficients between cAge and DNAmAge were 0.594 (P = 3.6 × 10−11) among women and 0.661 (P = 3.4 × 10−6) among men in the V90_2010 cohort. The corresponding values in V90_2014 were 0.925 (P = 5.4 × 10−24) among women and 0.903 (P = 1.2 × 10−10) among men. As shown in Table 1, Fig. 2a, b and Supplementary Figs. 3 and 4 (Additional File 2), the median DNAmAges were less than the corresponding cAges within most, but not all, categories: the median DNAmAges of the subjects aged 19–34 years in the V90_2014 cohort were higher than their cAges.
Fig. 2.
a, b The DNAmAge values in the V90 samples are visualized as boxplots. The V90_2010 sample (N = 143) is shown in panel a, and the V90_2014 sample (N = 81) is shown in panel b. The samples were categorized by cAge group (on the x-axis) and gender (box colours). A general trend of gender difference was observed in the DNAmAges of PBMCs in the nonagenarians and the young controls at both time points of the V90. The difference in the median DNAmAge values between nonagenarian men (N = 33) and women (N = 89) was significant (Mann-Whitney U test, P = 0.006) in the V90_2010 sample (panel a). However, in the categories with smaller sample sizes, in the younger cAge groups of the V90_2010 (panel a) and V90_2014 (panel b) samples and in the 94-year-old group of the V90_2014 (panel b) sample, the differences in the median DNAmAge values were not statistically significant between men and women (Mann-Whitney U test, P > 0.05). More detailed information can be found in Additional Files 1 and 2
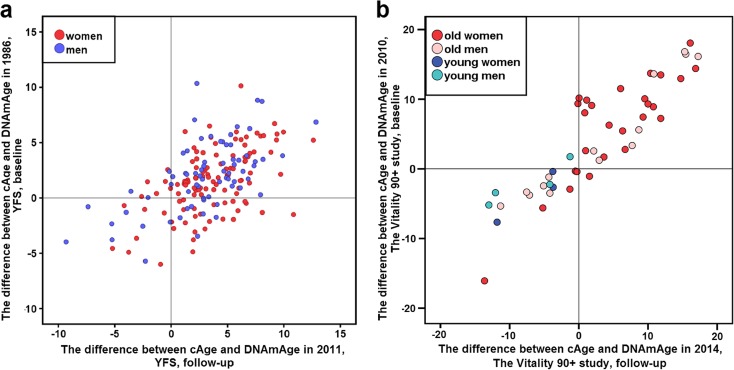
Fig. 3.
a, b The correlations between the Δ-cAge-DNAmAge values at different time points are shown in scatterplots. a Subjects participating in the YFS in 1986 and 2011. The dots corresponding to women (N = 113) are coloured red, and the dots corresponding to men (N = 70) are coloured blue. b Subjects participating in the V90 in 2010 and 2014 (N = 48). The colours of the dots for each subject are explained in the graph legend. The Spearman’s rank sum correlation coefficient between Δ-cAge-DNAmAge values was 0.535 (P = 6.1 × 10−15) for the YFS sample (N = 183, panel a) and 0.895 (P = 9.2 × 10−18) for all subjects in the V90 (N = 48, panel b). More detailed information can be found in Additional Files 1 and 2. (Colour figure online)
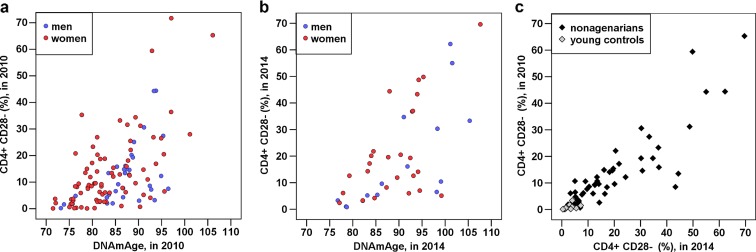
Fig. 4.
a–c The CD28−CD4+ cell counts of the participants examined in 2010 and 2014 correlate with their corresponding DNAmAge values and with one another. The associations are visualized as scatterplots. Each dot corresponds to one subject; the colours of the dots are explained in the graph legends. a The CD28−CD4+ proportions of 90-year-old subjects in V90_2010 correlated with their DNAmAge in 2010 (r = 0.540, P = 4.75 × 10−10, N = 115). b The CD28×CD4+ proportions of 94-year-old subjects in V90_2014 correlated with their DNAmAge in 2014 (r = 0.626, P = 1.20 × 10−5, N = 41). c The CD28−CD4+ proportions of all participants in V90_2010 correlated with their CD28−CD4+ proportions in V90_2014 (among all: r = 0.901, P = 9.48 × 10−24, N = 63; among elderly: r = 0.864, P = 3.33 × 10−16, N = 51; among young controls: r = 0.684, P = 0.014, N = 12). More detailed information can be found in Table 2 and Table 4 and Supporting Material in Additional File 1
To determine whether DNA methylomes of men, women, young, or old subjects aged differently during the follow-up period, the changes in DNAmAge from 2010 to 2014 (DNAmAge in 2014 − DNAmAge in 2010) were calculated. Over this period, the methylome of women aged 4.18 ± 3.04 years (N = 30) and that of men aged 6.31 ± 2.60 years (N = 18) (Mann-Whitney U test, P = 0.255, total N = 48). Within the cAge groups (categorized as nonagenarians and young controls), the changes of DNAmAge were 7.39 ± 2.11 years among the young controls (N = 7) and 4.53 ± 3.52 years among the nonagenarians (N = 41; Mann-Whitney U test, P = 0.020) during the 4-year follow-up period. There was no difference in the change in DNAmAge between nonagenarian men (N = 14) and women (N = 27) during the follow-up period (P = 0.582). The corresponding gender-wise analysis of young controls in the V90 cohort was discarded due to the small sample size of subjects participating at both time points.
Δ-cAge-DNAmAge in the YFS participants
The difference between calendar age and DNA methylome age (Δ-cAge-DNAmAge) in whole blood leukocytes (WBLs) was calculated for all study subjects. In the YFS, the Δ-cAge-DNAmAge was 3.24 ± 3.17 (median ± MAD) among women and 2.95 ± 3.20 among men in 1986 (baseline, 15–24-year-old subjects), and in 2011 (follow-up, 40–49-year-old subjects), the corresponding values were 1.50 ± 2.73 among women and 2.07 ± 2.77 among men. There was no statistically significant difference in the median Δ-cAge-DNAmAge between genders in either YFS_1986 (P = 0.314) or YFS_2011 (P = 0.654). The Δ-cAge-DNAmAge values are shown in further detail in Supplementary Figs. 1 and 2 (Additional File 2), in which the subjects were categorized by cAge group and gender. Figure 3a illustrates the relationship between the Δ-cAge-DNAmAge values in 1986 and those in 2011. The Spearman’s rank sum correlation coefficient between the Δ-cAge-DNAmAge values was 0.515 (P = 5.2 × 10−9) for women (N = 113), 0.567 (P = 3.1 × 10−7) for men (N = 70) and 0.535 (P = 6.1 × 10−15) for all participants (N = 183).
Δ-cAge-DNAmAge in the V90 participants
In the V90, the median Δ-cAge-DNAmAge values in 2010 (baseline) were 6.27 ± 6.99 among all 90-year-old subjects (N = 122) and −0.363 ± 2.83 among all 19–29-year-old subjects (N = 21). In 2014 (follow-up), the median Δ-cAge-DNAmAge values were 3.63 ± 8.74 among all 94-year-old subjects (N = 41) and −3.87 ± 3.34 among all 19–34-year-old subjects (N = 40). The Δ-cAge-DNAmAge values are shown in further detail in Supplementary Figs. 3 and 4 (Additional File 2), in which the subjects were categorized by cAge group and gender. The relationship between the Δ-cAge-DNAmAge values in 2010 and those in 2014 (N = 48) is illustrated in Fig. 3b; the corresponding Spearman’s rank sum correlation coefficient between the Δ-cAge-DNAmAge values was 0.895 (P = 9.2 × 10−18) for all subjects in the V90. Due to limited sample sizes, further statistical analyses in subgroups of small sample sizes were not performed.
Associations of DNAmAge with cell counts
The cell counts were determined using either genome-wide DNA methylation profiles together with a specific cell count estimation algorithm (Jaffe and Irizarry 2014) (YFS) or fluorescence-activated cell sorting (FACS) analysis (V90). Then, the associations between the cell counts and DNAmAge were determined using Spearman rank sum correlation analysis. First, the correlation of proportions of blood cell types at each time point with the corresponding Δ-cAge-DNAmAge values were examined (Tables 2 and 3). In the V90, the most significant peripheral blood mononuclear cell (PBMC) subtype correlate with Δ-cAge-DNAmAge was CD4+CD28− cells among nonagenarians: an increased proportion of these cells was associated with older DNAmAge. Other significant correlates in nonagenarians were CD4+ CD3+, CD8+ of CD3+, CD8+CD28− cells and the CD4+/CD8+ ratio (Table 2). In the young controls, the significant correlates were the CD4+/CD8+ ratio, CD3+ of live-gated, CD4+CD3+, CD4+CD28− and CD8+CD28− cells (Table 2). Among the YFS individuals aged 15–24 years, the significant WBL subtype correlate with Δ-cAge-DNAmAge was CD4+ T cells (Table 3). However, after 25 years, the significant correlates for the same individuals were the CD4+/CD8+ ratio and NK cells (Table 3).
Table 2.
The associations of PBMC subtype proportions with DNAmAge values at baseline and at follow-up among nonagenarians (born in 1920) and young controls (aged 19–34 years) in the V90 sample
| Nonagenarians | Young controls | |||||||
|---|---|---|---|---|---|---|---|---|
| Δ-cAge-DNAmAge at baseline in 2010, N = 115 | Δ-cAge-DNAmAge at follow-up in 2014, N = 41 | Δ-cAge-DNAmAge at baseline in 2010, N = 21 | Δ-cAge-DNAmAge at follow-up in 2014, N = 40 | |||||
| Cell type proportion (%) | r | P | r | P | r | P | r | P |
| CD3+ of live-gated cells | 0.106 | 0.261 | −0.251 | 0.114 | −0.065 | 0.780 | 0.404* | 0.010 |
| CD4+ of CD3+ | 0.316* | 0.001 | 0.311* | 0.048 | 0.505* | 0.019 | 0.382* | 0.015 |
| CD8+ of CD3+ | −0.310* | 0.001 | −0.326* | 0.038 | −0.233 | 0.309 | −0.232 | 0.150 |
| CD4+/CD8+ ratio | 0.309* | 0.001 | 0.346* | 0.027 | 0.396 | 0.075 | 0.332* | 0.036 |
| CD4+CD28− | −0.540* | 4.75e−10 | −0.626* | 1.20e−5 | −0.222 | 0.333 | −0.344* | 0.030 |
| CD8+CD28− | −0.269* | 0.004 | −0.327* | 0.037 | −0.369 | 0.100 | −0.348* | 0.028 |
| CD14+ of live-gated cells | −0.112 | 0.235 | 0.033 | 0.836 | −0.139 | 0.549 | 0.077 | 0.635 |
| CD19+ of live-gated cells | na | na | 0.246 | 0.122 | na | na | −0.025 | 0.879 |
The cell counts were determined using FACS. The associations were determined using Spearman rank-sum correlation analysis; larger and smaller Δ-cAge-DNAmAge values are referred to as ‘younger’ and ‘older’ DNA methylome age, respectively. Data not available are denoted as ‘na’. Associations with a P value less than 0.05 are denoted by asterisks. Additional young control subjects (N = 33) were recruited at follow-up in 2014
Table 3.
The associations of WBL subtype proportions with Δ-cAge-DNAmAge values at baseline in 1986 (cAge 15–24 years) and at follow-up in 2011 (cAge 40–49 years) of the subjects in the YFS
| YFS, Δ-cAge-DNAmAge in 1986, N = 183 | YFS, Δ-cAge-DNAmAge in 2011, N = 183 | |||
|---|---|---|---|---|
| Cell type proportion (%) | r | P | r | P |
| CD8+ T cells | −0.085 | 0.255 | −0.089 | 0.231 |
| CD4+ T cells | 0.150* | 0.044 | 0.093 | 0.211 |
| CD4+/CD8+ ratio | 0.106 | 0.160 | 0.224* | 0.003 |
| NK cells | −0.024 | 0.747 | −0.192* | 0.009 |
| B cells | 0.106 | 0.152 | 0.115 | 0.120 |
| Monocytes | −0.028 | 0.711 | −0.015 | 0.840 |
| Granulocytes | 0.029 | 0.700 | 0.054 | 0.465 |
The cell counts were determined using genome-wide DNA methylation profiles and a specific cell count estimation algorithm (Jaffe and Irizarry 2014). Associations were determined using Spearman rank-sum correlation analysis; larger and smaller Δ-cAge-DNAmAge values are referred to as ‘younger’ and ‘older’ DNA methylome age, respectively. Associations with a P value less than 0.05 are denoted by asterisks
Next, the fluctuations in cell subtype proportions in blood over the follow-up period were evaluated using correlation analysis. The proportion of each cell type and CD4+/CD8+ ratio at baseline were correlated against the corresponding variable at the follow-up time-point (Tables 4 and 5). During the 4-year follow-up period in the V90 participants, the cell proportions were stationary (Table 4, Fig. 4c). The most stabile cell types or ratios during the 25-year-follow-up period in the YFS individuals were NK and B cells and the CD4+/CD8+ ratio; the correlation coefficients for these variables were between 0.45 and 0.55 (Table 5).
Table 4.
Correlations of the PBMC subtype proportions of subjects (N = 63) in the V90 between 2010 and 2014
| Cell type proportion (%) | r | P |
|---|---|---|
| CD3+ of live-gated cells | 0.628* | 2.26e−06 |
| CD4+ of CD3+ | 0.830* | 5.34e−13 |
| CD8+ of CD3+ | 0.855* | 2.13e−14 |
| CD4+/CD8+ ratio | 0.811* | 7.93e−16 |
| CD4+ CD28− | 0.901* | 9.48e−24 |
| CD8+CD28− | 0.838* | 2.06e−13 |
| CD14+ of live-gated cells | 0.317* | 3.01e−02 |
The cell counts of the samples were determined using FACS. Associations were determined using Spearman rank-sum correlation analysis. Associations with a P value less than 0.05 are denoted by asterisks. Correlation analysis was performed on the total sample without further categorization due to a small sample size
Table 5.
Correlations of WBL subtype proportions for subjects in the YFS sample between 1986 and 2011
| YFS, all N = 183 | YFS, women N = 113 | YFS, men N = 70 | ||||
|---|---|---|---|---|---|---|
| Cell type proportion (%) | r | P | r | P | r | P |
| CD8+ T cells | 0.317* | 1.20e−05 | 0.356* | 1.35e−04 | 0.261* | 2.90e−02 |
| CD4+ T cells | 0.312* | 1.57e−05 | 0.349* | 1.86e−04 | 0.360* | 2.19e−03 |
| CD4+/CD8+ ratio | 0.453* | 1.97e−10 | 0.464* | 4.12e−07 | 0.511* | 1.00e−05 |
| NK cells | 0.398* | 2.18e−08 | 0.430* | 2.79e−06 | 0.312* | 8.55e−03 |
| B cells | 0.545* | 1.33e−15 | 0.484* | 8.69e−08 | 0.589* | 8.39e−08 |
| Monocytes | 0.547* | 9.73e−16 | 0.561* | 1.79e−10 | 0.521* | 3.71e−06 |
| Granulocytes | 0.206* | 4.92e−03 | 0.268* | 4.60e−03 | 0.080 | 0.509 |
The cell counts were determined using genome-wide DNA methylation profiles and a specific cell count estimation algorithm (Jaffe and Irizarry 2014). Associations were determined using Spearman rank-sum correlation analysis. Associations with a P value less than 0.05 are denoted by asterisks
Discussion
The human epigenetic clock, assessed as the DNAmAge estimate or DNAmAge acceleration (the difference between cAge and DNAmAge estimate), has recently gained much attention as a marker of human biological age. However, the utility and the behaviour of this marker in the course of the lifespan are incompletely understood. Here, we present a longitudinal evaluation of the characteristics of DNAmAge estimate (Horvath 2013) during adulthood and at advanced ages. The most notable finding in our study is that the estimate of DNA methylome age acceleration is remarkably stabile throughout adulthood and that the rate at which this trajectory proceeds appears to be set before adulthood. The data from the YFS demonstrated that already at the age of 15 years, the slope of DNA methylome ageing was already on an individual-specific level, such that over the 25-year follow-up period, very little deviation from this line was observed. A clear correlation in the scatterplot in Fig. 3a illustrates this finding. At old ages, the DNA methylome ageing rate was likewise steady (Fig. 3b). Previous findings provide support to our results. The creator of the DNAmAge calculator (Horvath 2013) reported first in his study using cross-sectional DNAmAge data that the epigenetic clock ticking rate is accelerated before adulthood and thereafter the rate slows down. In addition, the DNA methylomes of twins are identical at birth and begin to diverge thereafter, and by the late adulthood, the methylomes of twin pairs display only 39 % similarity (at median age of 63 years, all women) (Horvath 2013). Most importantly, a recent follow-up study (Simpkin et al. 2016) on children (baseline at birth, follow-ups at ages of 7 and 17 years) showed that epigenetic age acceleration (the difference between epigenetic and chronological age) is only minor at the time of birth when compared to later stages at childhood: the divergence in epigenetic age acceleration on those children was increased in association with chronological age. Simpkin et al. (2016) concluded also that within-subject correlation between epigenetic ages at different time-points increases with increasing chronological age. Thus, these results are well in line with our conclusion that the primary trajectory of epigenetic age is set before adulthood. Interestingly, Simpkin et al. (2016) found several phenotypes that associated with epigenetic age acceleration in the children: e.g. maternal alcohol consumption and smoking during pregnancy among others. Future long-term follow-up studies with full-scale phenotypic information from the beginning of life, through early childhood to later adulthood (e.g. middle-age) would provide more exact knowledge of epigenetic clock timing.
Several environmental factors, such as diet, physical and cognitive characteristics and toxin exposure, may influence the ticking rate of the epigenetic clock (Zampieri et al. 2015; Simpkin et al. 2016). Thus, in the current study, these and various other factors may underlie the observation that some of the individuals deviated from the diagonal line in the scatterplots of their Δ-cAge-DNAmAge values (Fig. 3a, b). In addition, some of the dispersion may be due to technical reasons, i.e. inaccuracies in the DNAmAge measurement. Although numerous factors may influence the DNAmAge estimate, surprisingly high correlation coefficients between inter-individual Δ-cAge-DNAmAge values at the two time-points were observed in both cohorts. Therefore, our results indicate that in the majority of individuals, ageing of the DNA methylome proceeds surprisingly steadily (Fig. 3a, b).
Another important finding in our study is that the DNAmAge estimate significantly paralleled the ageing-related shifts in leukocyte proportions (Table 2). The most significant PBMC subtype that correlated with Δ-cAge-DNAmAge in the V90 sample was CD4+CD28− cells; an increased frequency of these cells was associated with older DNA methylome age in the nonagenarians at both follow-up time points (Table 2, Fig. 4a, b). Moreover, CD4+CD28− cells were similarly associated with DNAmAge in the young controls (Table 2). The observation that increased proportions of immunosenescence-representing cell types (CD4+CD28− and CD8+CD28− cells) and a reduced CD4+/CD8+ cell ratio emerged as highly significant correlates of greater DNAmAge is of specific importance to ageing and age-related phenotypes. That is, the CD4+CD28− cell count can be a potential confounding factor in analyses of the associations between DNAmAge and ageing-related phenotypes. Interestingly, the association between DNAmAge and immune cell distribution is not restricted to ageing. It has been demonstrated that more rapid DNA methylome ageing estimate in HIV-1 patients is accompanied by increased frequencies of NK and CD28−CD34RA−CD8+ T cells and decreased frequencies of CD4+ granulocytes and naïve CD8+ T cells (Horvath and Levine 2015). Notably, HIV-1 patients are likely to prematurely develop ageing-related pathologies (Pathai et al. 2014). Based on the correlation between the DNAmAge estimate and the NK cell count in the YFS cohort follow-up when the participants were 40–49 years of age (Table 3), it seems that the NK cell count and DNAmAge estimate are associated only in later adulthood and not in adolescence and early adulthood. However, identifying the basis of this observation and elucidating the potential ageing- and immunosenescence-related methylomic changes in NK cells warrants further studies in purified NK cells.
As shown in our previous study (Marttila et al. 2015), CD4+CD28− cells appear to be an obvious confounding factor in analyses of age-related DNA methylation changes at genome-wide level in PBMCs; the proportion of CD4+CD28− cells explained a large part of the variation in DNA methylation levels of ageing-associated CpG sites. Although the mechanistic basis for the role of CD4+CD28− cells in ageing and immunosenescence is not completely clear, we speculate that certain intrinsic features of these cells, i.e. their cytokine secretion profile, shortened telomeres and resistance to apoptosis (Weiskopf et al. 2009; Arnold et al. 2011), might also be related to alterations in DNA methylation patterns in the ageing-associated sites as well as in the epigenetic clock sites. In addition to ageing, expansion of these cells has been reported in certain immune-related diseases, such as multiple sclerosis, rheumatoid arthritis and acute coronary syndromes (Broux et al. 2012). Hence, our findings may have implications on the methylomic analyses of these disorders as well.
Previous studies have shown that DNA methylomes of men age more rapidly than those of women (Hannum et al. 2013; Weidner et al. 2014). Thus, the results were interpreted according to gender. Although our subsample sizes of men and women were quite small for statistical analysis, a general trend of gender difference was observed in the DNAmAges of PBMCs in the nonagenarians and the young controls at both time points of the V90: men had older DNA methylomes than women (Fig. 2a, b); the difference was statistically significant in the V90_2010 cohort where the subsample sizes were sufficient. In the WBL samples from the YFS participants, no significant gender differences in the DNA methylome ages were observed in any cAge category (Table 1 and Figs. 1a, b). A reason for this negative result may have been the small sample size of the different cAge groups or variability of the blood sample types. However, in the YFS_1986 cohort, there was a slight trend in which 15–24-year-old men and women displayed unique, but constant trajectories of DNA methylome ageing estimate as a function of cAge (i.e. the slope of the trend line of this change was different between genders); women aged faster than men, as shown in Fig. 1a. Nevertheless, the overall ageing rate of DNA methylomes (i.e. how many years the DNA methylome had aged over the 4- or 25-year follow-up period) did not differ between women and men in either the V90 or the YFS. Further studies using larger samples are needed to clarify these observations.
Analogous to previous findings (Horvath 2013; Marioni et al. 2015a), the DNAmAge values correlated with cAge, showing high correlation coefficients (Spearman’s rho >0.5) and high statistical significance (P < 4 × 10−6) in both of our follow-up studies. Interestingly, as shown in Table 1 and Supplementary Table 1 (Additional File 2) as well as in Figs. 1 and 2, the median DNAmAges were often smaller than the corresponding median cAges; thus, the Δ-cAge-DNAmAge values were positive in the majority of samples, as shown in Figs. 3a, b and Supplementary Figs. 1–4 (Additional File 2). This result may be interpreted as an indicator of a ‘young’ DNA methylome age of blood cells (on both PBMC and WBL). Nevertheless, some of the young controls in the V90 exhibited older DNA methylomes in their PBMCs (Supplementary Fig. 4 in Additional File 2, Table 1). Analysis of the follow-up data revealed that the DNA methylomes in WBLs of the YFS subjects had aged by an average of ∼23 years during the 25-year follow-up period; this rate corresponds to 3.7 years of methylome ageing in 4 calendar years. During the 4-year-follow-up in the V90 cohort, the DNA methylomes in PBMCs of the individuals aged 19–34 years appeared to age faster than those of nonagenarians (∼7.4 years compared to ∼4.5 years). These results may refer to the slightly varying rates of DNA methylome ageing in PBMCs between different stages of the human lifespan. However, our results also indicate that the DNA methylome ageing rate is rather stabile after young adulthood, even at extreme ages (Figs. 3a, b).
Previous studies have shown that there are non-pathologic tissue-specific differences in DNAmAge. For example, the DNA methylome of the cerebellum ages more slowly (Horvath et al. 2015b) and the DNA methylome of sperm is considered much younger (Horvath 2013) than that in other parts of the human body. Studies of the within-individual differences in Δ-cAge-DNAmAge values between leukocytes and other healthy tissues using reasonable sample sizes are lacking. Few reports have assessed the estimated blood cell subtypes in their DNAmAge analyses (Horvath 2013; Marioni et al. 2015a) and concluded that DNAmAge is relatively independent of the fluctuation in the predominant blood cell subtypes. Nevertheless, these studies have assessed only the major blood cell categories, and most importantly, these studies have not examined very old individuals.
Our results demonstrate in both of our follow-up cohorts that (i) the DNAmAge is associated with the cell subtype distributions (Tables 2 and 3); (ii) there are highly significant intra-individual correlations of the blood cell subtype distributions during several years of follow-ups (Tables 4 and 5, Fig. 4c) and (iii) the difference between DNAmAge estimate and cAge is largely unchanged during the follow-ups (Fig. 3a, b). Our data do not permit us to elucidate why the proportions of the specific blood cell types are so strongly associated with the DNAmAge estimate, but the fact that the blood cell composition changes with age is also evident in our results (Table 1). This issue may be one of the major reasons for average DNA methylation differences in PBMCs over age. However, our results do not indicate a causal relationship between the ageing-associated changes in cell composition and the actual DNA methylation changes in specific loci per cell passage.
Our study contains evident limitations due to limited sample sizes. In addition, we did not obtain flow cytometry data for the proportions of minor cell subtypes aside from CD4+CD28− and CD8+CD28− cells. Furthermore, the cell counts of the YFS samples were determined using a prediction algorithm that relies on genome-wide cell-type-specific methylation profiles (Jaffe and Irizarry 2014). Moreover, the DNA samples were of slightly different origin: YFS sample DNA was extracted from WBLs, and V90 DNA was extracted from PBMCs. Nevertheless, we detected analogous results from the two independent follow-up cohorts examined in this study.
Conclusions
In summary, our results demonstrate that most of the differences in estimated DNAmAge arise before adulthood and that the estimated DNAmAge changing rate is rather stabile thereafter. We also provide evidence that DNAmAge estimate is tightly associated with blood cell subtype frequencies, especially with the immunosenescence-reflecting markers, the CD4+CD28− and CD8+CD28− cells and the CD4+/CD8+ ratio. However, it remains to be determined how and exactly when before adulthood the inter-individual differences in the DNAmAge arise. Moreover, other very important questions remain to be answered: to what extent is it possible to alter the ticking rate of the epigenetic clock by implementing lifestyle changes (diet, physical activity, etc.) during adulthood? Future studies are needed to address these questions.
Methods
Study populations
The Young Finns study
The YFS consisted of a series of cohorts representing the general population born in 1962, 1965, 1968, 1971, 1974 and 1977 from five cities with university hospitals in Finland (Helsinki, Kuopio, Oulu, Tampere and Turku) (Raitakari et al. 2008). A subsample of 183 individuals was randomly selected for this study from a follow-up study performed in 1986 (denoted as the YFS_1986 sample), and the same individuals were analysed again from a follow-up study performed 25 years later in 2011 (denoted as the YFS_2011 sample). The sample collections in 1986 and 2011 are described in further detail elsewhere (Akerblom et al. 1989; Nuotio et al. 2014). At baseline in 1986, the sample examined in the current study contained four age groups: 15-, 18-, 21- and 24-year-old subjects. The respective group sizes were 49, 44, 55 and 35, and the respective percentage of women was 59.2, 68.2, 56.4 and 60.0 %. All of the participants were of western European descent. This study followed the guidelines of the Declaration of Helsinki and was approved by the Ethical Review Committee of Turku University Hospital, and all participants provided informed consent.
The Vitality 90+ study
The Vitality 90+ study (V90) is an on-going prospective population-based study that includes both home-dwelling and institutionalized subjects aged 90 years or older who live in the city of Tampere, Finland. The recruitment and characterization of the subjects were performed as reported for previous V90 cohorts (Goebeler et al. 2003). In the DNA methylation analysis, the study population at baseline in 2010 (denoted as V90_2010) consisted of 122 subjects born in 1920 (women N = 89, men N = 33) and 21 young control subjects (19–28-year-old women N = 14, 20–29-year-old men N = 7). Of these subjects, 48 (control subjects N = 7 and 94-year-old surviving subjects N = 41; denoted as the V90_2014 sample) participated in a 4-year follow-up examination. In addition, 33 new control subjects (22–33-year-old men N = 9, 19–34-year-old women N = 24) were recruited together with the 4-year follow-up participants; therefore, the full sample size of the V90_2014 sample was 81. In total, 223 DNA samples from 176 subjects were used in the DNAmAge analysis. In addition, 136 subjects in the V90_2010 sample (95 % of the total sample size; young N = 21 and old N = 115) and 81 subjects in the V90_2014 sample (100 % of the total sample size; young N = 40 and old N = 41) were examined for PBMC subtype composition characterization via flow cytometry (FACS); PBMC samples from 63 subjects (51 nonagenarians and 12 young controls) were characterized using FACS at both time points, at baseline in 2010 and at follow-up in 2014. Further, of these subjects, 48 (the same as those mentioned above) were used for DNAmAge measurement at both time points. All study subjects were of Western European descent and had not suffered from any infections or received any vaccinations in the 30 days prior to blood sample collection. The study participants provided their written informed consent. This study has been conducted according to the principles expressed in the Declaration of Helsinki, and the study protocol was approved by the ethics committee of the city of Tampere (1592/403/1996; 765/13.03.01/2008, PSHP 7/2014, ETL R14002).
Sample preparation
WBL DNA of the YFS cohort at baseline and at the 25-year follow-up was obtained from blood samples stored in EDTA using a Wizard® Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions. The blood samples of the subjects in the V90 at baseline and at follow-up were collected into EDTA-containing tubes during a home visit. The samples were directly subjected to leucocyte separation on a Ficoll-Paque density gradient (Ficoll-Paque™ Premium, cat. no. 17-5442-03, GE Healthcare Bio-Sciences AB, Uppsala, Sweden). The PBMC layer was collected and suspended in 1 ml of a freezing solution (5/8 FBS, 2/8 RPMI-1640 medium, 1/8 DMSO) (FBS: cat. no. F7524, Sigma-Aldrich, St. Louis, MO, USA; RPMI: cat. no. R0883, Sigma-Aldrich, St. Louis, MO, USA; DMSO: cat. no. 1.02931.0500, VWR, Espoo, Finland) and stored in liquid nitrogen. DNA was extracted from PBMCs using the QIAamp DNA Mini Kit (Qiagen, CA, USA) according to the centrifugation protocol provided in the manufacturer’s instructions. The DNA was eluted in 60 μl of AE elution buffer and stored at −20 °C. The concentration and the quality of the DNA were assessed via the Qubit dsDNA HS Assay (Invitrogen, Eugene, OR, USA).
DNAmAge quantification
The genome-wide DNA methylation levels in WBLs from the YFS_2011 individuals and in PBMCs from the V90 participants were obtained using Illumina Infinium HumanMethylation450 BeadChips (Illumina, San Diego, CA, USA) (Bibikova et al. 2006; Bibikova et al. 2009; Bibikova et al. 2011) according to the manufacturer’s protocol at the Core Facility of the Institute of Molecular Medicine Finland (FIMM), University of Helsinki. The corresponding DNA methylation profiles of the YFS_1986 samples were measured using the same methodology at Helmholtz Zentrum, München, Germany. Samples were applied to the arrays in a randomized order. Aliquots of 1 μg of DNA were subjected to bisulphite conversion using the EZ-96 DNA Methylation Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions. A 4-μl aliquot of bisulphite-converted DNA was subjected to whole-genome amplification, followed by enzymatic fragmentation and hybridization to an Infinium HumanMethylation450 BeadChip. The BeadChips were scanned with the iScan reader (Illumina), and the measured probe intensities were transformed to β values using a standard equation in which β is the ratio of the methylated probe (m) intensities to the overall intensities (m + u + α, where α is the constant offset, 100, and u is the unmethylated probe intensity). Thus, in the heterogenic sample, the resulting β values ranged linearly from 0 (completely unmethylated, 0 %) to 1 (completely methylated, 100 %). The quality of the DNA samples and the methylation data was carefully ensured using standard examinations, such as principal component analysis (PCA), gender prediction and visualizations. The methylation intensity values and the PCA results were viewed using density plots, boxplots and dotplots. The gender prediction was based on the average levels of DNA methylation at CpG sites located in X chromosomes. Finally, the absolute methylation values (β values) of the selected probes (N = 28,587) were extracted from all data sets, and those values were used as the input for calculation of the DNAmAge (https://dnamage.genetics.ucla.edu/home) (Horvath 2013). The methylation data were inputted into the calculator in a blinded manner, i.e. without any preliminary background information concerning sample type, cAge or gender. Batch effects were normalized using the BMIQ function implemented in the DNAmAge algorithm.
FACS analysis of the V90 samples
The proportions of different leukocyte subtypes were determined with randomized sample orders using fluorescence-activated cell sorting analysis (FACS; BD FACSCanto II). In 2010, the results were analysed with BD FACSDiva, version 6.1.3 (BD Biosciences, Franklin Lakes, NJ, USA), and in 2014, the results were analysed with FlowJo software (Tree Star Inc., Ashland, OR, USA). The antibodies used were FITC-CD14 (cat. no. 11-0149), PerCP-Cy5.5-CD3 (45-0037), APC-CD28 (17-0289), PE-CD19 (12-0199) (eBioscience, San Diego, CA, USA), PE-Cy™7-CD4 (cat. no. 557852) and APC-Cy™7-CD8 (557834) (BD Biosciences). In 2010, CD19 antibody was not used. Staining was performed in phosphate-buffered saline (PBS) containing 1 % foetal bovine serum (FBS) after an incubation step with Fc Receptor Binding Inhibitor (cat. no 16-9161, eBioscience) to minimize non-specific staining of the cells.
Cell count estimates of the YFS samples
The leukocyte cell type distributions in the YFS samples were determined using R software (R>= 2.15.3) and the estimation algorithm implemented in the estimateCellCounts function of the minfi Bioconductor package (Jaffe and Irizarry 2014). This algorithm utilizes a subset of 600 control probes in the 450 BeadChip, which represent specific DNA methylation signatures of CD8+ and CD4+ T cells, monocytes, granulocytes, NK cells and B cells. The reference data used in the estimation algorithm are available in the FlowSorted.Blood.450K Bioconductor package.
Statistical analyses
Descriptive statistical analyses of the DNAmAges included calculations of medians and MADs. Between-group comparisons were conducted using non-parametric Mann-Whitney U tests (two-sided Wilcoxon’s test), and the correlation analyses were performed using non-parametric Spearman’s rank sum tests. The nominal P value threshold was set to 0.05. Analyses and visualizations were performed using R software (R>= 2.15.3) and IBM SPSS Statistics v.22 (IBM Corporation, Armonk, NY, USA).
Electronic supplementary material
Supporting data of this manuscript (DNAmAges and other fenotypes) (XLSX 90 kb)
More detailed summary information of the DNAmAges (PDF 354 kb)
Acknowledgments
The YFS has been financially supported by the Academy of Finland (grants 286284 (T.L), 132704 (M.H.), 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), 41071 (Skidi) and 250602 (Coctel to M.J.); the Social Insurance Institution of Finland; Kuopio, Tampere and Turku University Hospital Medical Funds (grant X51001 for T.L.); the Juho Vainio Foundation; the Paavo Nurmi Foundation; the Finnish Foundation of Cardiovascular Research (T.L.); the Tampere Tuberculosis Foundation (T.L., M.H., I.J.); the Emil Aaltonen Foundation (T.L., L.Ku.); the Finnish Cultural Foundation, Pirkanmaa Regional fund (N.M); and the Yrjö Jahnsson Foundation (T.L., M.H.) This work was also supported by grants from the Competitive Research Fund of Pirkanmaa Hospital District (9M017, 9N013 to M.H. and 9P002 to M.J.), Sigrid Juselius Foundation (I.J.), Finnish Medical Association (I.J.) and Competitive Research Fund of Fimlab Laboratories (X51409, I.J.). We thank Nina Peltonen, Sinikka Repo-Koskinen, Eeva Timonen, Janette Hinkka and Kristiina Lehtinen for their excellent technical assistance.
Authors’ contributions
LK processed the data, performed the analyses and was responsible for writing the manuscript. LK, SM, JJ, LKu, NM and TN were responsible for performing the experiments. MH, MK, IJ and TL provided reagents and materials. LK, SM, TL, JJ and MH contributed to the design of the study. MH, MJ, AH, OTR, MK and TL were responsible for recruiting the subjects for the study. All authors contributed to the writing of the manuscript and read and approved the final manuscript.
Abbreviations
- cAge
Calendar age
- DNAmAge
DNA methylome age
- FACS
Fluorescence-activated cell sorting analysis
- MAD
The median of the differences between DNAmAge and cAge
- PBMC
Peripheral blood mononuclear cell
- PCA
Principal component analysis
- WBL
White blood leukocyte
- YFS
Young Finns study
- V90
Vitality 90+ study
- Δ-cAge-DNAmAge
The difference between calendar age and DNA methylome age
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
References
- Akerblom HK, Viikari J, Rasanen L, Kuusela V, Uhari M, Lautala P. Cardiovascular risk in young Finns, results from the second follow-up study. Ann Med. 1989;21:223–225. doi: 10.3109/07853898909149938. [DOI] [PubMed] [Google Scholar]
- Arnold CR, Wolf J, Brunner S, Herndler-Brandstetter D, Grubeck-Loebenstein B. Gain and loss of T cell subsets in old age–age-related reshaping of the T cell repertoire. J Clin Immunol. 2011;31:137–146. doi: 10.1007/s10875-010-9499-x. [DOI] [PubMed] [Google Scholar]
- Benetos A, Okuda K, Lajemi M, Kimura M, Thomas F, Skurnick J, Labat C, Bean K, Aviv A. Telomere length as an indicator of biological aging: the gender effect and relation with pulse pressure and pulse wave velocity. Hypertension. 2001;37:381–385. doi: 10.1161/01.HYP.37.2.381. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, Gunderson KL. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocklandt S, Lin W, Sehl ME, Sanchez FJ, Sinsheimer JS, Horvath S, Vilain E. Epigenetic predictor of age. PLoS One. 2011;6:e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boks MP, van Mierlo HC, Rutten BP, Radstake TR, De Witte L, Geuze E, Horvath S, Schalkwyk LC, Vinkers CH, Broen JC, et al. Longitudinal changes of telomere length and epigenetic age related to traumatic stress and post-traumatic stress disorder. Psychoneuroendocrinology. 2015;51:506–512. doi: 10.1016/j.psyneuen.2014.07.011. [DOI] [PubMed] [Google Scholar]
- Broux B, Markovic-Plese S, Stinissen P, Hellings N. Pathogenic features of CD4+CD28- T cells in immune disorders. Trends Mol Med. 2012;18:446–453. doi: 10.1016/j.molmed.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Florath I, Butterbach K, Muller H, Bewerunge-Hudler M, Brenner H. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet. 2014;23:1186–1201. doi: 10.1093/hmg/ddt531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garagnani P, Bacalini MG, Pirazzini C, Gori D, Giuliani C, Mari D, Di Blasio AM, Gentilini D, Vitale G, Collino S, et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell. 2012;11:1132–1134. doi: 10.1111/acel.12005. [DOI] [PubMed] [Google Scholar]
- Goebeler S, Jylha M, Hervonen A. Medical history, cognitive status and mobility at the age of 90. A population-based study in Tampere, Finland. Aging Clin Exp Res. 2003;15:154–161. doi: 10.1007/BF03324494. [DOI] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan J, Gao Y, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Levine AJ. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis. 2015;212:1563–1573. doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schonfels W, Ahrens M, Heits N, Bell JT, Tsai PC, Spector TD, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci U S A. 2014;111:15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Garagnani P, Bacalini MG, Pirazzini C, Salvioli S, Gentilini D, Di Blasio AM, Giuliani C, Tung S, Vinters HV, et al. Accelerated epigenetic aging in Down syndrome. Aging Cell. 2015;14:491–495. doi: 10.1111/acel.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Mah V, Lu AT, Woo JS, Choi OW, Jasinska AJ, Riancho JA, Tung S, Coles NS, Braun J, et al. The cerebellum ages slowly according to the epigenetic clock. Aging (Albany NY) 2015;7:294–306. doi: 10.18632/aging.100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kananen L, Marttila S, Nevalainen T, Jylhava J, Mononen N, Kahonen M, Raitakari OT, Lehtimaki T, Hurme M. Aging-associated DNA methylation changes in middle-aged individuals: the Young Finns study. BMC Genomics. 2016;17:103. doi: 10.1186/s12864-016-2421-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LL, Emberly E, Fraser HB, Neumann SM, Chen E, Miller GE, Kobor MS. Factors underlying variable DNA methylation in a human community cohort. Proc Natl Acad Sci U S A. 2012;109(Suppl 2):17253–17260. doi: 10.1073/pnas.1121249109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, Gibson J, Redmond P, Cox SR, Pattie A, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015;44:1388–1396. doi: 10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttila S, Kananen L, Hayrynen S, Jylhava J, Nevalainen T, Hervonen A, Jylha M, Nykter M, Hurme M (2015) Ageing-associated changes in the human DNA methylome: genomic locations and effects on gene expression. BMC Genomics 16. doi:10.1186/s12864-015-1381-z [DOI] [PMC free article] [PubMed]
- Mather KA, Jorm AF, Parslow RA, Christensen H. Is telomere length a biomarker of aging? A review. J Gerontol A Biol Sci Med Sci. 2011;66:202–213. doi: 10.1093/gerona/glq180. [DOI] [PubMed] [Google Scholar]
- Nuotio J, Oikonen M, Magnussen CG, Jokinen E, Laitinen T, Hutri-Kahonen N, Kahonen M, Lehtimaki T, Taittonen L, Tossavainen P, et al. Cardiovascular risk factors in 2011 and secular trends since 2007: the Cardiovascular Risk in Young Finns Study. Scand J Public Health. 2014;42:563–571. doi: 10.1177/1403494814541597. [DOI] [PubMed] [Google Scholar]
- Pathai S, Bajillan H, Landay AL, High KP. Is HIV a model of accelerated or accentuated aging? J Gerontol Ser A Biol Med Sci. 2014;69:833–842. doi: 10.1093/gerona/glt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelec G, Larbi A, Derhovanessian E. Senescence of the human immune system. J Comp Pathol. 2010;142(Suppl 1):S39–S44. doi: 10.1016/j.jcpa.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Raitakari OT, Juonala M, Ronnemaa T, Keltikangas-Jarvinen L, Rasanen L, Pietikainen M, Hutri-Kahonen N, Taittonen L, Jokinen E, Marniemi J, et al. Cohort profile: the cardiovascular risk in Young Finns Study. Int J Epidemiol. 2008;37:1220–1226. doi: 10.1093/ije/dym225. [DOI] [PubMed] [Google Scholar]
- Simpkin AJ, Hemani G, Suderman M, Gaunt TR, Lyttleton O, Mcardle WL, Ring SM, Sharp GC, Tilling K, Horvath S, et al. Prenatal and early life influences on epigenetic age in children: a study of mother-offspring pairs from two cohort studies. Hum Mol Genet. 2016;25:191–201. doi: 10.1093/hmg/ddv456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegenga WT, Boekschoten MV, Lute C, Hooiveld GJ, de Groot PJ, Morris TJ, Teschendorff AE, Butcher LM, Beck S, Muller M. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age (Dordr) 2014;36:9648. doi: 10.1007/s11357-014-9648-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner CI, Lin Q, Koch CM, Eisele L, Beier F, Ziegler P, Bauerschlag DO, Jockel KH, Erbel R, Muhleisen TW, et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014;15:R24. doi: 10.1186/gb-2014-15-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Weinberger B, Grubeck-Loebenstein B. The aging of the immune system. Transpl Int. 2009;22:1041–1050. doi: 10.1111/j.1432-2277.2009.00927.x. [DOI] [PubMed] [Google Scholar]
- Zampieri M, Ciccarone F, Calabrese R, Franceschi C, Burkle A, Caiafa P. Reconfiguration of DNA methylation in aging. Mech Ageing Dev. 2015;151:60–70. doi: 10.1016/j.mad.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Zilbauer M, Rayner TF, Clark C, Coffey AJ, Joyce CJ, Palta P, Palotie A, Lyons PA, Smith KG. Genome-wide methylation analyses of primary human leukocyte subsets identifies functionally important cell-type-specific hypomethylated regions. Blood. 2013;122:e52–e60. doi: 10.1182/blood-2013-05-503201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting data of this manuscript (DNAmAges and other fenotypes) (XLSX 90 kb)
More detailed summary information of the DNAmAges (PDF 354 kb)