Summary
The association between the 6q25.1 locus and breast cancer risk may be mediated through SNPs that regulate expressions of the AKAP12 gene. SNP rs7763637, in strong LD with rs2046210, is a potential functional variant at the 6q25.1 locus.
Abstract
In a genome-wide association study conducted among Chinese women, we identified the single nucleotide polymorphism (SNP) rs2046210 at 6q25.1 for breast cancer risk. To explore a potential regulatory role for this risk locus, we measured expression levels of nine genes at the locus in breast cancer tissue and adjacent normal tissue samples obtained from 67 patients recruited in the Shanghai Breast Cancer Study. We found that rs2046210 had a statistically significant association with the expression levels of the AKAP12 and ESR1 genes in adjacent normal breast tissues. Women who carry the AA/AG risk genotypes had higher expressions of these two genes compared to those who carry G/G genotypes (P = 0.02 and 0.04 for the AKAP12 and ESR1, respectively). However, no significant differences of SNP rs2046210 with gene expression levels were found in tumor tissues. In The Cancer Genome Atlas samples, the AA/AG risk genotypes of SNP rs2046210 were associated with a significantly higher expression level of the AKAP12 gene and a lower level of the ESR1 gene in tumor tissue. Functional analysis using ENCODE data revealed that SNP rs7763637, which is in strong linkage disequilibrium with SNP rs2046210, is likely a potential functional variant, regulating the AKAP12 gene. Taken together, these results from our study suggest that the association between the 6q25.1 locus and breast cancer risk may be mediated through SNPs that regulate expressions of the AKAP12 gene.
Introduction
Genome-wide association studies have been instrumental in identifying common genetic variants associated with breast cancer risk. To date, nearly 100 novel genetic loci for breast cancer have been identified (1,2). We have previously identified several novel loci in Asian women (3–10), including a novel genetic susceptibility variant (rs2046210) at 6q25.1 (5). A nearly 60% elevated risk for breast cancer was found among women homozygous for the AA allele of rs2046210 (5). A similar association was found in other East Asian women including Japanese (10,11) and Koreans (12,13), while a weaker association was also found in women of European ancestry (5,10,14), and none for women of African ancestry (10,15,16). Single nucleotide polymorphism (SNP) rs2046210 is located in a non-coding region ~29kb upstream of the ESR1 gene and 6kb downstream of the CCDC170 (C6orf97) gene (5,10). Several previous studies have been conducted to identify potential causal and/or functional variants for this locus (10,17,18). However, the results are inconsistent. SNP rs2046210 and/or SNP(s) in strong linkage disequilibrium (LD) with SNP rs2046210 may affect breast cancer risk by regulating expressions of local genes in the locus. In this study, we investigated the expression levels of nine genes flanking a 1Mb region around SNP rs2046210 in breast cancer tissue and adjacent normal tissues from 67 patients, as well as data from The Cancer Genome Atlas (TCGA). We performed analyses to evaluate the association between expression of these genes and SNP rs2046210 and SNPs in strong LD with SNP rs2046210. Additionally, we performed functional annotation on local SNPs in strong LD with rs2046210 to identify potential functional variants in the locus.
Material and methods
Study subjects and tissues samples
The subjects in this study were a subset of patients of the Shanghai Breast Cancer Study (SBCS), a population-based case control study. Details of the study have been described elsewhere (19). The subjects in the current study were 67 randomly selected breast cancer patients who provided both tumor and adjacent non-tumor tissue samples (Table 1). All patients provided written, informed consent to participate in the study, and the study protocols were approved by the Institutional Review Boards of all institutions involved in the study. Tumor tissue samples were removed during surgery from the center of the lesion, whereas adjacent non-tumor tissue samples were obtained from the distal edge of the resection. These samples were snap-frozen in liquid nitrogen as soon as possible, typically within 10 minutes. Samples were stored at −80°C until the relevant assays were done. All patients were interviewed at the time of recruitment. Two senior pathologists reviewed all tissue slides to confirm the diagnosis.
Table 1.
Selected demographic characteristics and known breast cancer risk factors
| Breast cancer (n = 67) | |
|---|---|
| Age (mean) | 46.9 |
| Education ≥ high school (%) | 38.2 |
| Age at menarche (mean) | 14.2 |
| Post-menopause (%) | 32.7 |
| Body mass index (mean) | 24.4 |
| Waist-to-hip ratio (mean) | 0.79 |
| Regular physical activity (%) | 20.0 |
| Family history of breast cancer among first degree relatives | 7.3 |
| Estrogen receptor positive (%) | 63.0 |
| Progesterone receptor Positive %) | 67.9 |
Laboratory assays
Total RNA was extracted from tissue specimens by homogenization in TRIzol solution (Invitrogen, Carlsbad, CA), phase separation, precipitation and washing following the manufacturer’s instructions. The quality and quantity of RNA was measured by spectrophotometric analysis. RNA was reverse-transcribed (RT) using a High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) in a final volume of 15 μl containing 0.15 μg RNA and 1× RT polymerase chain reaction (RT-PCR) buffer, 5.5mM MgCl2, 500 μM each dNTP, 2.5 μM Random Hexamers, 0.4U/μl RNase inhibitor and 3.125U/μl MultiScribe reverse transcriptase. The mixture was incubated at 25°C for 10min, 37°C for 120min and 95°C for 5min.
We measured the mRNA expression levels of nine genes (PLEKHG1, MTHFD1L, AKAP12, ZBTB2, RMND1, ARMT1 (C6orf211), CCDC170 (C6orf97), ESR1 and SYNE1) located within the 1Mb region centered on SNP rs2046210. Real-time quantitative PCR reaction was carried out in an iCycler detection system (Bio-Rad, Hercules, CA) in a 25 µl volume. The reaction mixture consisted of 12.5 μl iQSYBR Green Supermix, 200nM of each primer and 1 μl of cDNA template. Reactions were performed for 45 cycles (95°C for 15s, 60°C for 30s and 72°C for 30s) after initial 3-min incubation at 95°C. Primers for the different genes amplified are presented in Supplementary Table 1, available at Carcinogenesis Online. All reactions were done in duplicates. The threshold cycle (Ct) was determined at 0.1 based on the amplification linear area of the target genes and the β-actin (ACTB) gene (an internal control). The normalized quantity of the target gene was calculated as 2−ΔCt, where ΔCt was obtained directly by subtracting Ct for the ACTB gene from Ct for the target gene. The final result was expressed as 2−ΔCt × 1000. In addition, in all experiments, appropriate negative controls containing no template were subjected to the same procedure to exclude or detect any possible contamination.
Genotyping data were obtained using the Affymetrix Genome-Wide Human SNP Array 6.0 as described previously (5).
Statistical analysis
We divided subjects into two groups based on the genotype of rs2046210 (i.e. AA/AG, GG). The average expression level of each gene for each group was measured with geometric means. The association significance for each gene was evaluated using the Student’s t-tests on log-transformed data. All P values are two-sided. SAS software was used for statistical analysis (v. 9.1; SAS Institute, Cary, NC).
Expression quantitative traitloci (eQTL) analysis
We downloaded the RNA-Seq V2 data (level 3) of 1006 breast cancer tumor tissues from the TCGA data portal (See URLs). DNA methylation data measured by the Illumina HumanMethylation450 BeadChip were also retrieved from TCGA level 3 data. We also downloaded level 3 SNP data genotyped using the Affymetrix SNP 6.0 array. Copy number variation data for the nine genes for TCGA samples were collected from the CbioPortal (see URLs) for tumor tissues. The eQTL analysis was performed in tumor tissue as previously described (3,18). Briefly, we transformed the RNA-Seq by the Expectation-Maximization (RSEM) value of each gene, and performed principal component correction in gene expression data to remove potential batch effects. Residual linear regression analysis was then used to detect eQTLs while adjusting for methylation and copy number variation, according to the approach proposed by Li et al. (3,18).
Functional annotation
For each SNP in strong LD with rs2046210 in Asians (r 2 > 0.8), we investigated its potential functional importance using data from the ENCODE (see URLs). First, we investigated whether the SNP was located in regulatory regions (i.e. promoter and enhancer) through chromHMM annotation tracks in ENCODE from the UCSC Genome Browser (see URLs) including nine cell lines: HMEC (breast normal cell line), GM12878, H1-hESC, K562, HepG2, HSMM, HUVEC, NHEK and NHLF. We also evaluated DNase I hypersensitive and transcription factor binding sites (TFBS) in all cell lines analyzed by ENCODE, including breast normal cell line, HMEC and breast cancer cell lines, T-47D and MCF-7. We assessed the histone modification markers H3K4Me1, H3K4Me3 and H3K27Ac in all cell lines analyzed by ENCODE through the layered histone tracks from the UCSC Genome Browser. Two publicly-available tools, RegulomeDB (20) (see URLs) and HaploReg V2 (21) (see URLs), were also used to evaluate candidate functional variants.
Results
Differential gene expression analysis
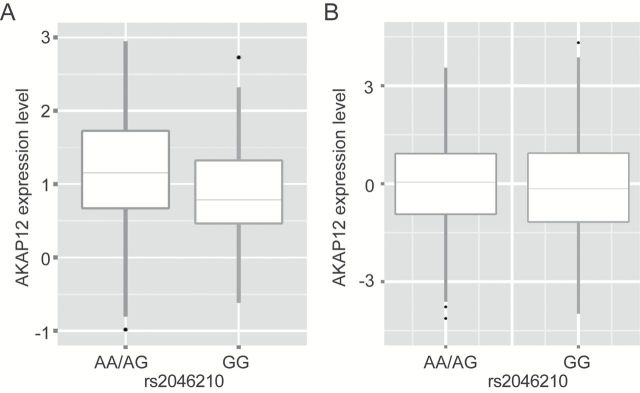
Geometric means and 95% confidence intervals (CI) for the expression levels of the nine genes in 67 SBCS breast cancer tumor and normal tissues are shown in Table 2. The RMND1 gene exhibited the highest expression level among nine studied genes, whereas the AKAP12, PLEKHG1 and ZBTB2 genes exhibited the lowest expression levels. SNP rs2046210 was significantly associated with expression levels of the AKAP12 and ESR1 genes in the adjacent normal tissues. Women who carry the rs2046210 AA/AG risk genotypes had substantially higher AKAP12 gene expression (3.35 vs. 1.36, P = 0.02) than those who carry the rs2046210 GG genotypes (Table 2; Figure 1A). This was also true for the expression level of the ESR1 gene (13.90 vs. 5.26, P = 0.04) (Table 2). The associations were seen in both estrogen receptor (ER)-positive and ER-negative samples (data not shown), although the associations were not statistically significant due to a smaller sample size. SNP rs2046210 was associated with MTHFD1L gene expression (4.37 vs. 2.11) with borderline significance (P = 0.08). No association between SNP rs2046210 and the other six genes was observed in the adjacent normal tissue. In the tumor tissues, however, expression levels of these nine genes did not differ significantly by rs2046210 genotype status (Table 2).
Table 2.
Comparison of gene expression levels (locus 6q25.1) by SNP rs2046210 genotypes in breast cancer tissues
| Marker | rs2046210 = ‘A/A’ and ‘A/G’ | rs2046210 = ‘G/G’ | P a | ||
|---|---|---|---|---|---|
| No. | Geometric means (95% CI) | No. | Geometric means (95% CI) | ||
| Adjacent normal tissue | |||||
| PLEKHG1 | 36 | 2.72 (1.79–4.13) | 24 | 2.56 (1.36–4.84) | 0.8657 |
| MTHFD1L | 35 | 4.37 (2.96–6.47) | 24 | 2.11 (0.93–4.81) | 0.0762 |
| AKAP12 | 35 | 3.35 (2.38–4.73) | 24 | 1.36 (0.60–3.11) | 0.0236 |
| ZBTB2 | 33 | 2.18 (1.67–2.86) | 23 | 1.96 (1.11–3.47) | 0.7011 |
| RMND1 | 35 | 55.33 (15.63–195.81) | 24 | 30.65 (7.99–117.53) | 0.5253 |
| ARMT1 | 37 | 11.07 (7.12–17.25) | 25 | 10.73 (6.37–18.07) | 0.9250 |
| CCDC170 | 37 | 5.62 (4.05–7.80) | 24 | 5.26 (2.78–9.97) | 0.8384 |
| ESR1 | 36 | 13.90 (7.88–24.54) | 24 | 5.26 (2.34–11.81) | 0.0423 |
| SYNE1 | 36 | 4.50 (3.26–6.22) | 25 | 6.05 (3.09–11.85) | 0.3766 |
| Tumor tissue | |||||
| PLEKHG1 | 39 | 1.60 (1.20–2.13) | 26 | 1.76 (1.25–2.49) | 0.6556 |
| MTHFD1L | 39 | 3.75 (3.08–4.56) | 28 | 3.49 (2.59–4.72) | 0.6785 |
| AKAP12 | 38 | 1.96 (1.14–2.75) | 27 | 2.05 (1.36–3.08) | 0.8802 |
| ZBTB2 | 35 | 2.31 (1.60–3.33) | 26 | 2.24 (1.62–3.11) | 0.9024 |
| RMND1 | 38 | 49.79 (15.69–158.07) | 27 | 35.02 (9.79–125.31) | 0.6820 |
| ARMT1 | 39 | 13.26 (9.48–18.56) | 28 | 12.73 (8.73–18.56) | 0.8698 |
| CCDC170 | 38 | 3.40 (2.27–5.10) | 26 | 3.95 (2.53–6.18) | 0.6198 |
| ESR1 | 39 | 10.17 (5.63–18.38) | 28 | 12.83 (6.23–26.40) | 0.6131 |
| SYNE1 | 39 | 3.91 (3.04–5.03) | 28 | 3.68 (2.70–5.02) | 0.7579 |
aFrom Student’s t-test.
Figure 1.
Association of SNP rs2046210 with AKAP12 gene expression levels. (A) SBCS adjacent normal tissue samples; (B) TCGA tumor tissue samples.
eQTL analysis using TCGA data
We also conducted an eQTL analysis to evaluate the association of SNP rs2046210 with expression of the AKAP12 and ESR1 genes in tumor tissue using data from 1,006 samples included in TCGA. SNP rs2046210 was found to be correlated with AKAP12 gene expression. The women who carry the AA/AG risk genotypes had a higher AKAP12 gene expression level than women who carry the GG genotypes (Figure 1B) (P < 0.05). The result was consistent with the observation in the adjacent normal tissue from the association analysis on 67 SBCS patients. We observed that SNP rs2046210 was significantly correlated with ESR1 gene expression level. Women who carry the AA/AG risk genotypes had a decreased expression level of ESR1 gene compared to women who carry the GG genotype in TCGA tumor tissues (P < 0.01). We also found a decreased expression in the SBCS tumor tissue samples among women who carry the AA/AG risk genotypes compared to women who carry the GG genotype, although the difference was not statistically significant (Table 2). However, this result was inconsistent with the observation in the adjacent normal tissue from the association analysis of the 67 SBCS patients. Nevertheless, our results suggest that the association between SNP rs2046210 and breast cancer risk may be mediated through its role in the regulating expressions of the AKAP12 and ESR1 genes.
Functional annotation
We further evaluated and annotated the functional significance of a total of 20 SNPs in strong LD with rs2046210 in an Asian population (r 2 > 0.8) using ChIP-Seq and DNase-Seq data from the ENCODE Project. In line with previous findings, no epigenetic signal was observed for rs2046210, indicating it may be a non-functional SNP (Supplementary Table 2, available at Carcinogenesis Online). Interestingly, SNP rs7763637, located 946bp away and in strong LD with SNP rs2046210 (r 2 = 0.92), was found to be potentially functional. The epigenetic signals of active promoter associated histone mark (H3K4Me3), enhancer associated histone marks (H3K4Me1 and H3K27Ac) and chromatin accessibility were enriched in the intervals containing SNP rs7763637 in multiple ENCODE cell lines. Functional annotation using chromatin states from nine ENCODE cell lines showed that this SNP is located in the active promoter region of the CCDC170 gene in two cell lines, including normal breast cell line HMEC. Using transcription factor (TF) ChIP-Seq data revealed that the variant is located in multiple TF binding sites, including several breast cancer-related TFs such as ZNF217, FOS, KAP1, JUND, FOSL2, JUN and MYC (Figure 2). In particular, TF FOSL2 and JunD were observed to harbor the variant in breast cancer cell line MCF-7 (Figure 2). Consistent with this observation, functional annotation using two publicly-available tools, RegulomeDB and HaploReg v2, revealed that this SNP exhibited potential functional significance (Supplementary Table 2, available at Carcinogenesis Online).
Figure 2.
Functional annotation of the potential functional SNP rs7763637 in 6q25.1. SNP rs7763637 is in strong LD with SNP rs2046210. Chromosome position was indicated by the dashed line. From top to bottom (1): Epigenetic landscape at 6q25.1, layered H3K4Me1, H3K4Me3, and H3K27Ac histone modifications are shown. The signals of different layered histone modifications from the ENCODE cell lines are shown (2). DNase I hypersensitivity sites (black) (3). Transcription factors binding sites in different cell lines by ChIP-Seq (4). Annotation using chromatin states on the ENCODE cell lines. Black and dark gray indicate promoter and enhancer, respectively (5). Transcription factors binding sites in breast cancer cell line MCF-7 by ChIP-Seq.
SNP rs7763637 was associated with breast cancer risk among 2867 breast cancer cases and 2,285 controls (P = 1.90×10–7), similar to that of rs2046210 (P = 2.54×10–7) in our Genome-wide association study samples. SNP rs7763637 was significantly associated with expression levels of the AKAP12 gene and borderline significant with the ESR1 gene in the adjacent normal tissues of SBCS patients (Supplementary Table 3, available at Carcinogenesis Online). Women who carry the rs7763637 AA/AG risk genotypes had substantially higher AKAP12 gene expression (3.62 vs. 1.26, P = 0.046) than those who carry the rs7763637 GG genotypes. This was also true for the expression level of the ESR1 gene (13.49 vs. 4.72, P = 0.06). No association between SNP rs7763637 and the other 7 genes was observed in the adjacent normal tissue (Supplementary Table 3, available at Carcinogenesis Online). Using the TCGA data, we found that SNP rs7763637 was associated with AKAP12 gene expression level. Women who carry the AA/AG genotypes had a higher AKAP12 gene expression level than women carrying the GG genotypes (P = 0.02). In addition, women who carry the AA/AG genotypes had lower expression levels of the ESR1 (P = 0.01), RMND1 (P = 0.003) and ZBTB2 (P = 0.03) genes than women carrying the GG genotypes in the TCGA tumor tissue samples. Taken together, these findings indicate that SNP rs7763637 is highly likely to be a functional variant at the 6q25.1 locus, and thus may affect the expression of target genes.
Discussion
SNP rs2046210, located upstream of the ESR1 gene, is associated with breast cancer risk among women of both European and East Asian ancestry (5,10). In this study, we investigated the mRNA expression levels of genes in the 1Mb region of SNP rs2046210 in breast tumor tissues and their adjacent normal tissues. We found that women who carry rs2046210 AA/AG risk genotypes had higher AKAP12 and ESR1 expression levels compared with women carrying G/G genotypes in the adjacent normal tissue. eQTL analyses using data from TCGA also showed that rs2046210 AA/AG risk genotypes were associated with higher level of AKAP12 gene expression, but associated with lower level of ESR1 gene expression in tumor tissue. Functional analysis revealed that SNP rs7763637, which is in strong LD with SNP rs2046210, is a potential functional variant at the 6q25.1 locus and may be associated with breast cancer risk mediated through the regulation of the AKAP12 gene.
Protein kinase A anchor protein 12 gene (AKAP12) lies ~26.8kb upstream of SNP rs2046210. It is a scaffold protein that plays important roles in cell proliferation, migration, apoptosis and angiogenesis (22). Studies have shown that AKAP12 functions as a metastasis suppressor (23). Downregulation or loss of AKAP12 was associated with malignancy and metastasis in many cancer types including breast cancer (22,24–28). In contrast, other studies have found that the expression of AKAP12 was increased in cancer tissue or cell lines (22,29,30), indicating multiple functions of the AKAP12 gene in carcinogenesis. However, no evidence to date has shown that AKAP12 affects primary tumor growth (23). Our study found that SNP rs2046210 may increase AKAP12 expression in both normal tissues and tumor tissues. This suggests that genetic variations in this locus may play a role in multiple stages of breast cancer development, including initiation, progression, and metastasis.
The ESR1 gene encodes estrogen receptor α (ER-α), which regulates signal transduction of estrogen, a sex hormone that plays a central role in the etiology of breast cancer. SNP rs2046210 is located 29kb upstream of the first untranslated exon and 180kb upstream of the transcription start site of exon 1 of the ESR1 gene (31). The associations of SNP rs2046210 with ESR1 gene expression differ between tumor tissues and normal tissues. Women in the SBCS who carry the rs2046210 AA/AG risk genotypes had a higher ESR1 gene expression level in adjacent normal tissues. However, women from TCGA who carry the AA/AG risk genotypes had a decreased expression level of the ESR1 gene in tumor tissues. We also found that ESR1 gene expression levels were lower in the AA/AG risk genotypes compared to the G/G genotype group in the SBCS tumor tissue samples, although the difference was not statistically significant. The reason for the different effect of rs2046210 on ESR1 gene expression in tumor and adjacent normal tissue is not clear. Genetic variations in this locus may affect allelic expression imbalance and/or DNA methylation, which may in turn affect ESR1 gene expression. Using TCGA RNA-seq data from 177 individuals, Li et al. found that rs2046210 genotypes were correlated with an allelic expression imbalance of the ESR1 gene. Experimental evidence also shows physical interaction between risk locus 6q25 and the promoter of ESR1 gene (18). Therefore, it is possible that SNP rs2046210 plays a regulatory role in ESR1 gene transcription.
SNP rs7763637, which is in strong LD with rs2046210, is located in multiple transcriptional regulatory regions such as the binding site of transcription factors FOS, FOSL2, and JunD. FOS and FOSL2 belong to the FOS gene family which can dimerize with proteins of the JUN family (including c-Jun, JunB and JunD) to form the transcription factor complex AP-1 (32,33). Several studies have suggested that the expression of AP-1 protein is related to breast cancer growth, progression and metastasis (34,35). Thus, SNP rs7763637 could be a potentially functional variant in breast cancer risk locus 6q25.1 that regulates the transcription of AP-1 proteins.
There are some limitations of our study. The sample size of SBCS tissue sample is not large. However, the results of the association of SNPs rs2046210 and rs7763637 with AKAP12 gene expression are consistent between SBCS samples and TCGA samples. In addition, we did not adjust for multiple testing in our analyses. Further studies with larger sample sizes are warranted to confirm our findings.
In summary, SNP rs2046210 and SNPs in strong LD with it may influence the expression of the AKAP12 gene in breast tissues. Future research is needed to elucidate the potential mechanism by which the AKAP12 gene is related to breast cancer risk.
Supplementary material
Supplementary Tables 1–3 can be found at http://carcin.oxfordjournals.org/
URLs
The Cancer Genome Atlas (TCGA), http://cancergenome.nih.gov/
1000 Genomes, http://browser.1000genomes.org
Minimac, http://genome.sph.umich.edu/wiki/Minimac
CbioPortal, http://www.cbioportal.org/public-portal/
ENCODE, http://genome.ucsc.edu/ENCODE/
UCSC Genome Browser, http://genome.ucsc.edu
RegulomeDB, http://regulome.stanford.edu/
HaploReg v2, http://www.broadinstitute.org/mammals/haploreg/haploreg.php
Funding
National Institutes of Health (R01CA064277, R01CA090899 and R01CA122756); Fogarty International Center training grant (D43 TW08313).
Supplementary Material
Acknowledgements
The authors wish to thank the study participants and research staff for their contributions and commitment to this project. We thank Regina Courtney and Jie Wu for laboratory assistance, and Nan Kennedy for assistance with editing and manuscript preparation. The sample preparation and gene expression assays was performed at the Survey and Biospecimen Shared Resource, which is supported in part by the Vanderbilt-Ingram Cancer Center (P30CA068485).
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- eQTL
expression quantitative traitloci
- LD
linkage disequilibrium
- SNP
single nucleotide polymorphism
References
- 1. Michailidou K., et al. (2013) Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet., 45, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fachal L., et al. (2015) From candidate gene studies to GWAS and post-GWAS analyses in breast cancer. Curr. Opin. Genet. Dev., 30, 32–41. [DOI] [PubMed] [Google Scholar]
- 3. Cai Q., et al. (2014) Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. Nat. Genet., 46, 886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng W., et al. (2013) Common genetic determinants of breast-cancer risk in East Asian women: a collaborative study of 23 637 breast cancer cases and 25 579 controls. Hum. Mol. Genet., 22, 2539–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zheng W., et al. (2009) Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet., 41, 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Long J., et al. (2010) Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet., 6, e1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai Q., et al. (2011) Genome-wide association study identifies breast cancer risk variant at 10q21.2: results from the Asia Breast Cancer Consortium. Hum. Mol. Genet., 20, 4991–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Long J., et al. (2012) Genome-wide association study in east Asians identifies novel susceptibility loci for breast cancer. PLoS Genet., 8, e1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Long J., et al. (2013) A common deletion in the APOBEC3 genes and breast cancer risk. J. Natl. Cancer Inst., 105, 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai Q., et al. (2011) Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of chinese, Japanese, and European ancestry. Cancer Res., 71, 1344–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mizoo T., et al. (2013) Effects of lifestyle and single nucleotide polymorphisms on breast cancer risk: a case-control study in Japanese women. BMC Cancer, 13, 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han W., et al. (2011) Common genetic variants associated with breast cancer in Korean women and differential susceptibility according to intrinsic subtype. Cancer Epidemiol. Biomarkers Prev., 20, 793–798. [DOI] [PubMed] [Google Scholar]
- 13. Kim H.C., et al. (2012) A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: results from the Seoul Breast Cancer Study. Breast Cancer Res., 14, R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han M.-R., et al. (2015) Evaluating 17 breast cancer susceptibility loci in the Nashville breast health study. Breast Cancer, 22, 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen F., et al. (2011) Fine-mapping of breast cancer susceptibility loci characterizes genetic risk in African Americans. Hum. Mol. Genet., 20, 4491–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hutter C.M., et al. (2011) Replication of breast cancer GWAS susceptibility loci in the Women’s Health Initiative African American SHARe Study. Cancer Epidemiol. Biomarkers Prev., 20, 1950–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stacey S.N., et al. (2010) Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet., 6, e1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Q., et al. (2013) Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell, 152, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao Y.T., et al. (2000) Association of menstrual and reproductive factors with breast cancer risk: results from the Shanghai Breast Cancer Study. Int. J. Cancer, 87, 295–300. [DOI] [PubMed] [Google Scholar]
- 20. Boyle A.P., et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res., 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ward L.D., et al. (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gelman I.H. (2010) Emerging roles for SSeCKS/Gravin/AKAP12 in the control of cell proliferation, cancer malignancy, and barriergenesis. Genes Cancer, 1, 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gelman I.H. (2012) Suppression of tumor and metastasis progression through the scaffolding functions of SSeCKS/Gravin/AKAP12. Cancer Metastasis Rev., 31, 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xia W., et al. (2001) The Src-suppressed C kinase substrate, SSeCKS, is a potential metastasis inhibitor in prostate cancer. Cancer Res., 61, 5644–5651. [PubMed] [Google Scholar]
- 25. Choi M.C., et al. (2004) AKAP12/Gravin is inactivated by epigenetic mechanism in human gastric carcinoma and shows growth suppressor activity. Oncogene, 23, 7095–7103. [DOI] [PubMed] [Google Scholar]
- 26. Jo U., et al. (2006) AKAP12alpha is associated with promoter methylation in lung cancer. Cancer Res. Treat. Off. J. Korean Cancer Assoc., 38, 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Su B., et al. (2006) SSeCKS metastasis-suppressing activity in MatLyLu prostate cancer cells correlates with vascular endothelial growth factor inhibition. Cancer Res., 66, 5599–5607. [DOI] [PubMed] [Google Scholar]
- 28. Hayashi M., et al. (2012) Identification of the A kinase anchor protein 12 (AKAP12) gene as a candidate tumor suppressor of hepatocellular carcinoma. J. Surg. Oncol., 105, 381–386. [DOI] [PubMed] [Google Scholar]
- 29. Lopez-Ayllon B.D., et al. (2014) Cancer stem cells and cisplatin-resistant cells isolated from non-small-lung cancer cell lines constitute related cell populations. Cancer Med, 3,1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yildirim M., et al. (2013) AKAP12/Gravin gene expression in colorectal cancer: clinical importance and review of the literature. J. BUON, 18, 635–640. [PubMed] [Google Scholar]
- 31. Koš M., et al. (2001) Minireview: Genomic Organization of the Human ERα Gene Promoter Region. Mol. Endocrinol., 15, 2057–2063. [DOI] [PubMed] [Google Scholar]
- 32. Matsui M., et al. (1990) Isolation of human fos-related genes and their expression during monocyte-macrophage differentiation. Oncogene, 5, 249–255. [PubMed] [Google Scholar]
- 33. Milde-Langosch K. (2005) The Fos family of transcription factors and their role in tumourigenesis. Eur. J. Cancer, 41, 2449–2461. [DOI] [PubMed] [Google Scholar]
- 34. Milde-Langosch K., et al. (2008) Role of Fra-2 in breast cancer: influence on tumor cell invasion and motility. Breast Cancer Res. Treat., 107, 337–347. [DOI] [PubMed] [Google Scholar]
- 35. Kharman-Biz A., et al. (2013) Expression of activator protein-1 (AP-1) family members in breast cancer. BMC Cancer, 13, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.