Abstract
Klf10, a member of the Krüppel-like family of transcription factors, is critical for osteoblast differentiation, bone formation and mineralization. However, whether Klf10 is involved in odontoblastic differentiation and tooth development has not been determined. In this study, we investigate the expression patterns of Klf10 during murine tooth development in vivo and its role in odontoblastic differentiation in vitro. Klf10 protein was expressed in the enamel organ and the underlying mesenchyme, ameloblasts and odontoblasts at early and later stages of murine molar formation. Furthermore, the expression of Klf10, Dmp1, Dspp and Runx2 was significantly elevated during the process of mouse dental papilla mesenchymal differentiation and mineralization. The overexpression of Klf10 induced dental papilla mesenchymal cell differentiation and mineralization as detected by alkaline phosphatase staining and alizarin red S assay. Klf10 additionally up-regulated the expression of odontoblastic differentiation marker genes Dmp1, Dspp and Runx2 in mouse dental papilla mesenchymal cells. The molecular mechanism of Klf10 in controlling Dmp1 and Dspp expression is thus to activate their regulatory regions in a dosage-dependent manner. Our results suggest that Klf10 is involved in tooth development and promotes odontoblastic differentiation via the up-regulation of Dmp1 and Dspp transcription.
Keywords: Klf10, Tooth development, Odontoblast, Dmp1, Dspp
Introduction
Tooth development is regulated by sequential and reciprocal interactions between the oral epithelial cells and the mesenchymal cells of the neural crest. Tooth morphology proceeds in three major stages: the bud, cap and bell. Mesenchyme cells form the dental pulp and the last division of pulp cells coming into contact with the basement membrane gives rise to odontoblasts. Odontoblasts have the ability to produce extracellular matrix components, such as dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (Dspp) and alkaline phosphatase (ALP) and have been implicated in dentin mineralization in response to caries, chemicals, or trauma (Mitsiadis and Rahiotis 2004). During this process, multiple signaling pathways converge to induce cell proliferation, differentiation and biomineralization (Thesleff and Mikkola 2002). However, the precise mechanism of tooth development is not completely understood.
Klf10, also known as transforming growth factor-β (TGFβ) inducible early gene-1 (TIEG1), is a member of the Krüppel-like family of transcription factors and was initially identified in normal human fetal osteoblasts (hFOB), following TGFβ treatment, by using the differential display polymerase chain reaction (PCR) method (Subramaniam et al. 1995). Klf10 encodes a 480-amino-acid protein, which contains several Src homology domains in the NH2-terminal region and three C2H2-type zinc finger DNA-binding motifs in the COOH-terminal domain (Subramaniam et al. 2010). Klf10 is expressed in a variety of cells and tissue types (Subramaniam et al. 1998, 2010) and directly regulates the TGFβ signaling pathway (Johnsen et al. 2002; Wara et al. 2011). Moreover, this gene functions as a tumor suppressor in cancer by inhibiting cell proliferation and inducing apoptosis (Chalaux et al. 1999; Ribeiro et al. 1999) and regulates a multitude of genes and biological processes (Subramaniam et al. 2007, 2010). Klf10 knockout (KO) mice display a gender-specific osteogenic phenotype with low bone mineral density, low bone mineral content and overall loss of bone strength in female mice (Subramaniam et al. 2005; Hawse et al. 2008, 2013). The molecular mechanism of Klf10 is to regulate osteoblast cell differentiation by stimulating osteoblast-specific gene expression such as that of Runx2 (Hawse et al. 2011). More importantly, allelic variations in Klf10 gene and altered Klf10 expression levels have been identified in patients with osteoporosis (Hopwood et al. 2009; Yerges et al. 2010). In addition, Klf10 is involved in angiogenesis and vascularization through the activation of cyclooxygenase 1 (COX-1; Wara et al. 2011; Yang et al. 2013).
Klf proteins contain similar structures and functionally compensate in biological activities (Heard et al. 2012; Spittau and Krieglstein 2012). Previously, we reported that Klf4 is specifically expressed in the polarizing and elongating odontoblasts during murine tooth development (Chen et al. 2009). Klf4 promotes the differentiation of odontoblasts via the up-regulation of Dmp1 (Lin et al. 2011, 2013). Since both Klf4 and Klf10 belong to the Krüppel-like factor family, they contain C-terminal DNA-binding domains (DBDs) consisting in three Cys2-His2 zinc fingers that are highly conserved (Knoedler and Denver 2014). In addition, Klf10 plays an important role in regulating osteoblast differentiation. Both osteoblasts and odontoblasts are derived from mesenchymal cells and mechanisms of osteogenesis and dentinogenesis resemble each other in critical steps. Factors involved in bone formation also play a critical role in dentinogenesis (Butler et al. 2003). Taking these data together, we hypothesized that Klf10 is involved in tooth development and odontoblast differentiation.
In this study, we evaluate the expression patterns of Klf10 in vivo from tooth bud formation to tooth eruption during mouse tooth development. Klf10 expression is also examined in an immortalized dental papilla mesenchymal cell line (iMDP-3). Klf10 induces iDMP-3 cell differentiation and mineralization and stimulates the expression of the Dmp1, Dspp and Runx2 genes. Furthermore, Klf10 controls Dmp1 and Dspp expression via the enhancement of their regulatory regions in iDMP-3 cells.
Materials and methods
Preparation of tissue sections
The Institution Review Board of the School and Hospital of Stomatology, Zhejiang University approved this study of animal use. Kun-ming mice were purchased from Zhejiang University (China). Noon of the day that a vaginal plug was found was considered as embryonic day 0.5 (E0.5), whereas noon of the day of birth was regarded as postnatal day 0.5 (PN0.5). At least three embryos and postnatal mice were killed for the collection of samples at various time points (E12.5, E14.5, E15.5, E16.5, E18.5, PN2, PN5, PN10 and PN20). The heads (E12.5-E16.5) and mandibles (E18.5-PN20) of embryonic and postnatal mice were dissected, fixed in 4 % formaldehyde and embedded in paraffin. Frontal serial sections (4 μm) were prepared.
Immunohistochemistry
Tissue sections were dewaxed in xylene for 15 min (3 times), hydrated in gradually decreasing ethanol concentrations for 2 min each and immersed in double-distilled H2O for 5 min (twice). Antigen retrieval was performed by treating the sections with 0.1 % (w/v) trypsin (Zhongshan, Beijing, China) at room temperature (RT) for 15 min. The sections were successively pretreated with 0.3 % hydrogen peroxide for 30 min, blocked in 2 % normal goat serum (Zhongshan) for 30 min and incubated with Klf10 antibody (a 1:100 dilution; Sigma-Aldrich, St. Louis, Mo., USA) at 4 °C overnight and subsequently with biotinylated goat anti-rabbit IgG and streptavidin-peroxidase conjugate (Zhongshan), respectively, at 37 °C for 30 min. The chromogen 0.1 % diaminobenzidine tetrahydrochloride (DAB) was used to visualize localization of the antigen. Finally, sections were counterstained with hematoxylin for 30 s. Negative controls were obtained by replacing primary antibody with mouse IgG I (Dakocytomation, Carpinteria, Calif., USA). For fluorescent immunohistochemistry, the tissue sections were dewaxed with xylene, rehydrated with ethanol and treated with H2O2. Then, the tissue sections were blocked with 10 % normal donkey serum (Sigma–Aldrich) and incubated with 10 mg/ml of either the primary goat or rabbit antibody, which was recognized by donkey anti-goat or anti-rabbit secondary antibody conjugated with Alexa Fluo 488 or Alex Fluo 568 (Molecular Probes, Eugene, Ore., USA) overnight at 4 °C. The sections were rinsed with phosphate-buffered saline, incubated with secondary antibody for 90 min and rinsed in H2O. For nuclear staining, the tissue sections were incubated with Hoechst (Sigma–Aldrich) for 5 min at RT. After being washed, the tissue sections were mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, Calif., USA). As a negative control, the primary antibody was replaced by mouse IgG I (Dakocytomation). Images of Alexa Fluo 488 and Alex Fluo 568 staining of the various proteins were captured by a Nikon Eclipse TE2000S microscope with a filter by means of a digital cooled camera connected to a PC computer and analyzed with NIS-Elements 3.2 software. Images of nuclear staining with Hoechst were obtained via filter UV-2E/C, C86826. For each experiment, all slides were simultaneously processed for a specific antibody, so that homogeneity in the staining procedure was ensured between samples. After the capture of the relevant images at the same magnification, the threshold was set with the same resolution and maintained for each slide in the experiment. The optical density was calculated by use of the morphometric analysis contained within the software package.
To determine the TGF-β1 effect on down-stream gene expression, mouse mandibles from E14.5 were isolated under stereomicroscopy. These mandibles were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco-BRL, Grand Island, N.Y., USA) supplemented with 0.5 % fetal bovine serum (FBS), 10 U/ml penicillin, 100 mg/ml streptomycin and treated with or without 10 ng/ml TGF-β1 (R&D Systems, Minneapolis, Minn., USA) for 2 days. The tissues were fixed in 4 % formaldehyde and embedded in paraffin. Serial sections were prepared. Fluorescent immunohistochemistry was performed by using antibodies directed against Dmp1 (gifts from Dr. Larry Fisher, NIDCR, USA), Klf5 (Santa Cruz Biotechnology, Santa Cruz, Calif., USA), Klf10 (Sigma-Al-drich) and Dsp (Alpha Diagnostic International, San Antonio, Tex., USA). Mouse IgG I was used as a negative control. Immunohistochemical assay was performed as described above with the corresponding secondary antibodies conjugated with Alexa Fluo 488 green fluorescent labeling (Molecular Probes). Images were observed under a Nikon Eclipse TE2000S microscope.
Cell culture
Immortalized dental papilla mesenchymal cells (iMDP-3) were generated as described previously (Wang et al. 2013). Cells were cultured in DMEM supplemented with 10 % FBS (Gibco-BRL), 100 U/ml penicillin and 100 mg/ml streptomycin in a humidified atmosphere of 5 % CO2 at 37 °C. For odontoblastic induction, iMDP-3 cells were seeded in six-well plates at an initial density of 2.5×104 cells/well and cultured in differentiation medium (DM; DMEM supplemented with 10 % FBS, 50 μg/ml ascorbic acid, 10 mM sodium β-glycerophosphate, 100 nM dexamethasone, 100 U/ml penicillin and 100 mg/ml streptomycin). The culture medium was replaced every 2 days during the incubation period. After induction for 0, 1, 3, 5, 7, 11 and 14 days, cells were prepared for ALP, alizarin red S (ARS), RNA extraction, Western blotting and immunofluorescent analyses.
Cell proliferation assay
Cell proliferation was determined by direct cell counting. Briefly, cells were plated into 6-well plates at 2.5×104 cells per well and cultured in DM for 0, 7 and 14 days at 37 °C. Cells were cultured in DMEM supplemented with 10 % FBS, 100 U/ml penicillin and 100 mg/ml streptomycin as a control. The cells were trypsinized and counted by using a hemocytometer under light microscopy. Cell counts were performed in triplicate and repeated in three cultures (n=5).
ALP staining
iMDP-3 cells were incubated for 0, 7 and 14 days in the differentiation medium as described above. Cells were fixed with 10 % formalin for 1 min and washed with PBS. In situ ALP staining was performed according to the manufacturer’s instructions (Bio-Rad Laboratory, Hercules, Calif., USA). Experiments were performed in triplicate and repeated in three cultures (n=5).
ARS staining
For cell mineralization assay, iMDP-3 cells were cultured in odontoblastic induction medium for 0, 7 and 14 days and stained by ARS staining (Sigma-Aldrich). Briefly, the cells were fixed in 10 % formalin for 30 min and rinsed with distilled water. Cells were then stained with 1 % ARS (pH 4.2) under gentle agitation. ARS staining was documented via a microscope (Carl Zeiss, Jena, Germany). The ARS staining was performed in triplicate and repeated in three cultures (n=5).
RNA extraction and quantitative real-time PCR
Total RNAwas isolated from whole tooth germs (E18.5, PN2, PN6, PN10 and PN20), the DM-induced iMDP-3 and control cells at 0, 1, 3, 5, 7, 11 and 14 days by using TRIzol Reagent (Invitrogen, San Diego, Calif., USA). The RNA was transcribed into cDNA by SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Specific primers for the PCR were synthesized as follows: Klf10, forward 5′-GTGACCGTCGGTTTATGAGG-3′, reverse 5′-ACTTCCATTTGCCAGTTTGG-3′; Dmp-1, forward 5′-CAGTGAGGATGAGGCAGACA-3′, reverse 5′-TCGATCGCTCCTGGTACTCT-3′; Dspp, forward 5′-AACTCTGTGGCTGTGCCTCT-3′, reverse 5′-TATTGACTCGGAGCCATTCC-3′; Runx2, forward 5′-TACAAACCATACCCAGTCCCTGTTT-3′, reverse 5′-AGTGCTCTAACCACAGTCCATGCA-3′; and Cyclo-A, forward 5′-GGTGACTTCACACGCCATAA-3′, reverse 5′-CATGGCCTCCACAATATTCA-3′. Quantitative real-time PCR was performed in an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, Calif., USA) with SYBR Green (Thermo Scientific, Waltham, USA). All samples were detected in triplicate in 96-well plates, with reactions being performed at 50 °C for 2 min and then at 95 °C for 10 min, followed by 35 cycles of 95 °C for 15 s, 60 °C for 40 s and 72 °C for 40s. Finally, the gene expression ratio of target genes and the reference gene was analyzed with a standard curve by ABI 7500 software 2.04 (Applied Biosystems). For data analysis, the expression levels of target genes were normalized to the expression of the reference gene, Cyclophilin A (Cyclo A). The expression levels of target genes, including Klf10, Dmp1, Dspp and Runx2, in DM-induced iMDP-3 cells were calculated by using the comparative cycle threshold method (ΔΔ CT) relative to the expression level in the control groups.
Protein extraction and Western blot analysis
iMDP-3 cells induced for various times (0, 1, 3, 5, 7, 11, and 14 days) were lysed with RIPA buffer (Santa Cruz Biotechnology) at 4 °C by vigorous shaking for 15 min. After centrifugation at 12,000g for 15 min, the supernatant was separated and stored at −80 °C until use. The protein concentration was measured by using a Bio-Rad protein assay kit (Bio-Rad Laboratory). An equal amount of proteins was loaded onto a 10 % SDS-polyacrylamide gel electrophoresis (SDS-PAGE) system and the bands were subsequently transferred to a membrane (Bio-Rad Laboratory). Rabbit anti-Klf10 (Sigma-Aldrich) and goat anti-actin (Santa Cruz Biotechnology) were used as primary antibodies. Transferred proteins were incubated with ECL substrate solution (Thermo Scientific) according to the manufacturer’s instructions and labeled bands were revealed on X-ray film.
Double immunofluorescence analysis
Cells were fixed in cold acetone and methanol (1:1), permeabilized in 0.2 % Triton X-100 at ice for 30 min and blocked with 10 % goat serum for 30 min at RT. For double-labeling, two primary antibodies were incubated simultaneously overnight at 4 °C at the following dilutions: rabbit anti-Klf10 (1:100; Sigma-Aldrich), goat anti-Dsp (1:100; Santa Cruz Biotechnology), goat anti-Runx2 (1:100; Santa Cruz Biotechnology) and mouse anti-Dmp1 (1:100; kind gift from Dr. Chunlin Qin, Texas A&M University, Baylor College of Dentistry, Tex., USA; Qin et al. 2006). After being washed, slides were incubated with the secondary antibody conjugated with Alexa Fluo 486 green and Alexa Fluo 568 red (1:500; Molecular Probes) for 1 h at RT. As a negative control, the primary Klf10 antibody was replaced by mouse IgG I. For nuclear staining, the cells were treated with Hoechst (Sigma-Aldrich). Slides were viewed and captured by using a Nikon inverted microscope. For double-fluorescent immunohistochemistry, the co-expression of Klf10 and PCNA (proliferating cell nuclear antigen) was analyzed in tissue sections from various mouse ages. Anti-PCNA antibody was purchased from Santa Cruz Biotechnology.
Plasmid constructions
Human Dmp1 promoter plasmids were generated as described previously (Chen et al. 2004a). The sequences between nucleotides (nt) −213 and +83, nt −656 and +83, nt −1187 and +83, nt −1656 and +83 and nt −2.6 kb and +83 were constructed into pGL-3 basic luciferase vector, respectively (Promega, Madison, Wis., USA). The mouse Dspp promoter plasmids were also constructed as described previously (Chen et al. 2004b, 2008). Briefly, the fragments between nt −591 and + 54, nt −1318 and +54, nt −1.5 kb and +54, nt −2.6 kb and +54 and nt −5.6 kb and +54 were subcloned into pGL-3 basic luciferase vector. The plasmid of Klf10 (pcDNA4-KLF10) was kindly provided by Dr. John R. Hawse (Department of Biochemistry and Molecular Biology, Mayo Clinic, Minn., USA; Hawse et al. 2011). All constructs were verified by DNA sequencing.
Transient transfection assay
iMDP-3 cells were cultured in six-well plates (nearly 80 % confluent) and maintained in α-minimal essential medium (Sigma-Aldrich) supplemented with 10 % FBS (Gibco-BRL Life Technologies, Paisley, UK). For transfection assay, the cells were transfected with Klf10 expression vector (pcDNA4-Klf10) or the respective empty vector by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The medium was removed after 5 h and replenished with media containing 10 % FBS. After 48 h, cells were harvested and used for real-time PCR, Western blot, ALP and ARS staining assays.
Luciferase assay
To detect transcriptional activity, the promoter activity of Dmp1 and Dspp was determined by using the dual-luciferase reporter assay kit (Promega). iMDP-3 cells were plated in 12-well plates and grown in culture to 80 % confluence. The pGL3-basic empty vector (Promega) was utilized as a baseline control. The cells were co-transfected with either pGL3-Dmp1-promoter or pGL3-Dspp-promoter and the TK-Renilla luciferase plasmid as an internal control by using the Lipofectamine 2000. For those experiments in which Klf10 was overexpressed, the plasmid pcDNA4-Klf10 was co-transfected with the luciferase reporter vectors. After 48 h of post-transfection, the cells were lysed for the promoter assay according to the manufacturer’s instructions. Promoter activity was equal to the ratio of Firefly and Renilla luciferase activities determined by using the Glomax Luminometer (Promega) and is presented as fold-change comparing empty vector pGL3 Firefly luciferase with Renilla luciferase. The promoter activity detection was repeated three times in parallel.
Statistical analysis
Statistical differences among groups were analyzed by using one-way analysis of variance within SPSS software (Version 10.0; SPSS, Chicago, Ill., USA). All values were calculated as means±standard deviation (S.D.). The statistical significance was determined by one-way analysis of variance at P<0.05.
Results
Expression of Klf10 during murine tooth development
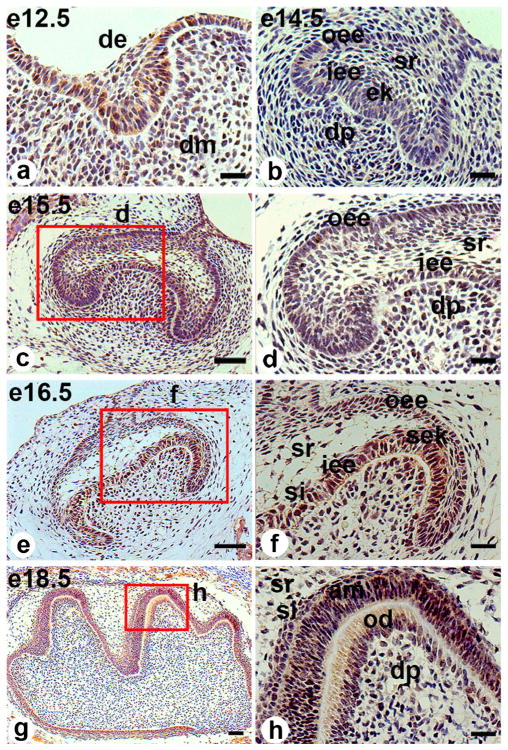
The morphogenesis and structure of the first molar from E12.5 to PN20 were determined by hematoxylin-eosin staining. The tooth germ underwent various development stages, including the bud, cap and bell stages and later stages including tooth root formation and tooth eruption. Immunochemistry revealed that Klf10 protein was expressed in various cell types of mouse molars from E12.5 to PN20.
At the bud stage (E12.5), the thickened epithelial buds arise from the dental lamina and the underlying mesenchyme cells condense. Both the epithelial and mesenchymal cells proliferate. Klf10 was slightly expressed in both the dental epithelium and the underlying mesenchyme (Fig. 1a). At the cap stage (E14.5), dental inner enamel epithelial cells proliferate and form the primary enamel knot at the central region. The inner and outer enamel epithelial cells are defined. The mesenchymal cells underlying the enamel knot proliferate and form the dental papilla (Fig. 1b). Klf10 expression was seen in the inner and outer enamel epithelial cells, the stratum reticulum and the mesenchymal cells and dental papillae (Fig. 1b). Moreover, the cells from the dental follicle adjacent to the enamel organ were positive for Klf10 immunostaining. Similar to E14.5, a condensed staining of Klf10 was observed in the above cells at E15.5 (Fig. 1c, d). At the early bell stage (E16.5), positive staining of Klf10 was found in the secondary enamel knot, inner and outer enamel epithelia, stratum intermedium, stratum reticulum and mesenchymal cells beneath the inner epithelia (Fig. 1e, f). At the late bell stage (E18.5), the dental epithelial cells begin to differentiate into ameloblasts and mesenchymal cells into odontoblasts (Fig. 1g, h). At this stage, the odontoblasts begin to polarize and become tall and columnar, with an increase in total organelles. The expression of Klf10 protein was clearly visible within both the cytoplasm and nuclei in the odontoblasts. Furthermore, strong Klf10 staining appeared in the differentiating ameloblasts, stratum intermedium, stratum reticulum and dental pulp cells (Fig. 1h).
Fig. 1.
Klf10 immunostaining in embryonic mouse molar germs. a At embryonic day (E) 12.5 (e12.5), Klf10 was expressed in both the dental epithelium and the underlying mesenchyme. b At E14.5 (e14.5), Klf10 appeared in the inner and outer enamel epithelia, stratum reticulum, primary enamel knot and dental papilla. c At E15.5 (e15.5), the Klf10 expression pattern was similar to that of E14.5. e By E16.5 (e16.5), Klf10 expression was found in the secondary enamel knot, inner and outer enamel epithelia, stratum intermedium and stratum reticulum and also in the mesenchymal cells beneath the inner enamel epithelia. g At E18.5 (e18.5), positive Klf10 staining appeared in the differentiating ameloblasts and odontoblasts, the outer enamel epithelia, the stratum intermedium, the stratum reticulum and the dental papilla cells. d, f, h Higher magnification images of red-boxed areas in c, e, g, respectively (de dental epithlium, dm dental mesenchyme, oee outer enamel epithelium, iee inner enamel epithelium, ek enamel knot, dp dental pulp, sr stratum reticulum, si stratum intermedium, sek secondary enamel knot, am ameloblast, od odontoblast). Bars 20 μM (a, b, d, f, h), 50 μM (c, e, g)
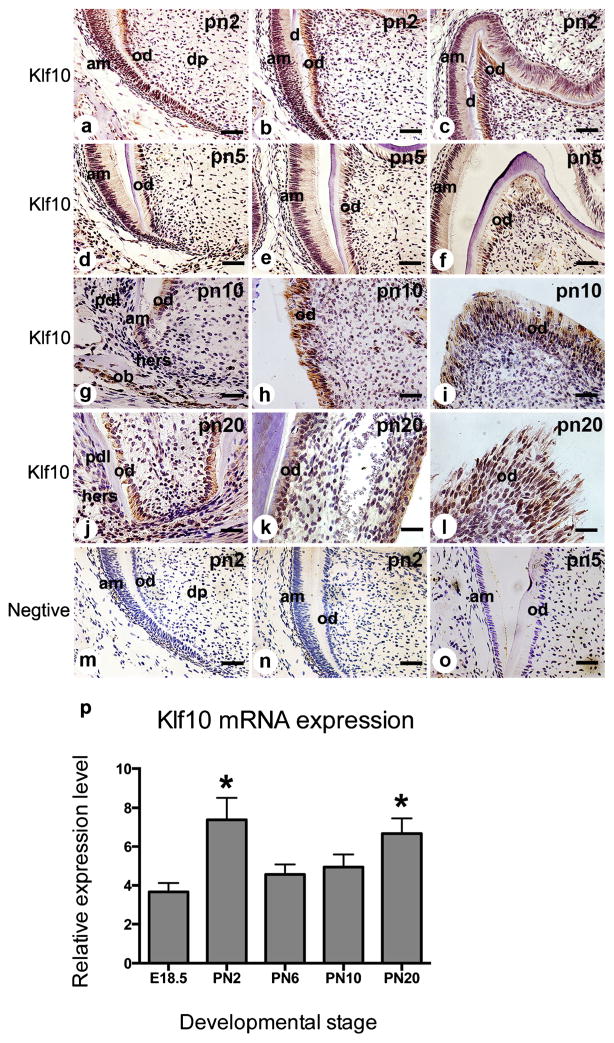
At PN2 (Fig. 2a–c), Klf10 was strongly expressed in preodontoblasts/preameloblasts and in polarizing, secretory and mature odontoblasts and ameloblasts. Immunoreaction of Klf10 was moderately observed in stratum intermedium and dental pulp cells (Fig. 2b, c). At PN5, the Klf10 expression pattern was similar to that in PN2 (Fig. 2d–f). By PN10, crown morphogenesis was basically completed. Klf10 expression was detected in the odontoblasts, ameloblasts and reduced dental epithelium (Fig. 2g–i); positive staining of Klf10 was also apparent in alveolar osteoblasts (Fig. 2g). By contrast, the cells of the Hertwig’s epithelium root sheath (HERS) and periodontal ligament (PDL) fibroblasts showed faint Klf10 staining (Fig. 2g). At PN20 (Fig. 2j–l), the first mouse molar erupts with hard-mineralized tissue and the tooth root undergoes advanced formation. Klf10 expression was seen in the odontoblasts both in the crown and in the root (Fig. 2j–l); weak staining of Klf10 was observed in HERS cells and PDL fibroblasts (Fig. 2j).
Fig. 2.
Klf10 expression in postnatal mouse molar development. a–c At PN2 (pn2), Klf10 was strongly expressed in preodontoblasts/ameloblasts and in polarizing, secretory and mature odontoblasts and ameloblasts and in several dental pulp fibroblasts. d–f At PN5 (pn5), the Klf10 expression pattern was similar to that at PN2. g–i At PN10 (pn10), Klf10 expression was apparent in ameloblasts, odontoblasts and osteoblasts, while its expression was moderately seen in dental pulp cells. Klf10 expression was barely visible in Hertwig’s epithelium root sheath (HERS) and periodontal ligament (PDL) cells. j At PN20 (pn20), Klf10 staining was observed in the odontoblasts in the root and faint staining of Klf10 was observed in HERS cells. k–l Klf10 was seen in the odontoblasts in the crown. m–o Tissue sections from PN2 and PN5 were stained with IgG as a negative control (am ameloblast, od odontoblast, ob osteoblast, pdl periodontal ligament fibroblast, hers Hertwig’s epithelial root sheath). Bars 20 μM (a–f, m–o), 50 μM (g–l). p Klf10 transcript was expressed in the first mandibular molars from E18.5 to PN20 (error bars means ± S.D.; *P<0.05 with significant difference versus E18.5)
To investigate Klf10 mRNA expression during murine tooth development, RNAs were isolated from various stages of mouse molars and Klf10 mRNA expression levels were analyzed by real-time PCR. Klf10 mRNA expression was significantly increased and had the highest level on PN2 and then maintained constant levels until PN20 (Fig. 2p). The Klf10 mRNA expression pattern coincided with that of Klf10 protein during mouse tooth development (Figs. 1, 2).
As Klf10 was expressed in primary and secondary enamel knots, the co-expression of Klf10 with cell proliferation marker, PCNA, was analyzed by using double-fluorescent immunohistochemistry. The results showed that Klf10 and PCNA were co-expressed in the primary and secondary enamel knots (Supplemental Fig. 1). Moreover, the expression of Klf10 and PCNA was seen in ameloblasts and odontoblasts in mouse teeth.
Expression of Klf10 and odontoblast marker genes during odontoblastic differentiation
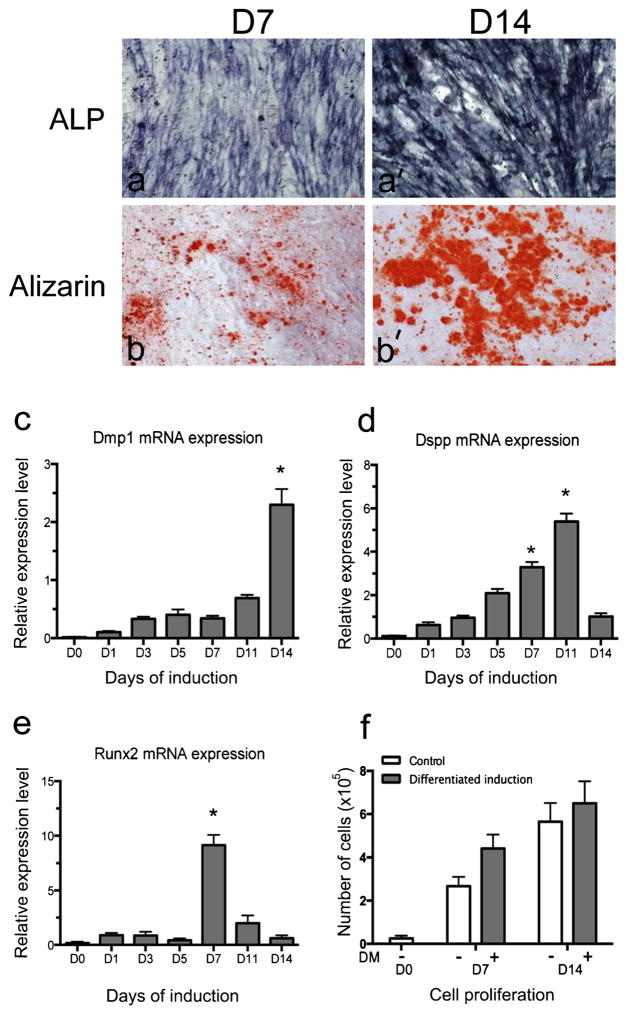
To examine odontoblastic differentiation, mouse dental papilla mesenchymal (iMDP-3) cells were cultured in differentiation medium for 7 and 14 days and the samples were collected and analyzed by ALP and ARS staining, as ALP and ARS are important markers during dental cell differentiation and biomineralization. The results demonstrated that ALP expression significantly increased in DM-induced iMDP-3 cells on day 7 and had a higher level on day 14 (Fig. 3a, a′). ARS staining showed the same trend as ALP staining. Mineralized nodules were detected on day 7 and the densities and sizes of the mineralized nodules increased with longer induction (Fig. 3b, b′).
Fig. 3.
Differentiation of dental papilla mesenchymal cells and odontoblast marker gene expression. (a, a′) Alkaline phosphatase (ALP) and (b, b′) Alizarin red S (Alizarin) staining of iMDP-3 cells on days 7 and 14 after differentiation induction. Messenger RNA of (c) Dmp1, (d) Dspp and (e) Runx2 in iMDP-3 cells after odontoblastic induction. Cell numbers (f) on days 7 and 14 after differentiation induction at the given time periods. *P<0.05 with significant difference versus day 0
To determine the gene(s) correlated with the differentiation and mineralization of mouse dental papilla mesenchymal cells, we examined the expression of the Dmp1, Dspp and Runx2 genes in this process, as these genes are involved in odontoblast differentiation and mineralization (Chen et al. 2004a, 2005). The messenger RNA level of Dmp1, Dspp and Runx2 was investigated by real-time PCR. Expression of Dmp1 and Dspp mRNAs gradually increased with time and the maximal level of Dmp1 expression was detected on day 14 (Fig. 3c), whereas the highest expression of Dspp mRNA was seen on day 11 (Fig. 3d). The messenger RNA level of Runx2 was increased significantly on day 7 (Fig. 3e).
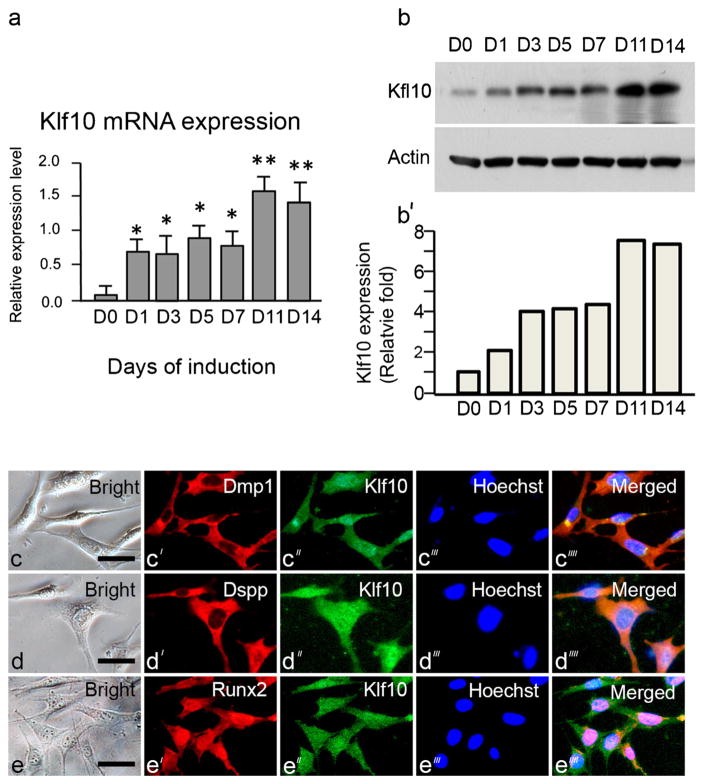
Similar to our in vivo study, the in vitro analysis showed that Klf10 expression increased during the odontoblast differentiation of iMDP-3 cells as analyzed by real-time PCR and Western blotting assays. Klf10 mRNA levels increased in a time-dependent manner following exposure to DM after the initiation of treatment and then significantly increased to higher peaks on days 11 and 14 (Fig. 4a). Furthermore, the Klf10 protein expression pattern in iMDP-3 cells was similar to that of Klf10 mRNA expression (Fig. 4b, b′).
Fig. 4.
Expression of Klf10 during odontoblastic differentiation of iMDP3 cells. a Messenger RNA of Klf10 during odontoblastic induction at various time intervals. *P<0.05 with significant difference versus day 0. b, b′ Protein level of Klf10 after odontoblastic induction was measured by Western blot assay by using anti-Klf10 antibody. Actin was used as the internal control. Expression of Klf10 and actin was quantitated by using image J software. Klf10 expression was normalized to actin expression. Klf10 protein on day 0 was arbitrarily considered to represent a one-fold increase. The expression level of Klf10 at various times was divided by the Klf10 expression on day 0. The co-expression of Klf10 (green, c″, d″ e″) with Dmp1 (red, c′), Dspp (red, d′) and Runx2 (red, e′) in iMDP-3 cells is shown in the merged images (c⁗–e⁗), respectively, after immunostaining with the relevant antibodies. Hoechst (blue) was used for nuclear staining (c‴–e‴). Bar 20 μm
As the expression of Klf10 and of Dmp1, Dspp and Runx2 increased during the differentiation of dental papilla mesenchymal cells, we next tested whether Klf10, Dmp1, Dspp and Runx2 were co-expressed in iMDP-3 cells. Using a double-fluorescent histochemistry assay, we found that the expression of Klf10 and these proteins is co-localized within the cytoplasm and nucleus (Fig. 4c′, c″, d′, d″, e′, e″). These results demonstrated that iMDP-3 cells could differentiate into odontoblast-like cells and that the differentiation of mouse dental papilla mesenchymal cells into odontoblast-like cells was relevant to odontoblast-relate gene expression including the expression of Klf10, Dmp-1, Dspp and Runx2. Moreover, DM-induced iMDP-3 cells displayed a more rapid growth rate than the non-induction cells as indicated by cell counting (Fig. 3f).
Effect of Klf10 on odontoblastic differentiation and odontoblast-relate gene expression
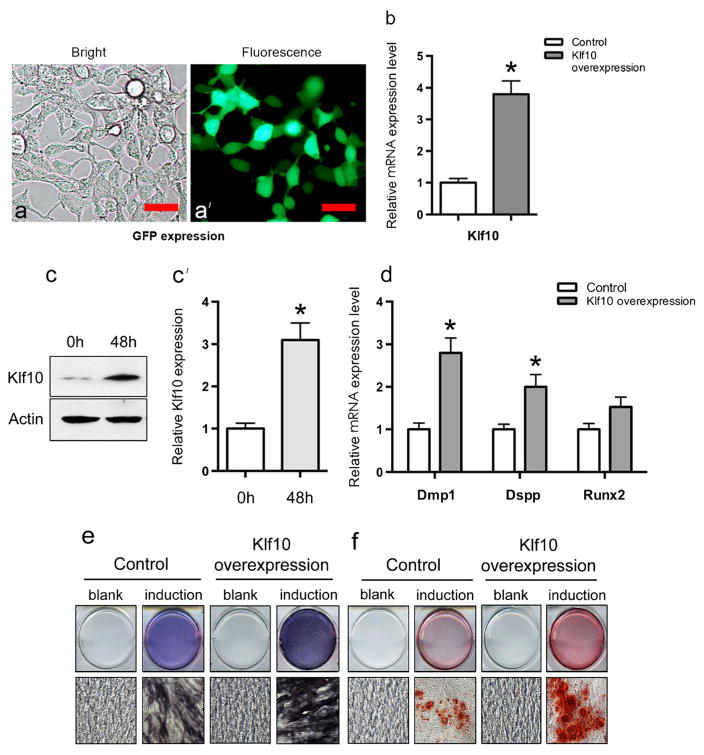
To gain further insight into the way that Klf10 affects odontoblastic differentiation, Klf10 was overexpressed in iMDP-3 cells. At first, in order to determine iMDP-3 cell transfection efficiency, the green fluorescent protein (GFP) gene was transiently transfected into iMDP-3 cells and GFP expression was observed by using reverted fluorescent microscopy after 48 h of transfection. Approximately 50 % GFP expression was seen in the transfected iMDP-3 cells (Fig. 5a, a′). This result indicated that iMDP-3 cells are a cell line suitable for functional study (Wang et al. 2013). Then, the Klf10 gene was transfected into iMDP-3 cells. We found that the mRNA level of Klf10 in the overexpression group was 3.8-fold higher than that in the control group (Fig. 5b). Consistent with the mRNA expression, the protein level of Klf10 also increased markedly in the overexpression group (Fig. 5c, c′). More importantly, the mRNA level of Dmp1, Dspp and Runx2 also increased in the Klf10 overexpression group as detected by real-time PCR (Fig. 5d). Moreover, Klf10 overexpression was able to induce cell differentiation and mineralization to a greater extent than in the control groups (Fig. 5e, f). These results indicated that Klf10 promoted odontoblastic differentiation and enhanced calcium deposition in iMDP-3 cells by regulating the Dmp1 and Dspp genes.
Fig. 5.
Effect of Klf10 on odontoblastic differentiation of iMDP-3 cells. a, a′ Green fluorescent protein (GFP) was present in approximately 50 % of iMDP-3 cells after transfection. b mRNA expression levels of Klf10 gene after Klf10 overexpression in iMDP-3 cells. Bars 20 μm. c, c′ Protein expression levels of Klf10 gene after Klf10 overexpression in iMDP-3 cells. d mRNA level of Dmp1, Dspp and Runx2 after Klf10 overexpression. e ALP staining in iMDP-3 cells detected on day 7 after Klf10 overexpression and differentiation induction. f Alizarin red S staining in iMDP-3 cells detected on day 7 after Klf10 overexpression and differentiation induction. *P<0.05 significant difference versus control
Klf10 expression is known to be induced by TGF-β1 (Johnsen et al. 2002; Wara et al. 2011). To determine whether TGF-β1 is able to up-regulate tooth-related gene expression via Kfl10, mandibles from E14.5 were treated with recombinant TGF-β1. The results demonstrated that TGF-β1 induced the expression of the Klf5, Klf10, Dmp1 and Dspp genes (Supplemental Fig. 2), indicating that Klf5, Klf10, Dmp1 and Dspp are downstream genes of TGF-β1 and that the expression of the Dmp1 and Dspp genes controlled by TGF-β1 might be further regulated through Klf10 and other Klf family members.
Klf10 promotes expression of Dmp1 and Dspp genes via their regulatory regions
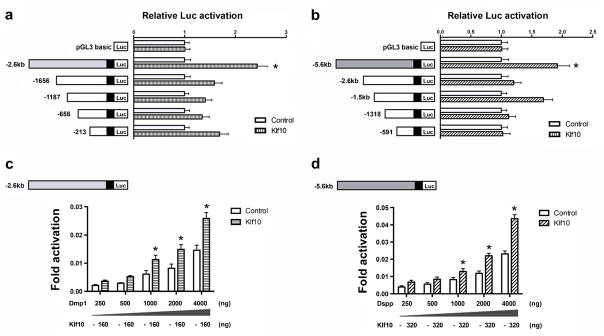
To elucidate further the mechanistic basis underlying the up-regulation of Dmp1 and Dspp expression by Klf10, we studied whether Klf10 regulated the expression of Dmp1 and Dspp by controlling their regulatory regions. Different constructs of Dmp1 and Dspp promoter regions were generated and subcloned into luciferase reporter vectors, respectively (Fig. 6a, b). These chimeric DNA constructs were co-transfected with Klf10 expression vector into iMDP-3 cells and the putative promoter activity was measured by a dual-luciferase assay. The luciferase assay showed that the variants of Dmp1 and Dspp promoter regions yielded different luciferase activities. For Dmp1 promoters, the responsible element(s) of Klf10 is located in pGL3-2.6 kb, showing a 2.4-fold increase in relative luciferase activity (Fig. 6a), whereas the pGL3-5.6 kb for the Dspp promoter appeared to increase luciferase activity approximately two-fold by Klf10 (Fig. 6b). These Dmp1 and Dspp reporter activities induced by Klf10 were dosage-dependent (Fig. 6c, d). Our results thus further confirmed that the induction of odontoblast differentiation and mineralization by Klf10 in part up-regulated the transcription of Dmp1 and Dspp in iMDP-3 cells.
Fig. 6.
Klf10 transactivates Dmp1 and Dspp genes in iMDP-3 cells. Mouse Klf10 expression plasmid or empty (−) expression vector were co-transfected into iMDP3 cells with (a) Dmp1 promoter constructs (pGL3-213, pGL3-656, pGL3-1187, pGL3-1656, or pGL3-2.6 kb) or with (b) Dspp promoter plasmids (pGL3-591, pGL3-1318, pGL3-1.5 kb, pGL3-2.6 kb, or pGL3-5.6 kb). Co-expression of Klf10 with either Dmp1 or Dspp reporter construct resulted in an increase of (c) Dmp1 pGL3-2.6 kb and (d) Dspp pGL3-2.6 kb activities in iDMP-3 cells in a dosage-dependent manner. Luciferase (Luc) activity was normalized to the control group. *P<0.05 significant difference versus control
Discussion
Klf10 is known to induce and repress the expression of multiple genes in several cell types and functions as a regulator of cell proliferation and apoptosis (Subramaniam et al. 2007). Immunohistochemical studies have demonstrated that Klf10 protein is expressed in numerous tissues, including the epithelial cells of the placenta, breast and pancreas, in osteoblast cells, in selected cells of the bone marrow (Subramaniam et al. 1998) and in the cerebral cortex (Kobori et al. 2002), myeloid cells (Noti et al. 2004), smooth muscle cells (Luo et al. 2005), fibroblasts (Mitsumoto et al. 2003), oligodendroglial cells (Bender et al. 2004) and glial cells (Yajima et al. 1997). In Klf10 KO mice, osteoblast cell differentiation and bone mineralization are impaired (Subramaniam et al. 2005). Like bone, dentin is derived from mesenchymal cells. In this study, we investigated whether Klf10 biological functions are relevant to odontoblast differentiation and dentin formation. We observed the expression patterns of Klf10 from early stages to later stages of murine molar development and determined the spatial-temporal expression of Klf10 at these stages. Furthermore, we found that Klf10 is co-expressed with odontoblast-relate genes Dmp1, Dspp and Runx2 in mouse dental papilla mesenchymal cells. The expression of these genes is increased during odontoblast differentiation. The forced expression of Klf10 promotes odontoblastic cell differentiation and mineralization via the regulation of Dmp1 and Dspp gene expression in mouse dental papilla mesenchymal cells.
One of the key steps in tooth development is the transition from the bud to cap stage. This is preceded by the condensation of the mesenchyme around the budding epithelium and the formation of the primary enamel knot (PEK) marking the position of the initiation of epithelial folding (Aberg et al. 2004). The PEK might act as an organizing center controlling tooth morphogenesis, causing the unequal growth of the enamel epithelium and inducing the formation of secondary enamel knots (SEKs) located at the tips of the forming cusps (Jernvall et al. 1994). Klf10 protein expression was found in both the enamel organ and the underlying mesenchyme at the bud and cap stages (Fig. 1a, b). Moreover, Klf10 is co-expressed with cell proliferation marker, PCNA, within PEK and SEKs in mouse teeth (Supplemental Fig. 1). This suggests that Klf10 is involved in the molecular regulation of tooth development.
At the bell stage, the enamel organ progressively delineates the dental papilla, cusps start to form and the size of the crown increases. At the same time, odontoblasts and ameloblasts occur showing cellular differentiation and polarization (Lesot and Brook 2009). At this stage, Klf10 expression was seen in the differentiating ameloblasts and odontoblasts. Furthermore, at postnatal days, Klf10 protein is also detectable in the preodontoblasts/preameloblasts and in polarizing, secretory and mature odontoblasts and ameloblasts. These consistent expression patterns are assumed to indicate that Klf10 is closely correlated with ameloblast and odontoblast differentiation.
By contrast, Klf10 shows weak expression in HERS and PDL fibroblasts during postnatal days. During tooth root development, the two possible fates of HERS are to differentiate toward PDL fibroblasts (Andujar et al. 1985) and cementoblasts (Sonoyama et al. 2007; Zeichner-David et al. 2003), whereas PDL fibroblasts maintain their potential to differentiate into osteoblasts and cementoblasts under certain conditions (Camilleri and McDonald 2006; Saito et al. 2014). Interestingly, we found the positive expression of Klf10 in osteoblasts but only faint staining in the progenitor cells. These results indicate that Klf10 expression is relevant to cell differentiation and cell cycles in certain cell types.
Klf10 has previously been reported to be predominantly located within the nucleus (Cook et al. 1998; Spittau et al. 2007; Subramaniam et al. 1998), although Klf10 expression has also been detected in the cytoplasm and this protein is inducible from the cytoplasm into the nuclei in human fetal osteoblast cells after bone morphogenetic protein-2 treatment (Hefferan et al. 2000). In this study, we observed that Klf10 is localized in both the cytoplasm and nuclei of odontoblasts and ameloblasts (Figs. 1, 2, 4c″). The expression of Klf10 within the cytoplasm and nucleus might be dependent on the cell type.
To improve the definition of the role of Klf10 during tooth formation and mineralization, we also monitored Klf10 expression in vitro and in vivo. Klf10 expression increases during odontoblastic cell differentiation (Figs. 1, 2, 4). In addition, Dmp1, Dspp and Runx2, as important markers of odontoblast differentiation, also increase during this process. Klf10 is one member of the family Sp1/Krüpped-like zinc finger transcriptional factors and shares a highly conserved COOH-terminal DNA-binding domain containing three C2H2 zinc finger motifs that facilitate binding to GC-rich regulatory regions in the target genes (Kaczynski et al. 2003); Klf10 functionally compensates for other Klf members such as Klf11 (Spittau and Krieglstein 2012). To assess the role of Klf10 in iMDP-3 cell differentiation, we overexpressed the Klf10 gene in iMDP-3 cells. The results show that the forced expression of Klf10 induces iDMP-3 cell differentiation and mineralization as assayed by ALP and ARS analyses (Fig. 5e, f). Moreover, Klf10 can up-regulate the expression of Dmp1, Dspp and Runx2 in iDMP-3 cells. These in vitro data are consistent with our in vivo results showing that Klf10 is apparently expressed in the differentiating and mature odontoblasts (Fig. 2). Furthermore, the strong expression of Klf10 in the osteoblasts of alveolar bone (Fig. 2g) is consistent with other studies (Hawse et al. 2011; Subramaniam et al. 1998).
Klf10 has been verified to play a role in regulating the expression of bone-related genes, including Runx2, OPG, BMP2 and RANKL, and in osteoblast maturation/differentiation and osteogenesis (Bensamoun et al. 2006). The factors involved in osteogenesis and bone repair, such as BMP2, RANKL and OPG, also play important roles in dentinogenesis (Feng et al. 2011; Kuntz et al. 2001). Previous studies have shown that growth factors such as TGF-β1 and BMP2 up-regulate downstream gene expression through Klf10 signaling (Bensamoun et al. 2006; Hawse et al. 2011). Our study revealed that TGF-β1 up-regulates the expression of the Klf10, Klf5, Dmp1 and Dspp genes in dental epithelial and mesenchymal cells (Supplemental Fig. 2) and we assume that TGF-β1 controls the expression of the Dmp1 and Dspp genes through Klf10 and other Klf family members. The molecular mechanisms of TGF-β1 in dental relate genes through Klf10 need to be further investigated in the future. Taken together, these data imply that Klf10 is closely correlated with odontoblast differentiation via the regulation of the expression of the downstream target genes.
Runx2 is known to be essential for tooth formation, being intimately involved in the development of calcified tooth tissue and it exerts an influence on the proliferation of the dental lamina (Camilleri and McDonald 2006). Runx2 participates in activating and suppressing Dspp expression, which is mainly involved in dentinogenesis (Chen et al. 2005; Li et al. 2011). Dspp mutations in humans and mice are associated with dentinogenesis imperfect a type II and type III (DGI-II and DGI-III) and with dentin dysplasia type II (DD-II; Shields et al. 1973). Moreover, Klf10 KO mice display reduced expression levels of Runx2 and exhibit significant delays in the rate of osteoblast mineralization (Hawse et al. 2011). In the present study, we have found that the overexpression of Klf10 stimulates Runx2 expression in iDMP-3 cells, indicating that Klf10 induces differentiation and mineralization in dental papilla mesenchymal cells via the Runx2 signal.
In this study, the forced expression of Klf10 resulted in the up-regulation of odontoblastic differentiation markers, Dmp1 and Dspp, in addition to Runx2 and enhanced ALP expression and the appearance of dense mineralized nodules. These results indicate that Klf10 is able to promote odontoblastic differentiation and increase calcium deposition in iMDP-3 cells by controlling the expression of Dmp1, Dspp and Runx2. In order to gain an understanding of the molecular mechanism of Klf10 in the regulation of Dmp1 and Dspp expression, either the Dmp1-luciferase reporter or the Dspp-luciferase reporter gene was co-transfected with Klf10 expression vector in iDMP-3 cells. We observed that Klf10 was able to significantly activate the promoter activity of the Dmp1 and Dspp genes in a dosage-dependent manner. These data suggested that Klf10 transcriptionally activates the promoters of the Dmp1 and Dspp genes. Although Klf10 KO mice exhibit the impairment of osteoblast differentiation and bone mineralization and of bone-relate gene expression, whether Klf10 regulates tooth development and formation in vivo remains unknown, as Klf family members functionally compensate with regard to their biological activities (Heard et al. 2012; Spittau and Krieglstein 2012). Use of the gain-or loss-of-function of Klf10 in transgenic mice will allow a detailed study of the biological roles of Klf10 in mouse tooth development and formation.
In conclusion, our results indicate that Klf10 is differentially expressed in the enamel organ and the underlying mesenchyme, ameloblasts and odontoblasts at early and later stages of murine molar formation. Additionally, we found that Kfl10 expression increases during odontoblastic differentiation in vitro. The expression pattern of Klf10 coincides with that of Dmp1, Dspp and Runx2 in this process. Thus, Klf10 seems to control odontoblast differentiation and tooth formation through the up-regulation of the expression of the Dmp1 and Dspp genes.
Supplementary Material
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (no. 81100727), the Education Department of Zhejiang Province (no. Y200909390) 2011 China State Key Clinical Department Grants and the National Institute of Dental and Craniofacial Research (NIDCR; RO1 DE019892).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00441-015-2260-2) contains supplementary material, which is available to authorized users.
Contributor Information
Hui Chen, Email: huic66@hotmail.com.
Shuo Chen, Email: chens0@uthscsa.edu.
References
- Aberg T, Wang XP, Kim JH, Yamashiro T, Bei M, Rice R, Ryoo HM, Thesleff I. Runx2 mediates FGF signaling from epithelium to mesenchyme during tooth morphogenesis. Dev Biol. 2004;270:76–93. doi: 10.1016/j.ydbio.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Andujar MB, Magloire H, Hartmann DJ, Ville G, Grimaud JA. Early mouse molar root development: cellular changes and distribution of fibronectin, laminin and type-IV collagen. Differentiation. 1985;30:111–122. doi: 10.1111/j.1432-0436.1985.tb00522.x. [DOI] [PubMed] [Google Scholar]
- Bender H, Wang Z, Schuster N, Krieglstein K. TIEG1 facilitates transforming growth factor-beta-mediated apoptosis in the oligodendroglial cell line OLI-neu. J Neurosci Res. 2004;75:344–352. doi: 10.1002/jnr.10856. [DOI] [PubMed] [Google Scholar]
- Bensamoun SF, Hawse JR, Subramaniam M, Ilharreborde B, Bassillais A, Benhamou CL, Fraser DG, Oursler MJ, Amadio PC, An KN, Spelsberg TC. TGFbeta inducible early gene-1 knockout mice display defects in bone strength and microarchitecture. Bone. 2006;39:1244–1251. doi: 10.1016/j.bone.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Butler WT, Brunn JC, Qin C. Dentin extracellular matrix (ECM) proteins: comparison to bone ECM and contribution to dynamics of dentinogenesis. Connect Tissue Res. 2003;44(Suppl 1):171–178. [PubMed] [Google Scholar]
- Camilleri S, McDonald F. Runx2 and dental development. Eur J Oral Sci. 2006;114:361–373. doi: 10.1111/j.1600-0722.2006.00399.x. [DOI] [PubMed] [Google Scholar]
- Chalaux E, Lopez-Rovira T, Rosa JL, Pons G, Boxer LM, Bartrons R, Ventura F. A zinc-finger transcription factor induced by TGF-beta promotes apoptotic cell death in epithelial Mv1Lu cells. FEBS Lett. 1999;457:478–482. doi: 10.1016/s0014-5793(99)01051-0. [DOI] [PubMed] [Google Scholar]
- Chen S, Inozentseva-Clayton N, Dong J, Gu TT, MacDougall M. Binding of two nuclear factors to a novel silencer element in human dentin matrix protein 1 (DMP1) promoter regulates the cell type-specific DMP1 gene expression. J Cell Biochem. 2004a;92:332–349. doi: 10.1002/jcb.20051. [DOI] [PubMed] [Google Scholar]
- Chen S, Unterbrink A, Kadapakkam S, Dong J, Gu TT, Dickson J, Chuang HH, MacDougall M. Regulation of the cell type-specific dentin sialophosphoprotein gene expression in mouse odontoblasts by a novel transcription repressor and an activator CCAAT-binding factor. J Biol Chem. 2004b;279:42182–42191. doi: 10.1074/jbc.M402476200. [DOI] [PubMed] [Google Scholar]
- Chen S, Rani S, Wu Y, Unterbrink A, Gu TT, Gluhak-Heinrich J, Chuang HH, Macdougall M. Differential regulation of dentin sialophosphoprotein expression by Runx2 during odontoblast cytodifferentiation. J Biol Chem. 2005;280:29717–29727. doi: 10.1074/jbc.M502929200. [DOI] [PubMed] [Google Scholar]
- Chen S, Gluhak-Heinrich J, Martinez M, Li T, Wu Y, Chuang HH, Chen L, Dong J, Gay I, MacDougall M. Bone morphogenetic protein 2 mediates dentin sialophosphoprotein expression and odontoblast differentiation via NF-Y signaling. J Biol Chem. 2008;283:19359–19370. doi: 10.1074/jbc.M709492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Couble ML, Mouterfi N, Magloire H, Chen Z, Bleicher F. Spatial and temporal expression of KLF4 and KLF5 during murine tooth development. Arch Oral Biol. 2009;54:403–411. doi: 10.1016/j.archoralbio.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Cook T, Gebelein B, Mesa K, Mladek A, Urrutia R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J Biol Chem. 1998;273:25929–25936. doi: 10.1074/jbc.273.40.25929. [DOI] [PubMed] [Google Scholar]
- Feng J, Yang G, Yuan G, Gluhak-Heinrich J, Yang W, Wang L, Chen Z, Schulze McDaniel J, Donly KJ, Harris SE, Macdougall M, Chen S. Abnormalities in the enamel in bmp2-deficient mice. Cells Tissues Organs. 2011;194:216–221. doi: 10.1159/000324644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawse JR, Iwaniec UT, Bensamoun SF, Monroe DG, Peters KD, Ilharreborde B, Rajamannan NM, Oursler MJ, Turner RT, Spelsberg TC, Subramaniam M. TIEG-null mice display an osteogenic gender-specific phenotype. Bone. 2008;42:1025–1031. doi: 10.1016/j.bone.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawse JR, Cicek M, Grygo SB, Bruinsma ES, Rajamannan NM, Wijnen AJ, van Lian JB, Stein GS, Oursler MJ, Subramaniam M, Spelsberg TC. TIEG1/KLF10 modulates Runx2 expression and activity in osteoblasts. PLoS One. 2011;6:e19429. doi: 10.1371/journal.pone.0019429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawse JR, Pitel KS, Cicek M, Philbrick KA, Gingery A, Peters KD, Syed FA, Ingle JN, Suman VJ, Iwaniec UT, Turner RT, Spelsberg TC, Subramaniam M. TGFbeta inducible early gene-1 plays an important role in mediating estrogen signaling in the skeleton. J Bone Miner Res. 2013;29:1206–1216. doi: 10.1002/jbmr.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard ME, Pabona JM, Clayberger C, Krensky AM, Simmen FA, Simmen RC. The reproductive phenotype of mice null for transcription factor Krüppel-like factor 13 suggests compensatory function of family member Krüppel-like factor 9 in the peri-implantation uterus. Biol Reprod. 2012;87:115. doi: 10.1095/biolreprod.112.102251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefferan TE, Subramaniam M, Khosla S, Riggs BL, Spelsberg TC. Cytokine-specific induction of the TGF-beta inducible early gene (TIEG): regulation by specific members of the TGF-beta family. J Cell Biochem. 2000;78:380–390. doi: 10.1002/1097-4644(20000901)78:3<380::aid-jcb4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Hopwood B, Tsykin A, Findlay DM, Fazzalari NL. Gene expression profile of the bone microenvironment in human fragility fracture bone. Bone. 2009;44:87–101. doi: 10.1016/j.bone.2008.08.120. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Kettunen P, Karavanova I, Martin LB, Thesleff I. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: non-dividing cells express growth stimulating Fgf-4 gene. Int J Dev Biol. 1994;38:463–469. [PubMed] [Google Scholar]
- Johnsen SA, Subramaniam M, Katagiri T, Janknecht R, Spelsberg TC. Transcriptional regulation of Smad2 is required for enhancement of TGFbeta/Smad signaling by TGFbeta inducible early gene. J Cell Biochem. 2002;87:233–241. doi: 10.1002/jcb.10299. [DOI] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoedler JR, Denver RJ. Kruppel-like factors are effectors of nuclear receptor signaling. Gen Comp Endocrinol. 2014;203:49–59. doi: 10.1016/j.ygcen.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori N, Clifton GL, Dash P. Altered expression of novel genes in the cerebral cortex following experimental brain injury. Brain Res Mol Brain Res. 2002;104:148–158. doi: 10.1016/s0169-328x(02)00331-5. [DOI] [PubMed] [Google Scholar]
- Kuntz KA, Brown CE, Jr, Legan JJ, Kafrawy AH. An immunohistochemical study of osteoprotegerin in the human dental pulp. J Endod. 2001;27:666–669. doi: 10.1097/00004770-200111000-00003. [DOI] [PubMed] [Google Scholar]
- Lesot H, Brook AH. Epithelial histogenesis during tooth development. Arch Oral Biol. 2009;54(Suppl 1):S25–S33. doi: 10.1016/j.archoralbio.2008.05.019. [DOI] [PubMed] [Google Scholar]
- Li S, Kong H, Yao N, Yu Q, Wang P, Lin Y, Wang J, Kuang R, Zhao X, Xu J, Zhu Q, Ni L. The role of runt-related transcription factor 2 (Runx2) in the late stage of odontoblast differentiation and dentin formation. Biochem Biophys Res Commun. 2011;410:698–704. doi: 10.1016/j.bbrc.2011.06.065. [DOI] [PubMed] [Google Scholar]
- Lin H, Xu L, Liu H, Sun Q, Chen Z, Yuan G, Chen Z. KLF4 promotes the odontoblastic differentiation of human dental pulp cells. J Endod. 2011;37:948–954. doi: 10.1016/j.joen.2011.03.030. [DOI] [PubMed] [Google Scholar]
- Lin H, Liu H, Sun Q, Yuan G, Zhang L, Chen Z. KLF4 promoted odontoblastic differentiation of mouse dental papilla cells via regulation of DMP1. J Cell Physiol. 2013;228:2076–2085. doi: 10.1002/jcp.24377. [DOI] [PubMed] [Google Scholar]
- Luo X, Ding L, Xu J, Chegini N. Gene expression profiling of leiomyoma and myometrial smooth muscle cells in response to transforming growth factor-beta. Endocrinology. 2005;146:1097–1118. doi: 10.1210/en.2004-1377. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Rahiotis C. Parallels between tooth development and repair: conserved molecular mechanisms following carious and dental injury. J Dent Res. 2004;83:896–902. doi: 10.1177/154405910408301202. [DOI] [PubMed] [Google Scholar]
- Mitsumoto M, Mitsumoto A, Demple B. Nitric oxide-mediated upregulation of the TGF-beta-inducible early response gene-1 (TIEG1) in human fibroblasts by mRNA stabilization independent of TGF-beta. Free Radic Biol Med. 2003;34:1607–1613. doi: 10.1016/s0891-5849(03)00211-9. [DOI] [PubMed] [Google Scholar]
- Noti JD, Johnson AK, Dillon JD. The zinc finger transcription factor transforming growth factor beta-inducible early gene-1 confers myeloid-specific activation of the leukocyte integrin CD11d promoter. J Biol Chem. 2004;279:26948–26958. doi: 10.1074/jbc.M310634200. [DOI] [PubMed] [Google Scholar]
- Qin C, Huang B, Wygant JN, McIntyre BW, McDonald CH, Cook RG, Butler WT. A chondroitin sulfate chain attached to the bone dentin matrix protein 1 NH2-terminal fragment. J Biol Chem. 2006;281:8034–8040. doi: 10.1074/jbc.M512964200. [DOI] [PubMed] [Google Scholar]
- Ribeiro A, Bronk SF, Roberts PJ, Urrutia R, Gores GJ. The transforming growth factor beta(1)-inducible transcription factor TIEG1, mediates apoptosis through oxidative stress. Hepatology. 1999;30:1490–1497. doi: 10.1002/hep.510300620. [DOI] [PubMed] [Google Scholar]
- Saito MT, Salmon CR, Amorim BR, Ambrosano GM, Casati MZ, Sallum EA, Nociti FH, Jr, Silverio KG. Characterization of highly osteoblast/cementoblast cell clones from CD105 enriched periodontal ligament progenitor cells population. J Periodontol. 2014;85:205–211. doi: 10.1902/jop.2014.130461. [DOI] [PubMed] [Google Scholar]
- Shields ED, Bixler D, el-Kafrawy AM. A proposed classification for heritable human dentine defects with a description of a new entity. Arch Oral Biol. 1973;18:543–553. doi: 10.1016/0003-9969(73)90075-7. [DOI] [PubMed] [Google Scholar]
- Sonoyama W, Seo BM, Yamaza T, Shi S. Human Hertwig’s epithelial root sheath cells play crucial roles in cementum formation. J Dent Res. 2007;86:594–599. doi: 10.1177/154405910708600703. [DOI] [PubMed] [Google Scholar]
- Spittau B, Krieglstein K. Klf10 and Klf11 as mediators of TGF-beta superfamily signaling. Cell Tissue Res. 2012;347:65–72. doi: 10.1007/s00441-011-1186-6. [DOI] [PubMed] [Google Scholar]
- Spittau B, Wang Z, Boinska D, Krieglstein K. Functional domains of the TGF-beta-inducible transcription factor Tieg3 and detection of two putative nuclear localization signals within the zinc finger DNA-binding domain. J Cell Biochem. 2007;101:712–722. doi: 10.1002/jcb.21228. [DOI] [PubMed] [Google Scholar]
- Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23:4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam M, Hefferan TE, Tau K, Peus D, Pittelkow M, Jalal S, Riggs BL, Roche P, Spelsberg TC. Tissue, cell type, and breast cancer stage-specific expression of a TGF-beta inducible early transcription factor gene. J Cell Biochem. 1998;68:226–236. [PubMed] [Google Scholar]
- Subramaniam M, Gorny G, Johnsen SA, Monroe DG, Evans GL, Fraser DG, Rickard DJ, Rasmussen K, Deursen JM, van Turner RT, Oursler MJ, Spelsberg TC. TIEG1 null mouse-derived osteoblasts are defective in mineralization and in support of osteoclast differentiation in vitro. Mol Cell Biol. 2005;25:1191–1199. doi: 10.1128/MCB.25.3.1191-1199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam M, Hawse JR, Johnsen SA, Spelsberg TC. Role of TIEG1 in biological processes and disease states. J Cell Biochem. 2007;102:539–548. doi: 10.1002/jcb.21492. [DOI] [PubMed] [Google Scholar]
- Subramaniam M, Hawse JR, Rajamannan NM, Ingle JN, Spelsberg TC. Functional role of KLF10 in multiple disease processes. Biofactors. 2010;36:8–18. doi: 10.1002/biof.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I, Mikkola M. The role of growth factors in tooth development. Int Rev Cytol. 2002;217:93–135. doi: 10.1016/s0074-7696(02)17013-6. [DOI] [PubMed] [Google Scholar]
- Wang F, Wu LA, Li W, Yang Y, Guo F, Gao Q, Chuang HH, Shoff L, Wang W, Chen S. Immortalized mouse dental papilla mesenchymal cells preserve odontoblastic phenotype and respond to bone morphogenetic protein 2. In Vitro Cell Dev Biol Anim. 2013;49:626–637. doi: 10.1007/s11626-013-9641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wara AK, Foo S, Croce K, Sun X, Icli B, Tesmenitsky Y, Esen F, Lee JS, Subramaniam M, Spelsberg TC, Lev EI, Leshem-Lev D, Pande RL, Creager MA, Rosenzweig A, Feinberg MW. TGF-beta1 signaling and Kruppel-like factor 10 regulate bone marrow-derived proangiogenic cell differentiation, function, and neovascularization. Blood. 2011;118:6450–6460. doi: 10.1182/blood-2011-06-363713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima S, Lammers CH, Lee SH, Hara Y, Mizuno K, Mouradian MM. Cloning and characterization of murine glial cell-derived neurotrophic factor inducible transcription factor (MGIF) J Neurosci. 1997;17:8657–8666. doi: 10.1523/JNEUROSCI.17-22-08657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DH, Hsu CF, Lin CY, Guo JY, Yu WC, Chang VH. Kruppel-like factor 10 upregulates the expression of cyclooxygenase 1 and further modulates angiogenesis in endothelial cell and platelet aggregation in gene-deficient mice. Int J Biochem Cell Biol. 2013;45:419–428. doi: 10.1016/j.biocel.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Yerges LM, Klei L, Cauley JA, Roeder K, Kammerer CM, Ensrud KE, Nestlerode CS, Lewis C, Lang TF, Barrett-Connor E, Moffett SP, Hoffman AR, Ferrell RE, Orwoll ES, Zmuda JM Osteoporotic Fractures in Men Study G. Candidate gene analysis of femoral neck trabecular and cortical volumetric bone mineral density in older men. J Bone Miner Res. 2010;25:330–338. doi: 10.1359/jbmr.090729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeichner-David M, Oishi K, Su Z, Zakartchenko V, Chen LS, Arzate H, Bringas P., Jr Role of Hertwig’s epithelial root sheath cells in tooth root development. Dev Dyn. 2003;228:651–663. doi: 10.1002/dvdy.10404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.