Abstract
Background
Pannexin 3 (PANX3) is a channel-forming protein capable of stimulating osteogenesis in vitro. Here we studied the in vivo roles of PANX3 in the chicken embryo using the RCAS retroviral system to over-express and knockdown expression during endochondral bone formation.
Results
In the limbs, PANX3 RNA was first detected in the cartilage condensations and became restricted to the prehypertrophic cartilage of the epiphyses, diaphysis and perichondrium. The increase in PANX3 was not sufficient to alter osteogenesis, however a virus containing an interference RNA construct caused a 20% reduction in bone volume. The control virus containing a shEGFP cassette did not affect development. Interestingly, the phenotype was restricted to later stages rather than to proliferation of the skeletogenic mesenchyme, formation of the cartilage condensation or creation of the hypertrophic zones. In addition, there was also no change in readouts of Hh, WNT, FGF or BMP signaling using either QPCR or radioactive in situ hybridization.
Conclusions
Based on the normal expression domains of PANX3 and the relatively late manifestation of the phenotype, it is possible that PANX3 hemichannels may be required to facilitate the transition of hypertrophic chondrocytes to osteoblasts, thereby achieving final bone size.
Keywords: Endochondral ossification, channel protein, retrovirus, chicken, skeletogenesis, limb bud, interference RNA
Introduction
Long bone development in the axial and appendicular skeleton occurs through the process of endochondral ossification (Berendsen and Olsen, 2015). Mesodermal precursor cells form the cartilage condensations. The cartilage elements are gradually replaced by bone, first in the centre or diaphysis and secondarily in the epiphyses. Chondrocytes become terminally differentiated and undergo hypertrophy in defined stacks of cells, leading to further longitudinal expansion. Osteoblasts differentiate from hypertrophic chondrocytes (Yang et al., 2014b; Zhou et al., 2014; Park et al., 2015) and perichondrium (Dirckx et al., 2013). The primary spongiosa is eventually remodeled into mature trabecular bone. In contrast, in parts of the skull and face, bone differentiates from mesenchyme directly using the process of intramembranous ossification (Berendsen and Olsen, 2015). Once the cartilage template is removed, similar transcription factors regulate both endochondral and intramembranous ossification including RUNX2 (runt-related transcription factor 2) and SP7 (Osterix) (Berendsen and Olsen, 2015).
We and others have previously demonstrated a novel connection between RUNX2 and the channel-forming protein Pannexin 3 (PANX3) in both osteoblasts and growth plate chondrocytes (Iwamoto et al., 2010; Bond et al., 2011; Ishikawa et al., 2011; Ishikawa et al., 2014). The Pannexins are a small family of chordate proteins homologous to the invertebrate gap junction proteins known as Innexins (Panchin et al., 2000). Unlike some of the Innexins, however, Pannexins do not form intercellular gap junctions under many physiologically relevant conditions (Sosinsky et al., 2011). Instead, they are understood to create large transmembrane channels that facilitate passage of ions and small molecules (such as Ca2+ and ATP) between the intercellular and extracellular spaces (Bond and Naus, 2014; Penuela et al., 2014). Overexpression and/or supression of Panx3 in cultured cell lines has been implicated in aberrant differentiation of keratinocytes (Celetti et al., 2010), osteoblasts (Ishikawa et al., 2014), and chondrocytes (Iwamoto et al., 2010). We therefore hypothesized that PANX3 was required for bone formation and wanted to test these ideas in vivo.
In this study, the chicken was chosen due to its long history as a model organism for skeletal development, and because it is amenable to localized manipulation of gene expression through retroviruses (Harpavat and Cepko, 2006; Chen et al., 2007; Chen et al., 2008; Gordon et al., 2009). Here, gallus PANX3 mRNA or PANX3-shRNA was delivered into early limb buds using the RCASBP vector (replication-competent ASLV long terminal repeat with a splice acceptor and Bryan polymerase; Gordon et al., 2009). The virus spreads vertically and laterally, leading to abnormal PANX3 expression starting approximately 24h from the time of infection. In our study we show that over-expression does not cause a phenotype, however effective knockdown causes a reduction in bone volume. We compare our findings to recent knockout studies carried out in mouse and zebrafish (Moon et al., 2015; Oh et al., 2015).
Results
PANX3 is expressed in intramembranous and endochondral bone in chicken
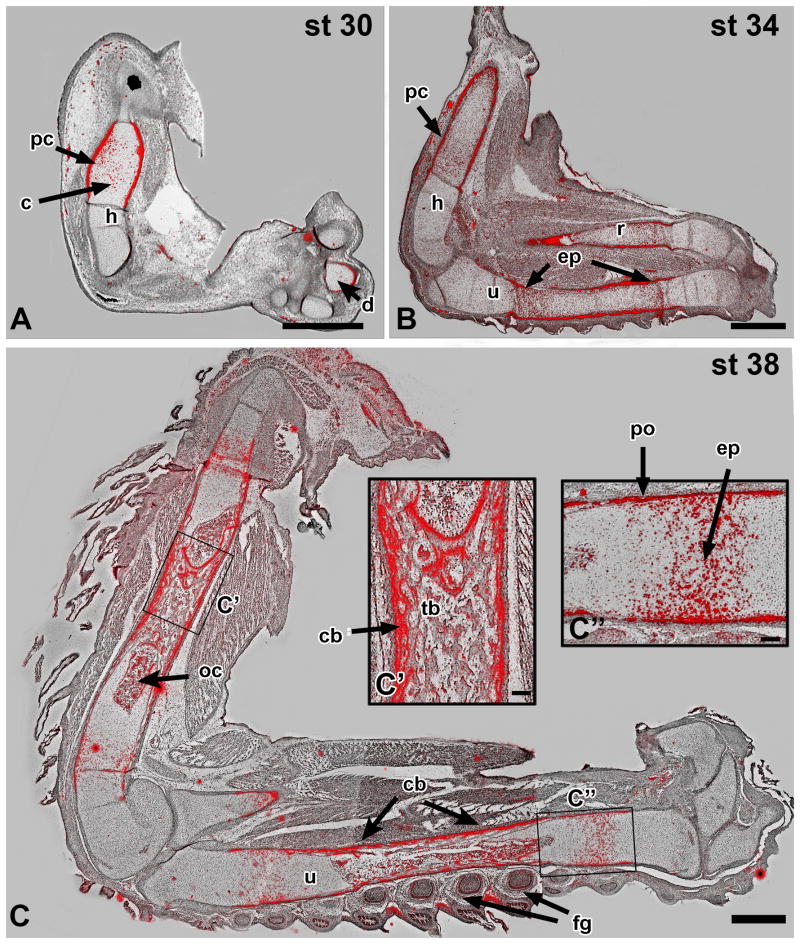
The presence of Panx3 has previously been reported in osteoblasts and hypertrophic chondrocytes in mouse (Bond et al., 2011; Kwon et al., 2014) where its expression is regulated by Runx2 (Bond et al., 2011). The chicken PANX3 promoter also contains a RUNX2 binding site (AACCACA (Ducy and Karsenty, 1995) at position 632, upstream of the start codon. The distribution of PANX3 transcript was assessed in the limbs of chicken embryos throughout condensation, formation of the growth plate and differentiation of endochondral bone using radioactive in situ hybridization (stages 28, 30, 34, and 38 (Hamburger and Hamilton, 1992). No expression was detected prior to stage 30. At stage 30 localized signal was seen in the cartilage condensations and perichondrium of the limb skeletal elements (Fig. 1A and data not shown). By stage 34, PANX3 transcript secondary sites of expression appeared in the epiphyseal plates of the humerus, radius, and ulna (Fig. 1B). Interestingly, there was a zone of lower expression in the centre of the diaphyses where ossification was initiating (Fig. 1B). By stage 38, the primary spongiosa (nascent trabecular bone) exhibited substantial expression, presumably in osteoblasts (Fig. 1C, C′). Expression of PANX3 in chondrocytes appeared to be highest in the pre-hypertrophic zone as well as in the bone collar, both sites of osteoblast generation (Figure. 1C′, C″). In addition to the skeletal system, PANX3 expression was also detected in the feather germs (Fig. 1C). Given the observed timing and distribution of PANX3 mRNA in developing bone we surmised that the translated protein is involved in osteogenesis rather than condensation formation.
Figure 1. Endogenous PANX3 expression during chicken limb development.
Radioactive in situ hybridization on sagittal sections of embryonic limbs was used to monitor the normal expression pattern of chicken PANX3. Silver deposits accumulate in regions of the tissue where radioactive probe has bound to complementary mRNA, and is visualized with a dark-field condenser. The signal has been false coloured (red) and overlaid upon bright field images. A) PANX3 is initially expressed in the centre of cartilage blastemas and is also elevated in the perichondrium. B) PANX3 is expressed in the epiphyseal plates and perichondrium by stage 34. There is relatively lower expression in the center of the diaphysis, the first site of chondrocyte hypertrophy (see centre of radius). C,C′: At stage 38 the ossification centers contain bone, and peripheral osteoblasts in the cortical bone express PANX3. The dermal papillae of the feather germs also express PANX3. C″: Prehypertrophic chondrocytes and the periosteum or bone collar express high levels of PANX3. Key: cb – cortical bone; d – digits; ep – epiphyseal plate; fg – feather germs; h – humerus; oc – ossification centre; pc – perichondrium; po – periosteum; r – radius; tb – trabecular bone; u – ulna. Scale bars A-C = 1 mm, C′, C″ = 100 μm.
Retroviral viral targeting to the limb covers the domains of PANX3 expression
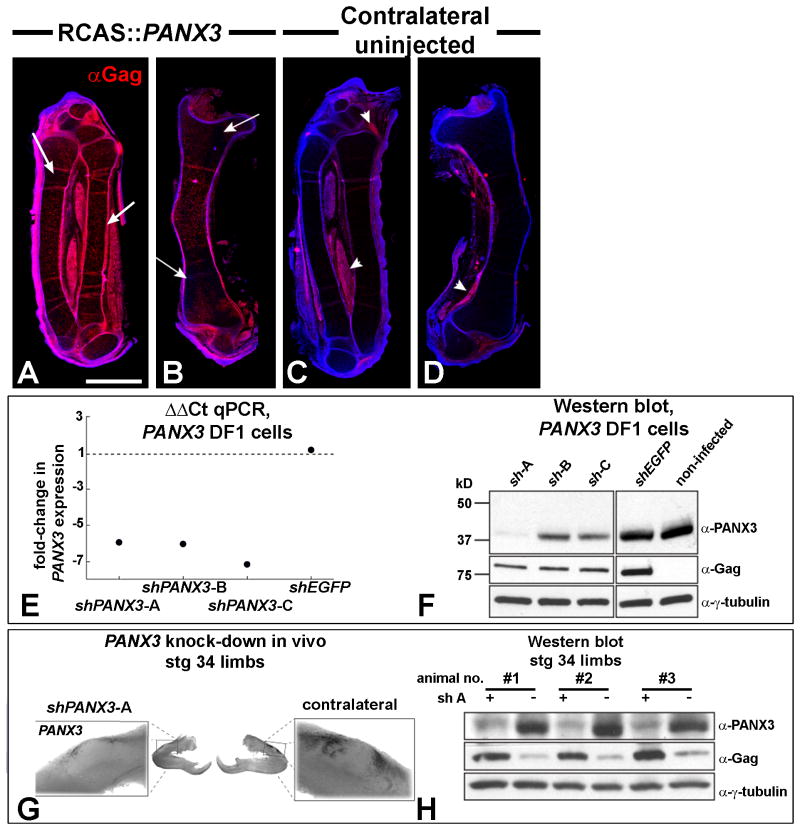
We used RCAS retroviral particles to deliver either wild-type PANX3, knockdown shPANX3 constructs or a control virus containing an shEGFP cassette. We confirmed that viral targeting was successful using immunofluorescence to detect the presence of the virus. The Gag antibody detected abundant expression on the treated side (Fig. 2A,B). In approximately 50% of treatments, the Gag antibody showed slight retrovirus spread to the contralateral side (Fig. 2C,D). The spread of virus is due to the replication-competent nature of RCAS virus. The targeting of viral injections was therefore very good and we expected that all viruses would be expressed over a similar region of the embryo.
Figure 2. Targeting of the virus and knockdown of PANX3 using RCASBP∷mir-30a shRNA constructs.
A-D) Embryos injected into the right forelimb at stage 20 with RCAS∷PANX3 and fixed 6 days later at stage 34. The red immunofluorescence signal from the anti-Gag antibody indicates presence of viral envelope protein. The virus is present throughout the skeletal elements of the injected limb and in the perichondrium (arrows). There is occasionally spread to the uninjected, contralateral side (C,D, arrowheads). E) DF-1 fibroblasts were transfected with a PANX3-expressing plasmid and then infected with the viruses expressing 3 different short hairpin constructs (A-C). RNA levels of PANX3 were measured 4 days later. Real-time qPCR shows a reduction of 6-fold in PANX3 compared to control cells infected with an shGFP virus. F) A subset of same DF1 cells were collected 4 days after viral infection and used to measure protein levels. shPANX3-A was the most effective. G) Whole mount in situ hybridization showed qualitatively lower endogenous PANX3 expression relative to the contralateral control limb. This embryo was injected at stage 20 with the virus and fixed 6 days later at stage 34. H) Cellular lysates were prepared from three RCAS∷shPANX3-A injected limbs (+) and their contralateral control limbs (-) 6 days after infection at stage 20. A Western blot showed a consistent knockdown of Panx3 across three biological replicates.
Effective knockdown of PANX3 in vitro and in vivo
We tested 3 different short hairpin sequences designed to reduce PANX3 expression (shPANX3-A,B,C starting at bp14, 347 and 620 respectively). Quantitative real-time PCR (qPCR, Fig. 2E) and Western blot confirmed (Fig. 2F) that the PANX3-shRNA constructs were able to reduced expression of plasmid-derived PANX3 in DF-1 cells. The control shEGFP virus had no effect on PANX3 expression (Figure 2E, F). Of the three PANX3-shRNA constructs, shPANX3-A caused the most significant knockdown at the RNA (Fig. 2G, Fig. 4) and protein level (Fig. 2H) in vivo, so the A version of the virus was chosen for all subsequent in vivo experiments.
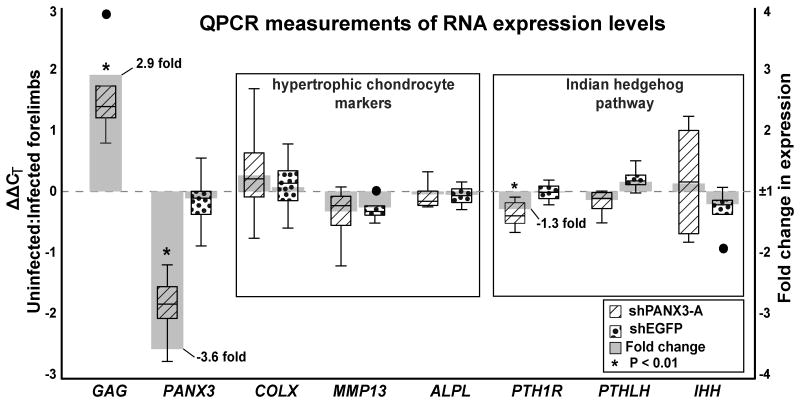
Figure 4. Real-time qPCR for genes involved in endochondral bone development.
Stage 37 forelimbs were injected with shPANX3-A or shEGFP. Comparisons were made to the contralateral limbs. Four pools of 4 RNA samples were made for each treatment. The axis on the left represents ΔΔCT (box plots), which is the log2 difference in mRNA concentration between injected and control, while the right axis represents fold change in expression. GAG, PANX3, and PTH1R demonstrated statistically significant changes in expression following shPANX3-A injection (1-sample t-test, H0 mean = 0.0, p-value ≤ 0.05).
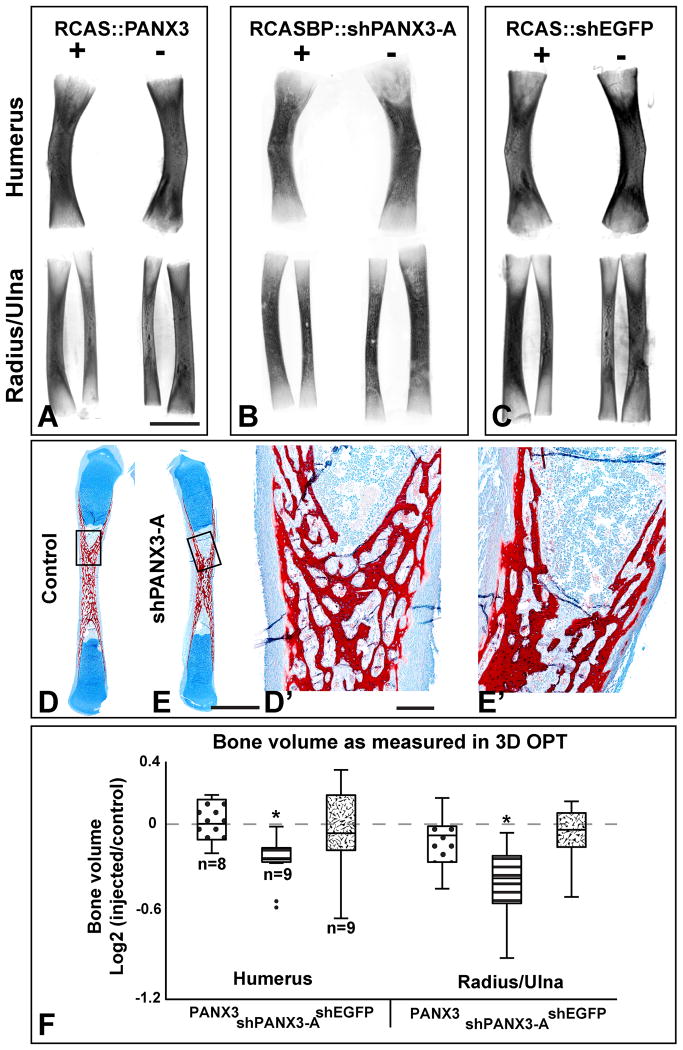
The skeletal effects of altering expression of PANX3 were investigated using 3D imaging and microscopy. Wings were collected 13-14 days following injection, and total volume of the Alizarin red-stained humerus, radius, and ulna were measured using OPT. The developmental stage of each embryo at the time of collection varied, so within-group sample variance was controlled by comparing each injected limb against its contralateral uninjected limb (I/C-score = log2[injected/uninjected]); significant deviation from a value of zero denotes a change in response to viral injection. PANX3 over-expression did not affect bone volume (I/C-score = -0.05 ±0.09; Fig. 3A, D) and values were not significantly different than the shEGFP controls (Fig. 3C,D). At the microscopic level, there were no changes in the arrangement of chondrocytes in the growth plate (data not shown). Therefore overexpression of PANX3 does not appear to affect chondrocyte stacking.
Figure 3. OPT scans of stage 40 forelimb bones.
Representative images of humerus (top) and radius/ulna (bottom) taken from the injected (right side, +) and contralateral (left, -) limb of the same animal. All limbs stained with Alizarin red, cleared in BABB and scanned on the red channel to reveal fluorescence. These are projections of the full 3-D scan. Limited information can be obtained about the trabecular pattern due to lack of penetration of the UV light. A) Wild-type RCAS∷PANX3 does not cause a change in volume. B) shPANX3 virus causes a reduction in size of forelimb bones. C) A virus containing a shEGFP cassette resulted in limbs of similar sizes. D-E′) Resin sections through the radii of stage 40 embryos, stained with Alcian Blue for cartilage and Alizarin red for bone. The red trabecular patterns in the diaphysis are indistinguishable.F) A whisker plot showing the bone volume of each sample comparing the injected (I) to its contralateral control (C; I/C score = log2[I/C]). An I/C-score of 0.0 indicates that the injected and control bones are the same size. PANX3 and shEGFP viral infection did not alter bone volume, while shPANX3 injection was correlated with a significant reduction of about 20%. Plus and minus symbols indicate the presence or absence of virus injection. Asterisks indicate p < 0.01, 1-sample, 2-tailed t-test. Dots represent valued that fall outside of the 95th percentile. Scale bar = 500 μm.
Alternatively, shPANX3-A retrovirus particles caused a significant reduction of the humerus, radius and ulna (19.9% ±5.8%; I/C-score = -0.34 ±0.11; P < 0.01; Fig. 3B, D). Thus, the 3.6-fold decrease measured with QPCR correlates with the decreased volume (Fig. 4). There were however no visible differences in histology of the cartilage or timing of replacement by bone (Fig. 3D-E′). Embryos treated with shPANX3-A appear to have smaller ossification centres but trabecular arrangement is normal (Fig. 3D,E).
One reason for the subtle difference in volume observed at stage 20 injections may be that peak viral expression did not coincide with the onset of PANX3 function. To determine whether a larger effect would be seen if virus expression initiated earlier, we performed injections at stage 15 (24 hours younger than stage 20). At stage 15 limb outgrowth is just beginning, however, injection of shPANX3-A failed to produce any phenotypic difference between knockdown and control limbs (injected with shEGFP; Table S2). Thus there is no evidence that PANX3 functions during cartilage formation but a later role in osteogenesis is likely.
We examined proliferation in animals fixed at both stage 35 (N = 7) and stage 37 (N = 14 shPANX3-A, N = 15 shEGFP) using BrdU labeling. There was no significant difference between treated and non-treated sides of the embryo at stage 35 (data not shown). Detailed examination of the epiphysis at stage 37 also did not show significant changes in proliferation, despite the fact that endogenous PANX3 is expressed in this region. Thus decreased bone volume is not due to an earlier drop in proliferation.
To further determine whether the knockdown of PANX3 had altered the molecular signaling involved in skeletogenesis, we measured the expression of bone development marker genes using qRT-PCR in stage 37 bones (Fig. 4). Matrix metallopeptidase 13 (MMP13), alkaline phosphatase (ALPL), and COLX are all classic markers for chondrocyte hypertrophy (Moog, 1944; Schmid and Conrad, 1982; D'Angelo et al., 2000). Rather unexpectedly, none of of these genes were differentially expressed in shPANX3-A treated versus shEGFP controls (Fig. 4). Thus terminal differentiation does not appear to be affected by PANX3 knockdown. We also examined ISBP and did not find significant expression differences (data not shown).
Altered expression of the Indian hedgehog pathway genes IHH, PTHLH, or PTH1R are known to disrupt the normal progression of chondrocyte maturation, usually leading to a reduction in final bone size (St-Jacques et al., 1999; Kronenberg, 2003). IHH and PTHLH expression remained unaffected by lower PANX3 levels (Fig. 4). On the other hand, PTH1R expression was reduced (1.3-fold). PTH1R is normally expressed at a low level throughout long bone cartilage, but is up-regulated in the pre-hypertrophic zone (Vortkamp et al., 1996; MacLean and Kronenberg, 2005; Chau et al., 2011). Increasing the activity of PTH1R in mouse chondrocytes delays hypertrophy (Weir et al., 1996; Schipani et al., 1997) by stimulating downstream protein kinase A (PKA; Li et al., 2004; Hsu et al., 2012), so the lower levels of PTH1R following treatment with shPANX3 would be predicted to lead to precocious chondrocyte differentiation. However as shown by histological analysis, the decrease in PTH1R at the RNA level was not sufficient to have a biological effect on the rate of bone differentiation.
Radioactive in situ hybridization was performed on serial sections of stage 35 embryos to determine (1) which genes overlapped in their expression with PANX3 and (2) whether expression of these genes were affected by knockdown of PANX3. We examined readouts and components of the Wingless pathway (LEF1, WNT5A, WNT5B), Hedgehog (Patched, PTCH1), Fibroblast growth factor (Sprouty, SPRY2) and Bone Morphogenetic protein (BMP2). In addition, markers of bone differentiation were examined (MMP13, RUNX2 and IBSP). Several genes were ubiquitously expressed (SPRY2, LEF1 and WNT5A, data not shown). PTCH1 was highly expressed in the joints and ends of the bones so it was not in the region of interest (data not shown).
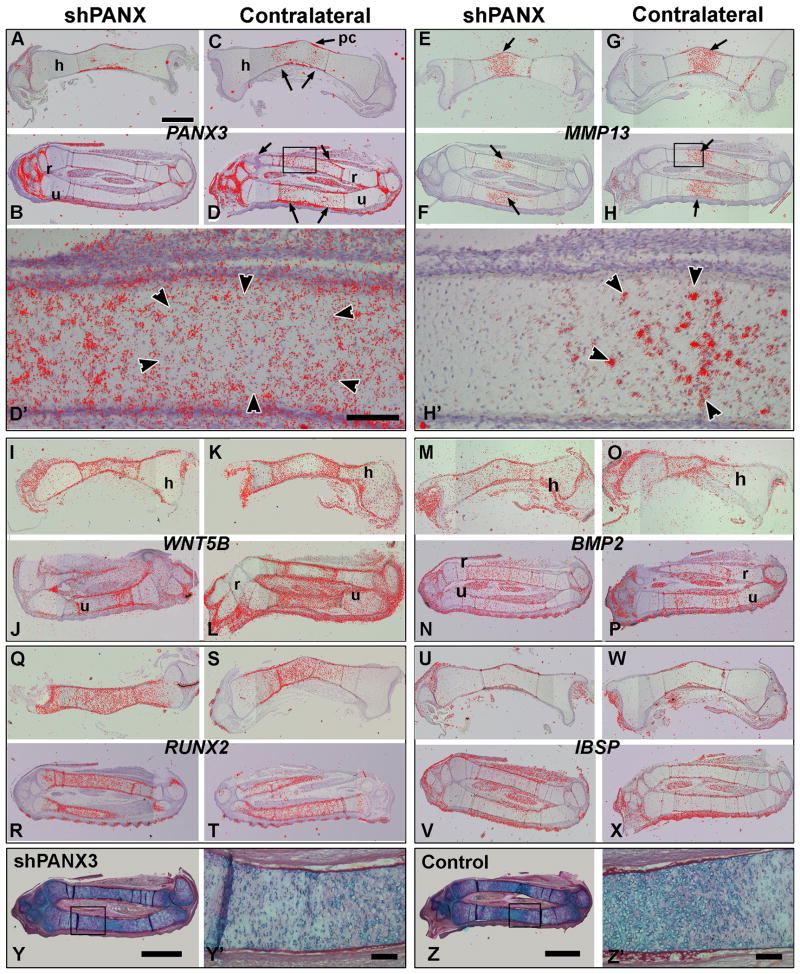
PANX3 was expressed in the perichondrium and flanking the diaphysis (Fig. 5C,D, D′). In the treated embryos there was visiblely reduced signal within the bones (Fig. 5A,B, N = 2). MMP13 had regionally restricted expression in the diaphysis, almost complementary to that of PANX3 (Fig. 5E-H,H′). MMP13 marked the future zone of chondrocyte hypertrophy prior to overt change in the cartilage (Fig. 5E-H, arrows). In higher power views there is some overlap between the MMP13 and PANX3 expression domains at the edges (Fig. 5D′,H′ arrowheads). In contrast, WNT5B (Fig. 5I-L) and BMP2 (Fig. 5M-P) transcripts closely overlapped the expression of PANX3, including the relatively low expression in the diaphysis. RUNX2 is expressed throughout the skeletal elements, extending beyond the PANX3 transcripts, indicating indicating that osteogenic commitment has been initiated by stage 35 (Fig. 5Q-T). IBSP (Integrin binding sialoprotein) is localized primarily to the perichondrium where initial ossification is beginning, overlapping with PANX3 expression (Fig. 5U-X). Although the levels of PANX3 were qualitative reduced, there was no change in distribution of all the genes we examined. In addition histodifferentiation of the cartilage was unaffected by the virus (Fig. 5Y-Z′). The lack of RNA expression differences at both stages 35 and 37 does not preclude a post-translational relationship between PANX3 and the products of these genes. It is also possible that other changes in expression may occur subsequent to stage 37.
Figure 5. Radiolabeled in situ hybridization for genes involved in endochondral bone development.
Sagittal sections of forelimb bones from two different stage 34 embryos. Both the right side injected with shPANX3 virus and the left side (uninjected control) are shown for comparison All sections were photographed with darkfield and brightfield illumination. Silver grains in the darkfield images were converted to red and overlaid on top of the brightfield images using Adobe Photoshop.A-D) Expression of PANX3 is reduced on the treated side (A,C) compared to the control side (B,D). On the control side, signal is present in the diaphysis (arrows in B,D) but a central region has relatively lower expression. E-H) The expression of MMP13 is highest in the centre of the diaphysis (arrows), complementary to the PANX3 signal. There is no qualitative difference in the levels of MMP13 on the treated side (E,G). I-L) WNT5B is expressed at high levels surrounding the diaphysis in both controls and experimentals. M-P) The expression of BMP2 is more ubiquitous that other genes and shows no difference between experimentals and controls. Q-T) RUNX2 is strongly expressed throughout the skeletal elements up to the epiphyses. There is no difference in expression in treated embryos. U-X) IBSP is mainly expressed in the perichondrium and again not affected by the knockdown of PANX3. Y-Z′) Adjacent sections from the same embryos used for in situ hybridization, stained with Alcian blue and picrosirius red. There are no differences in the density of chondrocytes in the knockdown limb (Y,Y′) as compared to the control limb (Z,Z′). Key: h – humerus, r – radius, pc – perichondrium, u - ulna. Scale bar in A = 1 mm and applies to all panels.
Discussion
Here, the chicken embryo was used to assess the importance of PANX3 during endochondral bone formation for the first time in vivo. Both over-expression and knockdown were achieved, but neither resulted in a major disruption of the developmental program. Misexpression of PANX3 outside its normal scope of osteoblasts and pre-hypertrophic chondrocytes appears to be completely benign, while suppressing endogenous expression causes a 20% reduction in bone volume, accompanied by reduced PTH1R expression.
Technical advances in the cloning of shRNA constructs into RCAS retroviruses
By combining the flexibility of restriction free cloning and the Gateway system, we have created a workflow to efficiently generate retroviral RNAi constructs for use in avian systems. This work builds upon the protocol originally described by Chen et al. (2007), who initially replaced the leader and trailer sequences immediately flanking the native chicken mir-30a hairpin with MluI and NcoI restriction sites to facilitate cloning custom hairpins into the target region. However, large 99 bp synthetic duplexes were required for each new construct. While the same group was later able to reduce the duplex size to 78 bp by utilizing SphI and NgoMIV restriction sites (Chen et al., 2008), our new PCR-based cloning method further optimizes the oligo sizes to 54 and 56 bp. Furthermore, adding a diagnostic KpnI site to the target pENTR-miR30a plasmid allows for rapid screening of clones. The ability to quickly and reliably create many new shRNA cassettes is very beneficial, because the effectiveness of any given cassette is difficult to predict as we have seen here.
Ectopic PANX3 expression is insufficient to stimulate precocious bone maturation in vivo
Exogenous PANX3 has previously been reported to enhance the differentiation rate of osteoblasts, chondrocytes, and osteoprogenitor cells in vitro, and possible mechanisms have been offered to explain these observations (Iwamoto et al., 2010; Ishikawa et al., 2011; Ishikawa et al., 2014). Using cultured ATDC5 and N1511 chondrocyte lines, one group proposed a model where ATP diffuses through Panx3 channels, preventing adenylyl cyclase from generating sufficient cAMP to activate pro-mitotic PKA (Iwamoto et al., 2010). This model suggests that ectopic PANX3 expression in proliferating chondrocytes should lead to increased ATP diffusion and a decrease in proliferation. We did not detect morphological changes in the RCAS∷PANX3 embryos. There was no evidence of decreased proliferation such as reduction of bone size and/or precocious differentiation. It seems likely that there are differences between the in vitro and in vivo overexpression. With the exception of our data in chicken, there are no other animal models in which over-expression of PANX3 has been studied. Our data suggests that PANX3 is not sufficient to alter channel function during embryogenesis.
Bone volume reduction in shPANX3 knockdown chickens is consistent with the phenotype observed in mouse knockouts
The reduction in bone size observed in stage 20 embryo injections is a real effect on the biology of bone formation. The question is why was this size difference so subtle? One possibility is that due to technical limitations of the technique (incomplete knockdown), the magnitude of the morphology changes was masked. Incomplete knockdown is unavoidable with a method such as retroviral infection. We did account for the time lag of about 18 hours between infection and expression of viral genes by injecting embryos at stage 20 which is at least 48h prior to the onset of endogenous PANX3 expression (Logan and Tabin, 1998; Gordon et al., 2009). We also treated embryos at younger stages but did not detect delays in mineralization or bone reductions. Even with high viral titre, a common problem with RNAi technology (Reynolds et al., 2004) is residual gene expression. We observed a 3.6 fold reduction in PANX3 RNA 7 days after infection with the virus so the remaining RNA would allow a certain level of wild-type PANX3 channel formation.
Recently two studies were published on the in vivo role of Panx3 in the mouse (Moon et al., 2015; Oh et al., 2015). These groups started with the identical DNA construct made available by the KnockOut Mouse Project consortium (KOMP2). Both used the C57/BL6 strain to generate the chimeras. However the groups employed different cre-deleter lines to delete exon 2 of Panx3. One group used CMV-cre mice (Moon et al., 2015) while the other (Oh et al., 2015) used Ella-Cre mice (Lakso et al., 1996). The reason to mention these details is that there was an ossification phenotype visible in the latter study using the Ella-cre germline deletion (Oh et al., 2015) but not in the study using the CMV-Cre mice (Moon et al., 2015). This appears to be the only technical difference between the two studies. Both studies confirmed that deletion was complete, that there was no aberrant protein expressed and that the mutated gene was still expressed in the correct distribution in the embryo. There is substantially increased prenatal lethality with the CMV-Cre Panx3 knockout mice however, the cause of which was not reported. The surviving mice are completely normal up to 6 weeks of age as determined with wholemount skeletal staining and micro-CT (Moon et al., 2015). If ossification defects were part of the lethal phenotype, they were not reported.
In the Ella-Cre Panx3-/- mice (Oh et al., 2015) the limbs are shortened at birth and there is delayed ossification in the parietal bones as shown in wholemount stained preparations. These phenotypes are not merely a developmental delay because 8 week-old Panx3-/- mice had similar decreases in bone length (Oh et al., 2015). Interestingly, only a 5-10% decrease in bone size was reported following knockout which is less than the 20% reduction we observed with the shPANX3 knockdown. Thus the magnitude of changes suggests that in our chicken experiment we are approaching the maximum phenotype for a loss-of-function experiment with PANX3.
Other similarities between our study and that of the study by Oh et al. (Oh et al., 2015) are that they investigated in detail the expression of genes characteristic of hypertrophic chondrocytes, including Col2a1, Mmp13, Ihh and Col10a1 in E14.5 and 16.5 mouse limbs. Similar to our findings they report no difference in expression patterns, although an increase in proliferation was detected in the proximal humerus at E15.5. At present it is difficult to reconcile the increased proliferation with the fact that the bones ended up being shorter. In our study we did not detect changes in proliferation. In summary, neither our study or the one by Oh et al. can provide a mechanism for the decrease in bone size.
Oh and coworkers also analyzed a panx3 morphant zebrafish (Oh et al., 2015), as a complement to the mouse studies. In the zebrafish, the main phenotypes were a delay in ossification of craniofacial bones with a minimal effect on pharyngeal arch cartilages. Thus in zebrafish there is a requirement for panx3 in the initiation of ossification that is not seen in amniotes.
Other membrane channels that could compensate for the loss of PANX3
Compensatory mechanisms may be at work in the chicken and mouse models. There are two other Pannxin genes, Panx1 and Panx2. There is some evidence that Panx1 and 3 are expressed in cartilage but no data provided on bone (Penuela et al., 2007). Panx2 is widely expressed in the mouse but again bone was not examined (Le Vasseur et al., 2014). We also lack sufficient information on the biochemical properties of PANX3 to confidently speculate about the likelihood of another pannexin acting in its place. Regardless, no bone defects have been reported in either the Panx1 or Panx2 knockout mice (Bargiotas et al., 2011). We also did not observe any changes in chicken PANX1 gene expression when PANX3 was knocked down (data not shown).
Other classes of membrane channels might compensate for the decrease in PANX3. Connexins form large aqueous channels between the intra- and extracellular compartment (i.e., hemichannels) with some similarity to PANX channels (Retamal, 2014). Connexin43 (CX43) coded by the gene Gap Junction protein, Alpha 1 (GJA1) is expressed in all tissues of the body including chondrocytes, osteoblasts and osteocytes (Plotkin, 2014; Zappitelli and Aubin, 2014). The disruption of Cx43 gap junction function or formation results in varying degrees of osteopenia in all the mouse models studied (Plotkin, 2014; Zappitelli and Aubin, 2014). However as pointed ou by Oh et al., there is no expression of Cx43 in hypertrophic chondrocytes (Oh et al., 2015) so it is unlikely that this channel protein is compensating for the loss of PANX3.
A potential role for PANX3 in the trans-differentiation of osteoblasts
There is yet another possible role for PANX3 that could explain the reduction in bone size. PANX3 could be required for the transformation of hypertrophic chondrocytes to osteoblasts. There are three studies in which the fate of hypertrophic chondrocytes was traced using genetic reporter mice (Yang et al., 2014b; Zhou et al., 2014; Park et al., 2015). A percentage of osteoblasts are derived from hypertrophic chondrocytes and that they persist after birth. The authors used ColXa1-Cre (Yang et al., 2014a; Yang et al., 2014b; Zhou et al., 2014; Park et al., 2015) or Aggrecan-Cre (Zhou et al., 2014) to drive fluorescence in hypertrophic chondrocytes. Importantly the promoter elements in the cre lines do not drive expression in the perichondrium which is the major source of osteoblasts for cortical and trabecular bone (Dirckx et al., 2013). In addition there does not seem to be aberrant expression of the reporter outside of the chondrocytes at any stage examined. The labeled cells formed about 20-30% of the osteoblasts as judged by co-expression of osterix (SP7) or osteocalcin (Park et al., 2015) in FACS analysis. Strikingly, the size of phenotypes we observed are similar to the proportion of osteoblasts derived from chondrocytes.
We currently have no means to perform lineage tracing of chondrocytes in chickens. However the negative data from our assays (lack of changes in proliferation and no quantitative changes in expression of MMP13 or COLXA1 at stage 37) points to a defect in osteogenesis that is unrelated to the earlier steps in endochondral bone formation. Instead, the bone phenotype caused by the PANX3 knockdown arises sometime between stage 37 and 40 which is likely when chondrocyte-osteoblast transition is taking place. The 5-10% reductions in bone size measured in the mouse knockout study by Oh et al. (Oh et al., 2015) are within the range that could be explained by a transdifferentiation defect. In the future, it will be fascinating to cross the reporter mice that mark hypertrophic chondrocytes with the Panx3-null mice. These studies may reveal roles for Panx3 channels in the transmission of specific signals to facilitate osteoblast transition.
Experimental Procedures
Animals and retroviral injection technique
Fertilized white leghorn eggs were purchased from the University of Alberta and incubated at 38°C until the desired stage (Hamburger and Hamilton, 1992). Stage 15 is 2.5 days of incubation and stage 20 is 3.5 days of incubation. The forelimb on the right-hand side of each animal was injected with concentrated virus + 0.05% Fast Green using a pulled glass pipette and a Picospritzer II (General Valve Corp., Fairfield, NJ). Embryos were allowed to continue developing at 38°C until the desired stage.
Plasmid construction
The chicken PANX3 (accession #XM_001231502) coding sequence was amplified from pooled embryonic cDNA from stage 24 embryos, and ligated into the SalI and EcoRI sites of pBluescript-SK+ (Agilent Technologies, Santa Clara, CA). PANX3 was then sub-cloned between the attL1 and attL2 sites of pENTR3C (Invitrogen, Carlsbad, CA) using BamHI and XhoI. PANX3 was also transferred from pENTR3C into a Gateway compatible variant of the RCASBP vector (Loftus et al., 2001) using the Gateway LR-clonase system (Invitrogen). It was necessary to add a canonical Kozak consensus sequence (GCCGCC) in front of the PANX3 start codon to achieve sufficient levels of over-expression from the RCASBP virus.
To create a restriction-free cloning system for producing short hairpin RNA (shRNA) delivery vectors, we replaced the native hairpin of the chicken mir-30a (miRbase accession #MI0001204) from a pENTR3C-mir-30a plasmid (Chen et al., 2007) with a KpnI site. In-house software was used to design 19-mer siRNA sequences based on previously described criteria (Reynolds et al., 2004; http://tiny.cc/naus_siRNA_pred; GitHub at http://tiny.cc/naus_siRNA_pred_script). The native 20 nucleotide (nt) mir-30a loop sequence was retained except for the first two and last two bases, which were changed from CT-to-TA and GG-to-AT respectively, because the CT and GG are expected to internally pair when converted to RNA (effectively increasing stem length with non-specific sequence). Based on these parameters, a 56-mer and 54-mer restriction-free cloning oligonucleotide was used to synthesize the constructs as follows; the forward primer consists of 17 nt leader sequence, 19 nt sense sequence, and 20 nt loop sequence, while the reverse primer consists of 15 nt trailer sequence, 19 nt anti-sense sequence, and 20 nt anti-sense loop sequence.
To incorporate the hairpin into pENTR3C-mir-30a, 25 ng of each primer was mixed with 100 ng of the empty plasmid in a standard 20 μl PCR reaction using iProof high fidelity DNA polymerase (Bio-Rad, Hercules, CA). The thermal cycling conditions were: denature 8s at 98° C, anneal 20s at 60° C, extend 75 s at 72° C, for 16 cycles. The reaction was treated with DpnI (to digest parental plasmid) and KpnI (to linearize any newly synthesized plasmid which failed to gain an insert) for 2 hours, and chemically competent bacteria were transformed with the un-purified reaction mix. Using super-competent bacteria (>108 CFU / μg pUC18) can be beneficial for this step, but sub-cloning grade (>106 CFU / μg pUC18) cells are usually sufficient. It should be noted that other loop sequences will probably yield equivalent knockdown (Miyagishi et al., 2004), allowing further reduction in synthetic oligonucleotide length should other investigators choose to explore this. Three 19-mer sequences from the PANX3 coding sequence, denoted shPANX3-A (5′ - ACACGGCTGCTGAGTACAT - 3′), shPANX3-B (5′ - TGGTGGCAGTGCTCATGTA - 3′), and shPANX3-C (5′ - TCATCTACCTCCTGAGGAA - 3′). A control shEGFP designed to knockdown EGFP expression (5′ - GGCACAAGCTGGAGTACAA - 3′) was also created. All cassettes were cloned into the hairpin region of pENTR3C-mir-30a for the current study.
Virus preparation and embryo injection
All RCASBP constructs used in this study contained the ‘A’ variant of the envelope protein that was converted to a Gateway compatible ‘Y’ format (Loftus et al., 2001). Viral particles were generated with a modified version of methods described previously (Logan and Tabin, 1998). Unlike the previously described method in (Logan and Tabin, 1998), cell debris was not pre-filtered from the media before centrifugation because a much higher final titre was recovered without this step. Pellets were drained and allowed to slowly resuspend in 100 μl of Opti-MEM (Invitrogen) on ice overnight. Debris was removed from the resuspension by transferring to a 1.5 ml tube and centrifuging for 5 minutes at 3000 g at 4° C, then 5 μl aliquots were flash frozen in liquid nitrogen and stored at -70°C.
Stage 15 embryos were injected with shPANX3 or shEGFP, while stage 20 embryos were injected with either wild-type PANX3, shPANX3 or shEGFP.
Validation of gene knockdown in cell culture using Western blots and qRT-PCR
Restriction-free cloning (Bryksin and Matsumura, 2010; Bond and Naus, 2012) was used to position PANX3 upstream of the internal ribosomal entry site of the pMES bicistronic expression plasmid (Swartz et al., 2001). The pMES plasmid vector contains the chicken beta-actin promoter, an IRES followed by the gene coding for GFP (Chen et al., 2004). Chicken fibroblasts (DF1) were transfected using FuGENE-6 reagent (Roche, Indianapolis, IN) according to the manufacturer's directions. Cells were cultured in Dulbecco's modified Eagle media, supplemented with 10% fetal bovine serum. There is no antibiotic resistance gene in pMES, therefore we enriched for cells expressing PANX3 through fluorescence-activated cell sorting (FACS) for 4 weeks. To assess the effectiveness of the shRNA constructs, shPANX3 virus was added to the culture media of the DF1 cell line and grown for 4 days. DF1 cell lysates were collected after 4 days and Western blotting was performed as described (Bond et al., 2011). The antibodies used were: Goat anti-Panx3 (N-20, Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-γ-tubulin (T6557, Sigma, St. Louis, MO), and mouse anti-viral GAG protein (Potts et al., 1987; 3C2, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA).
Separate cell cultures were prepared as above for RNA expression analysis. Total RNA was isolated using Trizol Reagent (Invitrogen) according to the manufacturer's directions. PANX3 expression was measured on a StepOnePlus qPCR machine (Life Technologies, Grand Island, NY). The final PCR mix included 10 ng of total RNA in 10 μl reactions, using the iScript One-Step RT-PCR kit with SYBR Green (Bio-Rad). Expression levels were standardized against eukaryotic 18S rRNA (Life Technologies, Carlsbad, CA) to generate Δct, and final relative expression (ΔΔct) was calculated as the ratio between the shPANX3 and shEGFP infected cultures.
Validation of gene knock-down and effects on target gene expression in vivo
Wholemount in situ hybridization was used to visualize expression of endogenous PANX3 following knockdown with the shPANX3 virus. Embryos were injected at stage 20 and then fixed 6 days later at stage 34. Embryos were hybridized to anti-sense PANX3 riboprobe as described (Song et al., 2004). A different set of embryos was collected for radioactive in in situ hybridization. Embryos were injected at stage 20 and grown to stage 28, 30 or 35. Following fixation, limbs were embedded in paraffin and sectioned. All probes were labeled with 35S-UTP and used at an activity level of 105 cpm/μl. Hybridizaton was carried out as described (Rowe et al., 1992). Sections were photographed under bright and dark field illumination and overlaid in Photoshop (Adobe) or presented without the overlay. Finally a third set of embryos was injected at stage 20. cell lysates collected at stage 34 and Western blots were carried out to assess level of PANX3 protein knockdown.
To quantify expression changes another set of animals was injected with shPANX3 or shEGFP and were terminated 8 days after viral injection (approximately stage 37). The forelimb long bones were manually cleaned of excess tissue and then RNA was extracted. The samples were pooled into four groups of four for each test condition, and these pools or biological replicates were used for subsequent qPCR measurement of PANX3, COLX, MMP13, ALPL, IHH, IBSP, PTHLH, PTH1R, and viral GAG. The primer sequences are given in Table S1.
Morphometric analysis of Skeletal Phenotype
Embryos injected at stage 20 were analyzed using 3D, Optical Projection Tomography (Sharpe et al., 2002). Forelimbs from stage 39 – 41 embryos were fixed directly in 100% ethanol for at least 48 hours, followed by Alizarin red staining of mineralized bone (Plant et al., 2000) however no cartilage staining was carried out. The specimens were cleared in 1% KOH for 2 – 4 days, and then any remaining soft tissue was manually dissected away. The humeri and combined radius/ulna were separated from one another for final analysis. The samples were dehydrated through graded methanol (to 100%) before final clearing in benzyl alcohol/benzyl benzoate (1:2). Each sample was directly pinned to a stub and scanned on the Texas Red channel. Rotational views were reconstructed into 3D models for volumetric measurement using NRecon and analyzed with CTan (SkyScan, Kontich, Belgium). Viral effects were determined by comparing the injected limb of each embryo against its uninjected contralateral limb. Direct comparison between the different viruses could not be carried out because the final developmental stage of the embryos at termination was not uniform (collections occurred 11-13 days post injection; N =9 shPANX3, N = 8 wtPANX3, N 9 = shEGFP).
Stage 15 injected embryos were collected 11 days post injection (stage 38) and cleared and stained using Alizarin red and Alcian blue (which stain bone and cartilage respectively; Plant et al., 2000). Subsequently, the limbs were photographed with a stereomicroscope. The 2D images were imported into ImageJ (National Institute of Mental Health, Bethesda, MD). The radius, ulna and humerus were measured for area, perimeter and Feret's diameter (the longest distance between any two points along a selection boundary; also known as the maximum caliper), comparing shRNA (n=11) to shEGFP injected right limbs (n=11).
Statistical analysis
All numerical results were trimmed of outlying data using the outlier labeling rule (Hoaglin and Iglewicz, 1987) and tested for a normal distribution (Shapiro-Wilk test, p-value ≥ 0.05) before graphing and applying further statistical tests. Log2 ratios of injected vs. contralateral uninjected limbs (I/C-scores; for OPT data and qPCR ΔΔCT values) were tested against a null mean of 0.0 using the 1-sample t-test. All confidence intervals (±) indicate the 95% confidence level. A 2-sample T-test was used to test for size differences in stage 15-injected limbs.
Supplementary Material
Key Findings.
PANX3 knockdown with an interference RNA retrovirus reduces size of endochondral bones in the chicken embryo
PANX3 overexpression is not sufficient to accelerate ossification
The late expression of the phenotype and extent of the volume decreases leave open the possibility PANX3 reduces a subset of osteoblasts
Acknowledgments
We would like to thank Dr. Sheri Holmen for kindly providing us with the pENTR3C-mir-30a plasmid. Preparation of this manuscript was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (SRB), NIH Ruth L. Kirschstein NRSA Postdoctoral Fellowship (JA), and the Canadian Institutes of Health Research (CCN and JMR). CCN holds a Canada Research Chair, and a Canadian Foundation for Innovation grant to the Faculty of Dentistry, UBC, funded the optical projection tomography scanner.
Footnotes
First two authors contributed equally to this work.
References
- Bargiotas P, Krenz A, Hormuzdi SG, Ridder DA, Herb A, Barakat W, Penuela S, von Engelhardt J, Monyer H, Schwaninger M. Pannexins in ischemia-induced neurodegeneration. Proc Natl Acad Sci U S A. 2011;108:20772–20777. doi: 10.1073/pnas.1018262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen AD, Olsen BR. Bone development. Bone. 2015;80:14–18. doi: 10.1016/j.bone.2015.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond SR, Lau A, Penuela S, Sampaio AV, Underhill TM, Laird DW, Naus CC. Pannexin 3 is a novel target for Runx2, expressed by osteoblasts and mature growth plate chondrocytes. J Bone Miner Res. 2011;26:2911–2922. doi: 10.1002/jbmr.509. [DOI] [PubMed] [Google Scholar]
- Bond SR, Naus CC. RF-Cloning.org: an online tool for the design of restriction-free cloning projects. Nucleic Acids Res. 2012;40:W209–213. doi: 10.1093/nar/gks396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond SR, Naus CC. The pannexins: past and present. Front Physiol. 2014;5:58. doi: 10.3389/fphys.2014.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryksin AV, Matsumura I. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques. 2010;48:463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celetti SJ, Cowan KN, Penuela S, Shao Q, Churko J, Laird DW. Implications of pannexin 1 and pannexin 3 for keratinocyte differentiation. J Cell Sci. 2010;123:1363–1372. doi: 10.1242/jcs.056093. [DOI] [PubMed] [Google Scholar]
- Chau M, Forcinito P, Andrade AC, Hegde A, Ahn S, Lui JC, Baron J, Nilsson O. Organization of the Indian hedgehog--parathyroid hormone-related protein system in the postnatal growth plate. J Mol Endocrinol. 2011;47:99–107. doi: 10.1530/JME-10-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Granger AJ, Vanbrocklin MW, Payne WS, Hunt H, Zhang H, Dodgson JB, Holmen SL. Inhibition of avian leukosis virus replication by vector-based RNA interference. Virology. 2007;365:464–472. doi: 10.1016/j.virol.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Chen M, Payne WS, Hunt H, Zhang H, Holmen SL, Dodgson JB. Inhibition of Marek's disease virus replication by retroviral vector-based RNA interference. Virology. 2008;377:265–272. doi: 10.1016/j.virol.2008.03.019. [DOI] [PubMed] [Google Scholar]
- Chen YX, Krull CE, Reneker LW. Targeted gene expression in the chicken eye by in ovo electroporation. Mol Vis. 2004;10:874–883. [PubMed] [Google Scholar]
- D'Angelo M, Yan Z, Nooreyazdan M, Pacifici M, Sarment DS, Billings PC, Leboy PS. MMP-13 is induced during chondrocyte hypertrophy. J Cell Biochem. 2000;77:678–693. [PubMed] [Google Scholar]
- Dirckx N, Van Hul M, Maes C. Osteoblast recruitment to sites of bone formation in skeletal development, homeostasis, and regeneration. Birth Defects Res C Embryo Today. 2013;99:170–191. doi: 10.1002/bdrc.21047. [DOI] [PubMed] [Google Scholar]
- Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CT, Rodda FA, Farlie PG. The RCAS retroviral expression system in the study of skeletal development. Dev Dyn. 2009;238:797–811. doi: 10.1002/dvdy.21907. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Harpavat S, Cepko CL. RCAS-RNAi: a loss-of-function method for the developing chick retina. BMC Dev Biol. 2006;6:2. doi: 10.1186/1471-213X-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoaglin DC, Iglewicz B. Fine-tuning some resistant rules for outlier labeling. J Am Stat Assoc. 1987;82:1147–1149. [Google Scholar]
- Hsu SH, Zhang X, Cheng S, Wunder JS, Hui CC, Alman BA. Suppressor of fused (Sufu) mediates the effect of parathyroid hormone-like hormone (Pthlh) on chondrocyte differentiation in the growth plate. J Biol Chem. 2012;287:36222–36228. doi: 10.1074/jbc.M112.382275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Iwamoto T, Fukumoto S, Yamada Y. Pannexin 3 inhibits proliferation of osteoprogenitor cells by regulating Wnt and p21 signaling. J Biol Chem. 2014;289:2839–2851. doi: 10.1074/jbc.M113.523241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Iwamoto T, Nakamura T, Doyle A, Fukumoto S, Yamada Y. Pannexin 3 functions as an ER Ca(2+) channel, hemichannel, and gap junction to promote osteoblast differentiation. J Cell Biol. 2011;193:1257–1274. doi: 10.1083/jcb.201101050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto T, Nakamura T, Doyle A, Ishikawa M, de Vega S, Fukumoto S, Yamada Y. Pannexin 3 regulates intracellular ATP/cAMP levels and promotes chondrocyte differentiation. J Biol Chem. 2010;285:18948–18958. doi: 10.1074/jbc.M110.127027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- Kwon TJ, Kim DB, Bae JW, Sagong B, Choi SY, Cho HJ, Kim UK, Lee KY. Molecular cloning, characterization, and expression of pannexin genes in chicken. Poult Sci. 2014;93:2253–2261. doi: 10.3382/ps.2013-03867. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Vasseur M, Lelowski J, Bechberger JF, Sin WC, Naus CC. Pannexin 2 protein expression is not restricted to the CNS. Front Cell Neurosci. 2014;8:392. doi: 10.3389/fncel.2014.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TF, Dong Y, Ionescu AM, Rosier RN, Zuscik MJ, Schwarz EM, O'Keefe RJ, Drissi H. Parathyroid hormone-related peptide (PTHrP) inhibits Runx2 expression through the PKA signaling pathway. Exp Cell Res. 2004;299:128–136. doi: 10.1016/j.yexcr.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Loftus SK, Larson DM, Watkins-Chow D, Church DM, Pavan WJ. Generation of RCAS vectors useful for functional genomic analyses. DNA Res. 2001;8:221–226. doi: 10.1093/dnares/8.5.221. [DOI] [PubMed] [Google Scholar]
- Logan M, Tabin C. Targeted gene misexpression in chick limb buds using avian replication-competent retroviruses. Methods. 1998;14:407–420. doi: 10.1006/meth.1998.0595. [DOI] [PubMed] [Google Scholar]
- MacLean HE, Kronenberg HM. Localization of Indian hedgehog and PTH/PTHrP receptor expression in relation to chondrocyte proliferation during mouse bone development. Dev Growth Differ. 2005;47:59–63. doi: 10.1111/j.1440-169x.2004.00781.x. [DOI] [PubMed] [Google Scholar]
- Miyagishi M, Sumimoto H, Miyoshi H, Kawakami Y, Taira K. Optimization of an siRNA-expression system with an improved hairpin and its significant suppressive effects in mammalian cells. J Gene Med. 2004;6:715–723. doi: 10.1002/jgm.556. [DOI] [PubMed] [Google Scholar]
- Moog F. Localizations of Alkaline and Acid Phosphatases in the Early Embryogenesis of the Chick. Biol Bull. 1944;86:51–80. [Google Scholar]
- Moon PM, Penuela S, Barr K, Khan S, Pin CL, Welch I, Attur M, Abramson SB, Laird DW, Beier F. Deletion of Panx3 prevents the development of surgically induced osteoarthritis. Journal of Molecular Medicine. 2015;93:845–856. doi: 10.1007/s00109-015-1311-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SK, Shin JO, Baek JI, Lee J, Bae JW, Ankamerddy H, Kim MJ, Huh TL, Ryoo ZY, Kim UK, Bok J, Lee KY. Pannexin 3 is required for normal progression of skeletal development in vertebrates. FASEB J. 2015;29:4473–4484. doi: 10.1096/fj.15-273722. [DOI] [PubMed] [Google Scholar]
- Panchin Y, Kelmanson I, Matz M, Lukyanov K, Usman N, Lukyanov S. A ubiquitous family of putative gap junction molecules. Curr Biol. 2000;10:R473–474. doi: 10.1016/s0960-9822(00)00576-5. [DOI] [PubMed] [Google Scholar]
- Park J, Gebhardt M, Golovchenko S, Perez-Branguli F, Hattori T, Hartmann C, Zhou X, deCrombrugghe B, Stock M, Schneider H, von der Mark K. Dual pathways to endochondral osteoblasts: a novel chondrocyte-derived osteoprogenitor cell identified in hypertrophic cartilage. Biol Open. 2015;4:608–621. doi: 10.1242/bio.201411031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penuela S, Bhalla R, Gong X-Q, Cowan KN, Celetti SJ, Cowan BJ, Bai D, Shao Q, Laird DW. Pannexin 1 and pannexin 3 are glycoproteins that exhibit many distinct characteristics from the connexin family of gap junction proteins. Journal of cell science. 2007;120:3772–3783. doi: 10.1242/jcs.009514. [DOI] [PubMed] [Google Scholar]
- Penuela S, Harland L, Simek J, Laird DW. Pannexin channels and their links to human disease. Biochem J. 2014;461:371–381. doi: 10.1042/BJ20140447. [DOI] [PubMed] [Google Scholar]
- Plant MR, MacDonald ME, Grad LI, Ritchie SJ, Richman JM. Locally released retinoic acid repatterns the first branchial arch cartilages in vivo. Developmental Biology. 2000;222:12–26. doi: 10.1006/dbio.2000.9706. [DOI] [PubMed] [Google Scholar]
- Plotkin LI. Connexin 43 hemichannels and intracellular signaling in bone cells. Front Physiol. 2014;5:131. doi: 10.3389/fphys.2014.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts WM, Olsen M, Boettiger D, Vogt VM. Epitope mapping of monoclonal antibodies to gag protein p19 of avian sarcoma and leukaemia viruses. J Gen Virol. 1987;68(Pt 12):3177–3182. doi: 10.1099/0022-1317-68-12-3177. [DOI] [PubMed] [Google Scholar]
- Retamal MA. Connexin and Pannexin hemichannels are regulated by redox potential. Front Physiol. 2014;5:80. doi: 10.3389/fphys.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Rowe A, Richman JM, Brickell PM. Development of the spatial pattern of retinoic acid receptor-beta transcripts in embryonic chick facial primordia. Development. 1992;114:805–813. doi: 10.1242/dev.114.3.805. [DOI] [PubMed] [Google Scholar]
- Schipani E, Lanske B, Hunzelman J, Luz A, Kovacs CS, Lee K, Pirro A, Kronenberg HM, Juppner H. Targeted expression of constitutively active receptors for parathyroid hormone and parathyroid hormone-related peptide delays endochondral bone formation and rescues mice that lack parathyroid hormone-related peptide. Proc Natl Acad Sci U S A. 1997;94:13689–13694. doi: 10.1073/pnas.94.25.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid TM, Conrad HE. A unique low molecular weight collagen secreted by cultured chick embryo chondrocytes. J Biol Chem. 1982;257:12444–12450. [PubMed] [Google Scholar]
- Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sorensen J, Baldock R, Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- Song Y, Hui JN, Fu KK, Richman JM. Control of retinoic acid synthesis and FGF expression in the nasal pit is required to pattern the craniofacial skeleton. Dev Biol. 2004;276:313–329. doi: 10.1016/j.ydbio.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Sosinsky GE, Boassa D, Dermietzel R, Duffy HS, Laird DW, MacVicar B, Naus CC, Penuela S, Scemes E, Spray DC, Thompson RJ, Zhao HB, Dahl G. Pannexin channels are not gap junction hemichannels. Channels (Austin) 2011;5:193–197. doi: 10.4161/chan.5.3.15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz ME, Eberhart J, Pasquale EB, Krull CE. EphA4/ephrin-A5 interactions in muscle precursor cell migration in the avian forelimb. Development. 2001;128:4669–4680. doi: 10.1242/dev.128.23.4669. [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- Weir EC, Philbrick WM, Amling M, Neff LA, Baron R, Broadus AE. Targeted overexpression of parathyroid hormone-related peptide in chondrocytes causes chondrodysplasia and delayed endochondral bone formation. Proc Natl Acad Sci U S A. 1996;93:10240–10245. doi: 10.1073/pnas.93.19.10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Zhu L, Hou N, Lan Y, Wu XM, Zhou B, Teng Y, Yang X. Osteogenic fate of hypertrophic chondrocytes. Cell Res. 2014a;24:1266–1269. doi: 10.1038/cr.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Tsang KY, Tang HC, Chan D, Cheah KS. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc Natl Acad Sci U S A. 2014b;111:12097–12102. doi: 10.1073/pnas.1302703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappitelli T, Aubin JE. The “connexin” between bone cells and skeletal functions. J Cell Biochem. 2014;115:1646–1658. doi: 10.1002/jcb.24836. [DOI] [PubMed] [Google Scholar]
- Zhou X, von der Mark K, Henry S, Norton W, Adams H, de Crombrugghe B. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 2014;10:e1004820. doi: 10.1371/journal.pgen.1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.