Abstract
Type 3 secretion systems (T3SSs) of bacterial pathogens translocate bacterial effector proteins that mediate disease into the eukaryotic cytosol. Effectors traverse the plasma membrane through a translocon pore formed by T3SS proteins. In a genome-wide selection, we identified the intermediate filament vimentin as required for infection by the T3SS-dependent pathogen Shigella flexneri. We found that vimentin is required for efficient T3SS translocation of effectors by S. flexneri and other pathogens that use T3SS, Salmonella Typhimurium and Yersinia pseudotuberculosis. Vimentin and the intestinal epithelial intermediate filament keratin 18 interact with the C-terminus of the Shigella translocon pore protein IpaC. Vimentin and its interaction with IpaC are dispensable for pore formation, but are required for stable docking of S. flexneri to cells; moreover, stable docking triggers effector secretion. These findings establish that stable docking of the bacterium specifically requires intermediate filaments, is a process distinct from pore formation, and is a prerequisite for effector secretion.
T3SSs are highly conserved structures used by >30 bacterial pathogens of humans, animals, and plants to establish infection through the delivery of effector proteins into host cells1, 2. T3SSs comprise a base that spans the bacterial membranes, a hollow needle that is anchored in the base and extends away from the bacterial surface, and a cap that prevents non-specific secretion1, 3, 4. Delivery of effectors into the host cytosol is tightly controlled5, 6. Contact of the cap with the plasma membrane activates the T3SS to secrete two proteins that form the translocon pore6, 7, which is required for delivery of effectors into the host cytosol. The timing of the delivery of effectors is likely regulated by signaling through the T3SS during interaction with the host. Loss of either cap protein, IpaD or IpaB, causes constitutive secretion5, and certain mutations of the needle8 or translocon proteins9, 10 alter the rate or selection of substrates secreted through the T3SS. A sorting platform that integrates these signals at the cytoplasmic membrane was identified in Salmonella enterica serovarTyphimurium11, 12; this platform is predicted to interact with bacterial effectors and their chaperones to coordinate loading of effectors into the T3SS. It is proposed that translocation of effectors into the host cell depends on docking of the T3SS needle on the translocon pore13, 14, which would allow passage of effectors from the bacterial cytosol to the host cytosol in a single step. Yet, whether docking occurs and which mechanisms regulate it are uncertain.
In a genome-wide selection designed to discover host factors involved in the pathogenesis of S. flexneri, we identified the intermediate filament vimentin. We demonstrate that intermediate filaments interact with the Shigella translocon pore protein IpaC. Vimentin and its interaction with IpaC are required for stable docking of S. flexneri on cells and for efficient translocation of T3SS effectors into cells, but not for T3SS translocon pore formation, indicating that stable docking of bacteria is a process that is distinct from pore formation. Moreover, docked bacteria, but not undocked bacteria, actively secrete effectors. These data establish that docking of the bacterium on the cell is required for efficient secretion and translocation of T3SS effector proteins.
Results
Host intermediate filaments are required for efficient S. flexneri infection
S. flexneri invasion of colonic enterocytes, which requires T3SS-mediated translocation of effector proteins into the host cytosol15, 16, is promoted by several functionally-independent host proteins17-21. To identify additional host genes required for S. flexneri infection, we performed a genome-scale selection in a loss-of-function library of human-derived haploid (Hap-1) cells22, 23. Selection of cells in the library was optimized by serial infection with S. flexneri that produced the E. coli adhesin Afa-124, which binds to CD55, a GPI-anchored human cell surface receptor. The adhesin enhanced S. flexneri attachment to Hap-1 cells and reduced Hap-1 survival from 20% to 5% (Fig. S1a). This approach, in conjunction with serial infections, enabled a highly stringent selection, with <0.1% Hap-1 cell survival (Fig. S1b). Hap-1 cell killing was dependent on S. flexneri invasion; infection with an invasion-deficient strain caused minimal cell death (Fig. S1a). S. flexneri infection lead to enrichment of 81 host genes (p value <0.05, Table S1), determined by sequencing of loss-of-function mutation sites within genomes of selected cells versus unselected cells. The most highly enriched genes included CD55 and genes associated with its biosynthesis, demonstrating specificity for the strain used in the selection. Among other genes that were enriched was ARF6, which encodes a small GTPase involved in endocytosis that we have since shown is required for efficient S. flexneri invasion21, further validating the selection.
Multiple host pathways contribute to S. flexneri infection, which raised the possibility that the relatively low level of enrichment of genes in the selection might be due at least in part to host redundancy. To prioritize the output, we screened for host pathways that contained more than one of the top 81 genes enriched by selection. A single multi-gene cluster was identified that included the enriched gene encoding the intermediate filament vimentin. Vimentin interacts directly with the enriched gene PKP1 (plakophilin I) and indirectly with the enriched genes PRKRIR (repressor of the inhibitor of serine/threonine protein kinase R), MID1 (midline-1), and NOL3 (nucleolar protein 3) (Fig. S1c). Intermediate filaments participate in a wide variety of cellular functions, including cell migration, signal transduction, and immune responses25-27. However, they are the least studied of the three major cellular cytoskeletal components, and their role during bacterial infection is poorly characterized. Since multiple enriched genes clustered with vimentin, and intermediate filament function in bacterial pathogenesis was poorly understood, vimentin was selected for further study.
Infection with S. flexneri lacking the Afa-1 adhesin caused 5.5-fold more cell death for mouse embryonic fibroblasts (MEFs) expressing vimentin (Vim+/+) than for MEFs lacking vimentin (Vim−/−) (Fig. 1a), confirming the results of the Hap-1 screen and indicating that vimentin was required for S. flexneri-induced cell death independently of the adhesin. Vimentin is the only cytosolic intermediate filament produced in these MEFs28, and cell death was partially rescued by expression of human vimentin in Vim−/− MEFs (Fig. 1a-b), indicating that cell death in the Vim+/+ cells was due to the presence of vimentin and that human vimentin functions in S. flexneri infection.
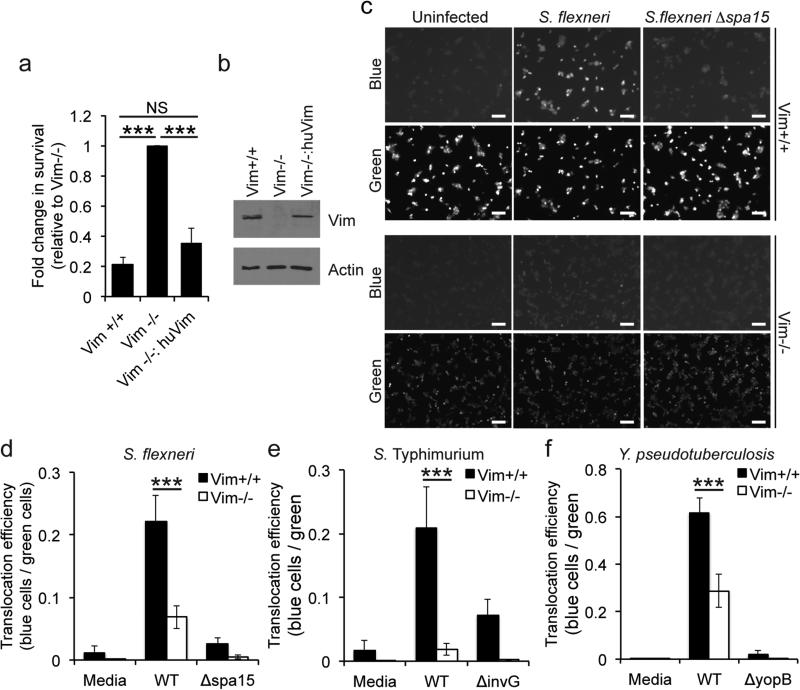
Figure 1. Efficient T3SS translocation depends on intermediate filaments.
(a) Cell viability of MEFs following infection with S. flexneri for 24 h. Viability determined by trypan staining and counting of recovered cells. Vim−/−:huVim, human vimentin expressed in Vim−/− MEFs. Data are mean ± SEM of three independent experiments for each group. *, P<0.05; one-way ANOVA with Tukey post hoc test. (b) Levels of vimentin in cells used for experiments in panel a, by western blot. Images shown are from one experiment, representative of three independent experiments. (c-f) Assessment of translocation efficiency by conversion of CCF4-AM fluorescence emission from green to blue upon infection of MEFs with bacteria that translocate a bacterial type 3 effector-β-lactamase reporter. (c) Images of fluorescence emission for S. flexneri expressing OspB-TEM. Images presented are representative of seven independent experiments. Scale bar, 100 μm. (d-f) Quantification of fluorescent images: (d) S. flexneri expressing OspB-TEM, mean ± SEM of seven independent experiments. (e) S. Typhimurium expressing SopE1-TEM, mean ± SEM of five independent experiments for media and WT and four independent experiments for ΔinvG. (f) Y. pseudotuberculosis expressing YopE-TEM, mean ± SEM of three independent experiments. Effector translocation was dependent on a functional T3SS apparatus, irrespective of the presence or absence of vimentin, as shown by Δspa15, ΔinvG, and ΔyopD mutants, which are deficient in T3SS activity.. ***, P<0.001; two-way ANOVA followed by Sidak post hoc test.
Intermediate filaments are required for efficient translocation by T3SSs
Intermediate filaments were required for effector protein translocation. Compared to Vim+/+ cells, the efficiency of translocation by S. flexneri into Vim−/− cells was reduced 62% ± 9% (Fig. 1c-d), as determined by cleavage of β-lactamase in cells loaded with the β-lactamase sensitive reporter CCF4-AM and infected with bacteria that deliver the T3SS effector OspB fused to β-lactamase. Similarly, efficient translocation of FLAG-tagged OspB, detected by differential fractionation of the cellular cytosol, depended on vimentin (Fig. S2).
The requirement for intermediate filaments to deliver T3SS effector proteins was conserved among other bacterial pathogens. Compared to Vim+/+ cells, translocation into Vim−/− cells was reduced by 94% ± 3% and 55% ± 7% for Salmonella Typhimurium (Fig. 1e) and Yersinia pseudotuberculosis (Fig. 1f), respectively. These results suggest that bacterial T3SS may generally require intermediate filaments to translocate effector proteins.
As might be expected from the observed defect in effector translocation, S. flexneri invasion was also defective in the absence of intermediate filaments (Fig. S3). The defect in effector translocation into Vim−/− MEFs correlated directly with defects in bacterial invasion (Fig. S3a-c). Natural infection by Shigella spp. occurs in the human colonic epithelium, in which keratins 8, 18, and 19 are expressed in the epithelial cells and vimentin is expressed in M cells and macrophages29, 30. Similar defects in invasion were observed in polarized human-derived Caco2 intestinal epithelial cells lacking keratin 8 or keratin 18 (Fig. S3d-e). Thus, for mammalian cell types relevant to natural infection, intermediate filaments were associated with events downstream of translocation, i.e., invasion of S. flexneri into cells.
The effect of vimentin on S. flexneri infection was independent of a general impact on the host cytoskeleton. Infection did not cause detectable rearrangements of intermediate filaments (Fig. S4). Moreover, although fewer actin-enriched entry sites were observed in Vim−/− cells compared to Vim+/+ cells (Fig. S5), among the subset of each that were infected, S. flexneri recruited actin to entry sites (Fig. S5b), indicating that actin rearrangements per se occur independently of vimentin.
T3SS translocon pore proteins interact with intermediate filaments
Intermediate filaments participate in functional interactions with the cortical actomyosin cytoskeleton31 and are also detected on the surface of cells32, 33. IpaC and its homologs are predicted to insert with the N-terminus extracellular and to contain either one or two hydrophobic transmembrane segments, placing the C-terminus either in the host cytosol or on the cell surface, respectively34-36 (Fig. S6a). The S. Typhimurium translocon pore protein SipC interacts with keratins in vitro37, 38, which raised the possibility that T3SS translocon proteins may interact with the intermediate filament cytoskeleton during infection. Human vimentin and keratins share substantial structural similarity26 and 30-36% amino acid sequence similarity. Using a distinct yeast-based cytoplasmic protein-protein interaction assay39, IpaC interacted specifically with vimentin (Fig. 2a). IpaC also interacted specifically with the intestinal intermediate filament keratin 18, but not with keratin 8 (Fig. 2b), suggesting that in the intestine, IpaC might interact with the keratin 18 component of the keratin 8-keratin 18 heterodimer. SipC, the S. Typhimurium homolog of IpaC, interacted with keratin 8 and keratin 18, consistent with previous reports37, 38, and interacted weakly with vimentin (Fig. 2c). YopD, the Y. pseudotuberculosis homolog of IpaC and SipC, interacted with vimentin and keratin 8 (Fig. 2d). Thus, like IpaC, homologs in two other enteric pathogens interact with intermediate filament proteins, indicating the interaction of the translocon pores with intermediate filaments is conserved among T3SSs.
Figure 2. The S. flexneri T3SS translocon pore protein IpaC and its homologs in other pathogens interact with intermediate filaments and this interaction is required for efficient translocation.
(a-f) Yeast based protein-protein interaction assay. The number of experimental replicates for each condition is included in Methods “Protein interaction assay”. *, P<0.05, **, P<0.01, ***, P<0.001; one-way ANOVA with Dunnett's post hoc test. (g) Infection of Vim+/+ MEFs with S. flexneri strains expressing OspB-TEM reporter to assess translocation efficiency by conversion of CCF4-AM fluorescence emission from green to blue upon type 3 translocation. The data are the mean ± SEM of three independent experiments. ***, P<0.001, one-way ANOVA with Dunnett's post hoc test.
The penultimate C-terminal residue of IpaC, R362, is conserved in S. Typhimurium (Fig. S6b) and required for S. flexneri invasion34, 35, 40. IpaC R362 was required for IpaC binding to intermediate filaments, as mutation to glutamine, leucine, or tryptophan markedly reduced IpaC interactions with vimentin or keratin 18, while not altering the level of protein synthesis, as determined by the intensity of the fluorescent signal from the IpaC construct (Fig. 2e-f and S7). Although YopD does not contain a residue homologous to IpaC R362, IpaC S349 is conserved across the three translocon homologs (Fig. S6b) and was required for interaction of IpaC with vimentin or keratin 18 (Fig. 2e-f). IpaC forms a hetero-oligomeric pore with IpaB7, yet co-expression of IpaB neither strengthened the interaction of IpaC with vimentin, nor rescued the interactions between the IpaC R362 mutants and vimentin (Fig. S8), suggesting that the interaction of IpaC with intermediate filaments is independent of IpaB.
The interaction of intermediate filaments with the translocon pore was required for efficient translocation. For S. flexneri producing IpaC mutants R362E, R362L, R362W, and S349A, the mutants showing reduced interaction with intermediate filaments, translocation efficiency was reduced by 73-90% compared to that of S. flexneri producing WT IpaC (Fig 2g). The translocation defect for IpaC R362W was also observed for translocation of OspB-FLAG into HeLa cells (Fig. S9), a common Shigella cell-based model that expresses vimentin, keratin 8, and keratin 18 (Fig. S4). It seemed unlikely that the defect in translocation efficiency of the IpaC mutants was due to a non-specific effect of the mutations on IpaC, as other functions of IpaC were normal, including secretion through the T3SS (Fig. S10a-b), as determined by chemical activation of T3SS activity with Congo red, and pore formation (Fig. S10c-d), as determined by hemoglobin release upon lysis of sheep erythrocytes resulting from translocon pore formation in the erythrocyte membrane7. Further, the C-terminus of IpaC polymerizes actin, yet mutation of R362 does not alter actin polymerization activity40. Thus, efficient translocation by T3SSs depends on the interaction of the C-terminal domain of IpaC with intermediate filaments.
Translocon pores form independently of intermediate filaments in nucleated cells
Sheep erythrocytes lack intermediate filaments, but Shigella forms pores in these cells (Fig. S10), which indicates that pore formation per se occurs in the absence of intermediate filaments. Supporting this, the abundance of IpaB and IpaC in the membranes of Vim−/− and Vim+/+ MEFs was similar (Fig. 3a).
Figure 3. Intermediate filaments are dispensable for translocon pore formation.
(a) Analysis of IpaC and IpaB incorporation in the plasma membrane using detergent fractionation of Vim+/+ and Vim−/− MEFs infected with S. flexneri. All panels are from the same experiment and are representative of three independent experiments. (b) Kinetics of translocon pore formation measured by BCECF dye release from Vim+/+ and Vim−/− bone marrow-derived macrophages infected with E. coli pSfT3SS at a sub-maximal multiplicity of infection. Data are the mean ± SD of one experiment, representative of three independent experiments. Independent wells were used for each experimental time point shown, media treatment was performed once, and infection with E. coli pSfT3SS was performed in triplicate wells. (c) BCECF release from Vim+/+ and Vim−/− bone marrow-derived macrophages infected with E. coli pSfT3SS as a function of the multiplicity of infection. The data presented are mean ± SD of one experiment performed in duplicate wells and are representative of three independent experiments. (d) Estimation of translocon pore diameter based on inhibition of BCECF dye release by osmoprotectants of varying diameters. E. coli pSfT3SS infection of Vim+/+ and Vim−/− bone marrow-derived macrophages. The data presented are mean ± SD of one experiment performed in duplicate wells and are representative of three independent experiments.
As the efficiency of pore formation and pore diameter impact bacterial effector translocation into the cytosol41, we tested whether the presence of intermediate filaments contributed to these traits. Unlike erythrocytes, in mammalian cells that normally contain intermediate filaments, pores formed by many T3SSs are not detected by lysis or dye release, presumably because they are blocked by the presence of effector proteins, since removal of effectors from Y. pseudotuberculosis enabled detection of pores41, 42. As cells naturally infected by Shigella contain intermediate filaments, we developed a system for analysis of translocon pore formation in these cells. As previously for WT Y. pseudotuberculosis43, pores were not detectable in cells infected with WT S. flexneri, which produces ~30 T3SS effectors, as minimal release of the small molecule dye BCECF (2’,7’-bis-(2-carboxyethyl)-5 (and 6)-carboxyfluorescein) was observed (Fig. S11). Of note, release of BCECF upon infection with WT S. flexneri was similar for Vim+/+ and Vim−/− macrophages (Fig. S12), suggesting that basal levels of translocation observed in Vim−/− cells (Fig. 1d) may be sufficient to block dye release or that other mechanisms contribute to blockage of pores formed by S. flexneri.
In contrast, E. coli pSfT3SS44, which is analogous to the effector-less Y. pseudotuberculosis system in that it carries the T3SS apparatus of S. flexneri and only 4 of the S. flexneri effectors, formed pores that were detectable by BCECF release (Fig. 3b-d). As for WT S. flexneri, efficient invasion and efficient translocation of the effector reporter OspB-TEM by E. coli pSfT3SS depended on vimentin (Fig. 4a-c), supporting use of this E. coli system as a model for the role of vimentin in S. flexneri T3SS function.
Figure 4. E. coli pSfT3SS reproduces S. flexneri phenotypes.
(a) Translocation of OspB-FLAG by E. coli pSfT3SS into the cytosol of Vim+/+ and Vim−/− MEFs. Western blot for FLAG, DnaK, a cytosolic bacterial protein, and actin. The two DnaK panels are from the same exposure of the same western blot, and all panels are from the same experiment, which is representative of three independent experiments. Within each experiment, translocation by E. coli pSfT3SS was assessed in triplicate. (b) Conversion of CCF4-AM fluorescence emission from green to blue upon translocation of the OspBTEM reporter by WT S. flexneri or E. coli pSfT3SS into Vim+/+ and Vim−/− MEFs. Data are the mean ± SEM of three independent experiments. **, P<0.01; two-way ANOVA with Sidak post hoc test. (c) Infection of Vim+/+ and Vim−/− MEFs by E. coli pSfT3SS, quantified by differential staining microscopy. Data are the mean ± SEM of three independent experiments. *, P<0.05; Student's t-test. (d) Pore size, as determined by the impact of osmoprotectants on lysis of erythrocytes by S. flexneri or E. coli pSfT3SS. Data are representative of two independent experiments. (e) Quantification of data from experiments depicted in panel d. Lysis is normalized to media alone in the presence of S. flexneri or E. coli pSfT3SS. Data are the mean ± SEM and of two independent experiments, each performed in triplicate. Means not significantly different by Student's t-test.
The percentage of cells from which dye was released and the kinetics of its release were similar for Vim+/+ and Vim−/− cells (Fig. 3b-c), indicating that the efficiency of pore formation is independent of vimentin. To test whether vimentin altered the diameter of pores that formed, we measured dye release in the presence of osmoprotectants of varying diameters, since the diameter of an osmoprotectant that blocks dye release from cells corresponds to the diameter of the pores that are formed in the plasma membrane41, 42. Pore diameter was similar in E. coli pSfT3SS-infected Vim+/+ and Vim−/− cells, as they each required PEG6000 to block infection-induced dye release (Fig. 3d). Moreover, in erythrocytes, the diameter of pores formed by E. coli pSfT3SS was similar to that of pores formed by WT S. flexneri, albeit their formation was less efficient (Fig. 4d-e).
Although pore formation in macrophage membranes activates the NLRP3 inflammasome45, it is unlikely the observed BCECF dye release was solely due to inflammasome activation in the ~90 min assays we used, since even at 4 hours of infection, E. coli pSfT3SS does not cause LDH release44, an indicator of loss of membrane integrity and cell viability. In HEK293T cells, which lack known inflammasome components46, infection with E. coli pSfT3SS induces dye release, whereas infection with WT S. flexneri does not (Fig. S11). Moreover, efficient activation of inflammasomes requires vimentin47, yet we observed no decrease in dye release in Vim−/− cells compared to Vim+/+ cells, further suggesting that the observed dye release was not due to inflammasome activation.
These results indicate that, whereas translocation of effectors depends on intermediate filaments, translocon pore formation occurs independently of them. Thus, pore formation in the plasma membrane is necessary but not sufficient for T3SS effector translocation. Moreover, in addition to participating in formation of the translocon pore, IpaC promotes bacterial effector translocation in a manner that depends on interactions between it and intermediate filaments.
Intermediate filaments are required for efficient S. flexneri docking to cells
To translocate effector proteins into host cells, the T3SS needle complex, which is anchored in the bacterial membrane, must either dock onto the translocon pore or secrete effectors nearby. For S. Typhimurium, the translocon pore itself is required for efficient docking of the bacterium to the host cell13, 14, suggesting that interaction of the needle complex with the translocon pore contributes to docking. We tested whether the interaction of IpaC with intermediate filaments promotes efficient docking by assessing, in a laminar flow chamber, binding of bacteria centrifuged onto cells. Bacterial entry was prevented by treatment of cells with cytochalasin D (Fig. S13a). S. flexneri docking to Vim+/+ cells was five-fold more efficient than to Vim−/− cells and was dependent on IpaC (Fig. 5a-b). Similar results were obtained in the absence of laminar flow (Fig. S13b). S. flexneri docking depended on the ability of IpaC to interact with intermediate filaments, as docking to Vim+/+ MEFs by bacteria producing WT IpaC was significantly greater than docking of S. flexneri producing IpaC R362W (Fig. 5c). Thus, intermediate filaments promote S. flexneri docking in a manner that is dependent upon their interaction with IpaC.
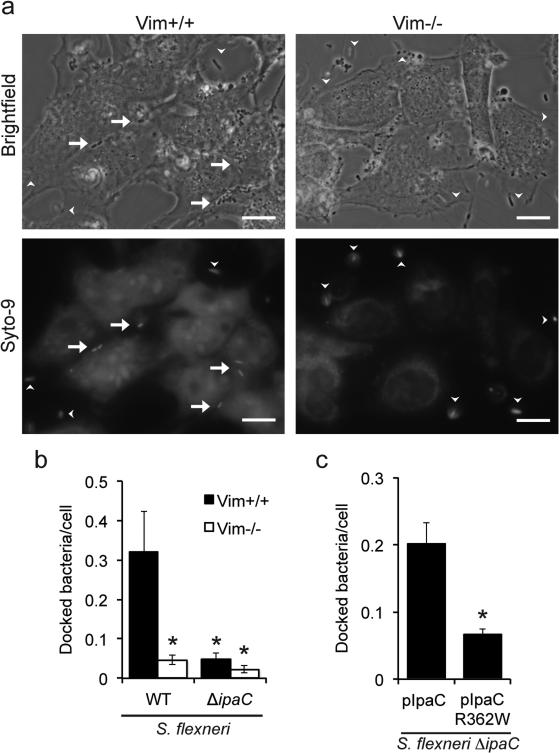
Figure 5. The interaction of intermediate filaments with the translocon pore is required for efficient docking of S. flexneri.
Bacterial docking to Vim+/+ and Vim−/− MEFs following a laminar flow of 0.125 dynes/cm2. (a) Representative images of S. flexneri-infected Vim+/+ and Vim−/− MEFs after centrifugation onto cells followed by exposure to laminar flow. Scale bar, 20 μm. Arrows, docked bacteria. Arrowheads, bacteria that were not docked, as indicated by their being outside the focal plane. (b) Quantification of docking of WT bacteria to Vim+/+ versus Vim−/− MEFs, as indicated. Data are the mean ± SEM of six independent experiments for WT infection of Vim+/+, five independent experiments for WT infection of Vim−/−, four independent experiments for ΔipaC infection of Vim+/+, and three independent experiments for ΔipaC infection of Vim−/−. *, P<0.05; two-way ANOVA with a Sidak post hoc test. (c) Quantification of docking of bacteria expressing WT or mutant IpaC to Vim+/+ MEFs. Data are the mean ± SEM of four independent experiments. *, P<0.05; Student's t-test.
For certain pathogenic E. coli, attachment to host cells is promoted by intermediate filaments. Tir, a T3SS effector of enteropathogenic E. coli interacts with keratin 18 in a complex with 14-3-3, which increases bacterial attachment at the pedestal48. The E. coli K1 surface protein IbeA promotes attachment to brain endothelial cells via interactions with vimentin33. The nature of these interactions is distinctly different from that described herein, in that for S. flexneri, they depend on the T3SS translocon pore.
Effector secretion occurs upon bacterial docking to cells
To determine whether bacterial docking is required for activation of effector secretion, we assessed T3SS activity within individual bacteria upon contact with Vim−/− versus Vim+/+ MEFs using TSAR (transcription based secretion activity reporter49). The percentage of bacteria displaying activation of T3SS was similar for bacteria docked on Vim+/+ MEFs and bacteria docked on Vim−/− MEFs (Fig. 6a-b), although as above, substantially more bacteria were docked to the Vim+/+ MEFs (Fig. 6a and c). For bacteria that had been brought into contact with cells but did not dock, essentially none displayed activation of T3SS (Fig. 6d-e). Complementation of Vim−/− MEFs with human vimentin restored the number of TSAR positive bacteria docked to cells (Fig. 6f), indicating that the absence of vimentin was responsible for the defect in docking and that human vimentin functions in this process, providing further support for the role of intermediate filaments in S. flexneri docking and translocation. These results indicate that stable docking of the bacterium to the cell is associated with activation of effector secretion.
Figure 6. Activation of effector secretion requires S. flexneri docking.
(a) Images of docked WT bacteria at 30 min of infection. Bacteria activated to secrete effectors through T3SS express GFP (TSAR, green), all bacteria express mCherry (red), DNA labeled with DAPI (blue). Arrows, TSAR positive bacteria. Scale bar, 10 μm. Images are representative of three independent experiments. (b-c) Quantification of cell-associated bacteria that have activated TSAR at 30 min or 60 min of infection. *P<0.05; Student's t-test. Data are the mean ± SEM of at least three independent experiments. (d) Absence of TSAR expression among bacteria centrifuged onto cells yet non-adherent at 30 min of infection. Images are representative of three independent experiments. Scale bar, 10 μm. (e) Quantification of data presented in d. Data are the mean ± SEM of three independent experiments. (f) Rescue of docking of bacteria by genetic complementation of Vim−/− MEFs with human vimentin. Vim−/−:huVim, human vimentin expressed in Vim−/− MEFs. Data are the mean ± SEM of three independent experiments. *P<0.05, **P<0.01; one-way ANOVA with Tukey post hoc test.
Discussion
A critical feature of T3SSs is the hierarchical secretion of substrates through the needle, such that the translocon pore proteins are secreted and inserted into the plasma membrane before secretion of effector proteins. By ensuring that the translocon pore is in place before effectors are secreted, this process guarantees that effector proteins are efficiently translocated into the host cytosol. Proper timing of effector secretion is facilitated by a sorting platform in the bacterial cytosol at the base of the T3SS apparatus that is loaded in a sequential manner with substrates11, 12. Data presented here establish that stable docking of S. flexneri on the host cell is independent of translocon pore formation per se, yet is dependent on the presence of intermediate filaments in the host cell and on their interaction with the translocon pore protein IpaC. Moreover, activation of effector secretion is associated with stable docking, but not with formation of the translocon pore.
These data support a model in which the tip of the T3SS transiently interacts with the host membrane to insert the translocon pore. We speculate that the interaction of IpaC with intermediate filaments, either in the host cytosol or at the cell surface, alters the extracellular aspect of the translocon pore complex in a manner that promotes stable docking of the needle apparatus (Fig. S14). Whether other proteins that were identified in our selection and which interact with vimentin also participate in bacterial docking is currently unknown. Our data demonstrate that pore formation and docking can be functionally separated (Fig. 3 and 5). In the context of infection, it is likely that these steps occur in concert, i.e., that as IpaC is inserted into the membrane, it is adjusted by interaction with intermediate filaments in a manner that allows the associated bacterium to stably dock. Consistent with a role of docking in the activation of effector secretion is the observation that mutations in MxiH8, the needle component of the S. flexneri T3SS, or in IpaD10, 50, 51, the cap protein on the needle complex, alter the selection of substrates and the rate of secretion through the T3SS, suggesting that these mutations are associated with conformational changes that promote the secretion of effectors3, 8, 51. Thus, we speculate that docking, enabled by the interaction of IpaC with intermediate filaments, leads to additional conformational changes that trigger effector secretion, resulting in effector translocation. These mechanisms are likely to be relevant to all the bacterial pathogens that utilize T3SSs during human infection.
Methods
Bacterial strains and plasmids
The strains and plasmids used in this study are listed in Table S2. The wild-type S. flexneri strain used in this study is serotype 2a strain 2457T52; all other S. flexneri strains used in this study are isogenic to it. BS103 is the virulence plasmid-cured derivative of 2457T53. 2457T ΔipaC was generated by P1-mediated transduction of the kanamycin-resistant locus from serotype 5a strain M90T ΔipaC::km54. E. coli pSfT3SS, which is DH10B carrying a portion of the S. flexneri 2457T virulence plasmid, has been described44. Of note, E. coli pSfT3SS does not produce IbeA, a protein that binds vimentin33. The sequences of primers used in PCR and sequencing are available from the authors upon request.
Cells
Vimentin +/+ (Vim+/+) and vimentin −/− (Vim−/−) mouse embryonic fibroblasts (MEFs) were kindly provided by Victor Faundez (Emory), and HeLa cells (ATCC, CCL2) and HEK-293T (ATCC, CRL-3216) were obtained from ATCC. These cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 0.45% glucose, 10% heat-inactivated fetal bovine serum under humidified air containing 5% CO2 at 37°C. Vim−/− MEFs were complemented using a human cDNA encoding vimentin (Thermo Scientific) cloned into the pBABE-puro retroviral vector. Retroviral particles packaging the vimentin gene or no insert were generated by transfection of 293T cells using pAdvantage (Promega), CMV-VSV-G and Gag-Pol, and were used to infect Vim−/− MEFs. Puromycin-resistant stable cell populations were generated, and clonal populations were isolated by limiting dilution. Knock-down of keratin 8 or keratin 18 in Caco-2 cells was by lentiviral transduction of control (sc-108080, Santa Cruz), keratin 8 (sc-35156-V, Santa Cruz), or keratin 18 (sc-35151-V, Santa Cruz) shRNA, with selection for puromycin-resistant, stable cell populations, performed by the Harvard Digestive Diseases Center. Bone-marrow derived macrophages were isolated from the bone marrow of the femurs and tibias of Vim+/+ and Vim−/− mice. The macrophages were differentiated from other bone marrow cells for 7 days in DMEM supplemented with 20% FBS, 25% L-cell supernatant, 25mM HEPES, 1% sodium pyruvate, 1% non-essential amino acids, 0.45% glucose and 1% glutamine55. Macrophages were used on days 7-21. All cells in the laboratory are periodically tested for mycoplasma; cells that test positive are treated or discarded.
Genome-wide S. flexneri selection of mutagenized HAP1 cells
A library of mutagenized HAP1 cells was generated as described previously22. Briefly, gene-trap retrovirus was produced in HEK-293T cells by transfection of pGT-GFP, pGT-GFP+1 and pGT-GFP+2 combined with appropriate packaging plasmids. Retrovirus was concentrated using ultracentrifugation before infecting 10×106 HAP1 cells in the presence of 8 μg/ml protamine sulfate (Sigma).
For selection with S. flexneri, cells were seeded in ten T175 flasks at 1 × 106 cells per flask in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% FBS, 0.45% glucose, and GlutaMAX, and were incubated at 37°C with 5% CO2. To achieve optimal selection of the Hap-1 cells, three serial infections with S. flexneri harboring pAfa-1, which contains a pilus adhesin of uropathogenic E. coli56, were performed. Each infection resulted in killing of approximately 95% of cells. Three infections were required to achieve killing of greater than 99.9% of cells (Fig. S1).
For each infection, cells were infected at a multiplicity of infection (MOI) of 100 using a small volume of media, so as to promote bacteria-cell contact. After 1 hour at 37°C, cells were washed 5 times and gentamicin was added to 25 μg/mL. Following incubation for an additional 20 hours at 37°C, gentamicin was washed out and two hours later, the next infection was carried out. After the third infection, cells were allowed to grow out for seven days, until the total number of surviving cells reached approximately 1×108.
Mapping of gene trap insertion sites
Insertion sites were identified as described previously23. Briefly, genomic DNA was isolated from 30×106 cells using a QIAamp DNA mini kit (Qiagen). Genomic DNA was digested with NlaIII, heat-inactivated and purified with a PCR purification kit (Qiagen). To generate the S. flexneri-selected dataset, insertion sites were identified by sequencing the genomic DNA flanking gene trap proviral DNA as reported previously23, 57. Deep sequencing was performed on an Illumina HiSeq2500 at the Biopolymers Facility at Harvard Medical School. The number of inactivating mutations (i.e. sense orientation or present in exon) per individual gene was determined, as well as the total number of inactivating insertions for all genes. A gene enrichment score was calculated for each gene by comparing the number of insertions in a gene per total number of insertions in the experimental dataset versus an unselected HAP1 control dataset that has been previously reported23. For each gene a p-value (corrected for false discovery rate) was calculated using the one-sided Fisher exact test, which yielded a total of 81 significantly enriched genes (p<0.05, Table S1). To prioritize the list of candidate genes, pathway analysis of these 81 enriched genes was performed using g:Profiler58. As a result of pathway analysis (Fig. S1c), 5 of the 81 enriched genes were prioritized, and vimentin was chosen for further study due to its poorly described role for bacterial infection and its physiological role as a component of the cytoskeleton.
Microscopy-based cell survival assay
Vim+/+ and Vim−/− MEFs, seeded at 4×105 cells per well in a 6-well tissue culture-treated plate, were infected with S. flexneri at a multiplicity of infection (MOI) of 100. Bacterial entry was synchronized by centrifugation at 800xg for 10 min at 25°C. Infected cells were incubated for 50 min at 37°C with 5% CO2, washed, and incubated in DMEM supplemented with 10% FBS and 50 μg/mL of gentamicin, which specifically kills the extracellular bacteria, for 23 hours at 37°C with 5% CO2. Cell viability was assessed by phase microscopy, and the number of viable cells was quantified by trypan staining and trypsinization of adherent cells, with enumeration using a hemocytometer.
Differential fluorescent microscopy
Differential staining to distinguish intracellular versus extracellular bacteria was performed as described previously21. Vim+/+ and Vim−/− MEFs were seeded at 3×105 cells per well in a 6-well tissue culture-treated plate and infected the next day at an MOI of 100 as described above (Microscopy-based cell survival assay). After one hour of infection, the cells were washed with PBS, and then fixed with 3.7% paraformaldehyde cytoskeleton F buffer59. Extracellular bacteria were stained with anti-S. flexneri (Difco Laboratories, 2883-47-3) and an Alexa Fluor 488 secondary antibody (Invitrogen, A11034). For experiments assessing E. coli pSFT3SS invasion, extracellular bacteria were labeled with rabbit anti-E. coli (Abcam, ab137967). The cells were then permeabilized with 0.5% Triton X-100 in cytoskeleton F buffer. Actin was stained with Alexa Fluor 568 phalloidin (Invitrogen), and cellular and bacterial DNA (both intracellular and extracellular bacteria) were stained with 4,6-diamidino-2-phenylindole (DAPI; Invitrogen).
Bacterial effector translocation and secretion assays
Fluorescence-based translocation assays were performed as described previously60. In brief, S. flexneri strains carrying an IPTG inducible OspB-TEM β-lactamase construct were grown to exponential phase at 37°C, and OspB-TEM production was induced with 1 mM IPTG for 30 min. Y. pseudotuberculosis were grown to exponential phase at 30°C, EGTA was added to 10μM, and the cultures were incubated an additional 1.5 hours at 37°C. S. Typhimurium carrying an IPTG inducible SopE1-TEM construct was grown to exponential phase at 37°C, and IPTG was added for 30 min at 1mM to induce SopE1-TEM production. Vim+/+ and Vim−/− MEFs, seeded at 2×104 cells per well in 96-well, flat-bottom, tissue culture-treated plates, were pre-loaded with CCF4-AM dye (Invitrogen) according to the manufacturer's protocol and infected at an MOI of 200. Bacterial entry was synchronized by centrifugation at 800xg for 10 minutes at 25°C. The infection continued for 50 min at 37°C with 5% CO2. Images were acquired with a Nikon Eclipse TE-300 inverted microscope using appropriate filters. The relative amount of translocation was determined using segmentation tools in iVision software (Biovision Technologies).
For quantification of intracellular bacterial effectors by western blot, HeLa cells or Vim+/+ and Vim−/− MEFs, seeded at 3×105 cells per well in 6-well tissue culture-treated plates, were infected with exponential phase S. flexneri pOspB-FLAG at an MOI of 200, as described above (Microscopy-based cell survival assay). After 50 min, the infected cells were lysed with RIPA buffer (50mM Tris, pH 8 containing 150mM NaCl, 1% nonidet-P40, 0.1% SDS, 10mM NaF, and EDTA-free protease inhibitor cocktail [Roche]), which releases soluble cytosolic components but does not lyse bacteria. Cellular debris and bacteria were removed by centrifugation. OspB-FLAG was detected by western blot using mouse antibody to FLAG (Sigma, F1804). Assessment of bacterial lysis was by western blot using mouse antibody to DnaK (Stressgen,SPA-880).
For measurements of T3SS activation using TSAR49, Vim+/+ and Vim−/− MEFs were seeded onto coverslips in 6-well plate and infected as described above (Microscopy-based cell survival assay). Cells and bacteria were co-cultured for 30 or 60 min. Non-adherent bacteria exposed to cells were collected from the infection supernatant and the washes of the cells; this fraction was washed once with PBS and fixed with 3.7% paraformaldehyde. Cells and associated bacteria were fixed with 3.7% paraformaldehyde, and the DNA was stained with DAPI. For measurement of TSAR activation in vimentin complementation experiments, after incubation of cells at 25°C for 45 min, the media on the cells was exchanged for warm DMEM supplemented with 10% FBS, the cells were incubated for 5 min at 37°C, and infections were performed as described above (Microscopy-based cell survival assay). Infected monolayers were incubated at 37°C for 60 min, washed once with PBS, and fixed with 3.7% paraformaldehyde. Images were acquired with a Nikon Eclipse TE-300 inverted microscope using appropriate filters. All bacteria expressed mCherry, and bacteria in which T3SS effector secretion was activated expressed GFP49.
Congo red secretion assay
Secretion through the T3SS by bacteria grown in vitro and in the absence of eukaryotic cells was induced by the addition to the growth media or to bacteria in phosphate-buffered saline of Congo red (to 10 uM), a commonly used activator of S. flexneri T3SS61.
Bacterial binding assays
For assays performed under laminar flow62-65, Vim+/+ and Vim−/− MEFs were seeded at 105 cells per chamber in a LuerI 0.4μm laminar flow chamber (Ibidi) in DMEM supplemented with 10% FBS. Cells were treated with 0.5μg/mL of cytochalasin D, from before infection and continuing through the infection. Exponential phase bacteria were stained with Syto-9 (Invitrogen, S34854) at 37°C for 20 min, washed with DMEM, and applied to cells at an MOI of 100. Infection was performed as above (Microscopy-based cell survival assay) and allowed to continue for 20 min at 37°C with 5% CO2. The bacteria were subjected to a shear force of 0.125 dynes/cm2 for 20 min at 37°C under a flow of DMEM supplemented with 10% FBS. To quantify bacteria associated with cells, ten random images were collected along the midline of the chamber.
Binding studies without laminar flow were performed as previously described14. 30 min prior to infection, Vim+/+ and Vim−/− MEFs were treated with cytochalasin D at 0.5 μg/mL and cytochalasin D was maintained for the duration of the infection, which followed the protocol for “Differential fluorescent microscopy” above and in supplemental methods.
Protein interaction assays
Yeast-based protein interact assays were performed as described previously39, 66, 67. Briefly, intermediate filament proteins were tagged with μNS, and the translocon proteins were tagged with mCherry (Table S2). Yeast were cultured overnight in the presence of 0.2% raffinose, diluted the next morning to OD600 of 0.5, and cultured for 2 hours at 30°C with 0.2% raffinose. Then, to induce protein synthesis, the media was changed to 0.2% galactose, and growth was allowed to proceed for 6-8 hours at 30°C. mCherry synthesis was monitored by fluorescence microscopy. Random images of ~100 yeast were collected for each condition in each experiment. Collection of images and analysis of data were performed in a blinded manner. Protein interaction was scored based on the presence or absence of fluorescent puncta among yeast cells that produced mCherry. Independent experimental replicates performed in Fig. 2: (a) Empty-IpaC, n=12; Vimentin-IpaC, n= 15; Empty-Vimentin, n=3. (b) Empty-IpaC, n=12; Keratin 8-IpaC, n=3; Keratin 18-IpaC, n=15. (c) Empty-SipC, n=8; Vimentin-SipC, n=9; Keratin 8-SipC, n=9; Keratin 18-SipC, n=11. (d) Empty-YopD, n=11; Vimentin-YopD, n=13; Keratin 8-YopD, n=7; Keratin 18-YopD, n=5. (e) Empty-IpaC, n=8; Vimentin-IpaC WT, n=11; Vimentin-IpaC R362E, n=8; Vimentin-IpaC R362L, n=4; Vimentin-IpaC R362W, n=3; Vimentin-IpaC S349A, n=4. (f) Empty-IpaC, n=8; Keratin 18-IpaC WT, n=11; Keratin 18-IpaC R362E, n=10; Keratin 18-IpaC R362L, n=5; Keratin 18-IpaC R362W, n=3; Keratin 18-IpaC S349A, n=5.
Quantification of translocon proteins in the plasma membrane
Membrane isolation and detection of translocon proteins was performed as described previously37. Briefly, Vim+/+ and Vim−/− MEFs, seeded at 6×106 cells per 10cm tissue culture-treated dish, were infected with early exponential phase bacteria in 2.5mL of DMEM supplemented with 10% FBS at an MOI of 100 for 1 hour at 37°C. For strains carrying an arabinose inducible construct, DMEM without glucose was used. The cells were washed 3 times with ice-cold PBS, scraped off the dish into ice-cold PBS containing protease inhibitors, and recovered by centrifugation at 225xg for 5 min at 25°C. The cell pellet was cooled on ice for 5 min and was resuspended in ice-cold 50mM Tris, pH 7.5 containing protease inhibitors and 0.2% saponin. After incubation on ice for 20 min and centrifugation at 16,100xg at 4°C for 30 min, the saponin-solubilized supernatant (cytosolic fraction) was collected, and the pellet was resuspended in 50mM Tris, pH 7.5 containing protease inhibitors and 1% Triton X-100. After additional incubation on ice for 30 min and centrifugation at 16,100xg at 4°C for 15 min, the Triton X-100-soluble supernatant (membrane fraction) was collected, and the pellet (detergent insoluble cellular debris and intact bacteria) was resuspended in 50mM Tris, pH 7.5 containing protease inhibitors and 1% Triton X-100. Western blots were performed using mouse anti-DnaK (Stressgen, SPA-880), rabbit anti-vimentin (Cell Signaling, 3932S), mouse anti-GAPDH (abcam, ab9484), rabbit anti-caveolin-1 (Sigma, C4490), rabbit anti-IpaC (gift from W. Picking), and rabbit anti-IpaB (gift of W. Picking).
Pore formation
Pore formation in sheep erythrocyte membranes was performed as previously described7. Briefly, 108 erythrocytes were washed with saline and infected with bacteria in stationary phase at an MOI of 100 in 30mM Tris, pH7.5. The bacteria were centrifuged onto the erythrocytes at 800xg for 10 min at 25°C and cultured at 37°C with 5% CO2 for 50 min. The sample was mixed and centrifuged at 800xg for 10 min at 25°C. As a control for lysis, erythrocytes alone were resuspended in 0.02% SDS prior to the last centrifugation step. The supernatants were transferred to a 96 well plate and OD570 measured using a Wallac 1420 Victor2 (Perkin Elmer). Pore diameters were estimated with osmoprotectants, as performed previously7, 41. For experiments measuring pore diameter, osmoprotectants were included at 30mM for the duration of the co-culture.
Pore formation in nucleated cells was performed as described previously41. Cells, seeded in 96 well flat-bottom plates at 5×104 (bone marrow-derived macrophages) or 1.5×104 (293T cells) per well, were preloaded with BCECF, AM [2’-7’-Bis-(2-Carboxyethyl)-5-(and-6)-Carboxyfluorescein, Acetoxymethyl Ester (Invitrogen, B1170)] and Hoechst (Invitrogen) in HBSS at 37°C at 5% CO2. The cells were infected with exponential phase bacteria at an MOI of 100 in RPMI; for MEFs, 4% FBS was included. Invasion was synchronized by centrifugation at 800xg for 10 min at 25°C, and the infection was continued at 37°C with 5% CO2 for indicated times. For assays to measure the translocon pore diameter, osmoprotectants of varying sizes were included at 60mM for the duration of the assay. Cells were stained with Hoescht (Invitrogen), and fluorescent microscopy was used to image the total number of cells (blue filter) and BCECF-positive cells (green filter). To quantify the percent of cells that were BCECF-positive, images were analyzed using Cell Profiler (Broad Institute).
Cytoskeletal assays
Cells were seeded at 3×105 cells per well on glass coverslips in a 6-well tissue culture-treated plate and infected the next day at an MOI of 400, as described above (Microscopy-based cell survival assay). After 30 min, the cells were washed with PBS, and then fixed with 3.7% paraformaldehyde and permeabilized with 0.5% Triton X-100, both in cytoskeleton F buffer59. Actin was stained with Alexa Fluor 568 phalloidin (Invitrogen, A12380), and cellular and bacterial DNA were stained with 4,6-diamidino-2-phenylindole (DAPI; Invitrogen). Actin rearrangements were identified by the presence of dense concentrations of actin21, 68. To visualize intermediate filaments HeLa cells were infected as above except the bacteria were added at an MOI of 100 and the infection carried out for 1 hour. Intermediate filaments were stained with rabbit anti-keratin 18 (abcam, 133263), rabbit anti-keratin 8 (Abcam, 53280), mouse anti-vimentin (Sigma, V6630, V-9 clone), goat anti-rabbit Alexa Fluor 488 (Invitrogen, A11034), and goat anti-mouse Alexafluor 488 (Invitrogen, A1101). Bacteria were visualized with DAPI.
Microscope and data analysis
Epifluorescence and phase-contrast microscopy were performed using a Nikon Eclipse TE300 or Nikon TE-2000-S microscope equipped with Chroma Technology filters and a Photometrics CoolSNAP HQ charge-coupled device camera (Roper Scientific) or a Q-Imaging Exi Blue camera (Q-Imaging). Images were acquired using iVision software (BioVision Technologies). Unless otherwise noted, for all other experiments involving microscopy, images were collected in a random manner across the coverslips. Color figures were assembled and digitally pseudocolored using Adobe Photoshop software.
For densitometry analysis of western blots, the chemoilluminescent signal was detected using film, films were scanned with an Epson Perfection 4990 Photo scanner, and band intensity was determined using ImageJ (NIH).
Unless otherwise indicated, for all experiments, a minimum of three independent experiments were conducted on independent days, using independent cultures, and where appropriate, independent plating of cells.
Statistical Analysis
All statistical analysis was performed using Graphpad Prism 6 (Graphpad Software). For experiments containing two groups, statistical significance was determined using a Student's t-test. Unless otherwise stated, for experiments containing three or more groups, statistical significance was determined using a one-way ANOVA followed by a Dunnett's post hoc test or a two-way ANOVA followed by a Sidak post hoc test. Experimental conditions were performed once per experiment, unless otherwise noted. Amino acid similarity among proteins was determined using clustalΩ69.
Supplementary Material
Acknowledgements
We thank Victor Faundez, Cammie Lesser, Wendy Picking and William Picking, Joan Mecsas, Victoria Auerbuch-Stone, Beth McCormick and Ana Maldonado-Contreras, and Claude Parsot for cell lines, bacterial strains, reagents, technical assistance and protocols, Alexandra Wiscovitch for technical assistance, and Matthew Brown for critical reading of the manuscript.
This work was funded by NIH RO1 AI081724 (to M.B.G.), NIH T32 AI007061 (to B.C.R.), NIH F32 AI092967 (to L.A.B.), NIH F32 AI114162 (to B.C.R.), a career development award through the New England Regional Center of Excellence in Biodefense and Emerging Infectious Disease and the ICCB-Longwood Facility at Harvard Medical School (NIH U54 AI057159, to L.M.S.), and the CAPES Foundation, Ministry of Education of Brazil, Brasilia-DF, 70040-020, Brazil (to A.L.Q.).
Footnotes
Author Contributions
B.C.R., M.B.G., and L.M.S. wrote the manuscript. B.C.R., M.B.G., L.M.S., M.R., S.P.W., and L.A.B. designed experiments and interpreted data. B.C.R., L.M.S., M.R., L.R.R., A.L.Q., B.B.H., L.A.B., C.M.K., S.B., G.M., E.K., L.R., Y.F., and N.M-V. performed experiments. All authors discussed the results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Galan JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol. 2014;68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 3.Cheung M, et al. Three-dimensional electron microscopy reconstruction and cysteine-mediated crosslinking provide a model of the type III secretion system needle tip complex. Mol Microbiol. 2015;95:31–50. doi: 10.1111/mmi.12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubori T, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 5.Menard R, Sansonetti P, Parsot C. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 1994;13:5293–5302. doi: 10.1002/j.1460-2075.1994.tb06863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veenendaal AK, Hodgkinson JL, Schwarzer L, Stabat D, Zenk SF, Blocker AJ. The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol Microbiol. 2007;63:1719–1730. doi: 10.1111/j.1365-2958.2007.05620.x. [DOI] [PubMed] [Google Scholar]
- 7.Blocker A, et al. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenjale R, et al. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J Biol Chem. 2005;280:42929–42937. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- 9.Meghraoui A, Schiavolin L, Allaoui A. Single amino acid substitutions on the needle tip protein IpaD increased Shigella virulence. Microbes Infect. 2014;16:532–539. doi: 10.1016/j.micinf.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Schiavolin L, Meghraoui A, Cherradi Y, Biskri L, Botteaux A, Allaoui A. Functional insights into the Shigella type III needle tip IpaD in secretion control and cell contact. Mol Microbiol. 2013;88:268–282. doi: 10.1111/mmi.12185. [DOI] [PubMed] [Google Scholar]
- 11.Stamm LM, Goldberg MB. Microbiology. Establishing the secretion hierarchy. Science. 2011;331:1147–1148. doi: 10.1126/science.1203195. [DOI] [PubMed] [Google Scholar]
- 12.Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE. A sorting platform determines the order of protein secretion in bacterial type III systems. Science. 2011;331:1188–1191. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misselwitz B, Kreibich SK, Rout S, Stecher B, Periaswamy B, Hardt WD. Salmonella enterica serovar Typhimurium binds to HeLa cells via Fim-mediated reversible adhesion and irreversible type three secretion system 1-mediated docking. Infect Immun. 2011;79:330–341. doi: 10.1128/IAI.00581-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lara-Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun. 2009;77:2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nhieu GT, Sansonetti PJ. Mechanism of Shigella entry into epithelial cells. Curr Opin Microbiol. 1999;2:51–55. doi: 10.1016/s1369-5274(99)80009-5. [DOI] [PubMed] [Google Scholar]
- 16.Ogawa M, Sasakawa C. Intracellular survival of Shigella. Cell Microbiol. 2006;8:177–184. doi: 10.1111/j.1462-5822.2005.00652.x. [DOI] [PubMed] [Google Scholar]
- 17.Tran Van Nhieu G, Ben-Ze'ev A, Sansonetti PJ. Modulation of bacterial entry into epithelial cells by association between vinculin and the Shigella IpaA invasin. EMBO J. 1997;16:2717–2729. doi: 10.1093/emboj/16.10.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mounier J, et al. Rho family GTPases control entry of Shigella flexneri into epithelial cells but not intracellular motility. J Cell Sci. 1999;112(Pt 13):2069–2080. doi: 10.1242/jcs.112.13.2069. [DOI] [PubMed] [Google Scholar]
- 19.Handa Y, et al. Shigella IpgB1 promotes bacterial entry through the ELMO-Dock180 machinery. Nat Cell Biol. 2007;9:121–128. doi: 10.1038/ncb1526. [DOI] [PubMed] [Google Scholar]
- 20.Hachani A, et al. IpgB1 and IpgB2, two homologous effectors secreted via the Mxi-Spa type III secretion apparatus, cooperate to mediate polarized cell invasion and inflammatory potential of Shigella flexenri. Microbes Infect. 2008;10:260–268. doi: 10.1016/j.micinf.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Garza-Mayers AC, Miller KA, Russo BC, Nagda DV, Goldberg MB. Shigella flexneri regulation of ARF6 activation during bacterial entry via an IpgD-mediated positive feedback loop. MBio. 2015;6:e02584. doi: 10.1128/mBio.02584-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 23.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernardini ML, Mounier J, d'Hauteville H, Coquis-Rondon M, Sansonetti PJ. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc Natl Acad Sci U S A. 1989;86:3867–3871. doi: 10.1073/pnas.86.10.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 26.Eriksson JE, et al. Introducing intermediate filaments: from discovery to disease. J Clin Invest. 2009;119:1763–1771. doi: 10.1172/JCI38339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mor-Vaknin N, et al. Murine colitis is mediated by vimentin. Sci Rep. 2013;3:1045. doi: 10.1038/srep01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colucci-Guyon E, Portier MM, Dunia I, Paulin D, Pournin S, Babinet C. Mice lacking vimentin develop and reproduce without an obvious phenotype. Cell. 1994;79:679–694. doi: 10.1016/0092-8674(94)90553-3. [DOI] [PubMed] [Google Scholar]
- 29.Chu PG, Weiss LM. Keratin expression in human tissues and neoplasms. Histopathology. 2002;40:403–439. doi: 10.1046/j.1365-2559.2002.01387.x. [DOI] [PubMed] [Google Scholar]
- 30.Jepson MA, Mason CM, Bennett MK, Simmons NL, Hirst BH. Co-expression of vimentin and cytokeratins in M cells of rabbit intestinal lymphoid follicle-associated epithelium. Histochem J. 1992;24:33–39. doi: 10.1007/BF01043285. [DOI] [PubMed] [Google Scholar]
- 31.Leube RE, Moch M, Windoffer R. Intermediate filaments and the regulation of focal adhesion. Curr Opin Cell Biol. 2015;32:13–20. doi: 10.1016/j.ceb.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 32.Mor-Vaknin N, Punturieri A, Sitwala K, Markovitz DM. Vimentin is secreted by activated macrophages. Nat Cell Biol. 2003;5:59–63. doi: 10.1038/ncb898. [DOI] [PubMed] [Google Scholar]
- 33.Zou Y, He L, Huang SH. Identification of a surface protein on human brain microvascular endothelial cells as vimentin interacting with Escherichia coli invasion protein IbeA. Biochem Biophys Res Commun. 2006;351:625–630. doi: 10.1016/j.bbrc.2006.10.091. [DOI] [PubMed] [Google Scholar]
- 34.Picking WL, Coye L, Osiecki JC, Barnoski Serfis A, Schaper E, Picking WD. Identification of functional regions within invasion plasmid antigen C (IpaC) of Shigella flexneri. Mol Microbiol. 2001;39:100–111. doi: 10.1046/j.1365-2958.2001.02210.x. [DOI] [PubMed] [Google Scholar]
- 35.Harrington A, et al. Characterization of the interaction of single tryptophan containing mutants of IpaC from Shigella flexneri with phospholipid membranes. Biochemistry. 2006;45:626–636. doi: 10.1021/bi0512593. [DOI] [PubMed] [Google Scholar]
- 36.Kuwae A, Yoshida S, Tamano K, Mimuro H, Suzuki T, Sasakawa C. Shigella invasion of macrophage requires the insertion of IpaC into the host plasma membrane. Functional analysis of IpaC. J Biol Chem. 2001;276:32230–32239. doi: 10.1074/jbc.M103831200. [DOI] [PubMed] [Google Scholar]
- 37.Scherer CA, Cooper E, Miller SI. The Salmonella type III secretion translocon protein SspC is inserted into the epithelial cell plasma membrane upon infection. Mol Microbiol. 2000;37:1133–1145. doi: 10.1046/j.1365-2958.2000.02066.x. [DOI] [PubMed] [Google Scholar]
- 38.Carlson SA, Omary MB, Jones BD. Identification of cytokeratins as accessory mediators of Salmonella entry into eukaryotic cells. Life Sci. 2002;70:1415–1426. doi: 10.1016/s0024-3205(01)01512-0. [DOI] [PubMed] [Google Scholar]
- 39.Schmitz AM, Morrison MF, Agunwamba AO, Nibert ML, Lesser CF. Protein interaction platforms: visualization of interacting proteins in yeast. Nat Methods. 2009;6:500–502. doi: 10.1038/nmeth.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terry CM, et al. The C-terminus of IpaC is required for effector activities related to Shigella invasion of host cells. Microb Pathog. 2008;45:282–289. doi: 10.1016/j.micpath.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwuan L, Adams W, Auerbuch V. Impact of host membrane pore formation by the Yersinia pseudotuberculosis type III secretion system on the macrophage innate immune response. Infect Immun. 2013;81:905–914. doi: 10.1128/IAI.01014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viboud GI, Bliska JB. Measurement of pore formation by contact-dependent type III protein secretion systems. Methods Enzymol. 2002;358:345–350. doi: 10.1016/s0076-6879(02)58100-3. [DOI] [PubMed] [Google Scholar]
- 43.Viboud GI, Bliska JB. A bacterial type III secretion system inhibits actin polymerization to prevent pore formation in host cell membranes. EMBO J. 2001;20:5373–5382. doi: 10.1093/emboj/20.19.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reeves AZ, Spears WE, Du J, Tan KY, Wagers AJ, Lesser CF. Engineering Escherichia coli into a Protein Delivery System for Mammalian Cells. ACS Synth Biol. 2015;4:644–654. doi: 10.1021/acssynbio.5b00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanaja SK, Rathinam VA, Fitzgerald KA. Mechanisms of inflammasome activation: recent advances and novel insights. Trends Cell Biol. 2015;25:308–315. doi: 10.1016/j.tcb.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 47.Dos Santos G, et al. Vimentin regulates activation of the NLRP3 inflammasome. Nat Commun. 2015;6:6574. doi: 10.1038/ncomms7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batchelor M, et al. Involvement of the intermediate filament protein cytokeratin-18 in actin pedestal formation during EPEC infection. EMBO Rep. 2004;5:104–110. doi: 10.1038/sj.embor.7400038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campbell-Valois FX, Schnupf P, Nigro G, Sachse M, Sansonetti PJ, Parsot C. A fluorescent reporter reveals on/off regulation of the Shigella type III secretion apparatus during entry and cell-to-cell spread. Cell Host Microbe. 2014;15:177–189. doi: 10.1016/j.chom.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Picking WL, et al. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect Immun. 2005;73:1432–1440. doi: 10.1128/IAI.73.3.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roehrich AD, Guillossou E, Blocker AJ, Martinez-Argudo I. Shigella IpaD has a dual role: signal transduction from the type III secretion system needle tip and intracellular secretion regulation. Mol Microbiol. 2013;87:690–706. doi: 10.1111/mmi.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Labrec EH, Schneider H, Magnani TJ, Formal SB. Epithelial Cell Penetration as an Essential Step in the Pathogenesis of Bacillary Dysentery. J Bacteriol. 1964;88:1503–1518. doi: 10.1128/jb.88.5.1503-1518.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maurelli AT, Baudry B, d'Hauteville H, Hale TL, Sansonetti PJ. Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect Immun. 1985;49:164–171. doi: 10.1128/iai.49.1.164-171.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Menard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo BC, et al. A Francisella tularensis locus required for spermine responsiveness is necessary for virulence. Infect Immun. 2011;79:3665–3676. doi: 10.1128/IAI.00135-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Labigne-Roussel AF, Lark D, Schoolnik G, Falkow S. Cloning and expression of an afimbrial adhesin (AFA-I) responsible for P blood group-independent, mannose-resistant hemagglutination from a pyelonephritic Escherichia coli strain. Infect Immun. 1984;46:251–259. doi: 10.1128/iai.46.1.251-259.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carette JE, et al. Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat Biotechnol. 2011;29:542–546. doi: 10.1038/nbt.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reimand J, Arak T, Vilo J. g:Profiler--a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res. 2011;39:W307–315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heindl JE, Saran I, Yi CR, Lesser CF, Goldberg MB. Requirement for formin-induced actin polymerization during spread of Shigella flexneri. Infect Immun. 2010;78:193–203. doi: 10.1128/IAI.00252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costa SC, Lesser CF. A multifunctional region of the Shigella type 3 effector IpgB1 is important for secretion from bacteria and membrane targeting in eukaryotic cells. PLoS One. 2014;9:e93461. doi: 10.1371/journal.pone.0093461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bahrani FK, Sansonetti PJ, Parsot C. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect Immun. 1997;65:4005–4010. doi: 10.1128/iai.65.10.4005-4010.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soyer M, Dumenil G. A laminar-flow chamber assay for measuring bacterial adhesion under shear stress. Methods Mol Biol. 2012;799:185–195. doi: 10.1007/978-1-61779-346-2_12. [DOI] [PubMed] [Google Scholar]
- 63.Mikaty G, et al. Extracellular bacterial pathogen induces host cell surface reorganization to resist shear stress. PLoS Pathog. 2009;5:e1000314. doi: 10.1371/journal.ppat.1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moreau-Marquis S, et al. The DeltaF508-CFTR mutation results in increased biofilm formation by Pseudomonas aeruginosa by increasing iron availability. Am J Physiol Lung Cell Mol Physiol. 2008;295:L25–37. doi: 10.1152/ajplung.00391.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muller NF, et al. Trimeric autotransporter adhesin-dependent adherence of Bartonella henselae, Bartonella quintana, and Yersinia enterocolitica to matrix components and endothelial cells under static and dynamic flow conditions. Infect Immun. 2011;79:2544–2553. doi: 10.1128/IAI.01309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yi CR, et al. Systematic analysis of bacterial effector-postsynaptic density 95/disc large/zonula occludens-1 (PDZ) domain interactions demonstrates Shigella OspE protein promotes protein kinase C activation via PDLIM proteins. J Biol Chem. 2014;289:30101–30113. doi: 10.1074/jbc.M114.595868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Costa SC, et al. A new means to identify type 3 secreted effectors: functionally interchangeable class IB chaperones recognize a conserved sequence. MBio. 2012;3 doi: 10.1128/mBio.00243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baxt LA, Goldberg MB. Host and bacterial proteins that repress recruitment of LC3 to Shigella early during infection. PLoS One. 2014;9:e94653. doi: 10.1371/journal.pone.0094653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sievers F, Higgins DG. Clustal omega. Curr Protoc Bioinformatics. 2014;48:3.13.11–13.13.16. doi: 10.1002/0471250953.bi0313s48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.