Abstract
Summary: The public availability of high throughput molecular data provides new opportunities for researchers to advance discovery, replication and validation efforts. One common challenge in leveraging such data is the diversity of measurement approaches and platforms and a lack of utilities enabling cross-platform comparisons among data sources for analysis. We present a method to map DNA methylation data from bisulfite sequencing approaches to CpG sites measured with the widely used Illumina methylation bead-array platforms. Correlations and median absolute deviations support the validity of using bisulfite sequencing data in combination with Illumina bead-array methylation data.
Availability and Implementation: https://github.com/Christensen-Lab-Dartmouth/methyLiftover includes source, documentation and data references.
Contact: brock.c.christensen@dartmouth.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
The public availability of high throughput molecular data provides new opportunities for researchers to advance discovery, replication and validation efforts. One common challenge in leveraging such data is the diversity of measurement approaches and platforms (Laird, 2010) and a lack of utilities enabling cross-platform comparisons among data sources for analysis. We introduce the methyLiftover utility, which allows simple and rapid remapping of DNA methylation data collected with bisulfite sequencing approaches to CpG sites measured with the widely used Illumina beadarray platforms. The methyLiftover utility was developed to increase the utility and comparability of the growing number of whole-genome bisulfite sequencing (WGBS) and reduced representation bisulfite sequencing (RRBS) data sets for the large number of researchers that collect and analyze methylation data from Illumina HumanMethylation 450K array. Bisulfite sequencing approaches to measure DNA methylation remain costly due to additional read depth and alignment issues associated with the reduced complexity of the bisulfite modified genome (Ziller et al., 2015). Though multiple large-scale epigenome characterization efforts using RRBS and WGBS continue to produce additional data, the research community will benefit from more readily available access and comparability of these data with much more widely used (and far less expensive) beadarray platforms from Illumina.
2 Methods
The methyLiftover utility provides two main functions. liftover450k accepts a user defined file input containing WGBS data (in BED or TXT format) and outputs an RData file containing sequencing data only from those CpG sites that are measured with the 450K array, selecting sites listed in the 450K annotation file via the IlluminaHumanMethylation450kanno.ilmn12.hg19 Bioconductor package (Hansen, 2015) (based on coordinates from Human Genome assembly hg19). The second function accepts two files (e.g. one containing WGBS or RBBS data, another containing 450K data) and joins them based on genomic position. Examples (with data) can be found with the code on the GitHub site. We note that the code can be used to reference the Illumina 850K annotation files using the function liftoverUserFiles. To test the validity of using whole-genome methylation values as a proxy for Illumina HumanMethylation 450K BeadChip methylation data, we filtered for known cross-hybridizing probes (Chen, 2013) calculated Pearson correlations (r) and median absolute deviation (MAD) between WGBS and 450K data collected from matched normal and tumor samples in The Cancer Genome Atlas (n = 22, Level = 3). For each biological specimen, we calculated Pearson correlations between whole-genome percent methylated values and 450K ‘beta values’ * 100. Correlations were mean-aggregated for each of five tissue/disease types: breast/BRCA, bladder/BLCA, lung/LUAD, stomach/STAD and uterine/UCEC. Illumina probe type normalization was done with beta-mixture quantile normalization (BMIQ) (Teschendorff et al., 2015).
3 Results
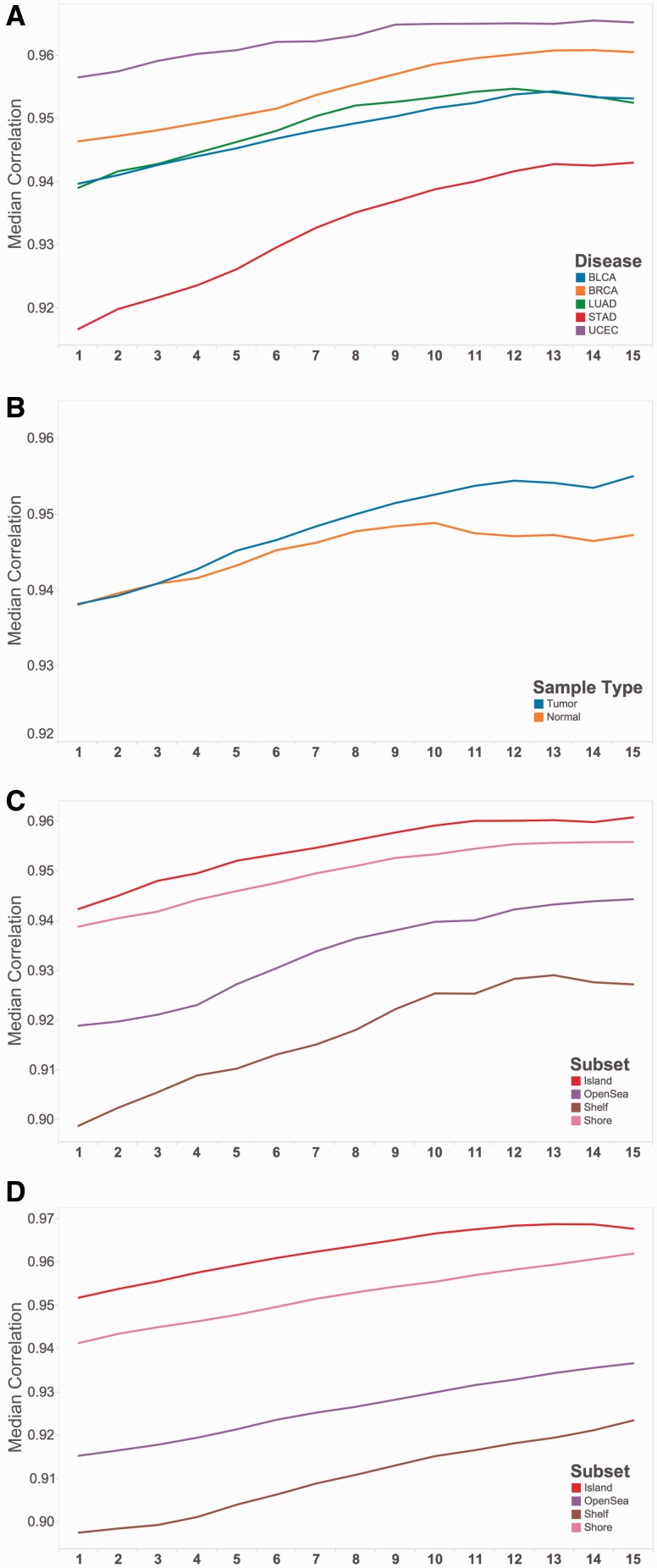
Running the methyLiftover utility on a local machine (8GB memory) is time efficient and low impact on computational resources. The tool imports 27M row WGBS BED files into memory in 12–15 s. Correlations within disease samples plateaued around a minimum read depth of 10 reads (Fig. 1A) in BRCA (r = 0.96, MAD = 5.46), BLCA (r = 0.95, MAD = 5.56), LUAD (r = 0.95, MAD = 5.79), STAD (r = 0.94, MAD = 6.52) and UCEC (r = 0.96, MAD = 4.41). The correlations were similarly strong with a minimum read depth of 10 reads in normal tissue (r = 0.95, MAD = 5.79, n = 5) and in tumor tissue (r = 0.95, MAD = 5.62, n = 17) (Fig. 1B). We identified an additional three matched samples (Heyn et al., 2012) in DNA from whole blood. Due to differences in WGBS and 450K sample labeling, we were able to confidently pair two of the WGBS/450K samples. Consistent with our TCGA results, the overall correlation between the 450K data and the methyLiftover WGBS subset in the two samples from Heyn et al. were 0.94 for a 103-year-old patient and 0.96 for a newborn patient. The data did not contain read depth (provided in original TCGA analysis) so we are unable to stratify by minimum read count and add them to Figure 1. Additionally, a beta value density comparison between BRCA normal and tumor samples is shown in Supplementary Figure 1.
Fig. 1.
Pearson correlation between whole-genome bisulfite sequencing and Illumina 450K array methylation data (n = 22) stratified by minimum read counts and (A) disease type, (B) tissue state (normal n = 5, tumor n = 17), and genomic context stratified by Infinium probes (C) type 1 and (D) type 2
For type I Illumina probes (MAD = 3.5), CpGs found in islands, shores, shelfs and open seas had correlations of 0.96, 0.952, 0.92 and 0.93, respectively (Fig. 1C). For type II probes (MAD = 6.5), CpGs found in islands, shores, shelfs, and open seas had correlations of 0.96, 0.95, 0.91 and 0.93, respectively (Fig. 1D). The upper quartiles of absolute deviation ranged from 10.5 to 17.5 and lower quartiles ranged from 1.9 to 3.63 across all samples. Our findings are consistent with previous comparisons of WGBS and 450K data in two paired samples (R2 = 0.95−0.96) (Bibikova, 2011). Further, correlation between methylation data from the Illumina 450K and the previous Illumina 27K array platforms was similar (R2 > 95) (Bibikova, 2011).
4 Conclusion
The use of WGBS and RRBS data in tandem with 450K methylation may help to expand the sample sizes available for cell-type specific and independent analyses. The methyLiftover utility will enhance the field of epigenomic research by expanding the comparability of DNA methylation data in the absence of the common Illumina 450K methylation data. The methyLiftover tool contains functions to subset and map WGBS and RRBS data to the CpG sites specific to the Illumina 450K array, both individually and as a whole directory, and create merged data sets from two user defined methylation data files (e.g. Illumina 850K annotation).
Funding
This work has been supported by P20GM104416/8189 (BCC), R01DE022772 (BCC), R01MH094609 (EAH) and R01ES024991 (EAH).
Conflict of Interest: none declared.
Supplementary Material
References
- Bibikova M. et al. (2011) High density DNA methylation array with single CpG site resolution. Genomics, 98, 288–295. [DOI] [PubMed] [Google Scholar]
- Chen Y. et al. (2013) Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics, 8, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K.D. (2015) IlluminaHumanMethylation450kanno.ilmn12.hg19: Annotation for Illumina’s 450k methylation arrays. R package, version 0.2.1.
- Heyn H. et al. (2012) Distinct DNA methylomes of newborns and centenarians. PNAS, 109, 10522–10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird P.W. (2010) Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet., 11, 191–203. [DOI] [PubMed] [Google Scholar]
- Teschendorff A.E. et al. (2012) A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics, 29, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller M.J. et al. (2015) Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat. Methods, 12, 230–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.