Abstract
The vast majority of chronic and recurrent bacterial diseases are attributed to the presence of a recalcitrant biofilm that contributes significantly to pathogenesis. As such, these diseases will require an innovative therapeutic approach. We targeted DNABII proteins, an integral component of extracellular DNA (eDNA) which is universally found as part of the pathogenic biofilm matrix to develop a biofilm disrupting therapeutic. We show that a cocktail of monoclonal antibodies directed against specific epitopes of a DNABII protein is highly effective to disrupt diverse biofilms in vitro as well as resolve experimental infection in vivo, in both a chinchilla and murine model. Combining this monoclonal antibody cocktail with a traditional antibiotic to kill bacteria newly released from the biofilm due to the action of the antibody cocktail was highly effective. Our results strongly support these monoclonal antibodies as attractive candidates for lead optimization as a therapeutic for resolution of bacterial biofilm diseases.
Keywords: Integration host factor (IHF), Histone like protein (HU), Nontypeable Haemophilus influenzae, Pseudomonas aeruginosa, Otitis media, Respiratory tract infection
Highlights
-
•
Monoclonal antibodies (MAbs) against protective epitopes of a DNABII protein disrupted diverse bacterial biofilms in vitro.
-
•
Delivery of these MAbs also provided therapeutic efficacy in two animal models of biofilm infection.
-
•
Bacteria newly released from the biofilm by the action of the MAbs were susceptible to host-mediated clearance and tobramycin.
Research In Context
The bacteria which cause the vast majority of chronic and recurrent diseases characteristically form ‘biofilms’. Biofilms are communities of bacteria with many unique properties, including being highly resistant to antibiotics and the body's immune response. To develop an effective therapeutic for biofilm infections, we targeted the DNABII proteins, a universal biofilm component that provides structural integrity. Monoclonal antibodies against domains of the DNABII proteins induced complete collapse of diverse biofilms with release of resident bacteria that were highly susceptible to killing by host immune effectors and traditional antibiotics. This therapeutic also mediated resolution of infection in two experimental animal models.
1. Introduction
The presence of a bacterial biofilm abrogates the ability of antibodies and antimicrobials to effectively eradicate the causative agent of many chronic and recurrent diseases. Given that biofilms are integral to the vast majority of these diseases (BROOK, 2016, DLUGASZEWSKA et al., 2015, SUBASHCHANDRABOSE and MOBLEY, 2015, DOU et al., 2016, SCALISE et al., 2015, HASSETT et al., 2014, SWORDS, 2012) knowledge of the key role of biofilms in pathobiology mandates the development of approaches for therapeutic disease resolution. The list of diseases wherein the ability to effectively disrupt bacterial biofilms would be beneficial is lengthy, and ideally this would involve eradication of both the microbes that induce its formation as well as the recalcitrant biofilm matrix. Towards our goal to develop a broad-based therapeutic approach for biofilm resolution, we focused on two common constituents of biofilms produced by multiple human pathogens, extracellular DNA (eDNA) and a family of bacterial DNA-binding proteins called the DNABII family.
Biofilms are communities of bacteria adhered to a surface with division of labor (e.g. altered gene expression), intercellular communication (e.g. quorum sensing), and the creation of a self-made extracellular polymeric substance (EPS) that enshrouds and protects resident bacteria (Flemming and Wingender, 2010). Often biofilms arise to weather stressful conditions, e.g. resistance to immune clearance and antibiotics (in excess of 1000-fold greater than needed to eliminate free-living bacteria). Key to the biofilm's protection is the extrapolymeric substance or EPS that constitutes the biofilm matrix. While the molecular makeup of the EPS varies among bacterial species, extracellular DNA (eDNA) is a common component (FLEMMING and WINGENDER, 2010, FONG and YILDIZ, 2015). Indeed, DNase can prevent biofilm formation by multiple pathogenic species, but does not effectively treat pre-formed biofilms despite the abundance of eDNA in mature biofilms (Flemming and Wingender, 2010). In association with eDNA is the DNABII family of proteins which serve as lynchpin proteins, positioned at the vertices of crossed strands of eDNA within the biofilm matrix, thus contributing to the structural stability of the biofilm matrix (GOODMAN et al., 2011, IDICULA et al., 2016, DEVARAJ et al., 2015, GUSTAVE et al., 2013).
The DNABII family is ubiquitous among eubacteria and has been studied for almost 40 years as an intracellular architectural element. This family is one of multiple nucleoid-associated proteins (NAPs) that maintain the structure and function of bacterial chromatin (Swinger and Rice, 2004). Recently, multiple labs showed that these proteins are also abundant extracellularly (GOODMAN et al., 2011, STINSON et al., 1998, LUNSFORD et al., 1996, GAO, 2000, BOLEIJ et al., 2009). The DNABII family members include integration host factor (IHF) which is a heterodimer of IHFA and IHFB and histone-like protein (HU), which is a hetero- or homodimer of each subunit. IHF and HU have a conserved sequence homology and as a result, a conserved architecture. This conserved architecture enables them to not only bind to and bend DNA (achieved by the insertion of two antiparallel β-ribbons into the DNA minor groove that cause the DNA to bend), but also show enhanced affinity to pre-bent DNA structures such as cruciforms or Holliday junctions (Swinger and Rice, 2004). These lynchpin proteins are present in the biofilms produced by multiple human pathogens (Goodman et al., 2011). Further, when biofilms are exposed to polyclonal rabbit antiserum directed against IHF isolated from Escherichia coli (anti-IHFE.coli), complete collapse of the biofilm occurs with release of resident bacteria (Brockson et al., 2014). These observations suggested that the DNABII proteins and eDNA might serve as universal biofilm constituents that not only contribute to structural integrity, but could also provide a mechanism for multispecies interaction and facilitate the development of mixed microbial biofilm consortia as typically exist in nature (Goodman et al., 2011).
Using nontypeable Haemophilus influenzae (NTHI) as a model organism to dissect the mechanism(s) responsible for the observed complete biofilm collapse, we have shown in vitro that anti-IHFE.coli captures DNABII proteins when they are in an ‘off’ state within the culture medium (e.g. when they are not in association with eDNA of the biofilm EPS) (Brockson et al., 2014). This action induces an equilibrium shift that results in removal of additional DNABII proteins from the biofilm matrix (e.g. those that are in an ‘on’ state or associated with eDNA of the biofilm EPS), resulting in structural collapse of the biofilm matrix with release of the resident bacteria. These newly released bacteria were not killed by the action of anti-IHFE.coli, however they were 4–8 fold more sensitive to the killing action of multiple antibiotics (Brockson et al., 2014). We have also already shown that this outcome is rapid, specific and dose-dependent but does not require direct contact between anti-IHFE.coli antibodies and the NTHI biofilm (Brockson et al., 2014). Moreover, this mechanism (which is characterized as ‘disruption’) was distinct from the ‘dispersal’ of an NTHI biofilm induced by exposure to antibodies directed against the Type IV twitching pilus which mediates a distinct ‘top-down’ dispersal of the biofilm that is linked to expression of the quorum signaling molecule AI-2 (Novotny et al., 2015b).
To then determine if antibodies with similar biofilm disruption functionality could be induced in vivo, we conducted a study which showed that active immunization with IHF [now isolated specifically from NTHI (IHFNTHI)], induced the formation of antibodies that disrupted biofilms formed by NTHI in the middle ears of chinchillas. This disruption leads to significantly more rapid resolution of experimental disease with eradication of mucosal biofilms (GOODMAN et al., 2011, BROCKSON et al., 2014). Via extensive epitope mapping efforts, combined with additional pre-clinical evaluation in the same chinchilla model of experimental otitis media (GOODMAN et al., 2011, BROCKSON et al., 2014), we found that DNABII proteins that are naturally associated with eDNA within the bacterial biofilm (as they are found in the disease state), do not induce a protective immune response, as binding to DNA obscures the protective epitopes within the DNABII protein. Pre-clinical studies using native protein (with no bound eDNA) versus that which was pre-complexed to DNA as comparative immunogens revealed that the typically obscured DNA-binding tip regions of the DNABII proteins served as the protective epitopes. We then showed that polyclonal rabbit antibodies directed against focused 20-residue peptides which mimicked these specific predicted protective domains within the DNA-binding tips of IHFNTHI, were equally effective as polyclonal antisera directed against the whole native IHFE.coli protein in terms of their ability to disrupt biofilms in vitro (GOODMAN et al., 2011, BROCKSON et al., 2014).
Having determined the mechanism of action, and shown the ability to utilize polyclonal antibodies to disrupt biofilms in vitro and also induce their formation via active immunization in vivo, it was important to ascertain the therapeutic potential of monoclonal antibodies in order to move closer to the ability to conduct human trials. Thereby, monoclonal antibodies directed against specific protective domains of the DNABII protein IHF were generated in an effort to develop a lead compound with a very specific epitope as a target. Ideally, this highly focused therapeutic would nonetheless provide broad protection against diverse bacterial pathogens which shared the same or similar targeted epitope. Thereby, in the present study, we extended our observations to determine if highly specific monoclonal antibodies (MAbs) directed against the DNA-binding tip regions of the alpha- and beta-subunits of IHFNTHI could individually disrupt biofilms formed in vitro by diverse strains of NTHI. In addition, we tested these MAbs against biofilms formed by four additional human pathogens: Staphylococcus aureus, Pseudomonas aeruginosa, Burkholderia cenocepacia and Moraxella catarrhalis. We also tested cocktails of these two MAbs both in vitro, to determine their ability to act synergistically, and in vivo using two distinct animal models of experimental upper and lower respiratory tract diseases to determine their ability to also resolve experimental infection. This outcome was achieved as evidenced by eradication of either mucosal biofilms formed by NTHI within the middle ears of chinchillas or classic multicellular aggregates of P. aeruginosa present within the lungs of mice.
2. Materials and Methods
2.1. Peptide Design and Synthesis
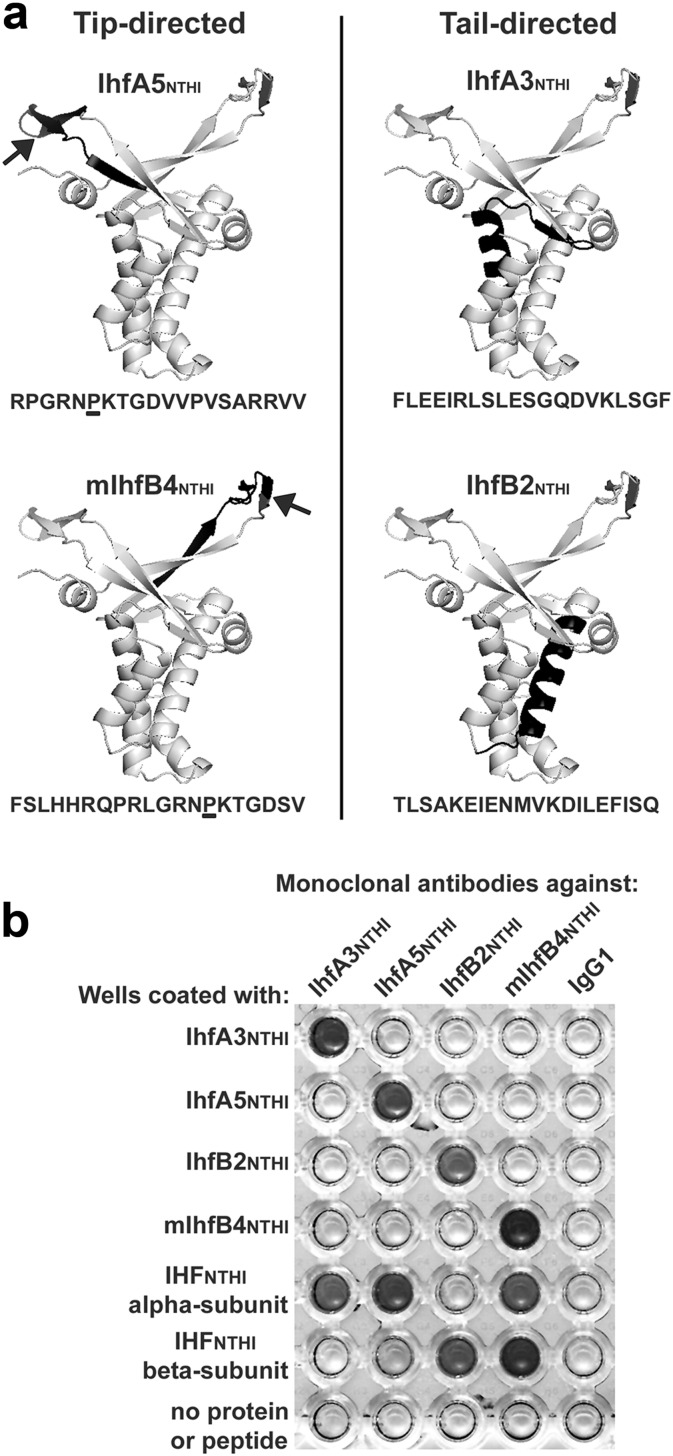
To identify regions of interest within the alpha and beta subunits of IHFNTHI, we used the known crystal structure of IHF from E. coli (Rice et al., 1996) along with the deduced amino acid sequence of IHFNTHI to approximate the location of the 20-mer synthetic peptides used for the generation of MAbs (Fig. 1A). Synthesis, purification and sequence confirmation of all synthetic peptides was performed by Ohio Peptide, LLC.
Fig. 1.
Design and validation of IHFNTHI tip- and tail-directed MAbs. (A) Structural models of the IHFNTHI dimer with the predicted location of each tip- and tail-directed synthetic peptide highlighted (black shading) as generated with PyMol software. The location of an essential proline residue within peptides IhfA5NTHI and mIhfB4NTHI are indicated by arrows in each model and are underlined within the respective amino acid sequences. (B) Specificity of IHFNTHI MAbs to the synthetic peptide and IHFNTHI protein subunit from which they were derived. Dark-colored wells within the scanned image of a representative ELISA plate displayed reactivity of MAbs to their respective peptide and protein subunit.
2.2. Generation of Hybridomas, Isolation, Purification and Validation of MAbs
Hybridoma cell lines were generated by Rockland Immunochemical, Inc. Briefly, spleen cells from mice immunized with synthetic peptides IhfA3NTHI, IhfA5NTHI, IhfB2NTHI or mIhfB4NTHI were fused with Sp2/0-Ag14 myeloma cells. To reduce the risk of overgrowth by non-producer cells and ensure antibody productivity, cell lines were subcloned one or more generations by limiting dilution. All MAbs were of the IgG1 kappa isotype except that to peptide IhfA5 which was IgG2a/IgM kappa isotype.
2.2.1. Culture of Hybridomas
Hybridomas were cultured in medium comprised of DMEM plus 4.5 g glucose/ml (Corning) supplemented with 2 mM l-glutamine (Corning), 100 U penicillin-streptomycin/ml (Corning) and 2 mM sodium pyruvate (Corning). For long-term culture, hybridomas were maintained at a density of 1 × 105 cells/ml in medium supplemented with 10% Superlow IgG fetal bovine serum (FBS; HyClone) and 1 × 106 cells/ml were transitioned to medium with 2.5% FBS for monoclonal antibody production. After 7–10 days in FBS-limited medium, hybridomas were removed from the cell culture supernatant by centrifugation and filtration. MAbs used herein were: IhfA3NTHI (clone 9B10·F2.H3); IhfA5NTHI (clone 14G8.F5.G6); IhfB2NTHI (clone 7 A4.E4.G11) and mIhfB4NTHI (clone 12E6.F8.D12.D5).
2.2.2. Purification of MAbs
Clarified cell culture supernatants were concentrated by centrifugation in Corning Spin-X UF columns (MWCO 30,000) with a final buffer exchange in 20 mM sodium phosphate, pH 7.0. The retentate was then applied to a Hi-Trap Protein G column (GE Healthcare), washed with 20 mM sodium phosphate, pH 7.0 and IgG eluted with 0.1 M glycine-HCl, pH 2.7 subsequently neutralized with 1 M Tris-HCl, pH 9.0. Antibody was dialyzed against Dulbecco's phosphate buffered saline, pH 7.0 for 24 h at 4 °C with d-tube dialyzer MAXI (MWCO 12–14 kDa; Novagen). The concentration of each monoclonal antibody was determined by Coomassie Plus Protein Assay (Thermo Scientific) versus a standard curve generated with BSA standards (Thermo Scientific). The purity of each preparation was confirmed by SDS-PAGE and gels stained with BioSafe Coomassie (BioRad) prior to imaging with Protein Simple FluoroChem M.
2.3. Cloning and Isolation of NTHI DNABII Proteins and Generation of Polyclonal Antisera Directed Against Each
Detailed protocol for cloning and isolation of NTHI DNABII proteins are described in Supplemental Experimental Procedures. Polyclonal antisera against purified DNABII proteins were generated at Spring Valley Laboratories (Sykesville, MD).
2.4. ELISA
To confirm specificity of each monoclonal antibody, an ELISA was performed. Synthetic peptides IhfA3, IhfA5, IhfB2 and mIhfB4 in addition to purified IHFNTHI α- and β-subunit proteins were suspended in carbonate-bicarbonate buffer (pH 9.6), 1.0 μg added to each well of MaxiSorp C-bottom microtiter plates (Nunc) and adsorbed for 18 h at 4 °C. Wells were then blocked with 2% bovine serum albumin (Sigma) in 10 mM phosphate buffered saline (pH 7.4; PBS), washed with 10 mM PBS and followed by addition of 0.1 μg monoclonal antibody per well. Wells were washed as before and goat anti-mouse IgG conjugated to HRP (Invitrogen; RRID AB_11180340) next added. Colour was developed with One-Step Ultra TMB (Pierce) and the reaction stopped with 2 M H2SO4. Assays were repeated a minimum of three times, on separate days. A representative assay is shown in Fig. 1B.
2.5. Biofilm Disruption Assay
Biofilms formed by NTHI strains 86-028NP, 1714, 1885 and 214; P. aeruginosa strain 27853; S. aureus strain 29213; Burkholderia cenocepcia strain K56; and M. catarrhalis strain 7169 were first established in 8-well chambered coverglasses (Lab-Tek) for 24 h prior to treatment (JURCISEK et al., 2011, BROCKSON et al., 2014) with 5 μg individual MAbs or a cocktail containing 2.5- or 5.0-μg each MAb (tail-directed cocktail IhfA3NTHI + IhfB2NTHI; tip-directed cocktail IhfA5NTHI + mIhfB4NTHI) for an additional 16 h. Non-specific murine IgG1 served as a negative control monoclonal antibody (eBiosciences; RRID AB_470112). Biofilms were then washed and stained with FM1-43FX bacterial cell membrane stain (Invitrogen) to characterize the contribution of bacteria themselves to the biofilm architecture (as opposed to other biofilm matrix materials, including eDNA) and fixed overnight at 4 °C in 16% paraformaldehyde, 2.5% glutaraldehyde, 4.0% acetic acid in 0.1 M phosphate buffer (pH 7.4) to prevent movement of biofilms during imaging. Biofilms were viewed on a Zeiss 510 Meta-laser scanning confocal microscope, images compiled with Zeiss Zen software and biofilm biomass calculated with COMSTAT2 software (HEYDORN et al., 2000, VORREGAARD, 2008). All assays were repeated a minimum of three times on separate days.
2.6. Chinchilla Study
Thirty-six adult chinchillas (Chinchilla lanigera; 659 ± 110 g) without evidence of middle ear disease or serum antibody to outer membrane proteins of NTHI 86-028NP were procured from Rauscher's Chinchilla Ranch (LaRue, Ohio) and were randomized to cohort by weight. All animals were challenged transbullarly with 1000 CFU NTHI strain #86-028NP per bulla delivered on a 0.3 ml volume of pyrogen-free sterile saline to induce experimental otitis media. Four days later, when it is known that biofilms typically fill ≥ 50% of the middle ear space of the vast majority of animals (in this model > 83% of ears will develop a biomass; > 67% of which will yield a score ≥ 2 on a 0 to 4 + scale) (NOVOTNY et al., 2011, NOVOTNY et al., 2013b) chinchillas were randomly grouped into cohorts of 3 animals each (6 total ears) and treatments (MAbs or IgG-enriched polyclonal rabbit sera) assigned as detailed in Table 1 (dosage selection process described in legend). A volume of 100 μl of each respective treatment diluted in sterile pyrogen-free sterile saline was delivered transbullarly and a total of three doses were administered at daily intervals. One day after receipt of the final dose, animals were sacrificed, middle ear fluids and mucosal biofilms collected, serially diluted and plated on to chocolate agar to quantitate the relative bacterial load within the planktonic and adherent biofilm populations, respectively. Mucosal biofilms were imaged with a Zeiss SV6 dissecting microscope, images compiled with Zeiss Zen software and then randomized. The relative amount of mucosal biofilm that remained within the middle ear was assessed by 5 blinded reviewers using an established mucosal biofilm scale (Novotny et al., 2011), whereby 0 = no mucosal biofilm visible, 1 ≤ 25% of middle ear space occluded by mucosal biofilm, 2 = ≥ 25–50% occluded, 3 = ≥ 50–75% occluded, 4 = ≥ 75–100% occluded. All chinchilla work was performed in accordance with state and institutional guidelines and under a protocol approved by Nationwide Children's Hospital Institutional Animal Care and Use Committee.
Table 1.
Cohort descriptions for three studies to assess efficacy of tip and tail derived polyclonal and monoclonal antibodies.
| Study | Host (n) | Treatment | Dose |
|---|---|---|---|
| Resolution of experimental NTHI-induced OM via treatment with polyclonal or monoclonal antibodies | Chinchilla (6) | Naive rabbit serum | 5.0 μg |
| Chinchilla (6) | Rabbit anti-IHFE. coli | 5.0 μg | |
| Chinchilla (6) | Rabbit anti-IHFNTHI | 5.0 μg | |
| Chinchilla (6) | Rabbit anti-HUNTHI | 5.0 μg | |
| Chinchilla (6) | MAb IhfA3NTHI | 5.0 μg | |
| Chinchilla (6) | MAb IhfA5NTHI | 5.0 μg | |
| Chinchilla (6) | MAb IhfB2NTHI | 5.0 μg | |
| Chinchilla (6) | MAb mIhfB4NTHI | 5.0 μg | |
| Chinchilla (6) | MAb IhfA3NTHI + IhfB2NTHI | 5.0 μg each | |
| Chinchilla (6) | MAbs IhfA3NTHI + IhfB2NTHI | 2.5 μg each | |
| Chinchilla (6) | MAbs IhfA5NTHI + mIhfB4NTHI | 5.0 μg each | |
| Chinchilla (6) | MAbs IhfA5NTHI + mIhfB4NTHI | 2.5 μg each | |
| Clearance of P. aeruginosa from the murine lung via delivery of MAb cocktails | Mouse (5) | None | None |
| Mouse (10) | Murine IgG1 | 10.0 μg antibody | |
| Mouse (10) | MAbs IhfA3NTHI + IhfB2NTHI | 5.0 μg each antibody | |
| Mouse (10) | MAbs IhfA3NTHI + IhfB2NTHI | 5.0 μg each antibody | |
| Clearance of P. aeruginosa from the murine lung via delivery of MAb cocktails ± tobramycin | Mouse (5) | None (sacrificed prior to treatment) | None |
| Mouse (3) | None | None | |
| Mouse (10) | Murine IgG1 + IgG2a | 5.0 μg each antibody | |
| Mouse (13) | Murine IgG1 + IgG2a + tobramycin | 5.0 μg each antibody + 60 mg tobramycin/kg | |
| Mouse (10) | MAbs IhfA3NTHI + IhfB2NTHI | 5.0 μg each antibody | |
| Mouse (13) | MAbs IhfA3NTHI + IhfB2NTHI + tobramycin | 5.0 μg each antibody + 60 mg tobramycin/kg | |
| Mouse (10) | MAbs IhfA5NTHI + mIhfB4NTHI | 5.0 μg each antibody | |
| Mouse (13) | MAbs IhfA5NTHI + mIhfB4NTHI + tobramycin | 5.0 μg each antibody + 60 mg tobramycin/kg | |
| Mouse (10) | Tobramycin | 60 mg tobramycin/kg |
In vitro, 5 μg MAb is typically added to each well of a chamberside to disrupt a 24 h biofilm which contains approx. 8 × 107 CFU NTHI per well of a chamberslide (includes both planktonic and biofilm-resident bacteria) (Brockson et al., 2014). In vivo, 4 days after TB challenge of a naive chinchilla there are approx. 108 CFU NTHI/ml MEF and ~ 106 CFU NTHI/mg mucosal biofilms (total of ~ 108 CFU/ml in the middle ear space) (Novotny et al., 2015a). Similarly, mice were challenged with 107 CFU of P. aeruginosa the day before administration of MAbs. Thereby, as a first approximation, we used what was known to be an effective dose in vitro to disrupt a biofilm that contained the approximate number of bacteria we expected to be within either the middle ear of chinchillas or lungs of mice and used this dose in our animal model systems.
2.7. Mouse Studies
2.7.1. Delivery of MAbs Only
Thirty-five male C57BL/6 mice (approximately 8 weeks of age) were obtained from Charles River Laboratories International, Inc. and were randomized to cohort by weight. Mice acclimated to their environment for 5 days prior to the start of study. All mice were inoculated with 1 × 107 CFU P. aeruginosa strain 27853 (ATCC) per 0.03 ml sterile pyrogen-free saline by intratracheal (IT) instillation using BioLite Intubation System (Braintree Scientific, Inc.). Twenty-four hours after IT challenge, 5 mice were sacrificed to obtain the baseline concentration of P. aeruginosa per lung prior to administration of treatment (SADIKOT et al., 2007, BJARNSHOLT et al., 2013, SINGH et al., 2000, BJARNSHOLT et al., 2009, BUCIOR et al., 2013, ALEMAYEHU et al., 2012, FOTHERGILL et al., 2014, MACHADO et al., 2011, FU et al., 2013). Lungs were aseptically collected and homogenized in 1.0 ml sterile saline using GentleMACs C-tubes (Miltenyi Biotec). Lung homogenates were then serially diluted in sterile saline and plated on to Trypic Soy agar (BBL). Due to the presence of lung tissue within homogenate aliquots, agar plates were incubated for 48 h to allow sufficient bacterial growth prior to enumerating the colony forming units per lung. The remaining 30 mice were randomly divided into 3 cohorts of 10 animals each and administered monoclonal antibody cocktails as described in Table 1 delivered IT in a total volume of 0.03 ml pyrogen-free sterile saline. To assess the ability of treatment with the IHFNTHI-targeted MAbs to reduce the bacterial load within the lung, five mice from each cohort were sacrificed 1 and 6 days after treatment (2 and 7 days after P. aeruginosa challenge, respectively). Lungs were collected, processed and homogenates plated as described. All mouse work was performed in accordance with state and institutional guidelines and under a protocol approved by Nationwide Children's Hospital Institutional Animal Care and Use Committee.
2.7.2. Delivery of MAbs in Combination With Tobramycin
Eighty-seven male C57BL/6 mice (approximately 8 weeks of age) were procured from Charles River Laboratories were randomized to cohort by weight and rested as before prior to IT challenge with 1 × 107 CFU P. aeruginosa strain 27853 in 0.03 ml pyrogen-free sterile saline. One day after IT challenge and prior to receipt of any treatment, 5 mice were sacrificed to obtain the pre-treatment bacterial concentration within the lungs as described. An additional 3 mice were sacrificed for histologic analysis as described below. The remaining 75 mice were randomly divided in 7 cohorts of either 10 mice for monoclonal antibody plus tobramycin treatment (60 mg tobramycin/kg) (Sabet et al., 2009) or 13 mice for monoclonal antibody treatment alone as described in Table 1. Murine IgG1 (eBioscience; RRID AB_470112) and IgG2a (eBioscience; RRID AB_470115) served as negative control MAbs. One day after receipt of treatment (two days after bacterial challenge), 5 mice from each cohort were sacrificed, lungs collected, homogenized and plated as described. Also at this time point, 3 additional mice from cohorts administered only MAb cocktails (no tobramycin) were sacrificed for histologic analysis as described below. The remaining 5 mice in each cohort were sacrificed six days after treatment (seven days after bacterial challenge), lungs collected and processed as described. Mice within each cohort were randomly selected for sacrifice. All mouse work was performed in accordance with state and institutional guidelines and under a protocol approved by Nationwide Children's Hospital Institutional Animal Care and Use Committee.
2.8. Histologic Analysis of Murine Lungs
At the time points described above, mice were sacrificed, lungs excised and inflated with 10% neutral buffered formalin, then embedded in paraffin and sectioned at 10 μm thickness. P. aeruginosa was detected using rabbit anti-whole Pseudomonas antibody (LifeSpan Biosciences; RRID AB_10570660) and revealed with goat anti-rabbit IgG conjugated to AlexaFluor 488 (Invitrogen). Lung sections were counter-stained with DAPI (Invitrogen) and visualized using a 10 × objective on a Zeiss Axiovert 200 M inverted microscope. Images were captured with Zeiss Axiovision software.
Immunolabled sections of lung were analyzed for the presence of bacterial aggregates. To do so, a minimum of three 10 × fields of view (FOV) from each lung were used to quantify the number of positively labeled P. aeruginosa aggregates per FOV. Evaluations of bacterial aggregates that remained in the lungs of mice following treatment with MAbs directed against DNABII proteins was conducted blindly.
To demonstrate the presence of bacterial eDNA, DNABII proteins and murine neutrophils within the lungs of mice challenged IT with P. aeruginosa, ten micron sections of paraffin embedded mouse lung were deparaffinized, hydrated to water and incubated with primary antibody cocktail followed by detection with fluorescently labeled secondary antibodies. Bacterial eDNA and DNABII proteins were detected using mouse anti-dsDNA (abcam; RRID AB_470907) and rabbit anti-IHFEcoli. Labeling by these primary antibodies was revealed with goat anti-mouse AlexaFluor 594 (Invitrogen) and goat anti-rabbit AlexaFluor 488 (Invitrogen) respectively. Images were captured using a 63 × objective on a Zeiss Axiovert 200 M inverted microscope and using Zeiss Axiovision software.
2.9. TEM of Murine Lungs
Ten µm serial sections from the paraffin-embedded lungs of each mouse were collected onto 22 mm coverglass, de-paraffinized, and infiltrated with LR White embedding medium (Electron Microcopy Sciences, Hatfield, Pa). Flat embedding in LR White was performed by placing a gelatin capsule (with the end removed) over the tissue section, filling with catalyzed LR White and allowed to polymerize. Thirty-five nanometer sections were cut using a Leica EM UC7 ultra microtome and collected on to a 100 mesh Formvar-coated Nickel grid (Electron Microscopy Sciences). Contrast staining with uranyl acetate and lead citrate was used to visualize tissue which was imaged using an Hitachi S-4800 TSEM.
2.10. Statistical Analysis
Statistically significant differences were calculated using GraphPad Prism 6 (GraphPad Software, Inc.). Differences in NTHI biofilm biomass after incubation with MAbs in vitro were determined by one-way ANOVA and Tukey's multiple comparisons test. Differences in CFU NTHI per mg mucosal biofilm and per ml middle ear fluid per cohort, and CFU P. aeruginosa per lung was calculated by Mann-Whitney test. Resolution of mucosal biofilm in vivo was analyzed by one-way ANOVA and Tukey's multiple comparisons test. For all analyses, a p-value ≤ 0.05 was considered significant.
3. Results
3.1. Design of Tip- and Tail-region Peptides
Prior epitope mapping efforts using a panel of synthetic peptides to mimic the deduced amino acid sequence of IHFNTHI as well as immune and non-immune chinchilla sera identified functional and protective domains positioned within the ends of the DNA-binding antiparallel β-ribbons (e.g. the DNA-binding ‘tips’) of each IHF subunit (GOODMAN et al., 2011, BROCKSON et al., 2014). To develop MAbs, two peptides derived from the tip regions of IHF (IhfA5NTHI and mIhfB4NTHI) and two derived from within the second long alpha-helix (IhfA3NTHI and IhfB2NTHI) that is, in part, involved in dimer-dimer interactions between the IHF subunits (e.g. the ‘tail’ regions of IHF) were used, the latter of which served as negative controls (Fig. 1A). Both tip peptides included an essential proline residue positioned approximately mid-sequence (Fig. 1A, arrows) that is predicted to support the native secondary structure of the turn in the protein architecture and appears to intercalate into DNA to facilitate or stabilize DNA bends (Rice et al., 1996).
3.2. Validation of MAb Specificity
By ELISA, all MAbs recognized the homologous peptide to which they were targeted as well as the isolated IHF subunit from which they were derived (Fig. 1B). Due to homology between the alpha- and beta-subunits of IHFNTHI (47% identical, 70% similar), MAbs directed towards one subunit often demonstrated some cross reactivity with the heterologous subunit. However, there was no cross reactivity between MAbs directed at tail peptides with tip peptides or vice versa.
3.3. Disruption of Diverse Bacterial Biofilms In Vitro
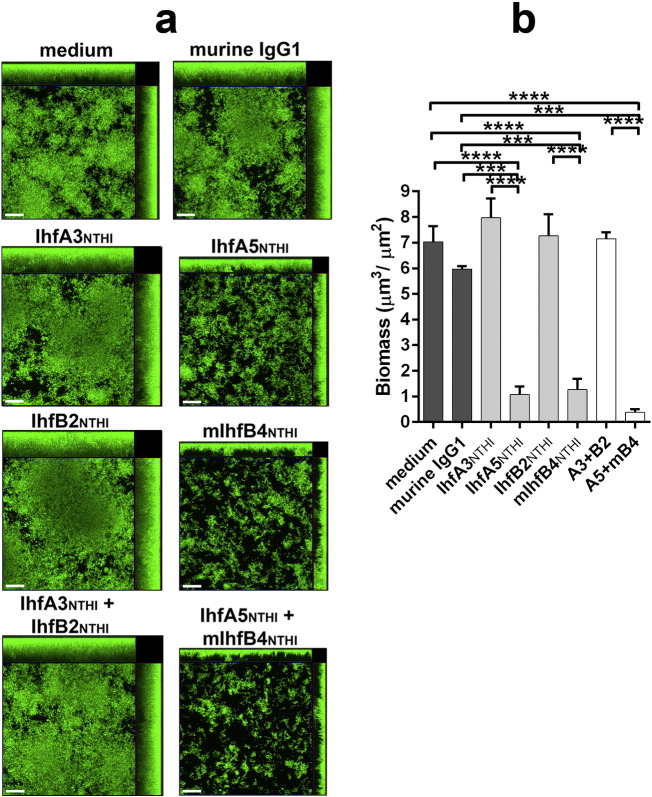
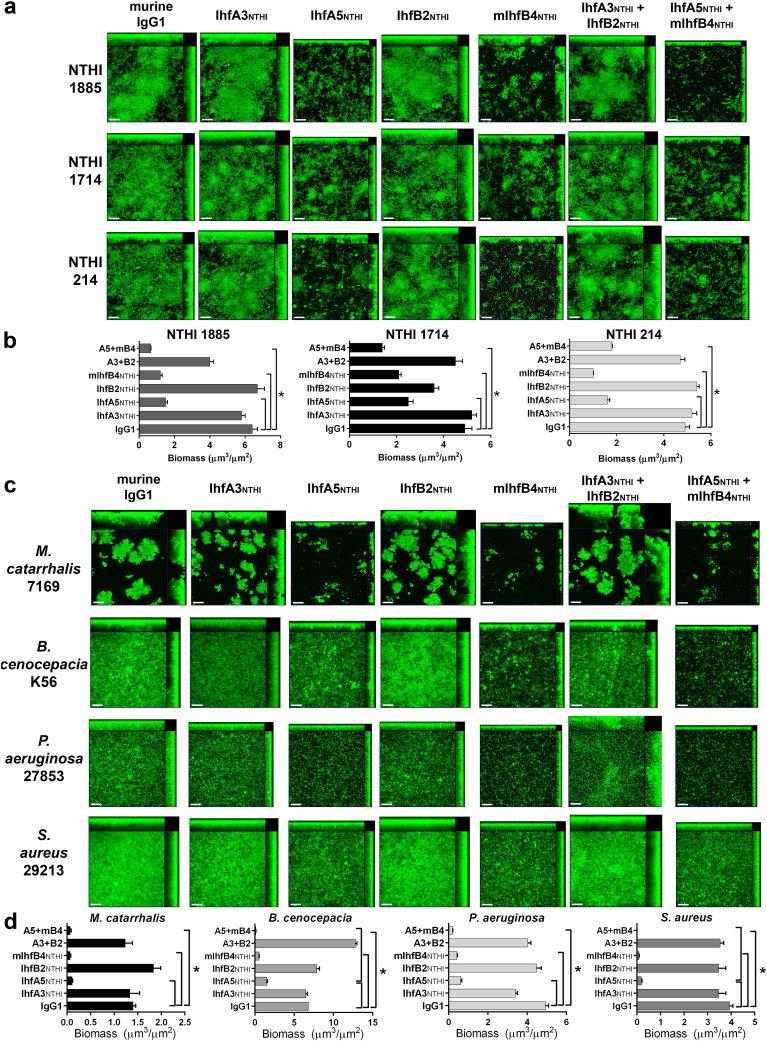
To assess the ability of the IHF-directed MAbs to disrupt pre-formed biofilms, 24-h NTHI biofilms were exposed to MAbs for 16 h in vitro. NTHI #86-028NP formed a robust biofilm that was undisturbed by treatment with non-specific murine IgG1 (Fig. 2A). Incubation with 5 μg of either of the two tip-directed MAbs (IhfA5NTHI and mIhfB4NTHI) induced a significant 6–7-fold reduction in biofilm biomass compared to either of the two tail-directed MAbs (IhfA3NTHI and IhfB2NTHI; p ≤ 0.0001; Fig. 2B). A cocktail of both tip-directed MAbs had an additive effect, this degree of disruption was significantly greater than that induced by treatment with a mixture of the tail-directed MAbs (p ≤ 0.0001) (Fig. 2A, bottom row). Comparable results were achieved with an additional three clinical isolates of NTHI (strains #1885, 1714 and 214) (Fig. 3A & B), thus demonstrating the breadth of efficacy of the IHF tip-directed MAbs against biofilms formed by diverse NTHI strains.
Fig. 2.
MAbs directed against the DNA-binding tip regions of IHFNTHI disrupted biofilms formed by NTHI 86-028NP in vitro. (A) Images obtained by confocal scanning microscopy demonstrating top-down and side views of representative 24-h biofilms formed by NTHI strain #86-028NP maintained in medium, or incubated for an additional 16 h with 5 μg murine MAbs as indicated. Bacteria within biofilms were stained with FM1-43FX membrane stain (green). Images are representative of three independent experiments. Scale bars, 20 μm. (B) Mean biofilm biomass ± SEM remaining after treatment as determined by COMSTAT2 analyses of three independent experiments. ***, p ≤ 0.001; ****, p ≤ 0.0001.
Fig. 3.
IHFNTHI tip-directed MAbs disrupted biofilms formed by clinically-relevant bacterial species. (A) Confocal scanning microscopy images of biofilms (green) formed by multiple clinical isolates of NTHI revealed disruption of pre-formed biofilms by incubation with the MAbs IhfA5NTHI and mIhfB4NTHI, a result not observed with the MAbs IhfA3NTHI and IhfB2NTHI. (B) This observation was further quantitated as a reduction in biofilm biomass by COMSTAT2 analyses. (C) Representative images showed disruption of biofilms formed by the bacterial pathogens M. catarrhalis, B. ceneocepacia, P. aeruginosa, and S. aureus by treatment with the IHFNTHI tip-directed MAbs and (D) quantitated as significant reduction in mean biofilm biomass. Images are representative of three independent experiments. Scale bars, 20 μm. *, p ≤ 0.05.
To determine if these MAbs could also disrupt biofilms formed by other human respiratory tract pathogens, we repeated biofilm disruption assays with M. catarrhalis (strain 7169), B. cenocepacia (strain K56), P. aeruginosa (strain 27853) and S. aureus (strain 29213). Biofilms formed by each bacterial species within 24 h varied greatly in architecture (Fig. 3C, 1st column), nevertheless tip-directed MAbs (IhfA5NTHI and mIhfB4NTHI) were highly effective against each of these pathogens, resulting in significant disruption (p ≤ 0.05) (Fig. 3C, columns 3, 5 & 7 and D) whereas those directed against the tails (IhfA3NTHI and IhfB2NTHI) were not (Fig. 3C, columns 2,4 & 6 and D).
3.4. Efficacy of the Tip-directed MAbs to Resolve Experimental NTHI-induced Otitis Media
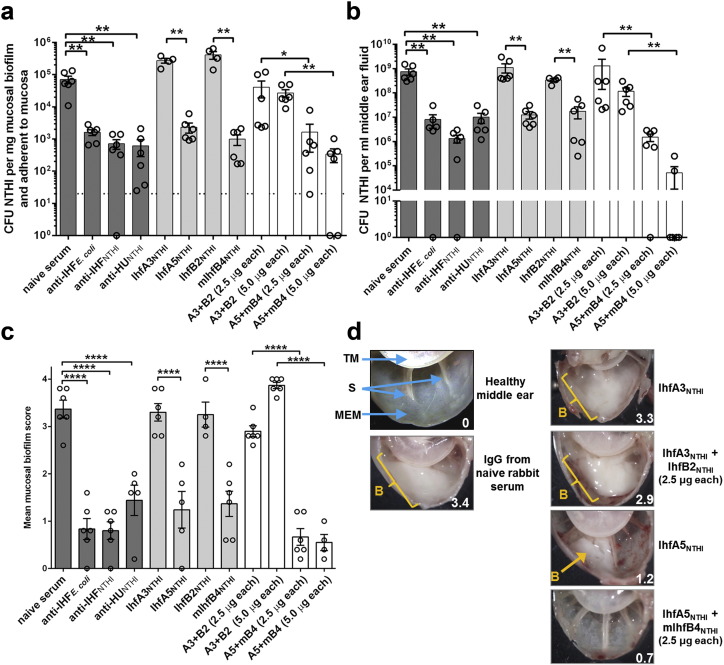
To test the ability of MAbs directed towards specific 20-mer epitopes of the alpha and beta subunit of IHFNTHI to render a therapeutic effect in vivo, we used a robust and highly reproducible chinchilla model of NTHI-induced otitis media (OM) (Bakaletz, 2009) wherein OM is first induced by inoculation of each middle ear with NTHI. Four days later, when it is known that NTHI biofilms typically fill ≥ 50% of the middle ear space of the vast majority of animals (in this model > 83% of ears will develop a biomass, > 67% of which will yield a score ≥ 2 on a 0 to 4 + scale) (NOVOTNY et al., 2011, NOVOTNY et al., 2013b), we instilled 100 μl of either: tip- or tail-directed MAb, IgG-enriched polyclonal rabbit antibody against a native DNABII protein or a cocktail of MAbs as detailed in Table 1 directly into the middle ear. This treatment was delivered daily for a total of three sequential days. Twenty-four hours after receipt of the final dose, animals were sacrificed and images of both middle ears were captured by light microscopy for blinded evaluation.
We found that for the subpopulation of NTHI that were either adherent to the middle ear mucosa or resident within mucosa-associated biofilms, IgG-enriched polyclonal rabbit antiserum to IHFE. coli, IHFNTHI or HUNTHI induced a significant decrease in bacterial load (1.6-, 2.0- and 2.1-log reductions, respectively) compared to naive rabbit IgG (p ≤ 0.01; Fig. 4A). No reduction in bacterial load was achieved with tail-directed MAbs IhfA3NTHI or IhfB2NTHI. There was however significant 1.5- and 1.9-log reductions in those cohorts that received MAbs to either IhfA5NTHI or mIhfB4NTHI, respectively (p ≤ 0.01). Receipt of a cocktail of tail-directed MAbs at either of two concentrations tested did not reduce the relative bacterial load within the middle ears. Conversely, a dose-dependent 0.7-log decrease in bacterial load was shown in the two cohorts that received cocktails of tip-directed MAbs with complete eradication of NTHI from two of these samples in the cohort that received the higher dose (5 μg of each MAb). This difference was statistically significant from those that received antibody against purified IHFE. coli (p ≤ 0.01) and also against those cohorts that received either of the two tail-directed MAbs individually (p ≤ 0.01), or an equivalent dose of a tail-directed MAb cocktail (p ≤ 0.05). For the planktonic bacterial sub-population present within middle ear fluids (Fig. 4B), the results were very similar to those seen for the adherent and mucosal biofilm sub-population. Again, we saw complete eradication of NTHI from the middle ear fluids of 4 ears in the cohort that received 5 μg of each MAb as a cocktail of tip-directed MAbs.
Fig. 4.
Clearance of NTHI from the middle ear in a chinchilla model of experimental otitis media. NTHI biofilms were established within the middle ears of chinchillas prior to infusion of the middle ear space with IgG-enriched polyclonal rabbit sera or murine MAbs. (A) CFU NTHI per mg mucosal biofilm and NTHI adherent to the middle ear mucosal epithelium. The limit of detection is indicated by a dashed horizontal line. (B) Clearance of NTHI from middle ear fluids. CFU NTHI in middle ear effusions after infusion of the middle ear space with IgG-enriched polyclonal rabbit sera or murine MAbs. (C) Resolution of established NTHI biofilms from the chinchilla middle ear. Images of each middle ear were blindly ranked to determine the relative quantity of mucosal biofilm that remained after infusion of the middle ear space with IgG-enriched polyclonal rabbit sera or murine MAbs. An established mucosal biofilm scale was employed (Novotny et al., 2011), whereby 0 = no mucosal biofilm visible, 1 ≤ 25% of middle ear space occluded by mucosal biofilm, 2 = ≥ 25–50% occluded, 3 = ≥ 50–75% occluded, 4 = ≥ 75–100% occluded. (D) Representative images of chinchilla middle ears after infusion of the middle ear space with IgG-enriched polyclonal rabbit sera or murine monoclonal antibodies. The mucosal biofilm score is indicated in the bottom right corner of each image. TM - tympanic membrane, S - bony septae, MEM - middle ear mucosa, B - mucosal biofilm. For bar graphs, values for each individual middle ear is shown (circles; 6 middle ears per cohort) and the mean ± SEM for the cohort presented (bars).*, p ≤ 0.05; **, p ≤ 0.01; ****, p ≤ 0.0001.
Importantly, we also quantitatively evaluated how much mucosal biofilm remained in chinchilla middle ears after receipt of treatment using a published scoring rubric that ranks the degree to which the middle ear space is filled with biofilm (where 1 + = < 25% filled and 4 + = ≥ 75–100% filled) (Novotny et al., 2011). As expected due to their demonstrated biofilm disruption capability in vitro, all IgG-enriched rabbit polyclonal antisera directed against a native DNABII protein were highly effective, significantly reducing NTHI biofilms within the middle ear compared to those that received naive rabbit IgG (p ≤ 0.0001) (Fig. 4C). Anti-IHFE. coli and anti-IHFNTHI appeared slightly more efficacious than anti-HUNTHI however this difference was not statistically significant. Following receipt of a single MAb, once again tail-directed MAbs IhfA3NTHI and IhfB2NTHI were not effective, whereas tip-directed MAbs IhfA5NTHI and mIhfB4NTHI induced significant resolution of mucosal biofilms (p ≤ 0.0001). In concordance with bacterial load data (Figs. 4A&B), no reduction in mucosal biofilm was observed in animals that received cocktails of tail-directed MAbs regardless of the dose received. Conversely, animals that received either the low or high dose cocktail of tip-directed MAbs demonstrated the greatest reduction in middle ear mucosal biofilm of any of the 12 cohorts (Fig. 4C).
Representative images of middle ears from selected chinchilla cohorts are shown in Fig. 4D. Robust biofilms remained in the middle ears of animals that received either naive rabbit IgG, tail-directed MAbs IhfA3NTHI (Fig. 4D) or IhfB2NTHI (not shown) or the cocktail of MAbs to the two tail peptides at either dose (lower dose shown). Conversely, there was little to no evidence of a mucosal biofilm remaining within the middle ears of animals treated with the tip-directed MAbs IhfA5NTHI (Fig. 4D) or mIhfB4NTHI (not shown) or the cocktail of tip-directed MAbs at both doses (lower dose shown). Intriguingly, there was also little or no evidence of any inflammation in the middle ears of these latter cohorts despite the fact that four days earlier this anatomical niche was filled with a Gram negative bacterial biofilm. Collectively, these data demonstrated the therapeutic efficacy of the tip-directed MAbs to resolve NTHI biofilms formed in the middle ear during experimental OM.
3.5. Efficacy of the Tip-directed MAbs to Resolve P. aeruginosa Lung Infection in a Murine Model
To determine if the outcome seen in the chinchilla model of experimental OM due to NTHI could be expanded and applied to diseases caused by another bacterial species, we then tested the cocktail of tip-directed MAbs in a mouse lung infection model. One day after mice were challenged with P. aeruginosa #27853 by intratracheal (IT) instillation, when the bacterial load in the lungs was ~ 8.4 × 103 CFU/lung (Fig. 5A), a single small volume dose (0.03 ml) of either nonspecific murine IgG1 (negative control) or a cocktail of tip-directed MAbs or tail-directed MAbs was administered IT. At both 1 and 6 days after this singular delivery of antibodies, mice were sacrificed and the lungs processed to determine relative bacterial load. Whereas at 1-day post-therapy there was no difference between cohorts that received nonspecific murine IgG1 and those that received the cocktail of tail-directed MAbs IhfA3NTHI and IhfB2NTHI, mice administered a cocktail of tip-directed MAbs IhfA5NTHI and mIhfB4NTHI showed a significant > 2-log reduction in bacterial load within this 24 h period (p ≤ 0.05) (Fig. 5A). This significant difference was maintained for 6 days despite the fact that both the control cohort of mice as well as those that had received the cocktail of tail-directed MAbs had also begun to clear P. aeruginosa from the lungs compared to pre-treatment. Nonetheless, the difference between these two latter cohorts and those that received the tip peptide-directed MAb cocktail remained statistically significant (p ≤ 0.05). Moreover, this latter cohort was the only cohort wherein several lungs yielded no bacterial counts.
Fig. 5.
Clearance of P. aeruginosa from the lungs of mice after treatment with MAbs directed against the DNA-binding tip regions of IHF. Mice were challenged intratracheally with P. aeruginosa followed one day later by instillation of a cocktail of (A) 10 μg tip-directed or tail-directed MAbs or (B) MAb cocktails ± 60 mg tobramycin per kg body weight. CFU P. aeruginosa per individual lung (5 mice per cohort) and mean CFU P. aeruginosa ± SEM for each cohort shown (bars). *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. Tobi: tobramycin.
Given that treatment with antibodies directed against a DNABII protein (or derived peptide) induced collapse of the biofilm with release of viable resident bacteria, we envision a combinatorial treatment for clinical evaluation wherein an effective antibiotic (or antiserum) is co-delivered to mediate killing of released bacteria. Thereby, we expanded the above-described study to add tobramycin to those MAb cocktails delivered to selected cohorts. One day after treatment, there was no reduction in CFU P. aeruginosa per lung homogenate in animals that received tobramycin alone despite the fact that this strain of P. aeruginosa is susceptible to the action of this antibiotic when grown in planktonic culture (Eckert et al., 2006). There was similarly no reduction in CFU P. aeruginosa in cohorts that received either a cocktail of nonspecific murine IgG1 and IgG2a with or without tobramycin, or a cocktail of tail-directed MAbs with or without tobramycin or tobramycin alone (Fig. 5B). Conversely, cohorts of mice that received a cocktail of tip-directed MAbs with or without tobramycin showed a significant reduction in lung bacterial load compared to mice administered the cocktail of murine IgG1 + IgG2a (p ≤ 0.01), those treated with tail-directed MAb cocktail of IhfA3NTHI + IhfB2NTHI (p ≤ 0.05) or those treated with tobramycin alone (p ≤ 0.05). The level of significance among these cohorts increased 6 days after treatment. The addition of tobramycin to a cocktail of tip-directed MAbs appeared to confer an added benefit although all cohorts that received this MAb cocktail generated very low or no CFU counts in lung homogenates, thereby this difference was not statistically significant. Again, only those cohorts that received tip-directed MAbs yielded lungs in which bacteria had been completely eradicated.
Histological evaluation of the murine lungs revealed multiple characteristic aggregates of P. aeruginosa (BJARNSHOLT et al., 2013, SINGH et al., 2000, LAFAYETTE et al., 2015) in sections of lung recovered prior to treatment (Fig. 6A; yellow, aggregates encircled), in mice treated with a cocktail of murine IgG1 + IgG2a (Fig. 6B) or those treated with the cocktail of tail-directed MAbs (Fig. 6C). Multicellular aggregates were observed dispersed throughout the lung with 7 to 15 aggregates observed in any single 10 × field of view in lungs from mice prior to treatment or in those cohorts treated with either murine IgG or with tail peptide MAbs. Upon examination at higher magnification, these aggregates appeared to contain a considerable number of bacterial cells along with positively labeled extracellular proteins (Fig. 6E-G). The number of bacterial cells within these aggregates was estimated to be 50 to 100. In lungs of mice treated with the cocktail of MAbs directed against IhfA5NTHI + mIhfB4NTHI, only 1 to 3 aggregates was observed within any 10 × field of view and most of these aggregates appeared to contain a single bacterium (Fig. 6D&H). Positively labeled extracellular proteins inhibited us from getting a more accurate count of bacterial cells per large aggregate via confocal microscopy, however when examined via TEM, > 100 bacterial cells were observed in large aggregates (Fig. 6I) in those cohorts of mice that did not clear P. aeruginosa (Fig. 6A–C). Individual bacteria or small aggregates of P. aeruginosa characteristic of mice treated with the tip-directed MAb cocktail (Fig. 6D & H) were too rare to be found by TEM. Four days after the IT challenge of mice with P. aeruginosa, bacterial eDNA with associated proteins and neutrophils were observed in the lungs of mice prior to treatment (Fig. 7).
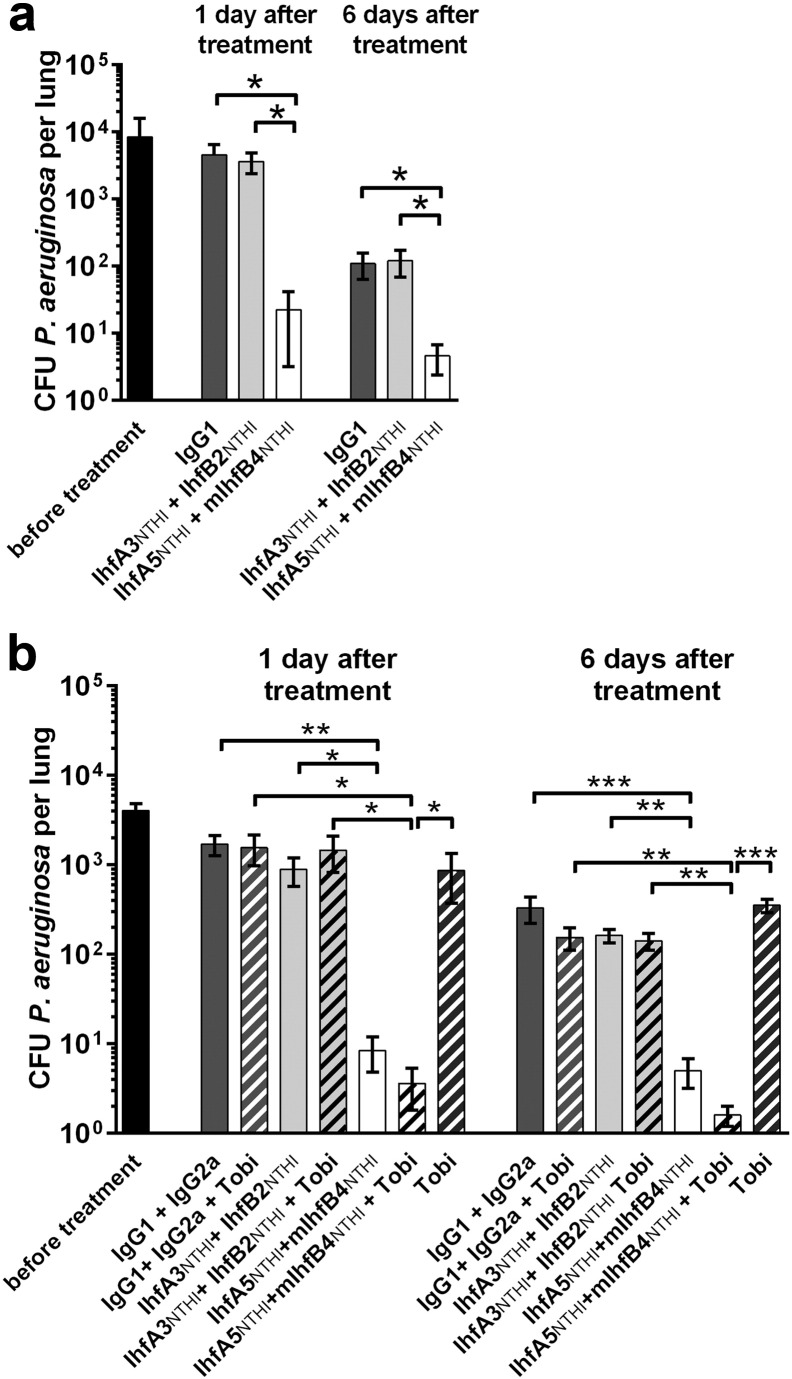
Fig. 6.
Reduction in both P. aeruginosa aggregate number and size within lungs treated with IHFNTHI tip-directed MAbs in an experimental murine model of lung infection. (A) Multiple bacterial aggregates were observed throughout the lungs of mice prior to treatment (yellow, aggregates encircled) and (B) in lungs after receipt of nonspecific murine IgG1 + IgG2a (C) or a cocktail of the tail-directed MAbs IhfA3NTHI + IhfB2NTHI. (D) In contrast, few P. aeruginosa were detected after delivery of tip-directed MAbs IhfA5NTHI + mIhfB4NTHI thus demonstrating the efficacy of this therapeutic strategy. DAPI counterstain (blue); scale bars, 50 μm. Panels E–H - higher magnification images of representative aggregates for each cohort showing either relative size of the aggregates or the presence of single bacteria [E- prior to treatment; F – treatment with nonspecific murine IgG1 + IgG2a; G – treatment with tail-directed MAbs IhfA3NTHI + IhfB2NTHI and H – treatment with tip-directed MAbs IhfA5NTHI + mIhfB4NTHI (P. aeruginosa aggregates were labeled with fluorescently tagged rabbit antibody to whole P. aeruginosa which labels both the bacteria themselves as well as bacterial proteins contained within the biofilm matrix hence the inability to readily resolve individual bacterial cells)]. Note that aggregates in Panels A, B & C and E, F & G are similar in size whereas those imaged in lungs treated with the cocktail of tip-directed MAbs appear as single bacterial cells (Panels D & H); scale bars, 5 μm. A representative TEM image of a P. aeruginosa aggregate as shown in Panels A, B & C or E, F & G is presented in Panel I. Bacterial cells are pseudo-colored green and mouse lung tissue is pseudo-colored blue to aid in visualization. Bacterial aggregates within lungs from mice that received murine IgG or tail-directed MAbs typically contained 50–100 bacteria. The relatively limited number of very small clusters or presence of only single bacteria resident within lungs from mice treated with tip-directed MAbs precluded visualization by TEM; Scale bar, 2 μm. Table: FOV – field of view.
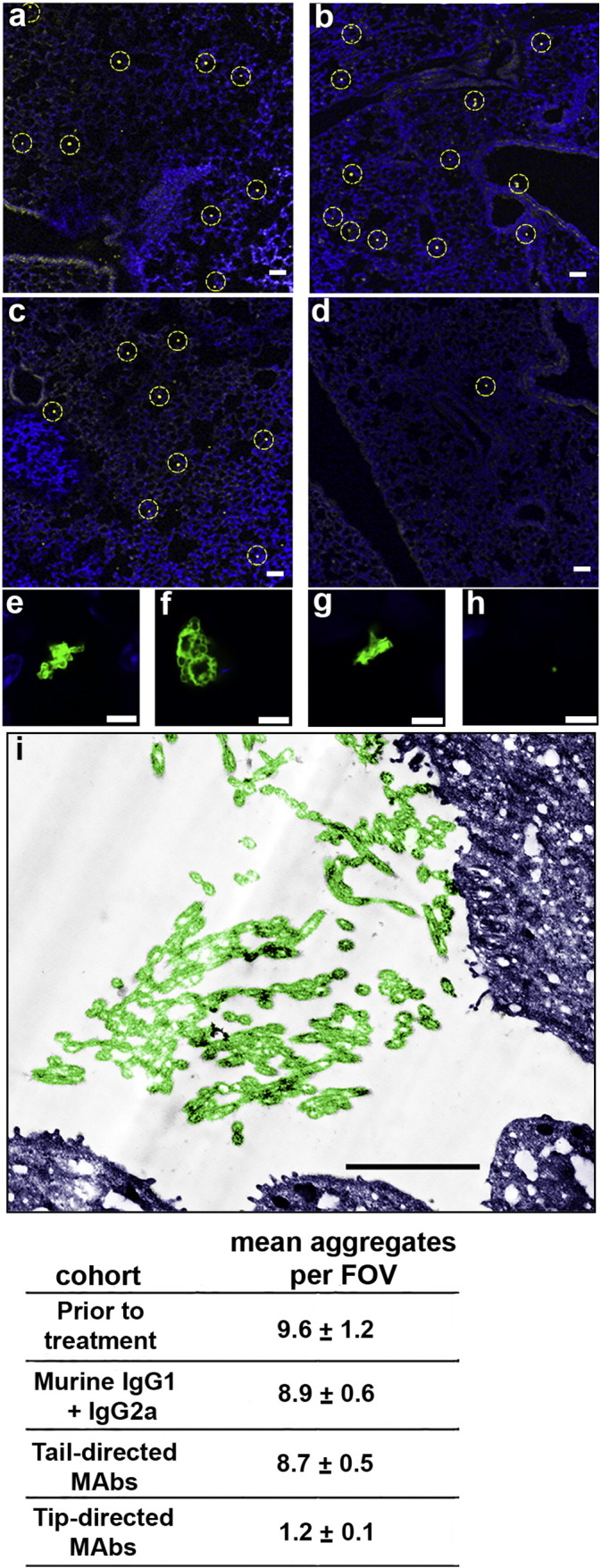
Fig. 7.
Detection of eDNA, DNABII protein and neutrophils within the lungs of mice after challenge with P. aeruginosa. Lungs collected from mice 4 days after IT challenge were embedded in paraffin and 10 μm sections were immunolabeled for the presence of eDNA, DNABII protein and neutrophils. Strands of eDNA (red, Panel A) and DNABII protein (green, Panel B) were present within the murine lung. Association between the eDNA and DNABII proteins is shown in the merged image (Panel C) where co-localization of eDNA and DNABII proteins appears yellow. Neutrophils appear white in these image (refer to arrowheads in panel A). Scale bar 5 μm.
4. Discussion
Extracellular DNA with associated DNABII proteins are common constituents of biofilms formed by a large number of human pathogens where they contribute significantly to the structural stability of these fortresses that house and protect resident bacteria from host immune effectors and the action of antimicrobials (Goodman et al., 2011). By targeting these lynchpin proteins that are positioned at the vertices of crossed strands of eDNA within the biofilm matrix, we've shown that we can induce complete collapse of the biofilm with release of resident bacteria without killing them (GOODMAN et al., 2011, BROCKSON et al., 2014). These newly released bacteria can then be cleared by either host innate and acquired immune effectors or antibiotics, to which they have significantly increased sensitivity (Brockson et al., 2014). This unique phenotype of newly released bacteria, which is distinct from those growing either planktonically or resident within a biofilm, has been reported by others (PETTIGREW et al., 2014, CHAO et al., 2014) and could provide an opportune treatment window for biofilm-associated diseases.
Whereas the efficacy of this DNABII protein-targeted approach had been demonstrated both in vitro (GOODMAN et al., 2011, GUSTAVE et al., 2013, BRANDSTETTER et al., 2013, NOVOTNY et al., 2013a, BROCKSON et al., 2014, DEVARAJ et al., 2015) and in vivo (JUSTICE et al., 2012, NOVOTNY et al., 2015b), the majority of the work published to date was conducted using polyclonal rabbit antiserum directed against native IHF that was isolated and purified from E. coli. To determine if this strategy was also effective when IHF or HU was isolated from another human pathogen, polyclonal antisera against these proteins as isolated and purified from the respiratory tract pathogen NTHI were used. Moreover, in an attempt to move closer to a lead clinical compound, peptides that targeted functional and protective domains of the alpha and beta subunit from IHFNTHI were designed to develop MAbs to these specific 20-mer domains. These MAbs were tested here both singly and admixed as cocktails in vitro against several human pathogens as well as in vivo, using two unique animal models: NTHI infection of the chinchilla middle ear or P. aeruginosa infection of the murine lung.
In vitro efficacy was shown against multiple clinical isolates of NTHI; the Gram negative cystic fibrosis (CF) and burn wound pathogen P. aeruginosa; the Gram negative respiratory tract pathogen M. catarrhalis; the organism responsible for deadly ‘cepacia syndrome’ in CF patients B. cenocepacia and Gram positive S. aureus which is responsible for a host of troublesome infections (Jansen et al., 2013). The ability to effectively disrupt biofilms formed by these diverse human pathogens is attributed to the relative conservancy of IHF among these five microbes. Members of the DNABII family have overall modest primary sequence identity despite a highly conserved secondary structure (Swinger and Rice, 2004). In the case of these four pathogens, compared to their closest DNABII orthologue in NTHI there is 38% to 62% overall sequence identity. However, within the DNA binding tips (from which the peptide epitopes were derived and the MAbs directed), there is overall greater sequence identity (e.g. 45% to 80%). Interestingly, Gram positive bacteria, including S. aureus have a single HU allele, their only DNABII gene. Despite its low primary sequence identity overall and between epitope tip regions with NTHI, there nonetheless appears to be sufficient structural recognition for the IHFNTHI-specific MAbs to readily resolve S. aureus biofilms in vitro. This outcome suggests that it is the combination of sequence and subsequent structure that is critical for antibody recognition and biofilm disruption.
We then used two models of biofilm infections to test the in vivo efficacy of these targeted MAbs and found that animals treated with MAbs derived against the tip peptides of the alpha and beta subunit of IHFNTHI resolved infection significantly earlier than other cohorts. Notably chinchillas treated with tip-directed MAbs demonstrated a marked reduction or complete eradication of pre-existing mucosal biofilms from the middle ear space with little to no residual signs of inflammation. Collectively these data suggested to us that in addition to the biofilm disruption effect mediated by delivery of IHF-directed antibodies into the middle ear, the host immune system also became engaged and contributed to the eradication of NTHI likely via the action of innate host defense peptides and phagocytosis.
Given the commonality of eDNA and associated DNABII proteins in biofilms formed by multiple and diverse human pathogens, the efficacy achieved by delivery of MAbs that target the DNA-binding tips of these proteins, in terms of reducing the bacterial load and/or eradicating mucosal biofilms, is strongly supportive of their continued development as a lead therapeutic compound. Whereas here, we used two models of respiratory tract diseases due to either NTHI (middle ear infection, the uppermost reach of the respiratory tract) or P. aeruginosa (lung infection, lower airway), the already demonstrated in vitro effectiveness of this approach against a diverse panel of pathogens suggests that the utility could be significantly broader with potential application to many diseases of man and animals. The transition to monoclonal antibodies directed against specific targeted epitopes is important as it allows the development of a significantly more focused therapeutic with the ability to humanize the monoclonal(s) for delivery in clinical trials; this is ongoing.
Funding Source
This work was supported by grant NIH/NIDCD R01 DC011818 to LOB and SDG. This funding source played no role in the conduct of the research or preparation of the manuscript.
Conflicts of Interest
LOB and SDG have equity in ProclaRx which has licensed this biofilm disruption technology. The authors declare no additional competing financial interests.
Author Contributions
Conceptualization, SDG and LOB. Methodology, JAJ and LAN. Investigation, LOB, JAJ and LAN. Writing – original draft, LOB. Writing – review and editing, LOB, LAN and SDG. Funding acquisition, LOB and SDG. Resources, SDG. Visualization, LAN and JAJ. Supervision, LOB and SDG.
Acknowledgments
We thank Jennifer Neelans for manuscript preparation, Hannah Wexner for generation and isolation of monoclonal antibodies, Lauren Warren for cloning NTHI IHFA and IHFB and John Buzzo for expression and purification of NTHI IHFA and IHFB. We are particularly indebted to Kenneth Brockman, Ph.D., Michael Ward, Jr., B.S. and Elaine Mokrzan, Ph.D. for outstanding assistance with chinchilla and mouse modeling studies and to Dr. Dan Wozniak for helpful advice regarding establishment of a murine model of P. aeruginosa lung infection.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.06.022.
Contributor Information
Laura A. Novotny, Email: Laura.Novotny@nationwidechildrens.org.
Joseph A. Jurcisek, Email: Joseph.Jurcisek@nationwidechildrens.org.
Steven D. Goodman, Email: Steven.Goodman@nationwidechildrens.org.
Lauren O. Bakaletz, Email: Lauren.Bakaletz@nationwidechildrens.org.
Appendix A. Supplementary data
Supplementary material.
References
- Alemayehu D., Casey P.G., Mcauliffe O., Guinane C.M., Martin J.G., Shanahan F., Coffey A., Ross R.P., Hill C. Bacteriophages phiMR299-2 and phiNH-4 can eliminate Pseudomonas aeruginosa in the murine lung and on cystic fibrosis lung airway cells. MBio. 2012;3:12. doi: 10.1128/mBio.00029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakaletz L.O. Chinchilla as a robust, reproducible and polymicrobial model of otitis media and its prevention. Expert Rev. Vaccines. 2009;8:1063–1082. doi: 10.1586/erv.09.63. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T., Alhede M., Eickhardt-SORENSEN S.R., Moser C., Kuhl M., Jensen P.O., Hoiby N. The in vivo biofilm. Trends Microbiol. 2013;21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T., Jensen P.O., Fiandaca M.J., Pedersen J., Hansen C.R., Andersen C.B., Pressler T., Givskov M., Hoiby N. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 2009;44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- Boleij A., Schaeps R.M., De Kleijn S., Hermans P.W., Glaser P., Pancholi V., Swinkels D.W., Tjalsma H. Surface-exposed histone-like protein a modulates adherence of Streptococcus gallolyticus to colon adenocarcinoma cells. Infect. Immun. 2009;77:5519–5527. doi: 10.1128/IAI.00384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstetter K.A., Jurcisek J.A., Goodman S.D., Bakaletz L.O., Das S. Antibodies directed against integration host factor mediate biofilm clearance from nasopore. Laryngoscope. 2013;123:2626–2632. doi: 10.1002/lary.24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockson M.E., Novotny L.A., Mokrzan E.M., Malhotra S., Jurcisek J.A., Akbar R., Devaraj A., Goodman S.D., Bakaletz L.O. Evaluation of the kinetics and mechanism of action of anti-integration host factor-mediated disruption of bacterial biofilms. Mol. Microbiol. 2014;93:1246–1258. doi: 10.1111/mmi.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook I. Microbiology of chronic rhinosinusitis. Eur. J. Clin. Microbiol. Infect. Dis. 2016 doi: 10.1007/s10096-016-2640-x. [DOI] [PubMed] [Google Scholar]
- Bucior I., Abbott J., Song Y., Matthay M.A., Engel J.N. Sugar administration is an effective adjunctive therapy in the treatment of Pseudomonas aeruginosa pneumonia. Am. J. Physiol. Lung Cell. Mol. Physiol. 2013;305:L352–L363. doi: 10.1152/ajplung.00387.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y., Marks L.R., Pettigrew M.M., Hakansson A.P. Streptococcus pneumoniae biofilm formation and dispersion during colonization and disease. Front Cell Infect. Microbiol. 2014;4:194. doi: 10.3389/fcimb.2014.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj A., Justice S.S., Bakaletz L.O., Goodman S.D. DNABII proteins play a central role in UPEC biofilm structure. Mol. Microbiol. 2015;96:1119–1135. doi: 10.1111/mmi.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugaszewska J., Leszczynska M., Lenkowski M., Tatarska A., Pastusiak T., Szyfter W. The pathophysiological role of bacterial biofilms in chronic sinusitis. Eur. Arch. Otorhinolaryngol. 2015 doi: 10.1007/s00405-015-3650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J.L., Jiang Y.W., Xie J.Q., Zhang X.G. New is old, and old is new: recent advances in antibiotic-based, antibiotic-free and ethnomedical treatments against methicillin-resistant Staphylococcus aureus wound infections. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17050617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert R., Brady K.M., Greenberg E.P., Qi F., Yarbrough D.K., He J., Mchardy I., Anderson M.H., Shi W. Enhancement of antimicrobial activity against Pseudomonas aeruginosa by coadministration of G10KHc and tobramycin. Antimicrob. Agents Chemother. 2006;50:3833–3838. doi: 10.1128/AAC.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H.C., Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Fong J.N., Yildiz F.H. Biofilm matrix proteins. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MB-0004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fothergill J.L., Neill D.R., Loman N., Winstanley C., Kadioglu A. Pseudomonas aeruginosa adaptation in the nasopharyngeal reservoir leads to migration and persistence in the lungs. Nat. Commun. 2014;5:4780. doi: 10.1038/ncomms5780. [DOI] [PubMed] [Google Scholar]
- Fu P., Mohan V., Mansoor S., Tiruppathi C., Sadikot R.T., Natarajan V. Role of nicotinamide adenine dinucleotide phosphate-reduced oxidase proteins in Pseudomonas aeruginosa-induced lung inflammation and permeability. Am. J. Respir. Cell Mol. Biol. 2013;48:477–488. doi: 10.1165/rcmb.2012-0242OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAO Q. Ph.D. Thesis. University of Southern California; Los Angeles, CA: 2000. S. mutans GtfB and GtfC gene expression. [Google Scholar]
- Goodman S.D., Obergfell K.P., Jurcisek J.A., Novotny L.A., Downey J.S., Ayala E.A., Tjokro N., Li B., Justice S.S., Bakaletz L.O. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011;4:625–637. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- Gustave J.E., Jurcisek J.A., Mccoy K.S., Goodman S.D., Bakaletz L.O. Targeting bacterial integration host factor to disrupt biofilms associated with cystic fibrosis. J. Cyst. Fibros. 2013;12:384–389. doi: 10.1016/j.jcf.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett D.J., Borchers M.T., Panos R.J. Chronic obstructive pulmonary disease (COPD): evaluation from clinical, immunological and bacterial pathogenesis perspectives. J. Microbiol. 2014;52:211–226. doi: 10.1007/s12275-014-4068-2. [DOI] [PubMed] [Google Scholar]
- Heydorn A., Nielsen A.T., Hentzer M., Sternberg C., Givskov M., Ersboll B.K., Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146(Pt 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Idicula, W. K., Jurcisek, J. A., Cass, N. D., Syed, A., Goodman, S. D., Elmaraghy, C. A., Jatana, K. R. & Bakaletz, L. O. 2016. Identification of biofilms in post-tympanostomy tube otorrhea. Laryngoscope, http://onlinelibrary.wiley.com/doi/10.1002/lary.25826/abstract, DOI: http://dx.doi.org/10.1002/lary.25826. [DOI] [PMC free article] [PubMed]
- Jansen K.U., Girgenti D.Q., Scully I.L., Anderson A.S. Vaccine review: “Staphyloccocus aureus vaccines: problems and prospects”. Vaccine. 2013;31:2723–2730. doi: 10.1016/j.vaccine.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Jurcisek, J. A., Dickson, A. C., Bruggeman, M. E. & Bakaletz, L. O. 2011. In vitro biofilm formation in an 8-well chamber slide. J. Vis. Exp., pii: 2481. doi: http://dx.doi.org/10.3791/2481. [DOI] [PMC free article] [PubMed]
- Justice S.S., Li B., Downey J.S., Dabdoub S.M., Brockson M.E., Probst G.D., Ray W.C., Goodman S.D. Aberrant community architecture and attenuated persistence of uropathogenic Escherichia coli in the absence of individual IHF subunits. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafayette S.L., Houle D., Beaudoin T., Wojewodka G., Radzioch D., Hoffman L.R., Burns J.L., Dandekar A.A., Smalley N.E., Chandler J.R., Zlosnik J.E., Speert D.P., Bernier J., Matouk E., Brochiero E., Rousseau S., Nguyen D. Cystic fibrosis-adapted quorum sensing mutants cause hyperinflammatory responses. Sci. Adv. 2015;1 doi: 10.1126/sciadv.1500199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunsford R.D., Nguyen N., London J. DNA-binding activities in Streptococcus gordonii: identification of a receptor-nickase and a histonelike protein. Curr. Microbiol. 1996;32:95–100. doi: 10.1007/s002849900017. [DOI] [PubMed] [Google Scholar]
- Machado G.B., De Oliveira A.V., Saliba A.M., De Lima C.D., Suassuna J.H., Plotkowski M.C. Pseudomonas aeruginosa toxin ExoU induces a PAF-dependent impairment of alveolar fibrin turnover secondary to enhanced activation of coagulation and increased expression of plasminogen activator inhibitor-1 in the course of mice pneumosepsis. Respir. Res. 2011;12:104. doi: 10.1186/1465-9921-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny L.A., Amer A.O., Brockson M.E., Goodman S.D., Bakaletz L.O. Structural stability of Burkholderia cenocepacia biofilms is reliant on eDNA structure and presence of a bacterial nucleic acid binding protein. PLoS One. 2013;8 doi: 10.1371/journal.pone.0067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny L.A., Clements J.D., Bakaletz L.O. Transcutaneous immunization as preventative and therapeutic regimens to protect against experimental otitis media due to nontypeable Haemophilus influenzae. Mucosal Immunol. 2011;4:456–467. doi: 10.1038/mi.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny L.A., Clements J.D., Bakaletz L.O. Kinetic analysis and evaluation of the mechanisms involved in the resolution of experimental nontypeable Haemophilus influenzae-induced otitis media after transcutaneous immunization. Vaccine. 2013;31:3417–3426. doi: 10.1016/j.vaccine.2012.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny L.A., Clements J.D., Bakaletz L.O. Therapeutic transcutaneous immunization with a band-aid vaccine resolves experimental otitis media. Clin. Vaccine Immunol. 2015;22:867–874. doi: 10.1128/CVI.00090-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny L.A., Jurcisek J.A., Ward M.O., Jr., Jordan Z.B., Goodman S.D., Bakaletz L.O. Antibodies against the majority subunit of type IV Pili disperse nontypeable Haemophilus influenzae biofilms in a LuxS-dependent manner and confer therapeutic resolution of experimental otitis media. Mol. Microbiol. 2015;96:276–292. doi: 10.1111/mmi.12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettigrew M.M., Marks L.R., Kong Y., Gent J.F., Roche-Hakansson H., Hakansson A.P. Dynamic changes in the Streptococcus pneumoniae transcriptome during transition from biofilm formation to invasive disease upon influenza A virus infection. Infect. Immun. 2014;82:4607–4619. doi: 10.1128/IAI.02225-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P.A., Yang S., Mizuuchi K., Nash H.A. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- Sabet M., Miller C.E., Nolan T.G., Senekeo-Effenberger K., Dudley M.N., Griffith D.C. Efficacy of aerosol MP-376, a levofloxacin inhalation solution, in models of mouse lung infection due to Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009;53:3923–3928. doi: 10.1128/AAC.00268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikot R.T., Zeng H., Azim A.C., Joo M., Dey S.K., Breyer R.M., Peebles R.S., Blackwell T.S., Christman J.W. Bacterial clearance of Pseudomonas aeruginosa is enhanced by the inhibition of COX-2. Eur. J. Immunol. 2007;37:1001–1009. doi: 10.1002/eji.200636636. [DOI] [PubMed] [Google Scholar]
- Scalise A., Bianchi A., Tartaglione C., Bolletta E., Pierangeli M., Torresetti M., Marazzi M., Di Benedetto G. Microenvironment and microbiology of skin wounds: the role of bacterial biofilms and related factors. Semin. Vasc. Surg. 2015;28:151–159. doi: 10.1053/j.semvascsurg.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Singh P.K., Schaefer A.L., Parsek M.R., Moninger T.O., Welsh M.J., Greenberg E.P. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- Stinson M.W., Mclaughlin R., Choi S.H., Juarez Z.E., Barnard J. Streptococcal histone-like protein: primary structure of hlpA and protein binding to lipoteichoic acid and epithelial cells. Infect. Immun. 1998;66:259–265. doi: 10.1128/iai.66.1.259-265.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subashchandrabose S., Mobley H.L. Virulence and fitness determinants of uropathogenic Escherichia coli. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.UTI-0015-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinger K.K., Rice P.A. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Swords W.E. Nontypeable Haemophilus influenzae biofilms: role in chronic airway infections. Front. Cell. Infect. Microbiol. 2012;2:97. doi: 10.3389/fcimb.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorregaard M. Master's Thesis. Technical University of Denmark (DTU); Kongens Lyngby, Denmark: 2008. COMSTAT2 - a modern 3D image analysis environment for biofilms. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.