Abstract
Inositol hexakisphosphate kinase 3 (IP6K3) generates inositol pyrophosphates, which regulate diverse cellular functions. However, little is known about its own physiological role. Here, we show the roles of IP6K3 in metabolic regulation. We detected high levels of both mouse and human IP6K3 mRNA in myotubes and muscle tissues. In human myotubes, IP6K3 was upregulated by dexamethasone treatment, which is known to inhibit glucose metabolism. Furthermore, Ip6k3 expression was elevated under diabetic, fasting, and disuse conditions in mouse skeletal muscles. Ip6k3−/− mice demonstrated lower blood glucose, reduced circulating insulin, deceased fat mass, lower body weight, increased plasma lactate, enhanced glucose tolerance, lower glucose during an insulin tolerance test, and reduced muscle Pdk4 expression under normal diet conditions. Notably, Ip6k3 deletion extended animal lifespan with concomitant reduced phosphorylation of S6 ribosomal protein in the heart. In contrast, Ip6k3−/− mice showed unchanged skeletal muscle mass and no resistance to the effects of high fat diet. The current observations suggest novel roles of IP6K3 in cellular regulation, which impact metabolic control and lifespan.
Inositol hexakisphosphate kinases (IP6Ks) are involved in diverse cellular signalling1,2, by generating inositol pyrophosphates and interacting with other cellular components. Inositol pyrophosphates are highly energetic molecules, and the phosphorylation of their already phosphorylated inositol hydroxyl group is catalysed by two different classes of enzymes: IP6Ks with 5-kinase activities3,4,5 and diphosphoinositol pentakisphosphate kinases (PPIP5Ks) with 1-kinase activity6,7,8. The IP6K 5-kinase activity pyrophosphorylates myo-inositol (1,3,4,5,6)-pentakisphosphate (InsP5) to 5-PP-InsP4, InsP6 to 5-PP-InsP5 (5-InsP7)9, and 1-PP-InsP5 to 1,5-PP2-InsP4 (InsP8), respectively. Among these, 5-InsP7 is possibly the most studied inositol pyrophosphate generated by IP6Ks, and shows diverse biological effects by target-protein binding and pyrophosphorylation, in which the energetic pyrophosphate group of 5-InsP7 donates its β-phosphate to a pre-phosphorylated serine residue of the target protein10. Although an additional naturally-occurring isomer of 5PPIP5K-generated InsP7 exists wherein a diphosphate group is placed at the 1-carbon (1-InsP7), most mammalian cellular InsP7 appear to be comprised of the 5-isomer (5-InsP7)3,11.
Mammals have three subtypes of IP6K: IP6K1–31. Recent studies using knockout mice have revealed the physiological importance of the first two IP6Ks in regulating diverse cellular functions. Ip6k1 gene deletion in mice resulted in growth retardation, reduced levels of circulating insulin from pancreatic beta-cells, and male sterility secondary to defective spermatogenesis12. In addition, knockout studies of Ip6k1 demonstrated its role in regulating platelet polyphosphate levels13 and neutrophil function in innate immunity14. A recent study also showed enhanced Akt signalling in the liver, fat, and muscle, thereby inducing increased insulin sensitivity wherein IP6K1-generated 5-InsP7 bound to the Akt-PH domain and inhibited insulin and insulin-like growth factor 1 (IGF1)-induced phosphatidylinositol (3,4,5)-trisphosphate signalling15. In contrast, Ip6k2 knockout mice did not exhibit growth, fertility, or insulin secretion defects but were resistant to ionizing radiation and their fibroblasts manifested up-regulated DNA repair as well as interferon-β resistance16. These mice were also more susceptible to squamous cell carcinoma in the oral cavity and oesophagus compared to wild-type mice when given water containing a carcinogen. Notably, IP6K2, via 5-InsP7 synthesis, represents a major mediator of cancer cell migration and tumour metastasis in cell culture and in intact mice17. These observations demonstrated that although both generate the same form of 5-InsP7, the functional redundancy between IP6K1 and IP6K2 is incomplete.
IP6K3 is less well characterized compared to IP6K1 and IP6K218. Northern blotting has shown abundant Ip6k3 mRNA in the rat cerebellum, and a GFP-IP6K3 fusion protein localized to both the cytoplasm and the nucleus when tested in HEK293 cells18. A study focusing on brain function demonstrated that IP6K3 physiologically regulates the morphology and synapse formation of cerebellar Purkinje cells through interacting with other proteins in the mouse brain19. Little is known, however, regarding the physiological role of IP6K3 in the whole body. Considering the known importance of the other two IP6K isoforms, we performed in vitro and in vivo analyses to determine the organismal role of IP6K3. The present study is the first to describe the role of IP6K3 in muscles. Here we show that Ip6k3 mRNA is highly expressed and responsive to energy and disease status in muscles and that Ip6k3 gene deletion induces metabolic changes and affects lifespan. We suggest that IP6K3 likely senses the energy status in this tissue and regulates downstream signalling that impacts metabolic control and lifespan in mice.
Results
Mouse and human IP6K3 mRNAs are highly expressed in skeletal muscles and Ip6k3 is induced by myotube formation in C2C12 cells
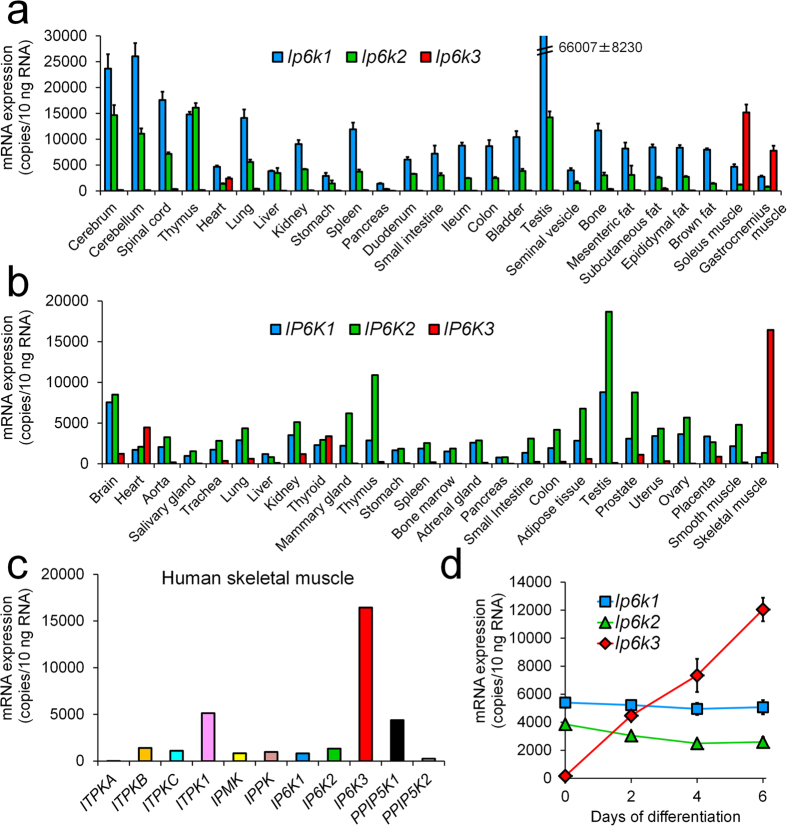
We first examined Ip6k3 mRNA tissue distribution in mice. Absolute mRNA level quantification revealed that Ip6k3 was highly expressed in murine skeletal muscles (the soleus and gastrocnemius muscles) followed by the heart (Fig. 1a). Ip6k3 was also detectable in the brain and other tissues at much lower levels. Similar results were obtained in human tissues (Fig. 1b), which also revealed the thyroid as an IP6K3-expressing site. Among the inositol kinases, IP6K3 showed the highest mRNA expression in human skeletal muscles (Fig. 1c). As IP6K3 reportedly functions in the brain19, the present study also characterized IP6K3 brain expression, revealing brain-region dependent IP6K3 expression in humans (Fig. S1). In contrast, the reported lack of Ip6k3 expression in mouse insulinoma MIN6 cells20 was reconfirmed in our study. In addition, neither the rat beta cell line INS-1 832/13 nor human islets, which contain non-beta endocrine cells, expressed IP6K3 (Fig. S2).
Figure 1. Mouse Ip6k3 and human IP6K3 mRNA are highly expressed in skeletal muscles, and Ip6k3 is induced by myotube formation in C2C12 cells.
Tissue distribution of each mRNA in C57BL/6J mice (a) and humans (b). Expression of inositol phosphate kinase family genes in human skeletal muscle tissues (c). Expression profiles of each mRNA during C2C12 myotube formation (d). Mouse Ip6k3 and human IP6K3 were highly expressed in skeletal muscles. Among the inositol kinase family members tested, IP6K3 showed the highest mRNA expression in human skeletal muscle. Ip6k3 mRNA was induced during myotube formation in C2C12 cells. Means ± SD of four mice consisting of two technical replicates for (a), means of two technical replicates for human pooled samples in (b,c), and means ± SD of three independent samples consisting of two technical replicates for (d). ITPKA-C, inositol-trisphosphate 3-kinase A-C; ITPK1, inositol-tetrakisphosphate 1-kinase; IPMK, inositol polyphosphate multikinase; IPPK, inositol-pentakisphosphate 2-kinase; PPIP5K1&2, diphosphoinositol pentakisphosphate kinase 1&2.
We next explored the Ip6k3 profiles in skeletal muscle cells in vitro. Quantitative PCR analysis of mouse C2C12 cells demonstrated that Ip6k3 mRNA in unfused myoblasts was almost undetectable whereas Ip6k1 and Ip6k2 were measurable and expressed equally. However, during myotube formation, Ip6k3 gene expression was robustly induced whereas Ip6k1 and Ip6k2 expression remained unchanged (Fig. 1d). Furthermore, when an AcGFP-human IP6K3 fusion protein construct was transfected into mouse C2C12 cells, the resultant AcGFP-IP6K3 was localized to both the cytoplasm and the nucleus (Fig. S3). Similar images were obtained from the human SJCRH30 rhabdomyosarcoma cell line, which expresses detectable IP6K3 mRNA baseline levels (Figs S3 and S4).
Ip6k3 mRNA expression is induced in diabetic, fasting, and disuse conditions in mouse skeletal muscles
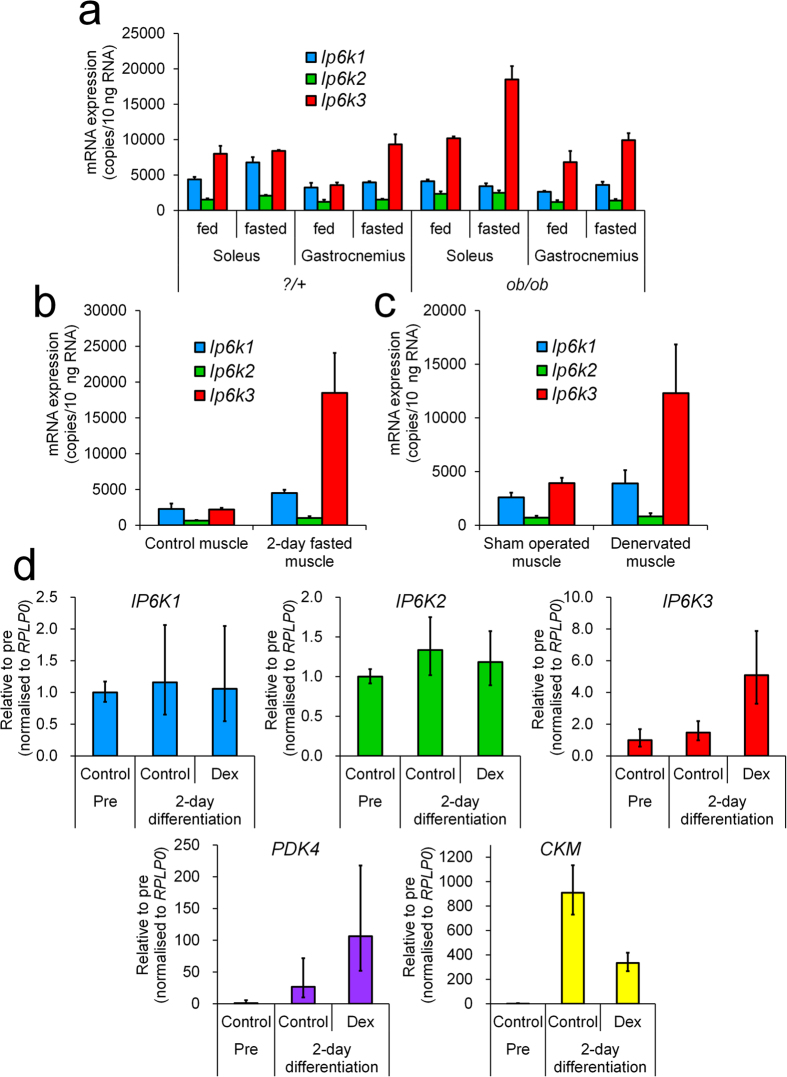
We investigated Ip6k3 expression changes in food and muscle-related disease conditions such as diabetes and muscle-disuse atrophy. In normal mice, fasting conditions increased Ip6k3 expression by 1.1- and 2.6-fold in the soleus and gastrocnemius muscles, respectively (Fig. 2a). In addition, Ip6k3 expression was elevated in the soleus and gastrocnemius muscles of non-fasting diabetic ob/ob mice by 1.3- and 1.9-fold, respectively, compared to normal mice (Fig. 2a); fasting further elevated this Ip6k3 expression by 2.2- and 1.1-fold (Fig. 2a). A longer 2-day fasting period increased Ip6k3 expression by 8.4-fold in the gastrocnemius muscle in C57BL/6J mice compared to the non-fasting condition (Fig. 2b). In addition, denervated gastrocnemius muscle (2 days after operation), which experiences impaired glucose metabolism and muscle atrophy, showed 3.1-fold increased Ip6k3 expression (Fig. 2c).
Figure 2. Ip6k3 mRNA is induced in diabetic, fasting, and disused conditions in mice, and IP6K3 mRNA is elevated by dexamethasone in human primary myotubes.
Ip6k1, Ip6k2, and Ip6k3 mRNA expression in the skeletal muscle from ?/+ littermates and ob/ob mice (a), in the gastrocnemius muscle of 2 days-fasted C57BL/6J mice (b), and in the denervated gastrocnemius muscle in C57BL/6J mice (c). Human primary skeletal myoblast cells were treated with 2% horse serum containing medium with or without dexamethasone and the respective mRNA levels were measured (d). Ip6k3 expression was induced by fasting and altered in the skeletal muscles of diabetic ob/ob mice. In addition, elevation of Ip6k3 mRNA was observed in 2 day-fasted gastrocnemius muscles and denervated gastrocnemius muscles at 2 days after operation. When human primary myotubes were treated with dexamethasone, which inhibits glucose metabolism, IP6K3 mRNA expression was elevated. Means ± SD of three ?/+ and ob/ob mice consisting of two technical replicates for (a), means ± SD of three mice consisting of two technical replicates for (b), means ± SD of six sham-operated and denervated mice consisting of two technical replicates for (c), means and range of three independent samples consisting of two technical replicates for (d). For (d), the comparative CT method was used and the ranges given were determined by evaluating the expression: 2−ΔΔCT with ΔΔCT + s and ΔΔCT − s, where s = the standard deviation of the ΔΔCT value. Dex, dexamethasone.
IP6K3 mRNA is induced in dexamethasone-treated human primary myotubes
We tested the effect of dexamethasone, a synthetic glucocorticoid that reportedly inhibits glucose metabolism in skeletal muscles21, in human primary myotubes. Myotube formation was observed following two days culture with medium containing 2% horse serum. Conversely, dexamethasone treatment resulted in inhibited myotube formation, attenuated CKM expression, and a 4.0-fold increase in pyruvate dehydrogenase kinase 4 (PDK4) mRNA expression, which inhibits glucose utilization and facilitates fat oxidation22. As observed with PDK4, IP6K3 expression was elevated 3.5-fold by dexamethasone treatment, whereas IP6K1 and IP6K2 expression was unchanged (Fig. 2d).
General observations of Ip6k3 −/− mice
As described in the Methods section, B6;129-Ip6k3−/− mice, B6.Cg-Ip6k3−/− mice, and their respective wild-type littermates were used in the present study. A test of five mating pairs (male Ip6k3−/− and female Ip6k3−/− mice, B6;129-Ip6k3−/− mice) resulted in 100% pregnancy rate and five females gave birth to 28 pups (14 males and 14 females), demonstrating normal fertility.
Food consumption, body composition, and blood parameters of Ip6k3 −/− mice
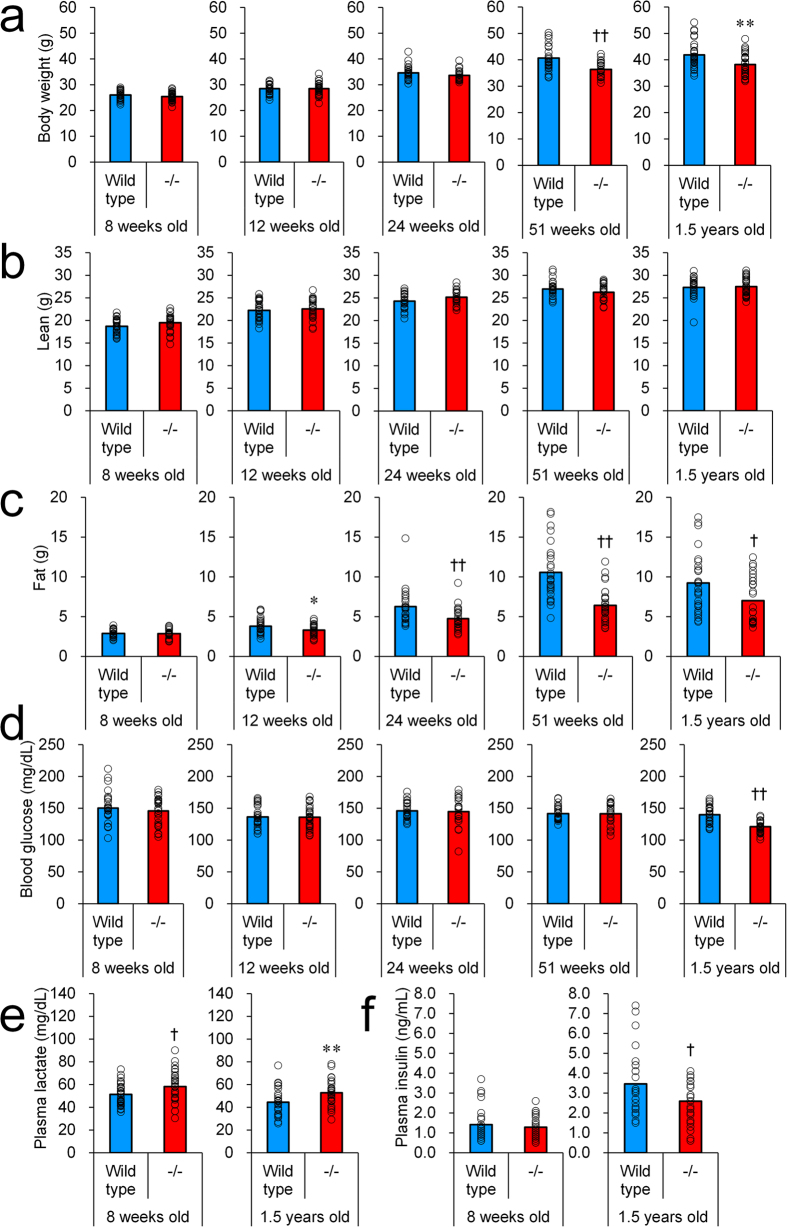
B6.Cg-Ip6k3−/− mice and their wild-type littermates were utilized in the present analysis. Wild-type and Ip6k3−/− mice showed similar levels of food intake (Table 1). Although body weight was unchanged in Ip6k3−/− compared to wild-type mice at 8 to 24 weeks of age, those of Ip6k3−/− mice were lower from 51 weeks to 1.5 years of age (9–11%) (Fig. 3a). Body composition of Ip6k3−/− mice determined using Echo-MRI (Fig. 3b,c) revealed unchanged lean mass and decreased fat mass (−13 to −39%) from 12 weeks to 1.5 years of age. Ip6k3−/− mice showed decreased levels of non-fasting blood glucose from 1.5 years of age (Fig. 3d). In contrast, plasma lactate levels were increased at 8 weeks and 1.5 years of age by 1.1–1.2-fold (Fig. 3e). Plasma insulin levels were unchanged at 8 weeks of age, whereas they were decreased by 25% in Ip6k3−/− mice at 1.5 years of age (Fig. 3f).
Table 1. Average food intake in Ip6k3−/− mice.
| 8 weeks old | 12 weeks old | 24 weeks old | 51 weeks old | 1.5 years old | |
|---|---|---|---|---|---|
| Wild-type | 3.5 ± 0.1 | 3.5 ± 0.2 | 3.6 ± 0.3 | 3.5 ± 0.2 | 3.4 ± 0.2 |
| −/− | 3.4 ± 0.1 | 3.6 ± 0.3 | 3.7 ± 0.1 | 3.5 ± 0.2 | 3.4 ± 0.2 |
B6.Cg-Ip6k3−/− and wild-type littermates were used for the analysis. Average daily food consumption per mouse is shown. The means ± SD are given. Cage number = 8 (total 24–30 mice).
Figure 3. Lower body weight, reduced fat mass, decreased blood glucose, higher plasma lactate, and reduced insulin in Ip6k3−/− mice.
Body weight (a), lean mass (b), fat mass (c), blood glucose (d), plasma lactate (e), and plasma insulin (f) of Ip6k3−/− mice and their wild-type littermates at the indicated ages. The observations became evident with ageing. B6.Cg-Ip6k3−/− and wild-type littermates were used for the analysis. The mean values and individual values are shown. *P < 0.05 and **P < 0.01 by the Student’s t-test, and †P < 0.05 and ††P < 0.01 by the Aspin-Welch test. n = 30 for 8 and 12 weeks old wild-type and Ip6k3−/− mice, n = 24 for 24 weeks old wild-type and Ip6k3−/− mice, n = 27 and 28 for 51 weeks old wild-type and Ip6k3−/− mice, n = 28 and 29 for 1.5 years old wild-type and Ip6k3−/− mice.
Lower glucose levels upon glucose and insulin tolerance testing in Ip6k3 −/− mice
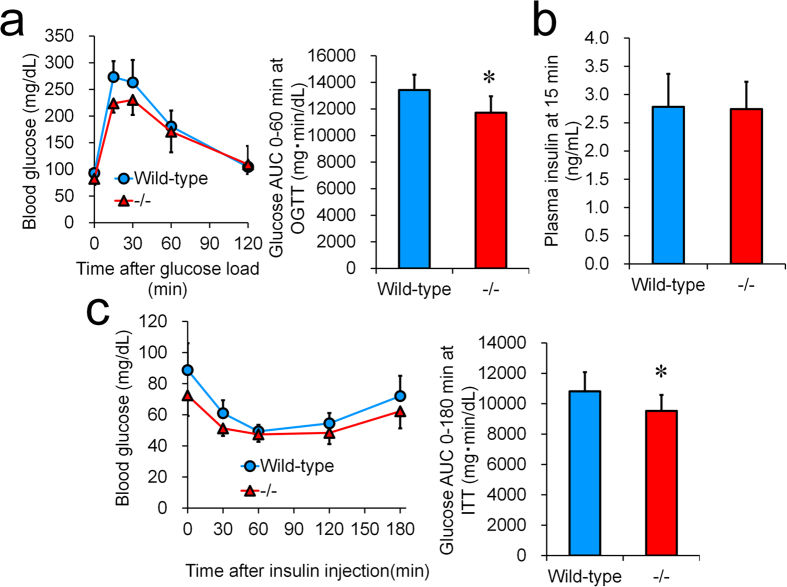
Glucose tolerance was investigated by applying an oral glucose tolerance test in B6;129-Ip6k3−/− mice and their wild-type littermates (Fig. 4a). After oral gavage of glucose (2 g/kg), Ip6k3−/− mice showed significant improvements in glucose tolerance (Fig. 4a). Plasma insulin levels at 15 min after an oral glucose load were unchanged (Fig. 4b). As observed for the oral glucose tolerance test, B6;129-Ip6k3−/− mice showed lower glucose levels compared to their wild-type littermates during an insulin tolerance test (Fig. 4c).
Figure 4. Lower glucose levels in glucose and insulin tolerance testing of Ip6k3−/− mice.
An oral glucose tolerance test was performed in 15 weeks old mice (a). Plasma insulin levels at 15 min after glucose loading are shown (b). Insulin (0.75 U/kg) was injected to 12 weeks old mice (c). B6;129-Ip6k3−/− and wild-type littermates were used for the analysis. Values represent the means ± SD. n = 8. *P < 0.05 by the Student’s t-test.
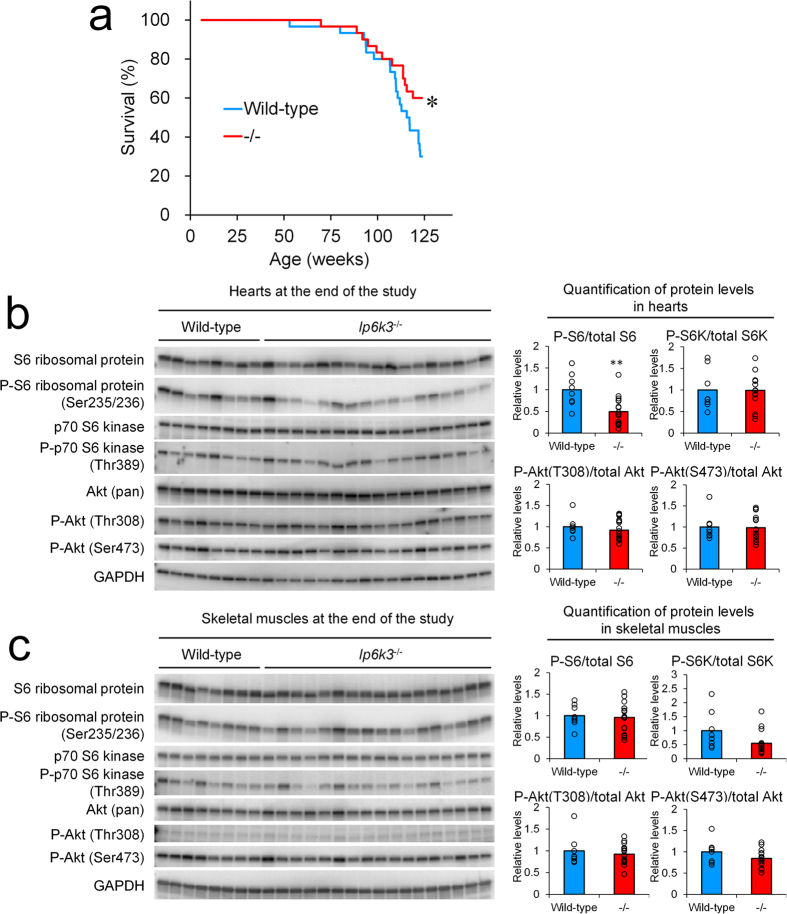
Prolonged lifespan of Ip6k3 −/− mice
Considering the roles of IP6Ks in regulating diverse cell signalling such as for metabolism control, we were interested to explore the possible association of IP6K3 with lifespan. As shown in Fig. 5a, the lifespan of Ip6k3−/− mice (B6.Cg-Ip6k3−/−) was significantly extended compared to wild-type littermates. The observed causes of death were general factors such as lymphoma and liver tumour, and we observed no differences in tumour formation in skeletal muscles or elsewhere in the Ip6k3−/− mice. As Ip6k3 is not expressed throughout the body but shows relatively restricted expression in the muscles, it was further of interest to explore its role in molecular signalling. We observed decreased phospho-S6 ribosomal protein in the heart of Ip6k3−/− mice at the end of the study period independent of the phosphorylation status of the upstream molecules S6 kinase and AKT23 (Fig. 5b). In contrast, decreased phospho-S6 ribosomal protein was not observed in the skeletal muscles (Fig. 5c). Although the phosphorylation status of S6K and AKT of skeletal muscles in Ip6k3−/− mice tended to be reduced, these changes did not reach statistical significance.
Figure 5. Extended lifespan of Ip6k3−/− mice.
Kaplan-Meier survival curves show a significant (log-rank χ2 = 4.2587 *P < 0.05) lifespan extension in Ip6k3−/− mice ((a) n = 30 for each group). Western blot analysis of signalling proteins and quantitation of proteins in the hearts and the skeletal muscles at the end of the lifespan study (b,c). Decreased phospho-S6 ribosomal protein was observed in the heart. B6.Cg-Ip6k3−/− and wild-type littermates were used for the analysis. The mean values and individual values are shown. **P < 0.01 by the Student’s t-test
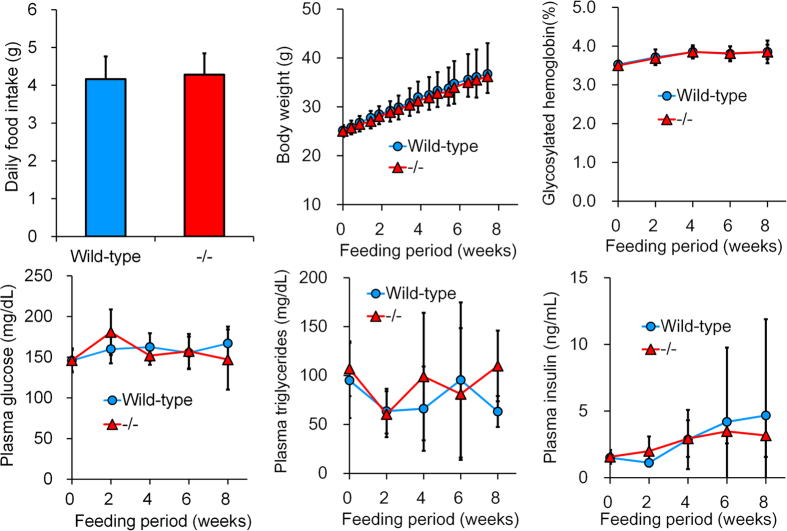
Similar levels of blood parameters and body weight in high fat diet fed Ip6k3 −/− compared to wild-type mice
The impact of Ip6k3 deletion was investigated under a high fat diet condition (B6;129-Ip6k3−/− mice). However, we observed no changes in food intake, body weight, plasma glucose, triglycerides, or insulin levels in Ip6k3−/− mice compared to wild-type littermates during the 8 weeks in which mice were fed the high fat D12451 diet (Fig. 6).
Figure 6. Ip6k3−/− mice show no resistance to high-fat diet feeding.
Ip6k3−/− mice were fed a high fat diet (45% kcal as fat) from 7 to 15 weeks old. Ip6k3−/− mice showed similar levels of the indicated parameters compared to wild-type littermates during the 8-week high fat diet feeding period. B6;129-Ip6k3−/− and wild-type littermates were used for the analysis. Values represent the means ± SD. n = 8.
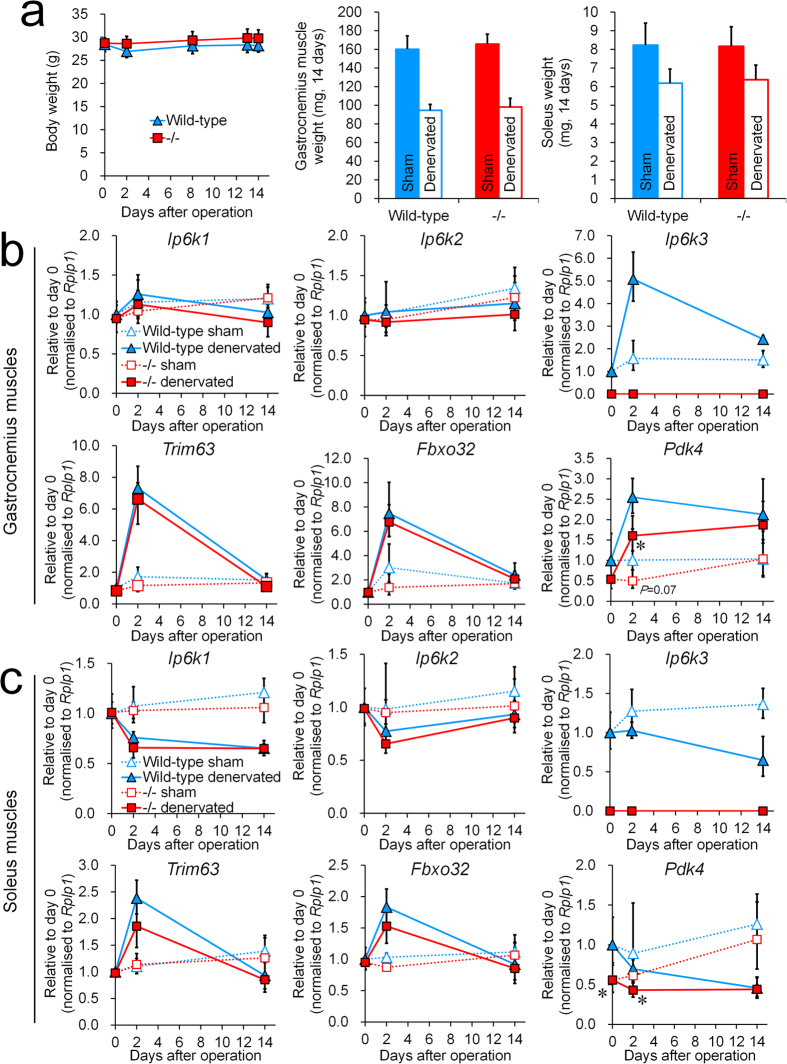
No impact on basal skeletal muscle mass and denervation-induced skeletal muscle atrophy but reduced levels of Pdk4 in Ip6k3 −/− mice
The impact of Ip6k3 deletion on basal skeletal muscle mass and muscle denervation was investigated using B6;129-Ip6k3−/− mice and their wild-type littermates. Body weight was unchanged in Ip6k3−/− mice during the study (Fig. 7a); the basal gastrocnemius muscle and soleus weights were unchanged as well (sham-operated muscles). After 14 days following the denervation operation, muscle weight was decreased by 41% and 22% compared to that following sham-operation in the gastrocnemius muscles and soleus in wild-type mice (Fig. 7a). Under this condition, similar decreases were observed in Ip6k3−/− mice as well (Fig. 7a). However, mRNA levels quantification showed that Ip6k3 mRNA was temporally induced at 2 days after the operation and then decreased in the gastrocnemius muscles but not in the soleus (Fig. 7b,c). Trim63 and Fbxo32, E3 ligases that ubiquitinate and degrade target proteins24, were transiently induced in both muscle types, and Ip6k3 deletion had no impact on their mRNA expression. In addition to facilitating muscle degradation, muscle denervation is known to induce metabolic disturbances in a fibre-type dependent manner25. Pdk4, which switches tissue from glucose catabolism to fatty acid utilization, was induced in denervated gastrocnemius muscles but not in the soleus (Fig. 7b,c), indicating separate conditions of metabolic disturbance between these muscles. Notably, we detected a down-regulation of Pdk4 mRNA levels in both muscles in Ip6k3−/− mice (Fig. 7b,c).
Figure 7. Ip6k3−/− mice have no impact on basal skeletal muscle mass and denervation-induced skeletal muscle atrophy but exhibit reduced levels of Pdk4.
Body, gastrocnemius muscle, and soleus weights (a), and gene expression in the gastrocnemius muscles (b) and soleus (c) after the denervation operation. Ip6k3 deletion had no impact on basal muscle mass and denervation-induced muscle loss, which was measured 14 days following the operation. Pdk4 was down-regulated in the muscles of Ip6k3−/− mice. B6;129-Ip6k3−/− and wild-type littermates were used for the analysis. *P < 0.05 by the Student’s t-test. Means ± SD of 6 mice for (a). Means and range are shown for (b,c). n = 4 and 6 for wild-type and Ip6k3−/− mice at day 0, n = 4 and 5 for wild-type and Ip6k3−/− mice at day 2, n = 6 for wild-type and Ip6k3−/− mice at day 14. For calculating gene expression in (b,c), the comparative CT method was used and the ranges given were determined by evaluating the expression: 2−ΔΔCT with ΔΔCT + s and ΔΔCT − s, where s = the standard deviation of the ΔΔCT value. Two technical replicates were performed per sample.
Discussion
There has been little research performed to elucidate the function of IP6K3 relative to IP6K1 and IP6K2. Here, we first confirmed that Ip6k3 was highly expressed in muscles. Muscle Ip6k3 expression was induced by diabetes, fasting, and disuse muscle conditions in mice. In addition, human primary myotubes treated with dexamethasone, which inhibits glucose metabolism, showed IP6K3 induction. Ip6k3−/− mice exhibited reduced glucose, decreased insulin, reduced fat mass, lower body weight, increased lactate, enhanced glucose tolerance, lower glucose during insulin tolerance testing, and longer lifespan compared to wild-type controls. However, in contrast to the changes in metabolism, no impact was observed on basal skeletal muscle mass and the skeletal muscle atrophy induced by denervation. Thus, the present study is the first to demonstrate novel roles of IP6K3 in regulating metabolism and lifespan.
Ip6k3 expression has been reported to be abundant in the rat brain18. Our detailed gene expression analysis revealed that mouse and human IP6K3 are comparably highly expressed in skeletal muscles, followed by the heart. Notably, IP6K3 levels were higher than those of any other inositol phosphate kinase in human skeletal muscles. In addition, Ip6k3 expression, which was undetected in C2C12 myoblasts, robustly increased during myotube differentiation. In contrast, generally available non-muscle cell lines such as HeLa and HEK293 cells did not express IP6K3 (Fig. S5). These expression profiles indicate that IP6K3 likely has a pivotal role in mature muscles. When tested under fasting conditions, muscle Ip6k3 was induced and higher Ip6k3 levels were observed in muscles obtained from diabetic ob/ob mice. In addition, denervation-induced disused muscles, which exhibit metabolic disturbance, also showed rapid Ip6k3 expression induction. Furthermore, dexamethasone inhibition of glucose metabolism also elevated IP6K3 mRNA in human myotubes. These data indicate that Ip6k3 levels might be controlled by energy and disease status in skeletal muscles.
Ip6k1−/− and Ip6k2−/− mice reportedly exhibit normal blood glucose levels under regular diet conditions12,16. In the present study, Ip6k3 gene deletion revealed its role in controlling glucose, insulin, fat mass, body weight, and lactate levels in mice. Enhanced glucose tolerance indicates a higher level of glucose disposal in the Ip6k3−/− mice. Furthermore, during an insulin tolerance test, glucose levels were lower in Ip6k3−/− mice compared to wild-type littermates. Considering the selective and elevated expression of Ip6k3 in muscle, this tissue is likely to represent the primary tissue responsible for the observed changes in the Ip6k3−/− mice. Taking into account that the glycolysis pathway is activated in Ip6k1−/− embryonic fibroblasts and in yeast and fungi mutated for KCS1, the IP6K ortholog26,27, Ip6k3 deletion might have resulted in increased glycolytic activity thereby increasing glucose uptake in muscles. Thus, the increased plasma lactate levels observed in Ip6k3−/− mice might be related to increased glycolytic activity. Notably, in the current study Ip6k3−/− mice showed no resistance to fat gain induced by 8 weeks of feeding with a high fat diet, in contrast to a previous IP6K1 study15. Ip6k1 is expressed in fat tissue and Chakraborty et al. revealed that the 5-InsP7 generated by IP6K1 interferes with Akt signalling, leading to insulin resistance and weight gain15. In addition, Szijgyarto et al. observed that yeasts devoid of KCS1 exhibit dysfunctional mitochondria, which are required for fatty acid synthesis27. These findings indicate that the Ip6k1 expressed in fat might have a direct role in fat accumulation via mitochondrial function. Thus, the absence of Ip6k3 expression in fat tissue might underlie the lack of impact observed on fat accumulation in Ip6k3−/− mice under a high fat diet condition. Furthermore, the decreased fat mass observed in Ip6k3−/− mice under regular diet conditions is likely to be secondary to the improved metabolic parameters.
The Ip6k3−/− mice exhibited prolonged lifespan under regular diet conditions, an intriguing effect as Ip6k3 is not expressed throughout the body but shows relatively restricted expression in the muscles. Furthermore, we observed decreased phospho-S6 ribosomal protein in the heart but not in the skeletal muscles of Ip6k3−/− mice at the end of the study period independent of the phosphorylation status of the upstream molecules S6 kinase and AKT, indicating the tissue dependent role of IP6K3. Recently, molecules affecting protein synthesis such as mTOR and S6K1 have been shown to regulate lifespan in mice28,29, and inhibiting protein synthesis pathways in mice has been demonstrated to prolong lifespan. In addition, a yeast KCS1 mutant was demonstrated to exhibit reduced protein synthesis, which is regulated by KCS1-produced inositol pyrophosphate30. Considering that S6 ribosomal protein phosphorylation physiologically regulates global protein synthesis31, this pathway might be related to the extended lifespan of Ip6k3−/− mice. Furthermore, Ip6k3 deletion may directly affect cardiac metabolism through reducing heart Pdk4 expression (Fig. S6); these hypotheses require further investigation.
A pathway governing protein degradation, mediated by the ubiquitin proteasome system, is activated in atrophying muscle through activation of the E3 ubiquitin ligases Trim63 and Fbxo3232,33. In the present study, hind limb denervation resulted in muscle mass reduction in the gastrocnemius and soleus muscles, wherein Trim63 and Fbxo32 were upregulated. Although Ip6k3 expression was induced in the gastrocnemius muscles, Ip6k3 deletion showed no impact on denervation-induced muscle atrophy. Considering that no change was observed in basal muscle weight, Ip6k3 is unlikely to control muscle mass and demonstrated no significant connection with the ubiquitin proteasome system, at least under the current tested conditions. In comparison, a previous study reported increased S6 ribosomal protein phosphorylation and tissue mass in gastrocnemius muscles in Ip6k1−/− mice under basal conditions15, which differs from the observations in Ip6k3−/− mice that showed unchanged S6 ribosomal protein phosphorylation and tissue mass in skeletal muscles. Considering, however, that reduced S6 ribosomal protein phosphorylation was observed in the heart of Ip6k3−/− mice, IP6K1 and IP6K3 might have independent roles in regulating S6 signalling. The development of selective inhibitors of IP6Ks that are effective in vivo would likely reveal the role of 5-InsP7 in S6 signalling and muscle mass regulation.
The switch from glucose catabolism to fatty acid utilization is regulated by phosphorylation and inactivation of the pyruvate dehydrogenase complex mediated by the kinase PDK434. Here, we found that Pdk4 baseline expression was decreased in the skeletal muscles and heart of Ip6k3−/− mice, indicating a metabolic shift toward glucose oxidation. Notably, Ip6k3 was upregulated under the conditions wherein Pdk4 was upregulated (human myotubes and the denervation study). Thus, there might be a biological connection between IP6K3 and PDK4 in muscles.
IP6K1 regulates insulin secretion as evidenced by the decreased serum insulin observed in Ip6k1−/−mice12 and the dysregulated insulin secretion in Ip6k1 siRNA-treated pancreatic beta cells20. We confirmed that Ip6k3 was undetectable in rodent pancreatic beta cell lines and that no IP6K3 was observable in human islets, indicating no direct role of IP6K3 in pancreatic endocrine cells. Consistent with this, plasma insulin levels at 15 min after glucose loading was unchanged in mice. However, we observed lowered plasma insulin levels in 1.5 years old Ip6k3−/− mice, suggesting that reduced plasma insulin is likely to be secondary to improved metabolic profiles.
It has been suggested that 5-InsP7 can act as a proactive metabolic messenger10,35, and that IP6K3-generated 5-InsP7 likely plays a role in regulating downstream signalling. In this study we were not able to determine the in vivo levels of 5-InsP7 in the muscle of Ip6k3−/− mice owing to methodological limitations; however, a new method applicable to tissue samples is under development in our laboratory. In addition, we failed to capture endogenous mouse IP6K3 protein using available antibodies under the current experimental conditions. A recent study has provided evidence that IP6K3 serves as a scaffold to link target proteins, a function not requiring the catalytic activity of IP6K3 demonstrated in mouse brain19. Thus, future studies exploring the protein-protein interactions in muscles are essential. Finally, IP6K3 is expressed outside the muscles (e.g. in the thyroid) and its function in those tissues needs to be determined.
In conclusion, the present study has revealed that Ip6k3 is highly expressed in muscles and that its mRNA levels are regulated in response to energy and disease status. The pivotal role of IP6K3 in metabolism and lifespan but not in the control of muscle mass was demonstrated using Ip6k3−/− mice. The current observations suggest the existence of novel roles of IP6K3 in sensing energy status and regulating downstream signalling to impact metabolic control and lifespan. Furthermore, the development of a selective inhibitor of IP6K3 might represent a new class of drug to treat diabetes, which might also have the enticing added benefit of potentially extending lifespan.
Methods
Reagents and cells
All reagents were purchased from Wako Pure Chemical Industries or Sigma-Aldrich, unless otherwise indicated. C2C12 cells were obtained from the American Type Culture Collection (ATCC). Human primary myoblasts were purchased from Thermo Fisher Scientific.
Cell culture
C2C12 cells were maintained in Dulbecco’s modified Eagle’s medium containing 25 mM glucose and 10% foetal calf serum (HyClone, GE Healthcare). Differentiation of C2C12 myoblasts was induced by replacing the medium with 2% horse serum (Thermo Fisher Scientific). Cells were cultured in a humidified atmosphere containing 5% CO2/95% air at 37 °C.
Human myotube study
Three independent lots of human skeletal myoblasts were treated with DMEM containing 2% horse serum with or without 1 μM dexamethasone for 2 days, and the collected samples were submitted to quantitative PCR analysis.
Mice
Male Lepob/Lepob (ob/ob; B6.V-Lepob/J) mice and their non-diabetic, untyped (?/+; B6.V-Lepob/J) littermates, and male C57BL/6J mice were obtained from Charles River Japan. The fasting duration was 16 hours for ob/ob and ?/+ mice, and a longer duration of 2-days was used for C57BL/6J mice. Tissue samples were collected at 8 weeks of age for C57BL/6J mice (for tissue mRNA distribution), 12 weeks of age for ?/+ and ob/ob mice, and 9 weeks of age for 2-days fasted C57BL/6J mice and denervated C57BL/6J mice. All animals were housed in cages in a room with controlled temperature (23 °C), humidity (55%), and lighting (lights on from 7:00 am to 7:00 pm) and were maintained on a laboratory chow diet (CE-2, CLEA). The care and use of the animals and the experimental protocols used in this research were approved by the Experimental Animal Care and Use Committee of Takeda Pharmaceutical Company, Ltd. All experiments were performed in accordance with the guidelines and regulations of Takeda Pharmaceutical Company, Ltd. (Shonan Research Center IACUC Guidelines). All animal studies were conducted after a more than 6-day acclimation period. Tissue samples were collected from anaesthetised mice.
Ip6k3 −/− mice
Ip6k3−/− mice lacking from the middle of exon 3 to exon 5 of Ip6k3 were obtained from Taconic Biosciences (TF1523). This strain lacks the lysine at position at 212 (exon 5) of mouse IP6K3 corresponding to the lysine at position 217 of human IP6K3, which is critical for its enzymatic activity18. Ip6k3 mRNA was undetectable in the muscle of Ip6k3−/− mice as determined using quantitative PCR detection of Ip6k3 sequence at the exon 3–4 boundary. In the present study, we used male Ip6k3−/− mice and their wild-type littermates with a mixed genetic background (B6;129-Ip6k3−/−) [C57BL/6 (75%) and 129SvEv (25%)] and C57BL/6 genetic background mice (B6.Cg-Ip6k3−/−), which was speed backcrossed five times to C57BL/6 mice.
General characterization of Ip6k3 −/− mice
Male B6.Cg-Ip6k3−/− mice and their wild-type littermates were used for the study. The mice were fed a CE-2 diet. Blood samples were collected at the indicated points from the tail vein (8:00 am, non-fasting condition) for metabolic parameters analysis. Body weight and food consumption were measured at regular intervals, and average daily food consumption per mouse was calculated. Blood glucose levels were measured using a LIFE CHECK (GUNZE). Plasma insulin levels were analysed with an enzyme-linked immunosorbent assay (Millipore). Plasma lactate levels were determined using a commercial kit (Determiner LA; Kyowa Medex). Body composition was quantitated by magnetic resonance imaging to directly measure total body fat mass and total body lean mass on unanaesthetised mice at the indicated ages (EchoMRI-900, Aloka).
Oral glucose tolerance test and insulin tolerance test
Male B6;129-Ip6k3−/− mice and their wild-type littermates at 15 weeks of age (n = 8/group) were fasted overnight and a glucose solution was orally administered (2 g/kg), then blood was collected from the tail vain and the glucose levels were measured by ACCU-CHEK (Roche). Plasma insulin levels were analysed with an enzyme-linked immunosorbent assay (Millipore). For insulin tolerance testing, male B6;129-Ip6k3−/− mice at 12 weeks of age (n = 8/group) were fasted overnight and insulin was injected intraperitoneally at 0.75 units/kg (Novo Nordisk), then blood was collected from the tail vain and the glucose levels were measured using LIFE CHECK (GUNZE).
Estimation of lifespan
Male B6.Cg-Ip6k3−/− mice and their wild-type littermates were used. The principal endpoint was the age at death (for mice found dead during daily inspections) or age at euthanasia (for mice deemed unlikely to recover or survive). Mice were examined at least daily for clinical signs, and were euthanized for humane reasons if they were so severely moribund that they were considered, by the researchers and veterinarians, unlikely to survive or recover. A mouse was considered severely moribund if it exhibited more than one of the following clinical signs: (a) rapid weight loss; (b) severe balance or gait disturbance; (c) tumour size over 10% of body weight; or (d) severely ulcerated skin. Tissue samples were collected from anaesthetised mice.
High fat diet feeding study
In the high fat diet study, male B6;129-Ip6k3−/− mice and their wild-type littermates (7 weeks old, n = 8/group) were fed on a standard high-fat diet (D12451, 45% kcal as fat; Research Diets) for 8 weeks, and the body weight and blood parameters were measured using an HLC-723 GHb G8 (Tosoh Bioscience) and a 7180 autoanalyser (Hitachi) at the indicated intervals. Plasma insulin levels were analysed with an enzyme-linked immunosorbent assay (Millipore).
Muscle denervation
Male B6;129-Ip6k3−/− mice and their wild-type littermates at 11 weeks of age were used (n = 4–6/group). Denervation was performed by excising the smaller section of the sciatic nerve (2–3 mm) and the non-denervated hind limb served as a sham-operated control in subsequent analyses, which were carried out 2 and 14 days after the operation. This procedure does not affect the ability of the animal to ambulate. Tissues from mice before the operation were used as pre-control samples. Tissue samples were collected from anaesthetised mice.
Total RNA and cDNA preparation
Tissues were dissected and stabilized using RNAlater RNA Stabilization Reagent (Thermo Fisher Scientific). Stabilized tissues were homogenised in QIAzol Lysis Reagent, followed by RNeasy kit purification (Qiagen). Cellular RNA samples were prepared using an RNeasy kit. First strand cDNA was synthesized from up to 1.5 μg total RNA per 20 μL reaction using the High-Capacity cDNA Reverse Transcription Kit reverse transcriptase (Thermo Fisher Scientific). Total RNA from various human tissues and brain regions was purchased from TaKaRa Bio Inc. (Premium RNA) and cDNA was synthesized from up to 1 μg total RNA per 20 μL reaction using Superscript II reverse transcriptase (Thermo Fisher Scientific).
Gene expression analysis
To determine the absolute copy numbers of the target genes, a standard curve was generated by amplifying known concentrations of synthetic oligonucleotides, and the target gene copy number was calculated using this curve. All oligonucleotide primers and dual-labelled (FAM, VIC, HEX - TAMRA, BHQ, MGB) oligonucleotide probes were synthesized by Sigma-Aldrich or Thermo Fisher Scientific (Table S1). The TaqMan gene expression assays used are described in Table S2. Relative mRNA quantification was performed in combination with an internal control gene, RPLP0 or Rplp1. Amplification was performed under the following conditions: initial denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing and extension at 60 °C for 60 s on the ABI PRISM 7900HT Sequence Detector (Thermo Fisher Scientific) using the EXPRESS qPCR supermix (Thermo Fisher Scientific).
Antibodies
The antibodies used were as follows: phospho-S6 ribosomal protein (Ser235/236) (#4858), S6 ribosomal protein (#2217), phospho-p70 S6 kinase (Thr389) (#9234), p70 S6 kinase (#2708), Akt (pan) (#4691), phospho-Akt (Thr308) (#2965), and phospho-Akt (Ser473) (#4060)(all from CST). Anti-rabbit IgG, HRP-linked antibodies were from CST (#7074) or Jackson Laboratories (111-035-144). GAPDH antibodies were from MBL (M171-7) or CST (#5174).
SDS-PAGE and western immunoblotting
Sample lysates were prepared by homogenizing in extraction reagent (T-PER, 78510, Thermo Fisher Scientific) with protease (05 892 970 001, Roche) and phosphatase (04 906 837 001; Roche) inhibitor cocktails. Concentration-adjusted protein (Pierce 660 nm Protein Assay) was heated at 100 °C for 5 min in Laemmli sample buffer (Bio-Rad) followed by SDS/PAGE (TGX gel, Bio-Rad). Samples were transferred to a PVDF membrane, blocked in 5% skim milk, and incubated overnight with primary antibodies following by the respective secondary antibodies. Membranes were developed using ECL reagent (RPN2232; GE Healthcare) and immunoreactive bands were quantified with Quantity One Software (Bio-Rad).
Statistical analyses and calculations
Statistical significance was analysed using the F test, which was used for testing the homogeneity of variances, followed by the Student’s t test (P > 0.2 by the F test) or the Aspin-Welch test (P ≤ 0.2 by the F test). Student’s t tests and Aspin-Welch tests were conducted at the two-tailed significance levels of 5% (0.05). Cumulative survival curves were constructed using Kaplan-Meier survivorship methods. Differences between the curves were tested for significance by the log-rank test. All data were presented as the means and SD, except for qPCR data generated using the comparative CT (ΔΔCT) method (User Bulletin #2, Applied Biosystems). For the comparative CT method, the ΔCT value was determined by subtracting the average internal control CT value from the average target CT value. The calculation of ΔΔCT involves subtraction by the ΔCT calibrator value. The range given for each mRNA level relative to the control was determined by evaluating the expression: 2−ΔΔCT with ΔΔCT + s and ΔΔCT − s, where s = the standard deviation of the ΔΔCT value.
Additional Information
How to cite this article: Moritoh, Y. et al. Inositol Hexakisphosphate Kinase 3 Regulates Metabolism and Lifespan in Mice. Sci. Rep. 6, 32072; doi: 10.1038/srep32072 (2016).
Supplementary Material
Acknowledgments
The authors would like to thank Jun-ichi Miyazaki (Osaka University, Japan) for providing the MIN6 cells, and Christopher B. Newgard (Duke University) for providing the INS-1 832/13 cells. We also thank Drs. Minoru Maruyama, Masako Matsuo, and Shoichi Okubo for providing materials. The present study was conducted with financial support from Takeda Pharmaceutical Company Ltd.
Footnotes
Author Contributions Y.M. conceived the project, and designed, performed, and analysed the experiments. M.O., Y.Y., H.H. and K.I. performed and analysed the experiments. H.F. and R.T. conceived the project and analysed the experiments. Y.M. and R.T. wrote the paper. All authors commented on and approved the paper.
References
- Barker C. J., Illies C., Gaboardi G. C. & Berggren P. O. Inositol pyrophosphates: structure, enzymology and function. Cell. Mol. Life Sci. 66, 3851–3871 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C., Szijgyarto Z. & Saiardi A. The signaling role of inositol hexakisphosphate kinases (IP6Ks). Adv. Enzym. Regul. 51, 74–82 (2011). [DOI] [PubMed] [Google Scholar]
- Lin H. et al. Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J. Biol. Chem. 284, 1863–1872 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A., Erdjument-Bromage H., Snowman A. M., Tempst P. & Snyder S. H. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 9, 1323–1326 (1999). [DOI] [PubMed] [Google Scholar]
- Draskovic P. et al. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem. Biol. 15, 274–286 (2008). [DOI] [PubMed] [Google Scholar]
- Choi J. H., Williams J., Cho J., Falck J. R. & Shears S. B. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J. Biol. Chem. 282, 30763–30775 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridy P. C., Otto J. C., Dollins D. E. & York J. D. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J. Biol. Chem. 282, 30754–30762 (2007). [DOI] [PubMed] [Google Scholar]
- Wang H., Falck J. R., Hall T. M. & Shears S. B. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol. 8, 111–116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears S. B. Inositol pyrophosphates: why so many phosphates? Adv. Biol. Regul. 57, 203–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. S., Livermore T. M. & Saiardi A. Inositol pyrophosphates: between signalling and metabolism. Biochem. J. 452, 369–379 (2013). [DOI] [PubMed] [Google Scholar]
- Albert C. et al. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem. J. 327, 553–560 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari R., Juluri K. R., Resnick A. C. & Snyder S. H. Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc. Natl. Acad. Sci. USA 105, 2349–2353 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S. et al. Inositol hexakisphosphate kinase 1 maintains hemostasis in mice by regulating platelet polyphosphate levels. Blood 122, 1478–1486 (2013). [DOI] [PubMed] [Google Scholar]
- Prasad A. et al. Inositol hexakisphosphate kinase 1 regulates neutrophil function in innate immunity by inhibiting phosphatidylinositol-(3,4,5)-trisphosphate signaling. Nat. Immunol. 12, 752–760 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A. et al. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell 143, 897–910 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison B. H. et al. Gene deletion of inositol hexakisphosphate kinase 2 predisposes to aerodigestive tract carcinoma. Oncogene 28, 2383–2392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F. et al. Inositol pyrophosphates promote tumor growth and metastasis by antagonizing liver kinase B1. Proc. Natl. Acad. Sci. USA 112, 1773–1778 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A., Nagata E., Luo H. R., Snowman A. M. & Snyder S. H. Identification and characterization of a novel inositol hexakisphosphate kinase. J. Biol. Chem. 276, 39179–39185 (2001). [DOI] [PubMed] [Google Scholar]
- Fu C. et al. et al. Inositol hexakisphosphate kinase-3 regulates the morphology and synapse formation of cerebellar purkinje cells via spectrin/adducin. J. Neurosci. 35, 11056–11067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illies C. et al. Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic beta cells. Science 318, 1299–1302 (2007). [DOI] [PubMed] [Google Scholar]
- Kuo T., Harris C. A. & Wang J. C. Metabolic functions of glucocorticoid receptor in skeletal muscle. Mol. Cell. Endocrinol. 380, 79–88 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Hulver M. W., McMillan R. P., Cline M. A. & Gilbert E. R. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr. Metab. (Lond). 11, 10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han E. K. et al. Akt inhibitor A-443654 induces rapid Akt Ser-473 phosphorylation independent of mTORC1 inhibition. Oncogene 26, 5655–5661 (2007). [DOI] [PubMed] [Google Scholar]
- Bonaldo P. & Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis. Model. Mech. 6, 25–39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans O. B. Muscle pyruvate oxidation following denervation and reinnervation. Muscle Nerve 6, 557–560 (1983). [DOI] [PubMed] [Google Scholar]
- Lev S. et al. Fungal inositol pyrophosphate IP7 is crucial for metabolic adaptation to the host environment and pathogenicity. MBio 6, e00531–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szijgyarto Z., Garedew A., Azevedo C. & Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science 334, 802–805 (2011). [DOI] [PubMed] [Google Scholar]
- Harrison D. E. et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C. et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326, 140–144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thota S. G., Unnikannan C. P., Thampatty S. R., Manorama R. & Bhandari R. Inositol pyrophosphates regulate RNA polymerase I-mediated rRNA transcription in Saccharomyces cerevisiae. Biochem. J. 466, 105–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O. Ribosomal protein S6 phosphorylation: four decades of research. Int. Rev. Cell Mol. Biol. 320, 41–73 (2015). [DOI] [PubMed] [Google Scholar]
- Gomes M. D., Lecker S. H., Jagoe R. T., Navon A. & Goldberg A. L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA 98, 14440–14445 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine S. C. et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708 (2001). [DOI] [PubMed] [Google Scholar]
- Kwon H. S. & Harris R. A. Mechanisms responsible for regulation of pyruvate dehydrogenase kinase 4 gene expression. Adv. Enzyme Regul. 44, 109–121 (2004). [DOI] [PubMed] [Google Scholar]
- Shears S. B. Diphosphoinositol polyphosphates: metabolic messengers? Mol. Pharmacol. 76, 236–252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.