Abstract
Myosins are actin-based motor proteins involved in many cellular movements. It is interesting to study the evolutionary patterns and the functional attributes of various types of myosins. Computational search algorithms were performed to identify putative myosin members by phylogenetic analysis, sequence motifs, and coexisting domains. This study is aimed at understanding the distribution and the likely biological functions of myosins encoded in various taxa and available eukaryotic genomes. We report here a phylogenetic analysis of around 4,064 myosin motor domains, built entirely from complete or near-complete myosin repertoires incorporating many unclassified, uncharacterized sequences and new myosin classes, with emphasis on myosins from Fungi, Haptophyta, and other Stramenopiles, Alveolates, and Rhizaria (SAR). The identification of large classes of myosins in Oomycetes, Cellular slime molds, Choanoflagellates, Pelagophytes, Eustigmatophyceae, Fonticula, Eucoccidiorida, and Apicomplexans with novel myosin motif variants that are conserved and thus presumably functional extends our knowledge of this important family of motor proteins. This work provides insights into the distribution and probable function of myosins including newly identified myosin classes.
Keywords: family, motor proteins, phylogeny
Introduction
Molecular motor proteins are mechanoenzymes associated with the cytoskeleton that powers much of the movement performed by living organisms. There are different motor proteins that coexist in every eukaryotic cell and they differ in the kind of filament they bind to, the direction in which they move along the filament, and their “cargo”.1 There are three major groups of motor proteins: kinesins and dyneins (that move along microtubules) and myosins (that move along actin filaments). The myosins are actin-based molecular motors and the amino acid sequences of myosin protein families are quite divergent in eukaryotes. The sequence identity of the motor domain could be lower than 20% among some members of the myosin family. In spite of the low sequence identity, the core tertiary structures of myosins are strongly preserved.2 They are characterized by a motor domain that binds to actin in an ATP-dependent manner, a neck domain consisting of varying numbers of IQ motifs, and an amino-terminal and carboxy-terminal domain of various lengths and functions. The known conserved sequence motifs, called structurally conserved regions (SCRs), were used as a prerequisite for a protein to be considered as a myosin.3
Abundant information is available about the structural, biochemical, and functional aspects of the motor domain of myosins.4 However, fewer studies have been performed on the phylogenetic relationships and evolutionary history of the myosin family.5,6 Some of these phylogenetic studies have been carried out by choosing exemplars of the functionally different classes of myosins, while others have focused on the phylogenetic relationships of restricted myosin clades. The structurally variable features of the myosins, such as sequence divergence, gene duplications, and length and domain architectures, might account for the continuous need for a comprehensive phylogenetic analysis of such a prolific family.7
Given the range of functions, structures, and interactions mediated by myosins, we report a survey of the abundance, protein architecture, conservation, and structure of gene products that contain sequence signatures of myosin domains across the entire proteome and myosin classes. An earlier study is consistent with many results presented here, and we now contribute an expanded structure and motor domain–centric analysis of myosins encountering new myosin families, with an emphasis on the scope of comparative analysis in these protein families.8 Our results should be useful for the structural and functional prediction by analogy for less well-characterized myosin classes.
Myosins have undergone marked diversification and domain rearrangements; the comparative study of these molecular motors offers great potential to untwine early eukaryote evolution.9 Other studies have attempted to infer inter-specific relationships specifically within the myosin family, though often with limited taxon sampling, and such analyses of few taxa can be subjected to strong systematic biases, which in turn produce high measures of repeatability (such as bootstrap proportions).10 In this article, however, we report a large-scale study in the entire non-redundant sequence database to present an unprecedented number of 4,064 putative myosins from different phyla, after certain filters such as the presence of SCRs and motor domain length, in agreement with known classical myosins. In addition, the myosin families are examined in the context of domain architectures and report discovery of ten novel and distinct classes of myosins, mainly from Oomycetes, Cellular slime molds, Choanoflagellates, Pelagophytes, Eustigmatales, Fonticula, and Eucoccidiorida. Due to the availability of large datasets, most of the discussion will focus on the fungal and novel myosin classes.
Results
Identification of myosin genes
To sketch the evolutionary history of the myosin gene family, we surveyed in those eukaryotic genomes for which complete or near-complete genomic data were publicly available. These organisms represent a wide taxonomic diversity of eukaryotes and encompass the major eukaryotic “supergroups.” To survey for myosins, we employed a Hidden Markov Model (HMM)-based sequence search strategy, starting from the head domain of classical myosins (please see Methods for details), to identify myosins using an E-value cutoff of 1e-57. This approach identified 6,044 encoded myosin-like protein sequences, which included sequence fragments (Supplementary File 1). To improve phylogenetic resolution, we considered only 4,064 of them that retain conserved myosin motifs in ATPase domain and the length of the domain was in agreement with the previously known myosin ATPase (head) domains.
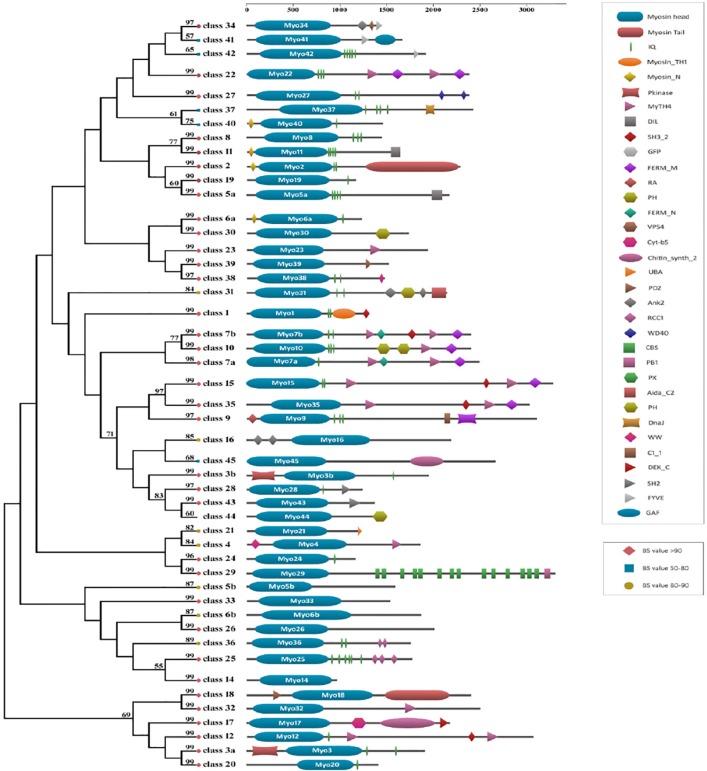
We aligned the 4064 head domains of the myosins and calculated a maximum likelihood phylogeny using RAxML to construct a global tree (Supplementary Fig. 1). We considered local bootstrap probability (LBP) and literature information of different functional groups of myosins to divide the global tree into different subtrees, and further, the subtrees were phylogenetically evaluated. The phylogeny for the myosin repertoire thereby includes members from 340 diverse eukaryote species. Members from each of the 35 myosin classes, defined previously by Odronitz and Kollmar (ie, myosin 1 to myosin 35), were also retrieved and included in phylogeny with strong bootstrap support (>50% BS value). A simplified phylogeny showing representative members from each class, along with their domain architecture, has been provided (Fig. 1).
Figure 1.
The maximum-likelihood trees were constructed based on the representative motor domain sequences selected from the global tree (Supplementary Fig. 1); bootstrap values (BS) above 50 are shown near the tree nodes with shapes and color.
Notes: Diamond node indicates BS value >90, circle node indicates BS value in the range of 80 to 90, and square node indicates BS value in the range of 50 to 80. The scale bar on the top represents the number of amino acids in each gene product as defined by the primary structure of the gene. The schematic representation of consensus domain architecture for each myosin class is mapped onto the tree.
Nomenclature and population of myosin classes
The previous study for the myosin superfamily reported 35 class, named myosin 1 to myosin 35.8 The phylogeny presented here confirms the monophyly of most of the previously known myosins to a large extent. Few myosin classes, like myosin 3, 5, and 6, have been split into multiple clades, with adjacent clusters consisting of members of other classes. Our inclusion of newly available sequences, from a more diverse range of organisms, undoubtedly also expands several preexisting families (Fig. 2). For example, the cluster corresponding to myosin 7 class, has now expanded to include previously unclassified sequences. More importantly, greater taxon sampling identifies new myosin groups, including ten novel myosin classes. In the following, by extension to the standard myosin nomenclature, we refer to these new families as myosin 36 to myosin 45. The sequence similarity between myosins of different classes was also plotted in the form of a heatmap (Fig. 3). These observations support the previously known details, like the similarity between classes 15 and 35, 8 and 11 and 7, 10, and 15. We also observe similar but new mega clusters involving classes 3b-9-16-28-43-43-45 and 22a-22b-34-41-42 (Supp. Fig. 1).
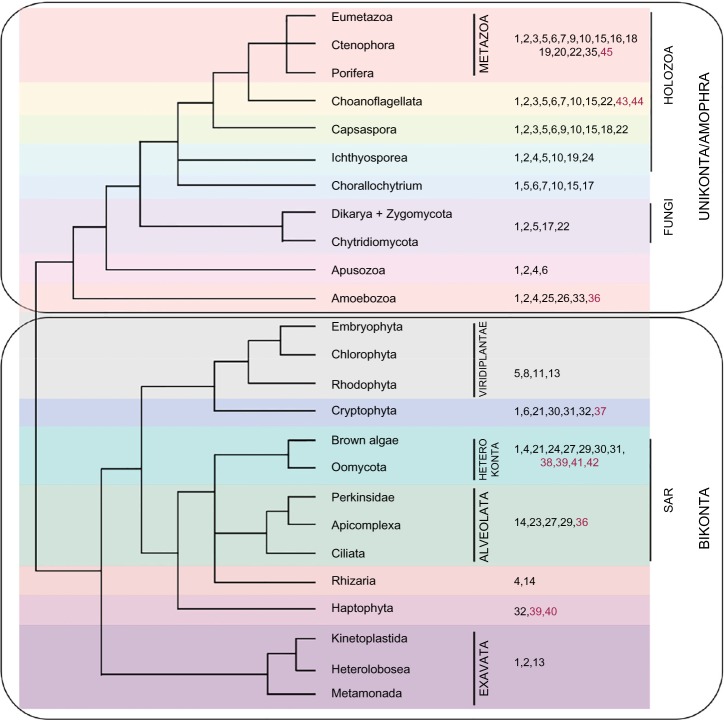
Figure 2.
Taxonomic distribution of myosin classes in eukaryotic genomes.
Notes: The numbers to the left indicate the myosin classes and the leaves of the tree represent the taxonomic clades. The kingdoms within Eukaryota have been represented in different colors. The previously known myosin classes have been colored in black and novel myosin classes are colored in brown. Tree topology has been adapted from the recent phylogenomic study (Derelle and Lang 2012).
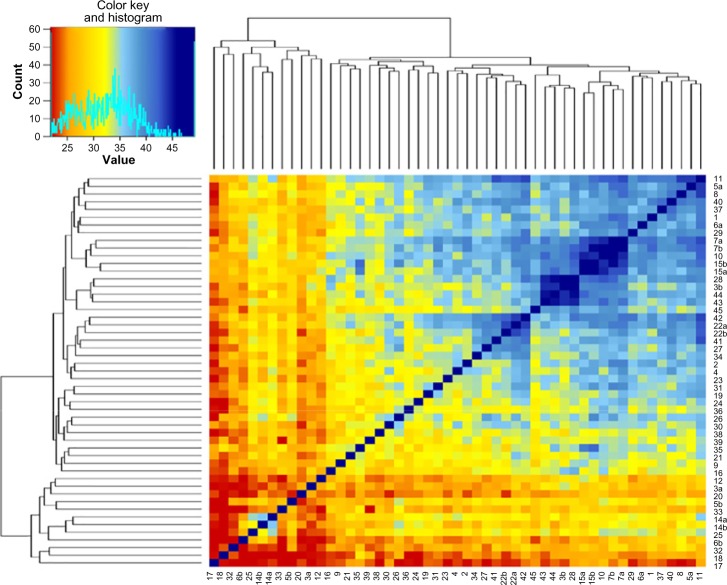
Figure 3.
A heat map showing the sequence identity between the motor domains of the myosin classes.
Notes: The color convention is indicated on the top left panel. The dendograms are derived from average sequence identity among the myosin classes. In the top left panel, the X axis represents the percentage identity among the 45 myosin classes and the Y axis indicates the counts of myosin classes, which fall in these sequence identity range. The cyan line indicates the actual average sequence identity value shown by the myosin classes.
Conservation of catalytically important residues in the motor domains of novel myosins
Sequence analysis within core motor domains of myosins classes shows class-specific insertions at nucleotide-binding regions analogous to other ATPases -the P-loop or Walker A and a Switch-II motif, which undergo large conformational changes in the contractile cycle and are highly conserved across all myosin classes and a Switch-I region, which are further variable.11 The phosphate-binding loop (P-loop) motif GESGAGKT is well conserved across the novel myosins, except for myosin Class-40 where it is replaced by GESGSGKT, similar to myosin classes 9, 12, 15, 16, 17, 20, and 21 (Supplementary Fig. 2). The switch-I region, which has a consensus LEAF-GNAKTIRNNNSSRFGK in conventional myosins, is more variable. In the switch-II region, LDIYGFE, is replaced with LDIFGFE (which occurs in most of the unconventional myosins, 4, 7, 9, 10, 14, 16, 21, 22, 26, 27, 28, 30, 31, 33, and 34) in almost all of the novel classes, except classes 41 and 45, which show a consensus of VDIFGFE and LDLS-GFE, respectively (Supplementary Fig. 3). Nucleotide state is communicated to the actin-binding region, and the lever arm and differences in the consensus sequences could indicate subtle variations that have evolved in nucleotide binding and dissociation affecting the kinetics of myosin motor movement across different classes.11
Diversification of myosins are achieved by diverse domain architecture of the genes that contain them
Motor proteins are generally composed of a motor head domain that converts chemical energy to force, and a range of additional domains that bind cargo, filaments or accessory proteins.12 Since regions outside of the motor head domain direct many interactions, considerable functional diversification might be achieved through the evolution of the protein domain combinations. To further investigate the diversification of the myosin family, we identified putative domain architectures for the initial dataset, of all 4064 identified, myosin proteins using Pfam,13 SMART,14 CDD,15 and Paircoil216 database searches. In total, we found 218 different myosin protein domain architectures for all identified myosins (individual domain architectures of each of the genes under analysis are available in (Supplementary Fig. 4).
Surprisingly, most such coexisting domains were specific only to particular taxa in our analysis. For instance, Membrane Occupation and Recognition Nexus (MORN) repeats in Haptophytes, actin-binding calponin homology domains were observed only in Oomycetes, and ubiquitin-associated domain (UBA) in Kinetoplastids, indicating that these domain combinations were relatively recent acquisitions (Supplementary Fig. 5). It is also noteworthy that few myosins in our analysis, like in myosin classes 14a, 33, and 26, possess no identifiable protein domains outside of the motor domain. This implies that the great majority of the interactions between these motors and other proteins is controlled either by poorly conserved stretches of peptide or protein domains that are not yet described in protein domain databases, for example, in myosin 14.17 It is possible to identify myosin-containing genes of unusual architectural forms by annotating the motor domain phylogeny with the protein domain architectures. The unusual domain architectures could be due to the result of secondary loss and gain of domains (Supplementary Figure 6). Accounting for these secondary loss events, 218 protein domain architectures, which were observed in multiple genomes, were specific to a paralogue or family on the myosin phylogeny, suggesting that they represent synapomorphic character state.
Myosin functional diversity
Our data show that there are robust phylogenetic patterns in terms of the abundance of members and varied domain architecture across myosin classes (Supplementary Fig. 4). Even within the same class, there is a diversity in coexisting domains, which might have major evolutionary significance and influence the topology of protein–protein interaction network (Fig. 4A). Some of the unusual domain architectures in the previously known myosin classes have been discussed in Supplementary Data.18–23 The former classes, 3, 5, 6, 7, 14, 15, and 22 members, have split into two clades and have distinct conserved motifs from each other. Myosin 3a is more similar in sequence to myosins 17, 33, 22, and 20 and myosin 3b to classes 9, 43, 44, 45, 28, and 16. Class-5b is closer to 6b than 5a. Class-7a and 7b are as closer to each other as they are to classes 10, 35, and 15. Class-14a is more similar in sequence to Class-25, and Class-14b is more similar to Class-12. Class-15a is closer to Class-35 than Class-15b. Classes 22a and 22b cluster with classes 27, 41, 34, and 42. Alignments of motor domains of such split myosin classes are provided in the link http://caps.ncbs.res.in/download/myosins/split_myosin_alignments.
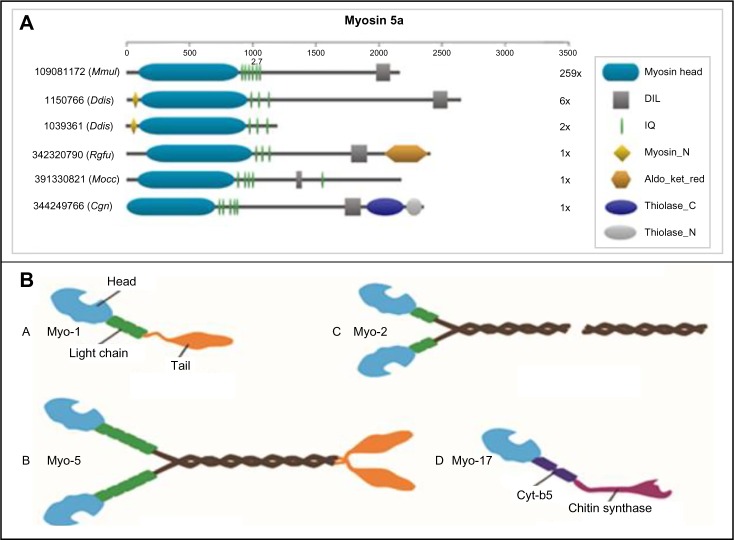
Figure 4.
(A) Diverse domain architecture observed in myosin Class-5a. The four-letter code refers to the following organisms – Mmul (Macaca mulatta), Ddis (Dictyostelium discoideum), Rglu (Rhodotorula glutinis), Mocc (Metaseiuluso ccidentalis), and Cgri (Cricetulus griseus). The gene identifiers are also indicated in the figure. The numbers next to the domain architectures indicate the number of sequences with that domain architecture. (B) Cartoon representation of fungal myosin classes. All myosins share a motor domain on their heavy chains at the amino-terminus (the “head” domain), but they differ in IQ motifs considerably and in domain composition. Certain myosins are known to exist as dimers. The Class-2 fungal myosins have a long coiled-coil region, so they have been represented using a break. Please refer to Supplementary Figure 5 for representative domain architectures for fungal myosins belonging to each of these classes.
Fungal myosins
A total of 1,365 myosins belonging to the four myosin families 1, 2, 5, and 17 were identified in the 244 fungal genomes for which complete or near-complete genomic data are available. Figure 4B shows a cartoon representation of these fungal myosins showing their domains and dimerization status.
Microsporidia are obligate, spore-forming unicellular parasites with a wide host range and they are most closely related to Fungi, despite obvious differences in existence and biology.19 Although other fungal myosins belong to classes 1, 2, 5a, and 17, the distantly related Microsporidia taxa reveal only two types of myosins: Class-2 and the subclass 5b. The subclass was named as 5b because of co-clustering of few members of the previous myosin Class-5 and similar domain architecture (Mysc, IQ repeats, coiled-coil, and C-terminal DIL domain). However, we also observe less diverse domain architecture in fungal myosins; even though the core domain content is conserved, the data clearly show progressive changes and expansions of individual myosin class domain architectures (Supplementary Fig. 7). Foth et al had identified orphan myosin sequences in Encephalitozoon cuniculi and Encephalitozoon intestinalis. Now the class has expanded to include sequences from Encephalitozoon hellem, Encephalitozoon romaleae, Antonospora locustae, Nosema ceranae BRL01, Vittaforma corneae ATCC 50505, Enterocytozoon bieneusi H348, and Spraguea lophii 42_110. In our maximum likelihood analysis, we have observed this monophyletic association and the unclassified sequences of 5b fall in close proximity to myosin 1, 2, and 5a, compared to myosin 17 with good statistical support (Fig. 5).
Figure 5.
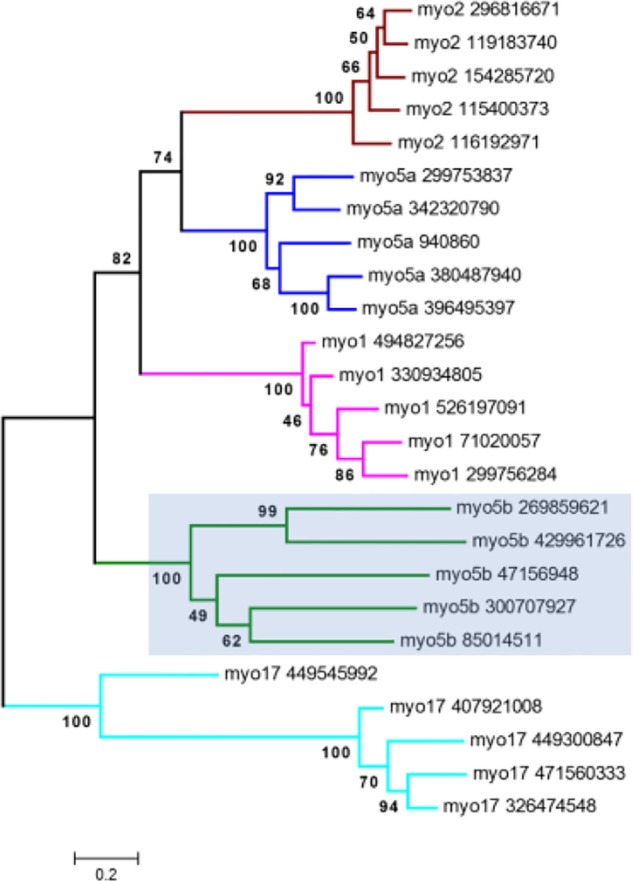
A phylogram of representative fungal myosins created by the maximum-likelihood method based on 1,000 replicates. Bootstrap values are given at the nodes.
Excavate myosins
Myosin 21 was specific to metazoans and retain the Pkinase domain and IQ repeats as coexisting domains. This myosin class now has 49 sequences, is also seen in Excavates, Kinetoplastida, and Heterolobosa and shows additional domains like the C-terminal UBA domain, IQ repeats, and an N-SH3 domain. Myosin 33 is Trypanosoma specific and does not have any associated domains.
Apicomplexan myosins
Myosin 14 has a single-motor domain. Class-14a is found in Apicomplexans and has a GESGAGKT motif, while myosin 14b is found in Plasmodium and Paramecium and has a GESGSGKT motif. One sequence (Paramecium tetraurelia) has IQ repeats and the RCC_1 domain; another sequence has a Vsp54 along with these two domains.
Yusuf et al, 2015 studied the PfMYOB isoform of Class-14 myosin in Plasmodium falciparum. PfMYOB associates with a light chain called MyoB light chain (MLC-B). The additional light chain ELC (predicted)- and MLC-B (experimentally verified)-binding regions have been identified in the neck regions of the myosin sequences, PfMYOA, PfMYOB, and TgMYOA. No domains have been identified in the neck region. The authors proposed that for the short Class-14 myosins that lack a tail region, atypical myosin light chains like MLC-B may fulfill that role.17
Classes 23 to 27 were represented only by Apicomplexan species. Myosin 23 has a MyTH4 domain. Apart from Apicomplexa, myosin 24 sequences are found in Oligohymenophorea, Spirotrichea, and Rattus norvegicus and consist of IQ repeats. Myosin 25 has IQ and RCC1 (related regulator of chromosome condensation 1) repeats. One sequence from Theileria orientalis has an N-terminal SH3 domain. Myosin 26 has no additional (coexisting) domains, while myosin 27 has IQ and WD40 repeats. WD40 domains may target to centrosomes or the nucleolus or bind phosphoinositides. Some sequences have additional domains like Ras (Perkinsus marinus) and Miro-mitochondrial Rho-GTPase (Plasmodium yoelii).
Identification of novel myosin classes and their putative function
The novel class myosin 36 form a separate cluster with a bootstrap value of 75 next to myosin classes 14, 25, and 11 in the phylogenetic tree (Supplementary Fig. 1). While the Class-25 cluster has Apicomplexan myosins, Class-21 is specific to metazoans and excavates; this new class (myosin 36) is found in members of Amoebozoal phyla Dictosteliida, and the ciliates Oligohymenophorea (Tetrahymena and Paramecium) and Spirotrichea (Supplementary Fig. 1). Some of the sequences from Dictyostelium, Paramecium, and Tetrahymena were reported as orphan myosins by Odronitz and Kollmar (2007). The neighboring Class-14 has only a single motor domain, Class -25 has RCC1 repeats, and Class-21 has Pkinase and IQ repeats. The domain architecture in Class-36 is diverse – two IQ repeats followed by two to four RCC1 repeats (similar to Class-25), MyTH4 domain followed by a FERM domain or RhoGEF alone or co-occurring with IQ, PH, or N-terminal SH3 domain. Both RCC1 and RhoGEF are guanidine exchange factors and could be involved in the regulation of the cellular signaling events. The consensus P-loop motif sequence is GESGAGKT. Some Dictyostelium sequences have GESGSGKS instead of the consensus P-loop motif and a Tetrahymena thermophila sequence shows the GESGTGKT (Supplementary Table 1) motif in that position.
The members in the newly found class, myosins 37, cluster with myosin Class-4 (seen in lower eukaryotes) and Class-31 (specific to Oomycete, Pelagophyte, and Phaeophycae) with a bootstrap value of 55. They are specific to the Cryptophytes Guillardia theta CCMP2712 (5 sequences) and the red alga Galdieria sulphuraria. Myosin 4 has the consensus domain architecture of the motor domain, sometimes followed by MyTH4 and SH3 or Ank and preceded by N-SH3 or WW domains. In the case of Class-31 myosins, IQ motifs, and two Ankyrin repeats interrupted by a PH domain, an Aida_C2 (axin interaction dorsal-associated) domain in the carboxy terminus could be identified. In myosin 37, however, the Guillardia sequences consist of only the motor domain, perhaps due to incomplete gene models. The Galdieria sequence has five IQ repeats and a DnaJ domain and was reported as an orphan sequence in the study by Odronitz and Kollmar. Since Galdieria is a thermoacidophile, living in acidic spring habitats, the chaperone DnaJ domain, which associates with Hsp 70, could help in the folding of unfolded proteins at high temperatures. Out of the six sequences, three have GESGAGKT as the P-loop motif and other three have GESGSGKT instead.
The myosins of Class-38 cluster next to myosin Classes 6a, 30, and another new myosin class, (Class-39) with a significant bootstrap value of 100. They are found in Oomycetes, Bacillariophyta, Pelagophyceae, and Phaeophyceae, sharing a similar phylogenetic distribution as Class-30 myosins, whereas Class-6a myosins are seen only in Apicomplexans. Myosin 6a has the motor domain only, while some sequences have N-terminal SH3 or IQ domains. Myosin 30 sequences have either of the Phosphatidylinositol-binding domains PH (Pleckstrin homology) domain or PX (phagocytic oxidase) domain, sometimes with IQ repeats. One sequence from the alga (Aureococcus anophagefferens) has the protein-binding WW domain and the PH domain. The Class-38 sequences show various domain compositions. The domain architecture, containing up to 11 IQ repeats, is sometimes characterized by the WW domain or PX domain or an N-terminal CH domain and is similar to myosin 30. Two sequences show only motor and Tub domains, one sequence has an N-terminal PcF and a PX domain, whereas another sequence has an N-terminal SH3-like domain and an IQ motif (Supplementary Fig. 2). A Thalassiosira pseudonana myosin shows a variant Walker motif (GILGAGKS, different from the consensus GESGAGKT) in its sequence.
Myosin Classes 39 and 40 are species-specific myosins. Class-39 myosins occur in the Isochrysidale (Emiliania huxleyi CCMP1516) and cluster closer to classes 38 and 30 at a bootstrap of 62. One sequence has a Tub domain, one has an IQ motifs, and two have PDZ domains. All sequences have GESGAGKT at the P-loop site. Myosins belonging to Class-40 cluster separately (at a bootstrap value of 100) next to myosin class 28 and the classes 38, 30, 39, and 6a share a common origin in the phylogenetic tree and are found only in the Cryptophytes-Guillardia theta CCMP2712. There are five sequences, two of which have a single IQ motif and one has an N-terminal SH3-like domain. This class has a variant Walker motif (GDSGSGKT), whereas one sequence has the exact GESGAGKT at the site.
Myosins in 41 and 42 classes are phylogenetically closer to myosin 22 and 34 (with bootstrap value 85 and 92, respectively). While myosin 23 is found in Phaeophyceae and Bacillariophyta, myosins of classes 22, 41, and 42 occur in Oomycetes. Myosin 34 contains IQ and Ank repeats followed by FYVE as coexisting domains and myosin 22b has IQ and other associated domains like Ank, Pkinase, and adh short C2 domains. The novel class domain architectures are similar to those of Class-34 myosins. Class-41 has two distinct domain architectures – Myosin head followed by FYVE and GAF domain and the other with the myosin head followed by two to four IQ repeats, with or without PDZ domain. Five sequences have GESGAGKT, four have GESGSGKT and one has GESGTGKT in the P-loop ATP-binding site. Class-42 contains the myosin motor domain, followed by FYVE and sometimes interspersed with IQ motifs. Since all the associated domains are known to interact with cell-signaling components, these myosins could be localized in the cell membrane and they have a role in cell signaling. All sequences in Class-42 have a GESGAGKT motif, except two Phytophthora sequences, which have GESGSGKT instead.
The myosin classes 43, 44, and 45 cluster with classes 3b, 28 and 16 with bootstrap values of 58, 60, and 87 respectively. Some of the sequences are among the Choanoflagellate orphans reported by Odronitz and Kollmar.8 Myosins belonging to classes 3b, 9, 16, and 28 are seen only in metazoans. Myosins of classes 43 and 44 are unique to Choanoflagellates and the former has SH2 domain, which is similar to myosin 28 domain architecture (IQ and SH2), whereas the latter has PH, SH2, or Mcp5_PH domains co-occurring with the motor domain. Since PH is involved in cell signaling and SH occurs in membrane kinases, these myosins could also be localized to cell membrane and be involved in cell signaling. The other myosins in the cluster have different domain architectures than the ones seen in the two novel classes: N-terminal Pkinase-IQ (Myosin 3); RA-IQ- C1-RhoGAP (Myosin 9); N-terminal Ank -NYAP (Myosin 16) and IQ-SH2 (Myosin 28). All the sequences in both these classes have a consensus P-loop motif of GESGAGKT. Class-45 myosins are found in molluscs and annelids and are associated with Glucosyl transferase and chitin synthase domains like the fungal myosins. These animals have either a chitinous exoskeleton (Mollusca) or chitin bristles in the exterior surface (Capitella telata). One of these sequences is from the mollusc, Atrina rigida and was reported in the Odronitz and Kollmar (2007) paper8 as an orphan myosin. One sequence from the mollusc, Crassostrea gigas, has neither of the fungal myosin-associated domains but has a MH2 domain instead. All the sequences have a Walker motif of GESGAGKT, with the exception of one (Mizuhopecten yessoensis myosin that contains an equivalent – GNSGAGKT motif).
Orphan myosins
The orphan myosins could not be classified since they were less in number (from single to five sequences) and clustered separately in the phylogeny with high bootstrap values. There are 38 myosin sequences that are not affiliated to any of the 45 classes. They are 17 from Choanoflagellates (Salpingoeca rosetta and Monosiga brevicollis MX1), nine from Haptophytes (all from Emiliania huxleyi), three from animals (Amphimedon queenslandica, Nematostella vectensis, and Clonorchis sinensis), two from Pelagophytes (Aureococcus anophagefferens), two from Cryptomonads (Guillardia theta CCMP2712), two from Capsaspora (Capsaspora owczarzaki ATCC 30864), one from brown algae (Ectocarpus siliculosus), one from Oomycetes (Saprolegnia diclina VS20), and one is found in Longamoebia (Acanthamoeba castellanii str. Neff). One of the M. brevicollis myosins was identified previously as an orphan. The N. vectensis orphan was previously classified as a Class-15 myosin.4 The E. huxleyi myosins are interspersed between myosins from classes 6a, 38, 39, 30, and 40 in the phylogenetic tree.
The motor domains of Choanoflagellate and Haptophyte orphan myosins have distinct sequences from the other myosins. This illustrates the sequence diversity and evolutionary divergence of myosins from these phyla. As lower extant genomes are analyzed, further divergent myosins can be expected. Some orphan myosins have unusual domains like Ribonuc_l-PSP (E. huxleyi), DUF4339 (S. diclinica), Y_phosphatase (M. brevicollis), Mcp5_PH (E. huxleyi), Rap_ GAP (C. owczarzaki), and MH2 (S. rosetta). The detailed diverse domain organization of orphan myosins is shown in Supplementary Fig. 8.
Conclusions
Recently, genome sequencing projects have allowed the identification of the precise number of myosin genes in major eukaryotic species and have remarkably expanded the taxonomic range of known myosins. With more than 4,000 proteins analyzed and a reasonable representation of the different myosin paralogues in the key groups of living organisms, the recovered phylogeny confirms and generalizes previous analyses that were carried out with decreased and taxonomically more limited data sets. In the present study, we reconstructed a myosin family phylogeny taking into account the new genomic information. Starting from 378 classical myosin sequences and their ATPase domains, sensitive computational searches using HMMs against the entire nonredundant database of sequences, gave rise to a broader understanding of putative myosin types in the whole taxa of life. The hits obtained from the initial search, followed by computational validations, permitted us to accept 4,064 myosin-containing gene products for our starting analysis. In our analysis, the most primitive myosins are from Dictyostelium. The classification and composition of the known myosin families from our analysis agree very well with previously reported classifications.17
Various analyses such as taxanomic distribution, expansions of existing myosin families, and the realization of new myosin families were possible. We can reiterate about the new myosin families here. Unlike previous analyses reported by other groups, our searches were not taxon-restricted. Our analysis of domain architectures of myosin-containing genes also did not assume one domain architecture for each myosin family. Indeed, this provides a perception of gain or loss of domain architectures across the eukaryotic kingdoms. Certain domain architectures are highly preferred. The classical domain architecture of a myosin containing a head-domain, few IQ repeats, followed by a coiled-coil and cargo-binding domain is too idealistic and examples are slowly emerging in the literature of exceptions to such a domain architecture. Our data show considerable diversity of domain architecture among the classes, for example, myosin 5a reinforcing the notion that individual myosin classes have undergone dynamic evolution. This also indicates that domain-level classification, which secludes the information encoded in the accessory domains, might not be the appropriate approach to understand evolutionary relationship for such a diverse motor family and one should take into account the full-length sequence, consolidating the complete information to comprehensively understand the evolutionary forces that shape the existence of these protein functional domains and domain combinations in each species. For example, myosin 6 does not contain a long-length, coiled-coil domain.24 It is likely that the presence of a small fraction of coexisting domains and motifs has remained unnoticed, since the large analysis and automatic searches require stringent thresholds. Our report is attempting to aim a broad bioinformatics overview of all possible putative myosins in the entire universe so that several new, hitherto uncharacterized interesting variations of myosin families and myosin family members can be adopted for detailed structure-function characterization. This work also provides a framework and insights into the distribution and likely function of myosins including newly identified myosins supported by phylogenetic inferences and available experimental data, for revealing the relationships and functions.
Materials and Methods
Sequence searches and alignments
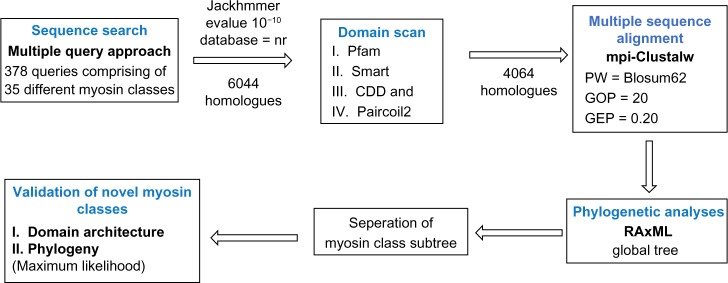
Sequence search for myosins was performed using the standalone JACKHMMER25 program at an E-value of 10−10 against the complete non-redundant protein database (NCBI) as of March 13, 2013, release 58. We started our search with 378 myosin queries that belong to functionally different groups (to find similar sequences in the database).8 The sequence corresponding to only the ATPase domain was considered for sequence searches, as per previous work.19 Hits obtained from such sequence searches were included in the myosin family for further analysis if the sequence identities with the query were above 20%, key conserved myosin motifs in ATPase domain could be observed, and the length of the head domain of the hit was in agreement with previously known myosins (Fig. 6).26 Only those sequences that had a motor domain as verified using standalone PfamScan at E-value of 10−2 were considered.
Figure 6.
Workflow of sequence search and validation. In all, 378 myosins from 35 classes (Odronitz and Kollmar et al., 2007) were taken as queries for a Jackhmmer search against the NR database. The hits were filtered for the presence of functional (ATP-binding) motifs to obtain 4,064 sequences. The hits were aligned using the indicated parameters in Clustal-W and a phylogenetic tree was constructed using RAxML.
Protein sequences were aligned using CLUSTALW version 1.7, BLOSUM series scoring matrix and a gap penalty mask based on the aligned secondary structures of the myosins whose crystal structures were known.27 Where required, minor manual corrections were performed on the alignment based on the knowledge of myosin motifs. The same strategy was employed for each separate alignment of the different myosin groups (see below).
Phylogenetic analyses
Phylogenetic analyses based on protein sequences were carried out using the maximum-likelihood method with the RAxML software (Version 7.4.2) with 1,000-bootstrap replicates.28 The number of sequences is the most important limiting factor when an exhaustive phylogenetic analysis is attempted under the maximum-likelihood principles. Since the data size of the aligned set of sequences was large, we first reconstructed a global tree based on the protein sequences with the kinesin motor domain as the outgroup. The steps for obtaining the global tree were (1) selection of best-fit models of amino acid replacement for the data (2) calculation of a maximum-likelihood distance matrix with the RAxML program under the BLOSUM model, and (3) the use of the resulting tree topology as a seed to search for a topology with a higher likelihood value using the same amino acid substitution model.
We then divided the global tree into subtrees following two criteria: an LBP greater than 50% and literature information on different functional groups of myosins. The sequences composing each group were aligned using MUSCLE (with iterative refinement).29 We selected the myosin 1 sequence, which is widely distributed across eukaryotes as an outgroup for the purpose of tree representation in other myosin class–specific trees. In groups of more than five sequences, we followed the same strategy as that used in estimating the global tree. A majority rule of LBP 50% was established for each node in every subtree; unsupported nodes were excluded and their branches forced to yield polytomies.
Domain architecture
All the gene products, containing at least one putative myosin head domain, were then searched against the protein conserved database CDD,15 SMART,14 and Pfam database13 at an E-value of 10−2, to identify and classify coexisting domains. The prediction of coiled-coils was inferred separately using the PAIRCOIL2 program with P-score cutoff of 0.025.16 Protein domain identification is limited by the sensitivity of the search and the diversity of protein domains in the database. CDD, SMART, and Pfam were used in combination to increase both the sensitivity and the protein diversity.
Gene ontology (GO) annotation
Sensitive sequence searches revealed 134 novel myosins with signatures of functional motifs. These sequences were further validated for the presence of functional motifs and GO annotations using Blast2GO30 with default parameters. The details are described in Supplementary Table 2.
Supplementary Material
The consensus domain architecture, taxonomic distribution and expansion, and unusual domain architecture of the classical myosins have been discussed in detail.
Supplementary Figure 1. ML tree of myosin head domains. The tree is collapsed at key nodes and rooted using the midpoint-rooted tree option. Myosin classes are indicated. Nodal support was obtained using RAxML with 1,000 bootstrap replicates. Different colors have been used to indicate different myosin classes. The arrows indicate the direction in which the phylogenetic tree has to be read.
Supplementary Figure 2. Alignments of the ATP-binding conserved regions in the different myosin classes. The residues that are completely conserved are shaded in black. The green box in the alignment indicates the motif region. The consensus sequence is based on MultAlin31: uppercase is identity, lowercase is consensus level >0.5, ! is any one of the amino acids I or V, $ is anyone of L/M, % is anyone of F/Y, and # is anyone of NDQEBZ. Lowercase is consensus level > SimilarityGlobalScore if S, M, or E are used as Similarity Type. Please see Supplementary Fig. 3 for alignment of all the novel myosin classes.
Supplementary Figure 3. Alignments of the ATP- binding conserved regions within the novel myosin classes. The residues that are completely conserved are shaded in black. The consensus sequence is based on MultAlin: uppercase is identity, lowercase is consensus level >0.5, !is anyone of IV, $ is anyone of LM, % is anyone of FY, and # is anyone of NDQEBZ. Lowercase is consensus level >SimilarityGlobalScore if S, M, or E are used as Similarity Type
Supplementary Figure 4. Diversity in domain architectures observed among each myosin classes. Pfam, CDD, and SMART searches were used to identify putative gene architectures. The gene identifier and four-letter code are elaborated at the end.
Supplementary Figure 5. Diagram of protein domain composition of myosins in different eukaryotic groups. The lineage-specific Pfam domains are illustrated. Darker circles denote a frequency of occurrence in more than five sequences and lighter circles indicate a lesser frequency. Detailed information is provided in Supplementary Figure 6.
Supplementary Figure 6. Detailed diagram of protein domain composition of myosins in different eukaryotic groups. The lineage-specific Pfam domains are illustrated. Darker circles denote a frequency of occurrence in more than five sequences and lighter circles indicate a lesser frequency.
Supplementary Figure 7. The domain-tree view of diverse domain architectures observed in different fungal myosin classes. Schematic representation of domain architecture for each myosin is mapped onto the tree. The tree is based on the distance matrix computed by Jaccard similarity coefficient using the DoMosaic tool. Values marked above the branches refer to Jaccard index.
Supplementary Figure 8. Domain Architecture of the orphan myosins identified in our study. The four-letter code refers to the following organisms: Nvec (Nematostella vectensis), Mbre (Monosiga brevicollis MX1), Esil (Ectocarpus siliculosus), Csin (Clonorchis sinensis), Cowc (Capsaspora owczarzaki ATCC 30864), Acas (Acanthamoeba castellanii str. Neff), Ehux (Emiliania huxleyi CCMP1516), Sros (Salpingoeca rosetta), and Sdic (Saprolegnia diclina). The gene identifiers are also indicated in the figure.
Supplementary File 1. The fasta file of myosin head domains used to compute the phylogenetic tree shown in Supplementary Figure 1.
Supplementary Table 1. The gene identifier, accession Id, organism name, taxa Id, and P-loop motif for all the novel myosin sequences are provided. The sequences having differences in the GESGAGKT motif are highlighted.
Supplementary Table 2. Mapping of Gene Ontology terms (cellular component, molecular function, and biological process) for all the novel myosin domains.
Footnotes
ACADEMIC EDITOR: Liuyang Wang, Associate Editor
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 2,943 words, excluding any confidential comments to the academic editor.
FUNDING: The work was supported by the Human Frontier Science Program (Grant No: HFSP/RGP0054/2009-C). NSP is currently supported by Centre of Excellence (Grant No. BT/01/COE/09/01) funded by Department of Biotechnology, India. We thank the Tata Institute of Fundamental Research (TIFR) for funding IM. The authors thank NCBS (TIFR) for infrastructural facilities. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived the study: RS. Constructed the workflow: SNP. Performed the sequence searches: SNP. Performed analysis: SNP, IM. Contributed to the writing of the manuscript: SNP, IM, RS. Made critical revisions and approved the final version: RS. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496(1):3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 2.Cope MJ, Whisstock J, Rayment I, Kendrick-Jones J. Conservation within the myosin motor domain: implications for structure and function. Structure. 1996;4(8):969–87. doi: 10.1016/s0969-2126(96)00103-7. [DOI] [PubMed] [Google Scholar]
- 3.Huang IK, Pei J, Grishin NV. Defining and predicting structurally conserved regions in protein superfamilies. Bioinformatics. 2013;29(2):175–81. doi: 10.1093/bioinformatics/bts682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson RF, Langford GM. Myosin superfamily evolutionary history. Anat Rec. 2002;268(3):276–89. doi: 10.1002/ar.10160. [DOI] [PubMed] [Google Scholar]
- 5.Goodson HV, Spudich JA. Molecular evolution of the myosin family: relationships derived from comparisons of amino acid sequences. Proc Natl Acad Sci U S A. 1993;90(2):659–63. doi: 10.1073/pnas.90.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sebé-Pedrós A, Grau-Bové X, Richards TA, Ruiz-Trillo I. Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome Biol Evol. 2014;6(2):290–305. doi: 10.1093/gbe/evu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allaby RG, Woodwark M. Phylogenetics in the bioinformatics culture of understanding. Comp Funct Genomics. 2004;5:128–46. doi: 10.1002/cfg.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8(9):R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436(7054):1113–8. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- 10.Hedtke SM, Hillis DM. Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol. 2008;46:239–57. [Google Scholar]
- 11.Sweeney HL, Houdusse A. Structural and functional insights into the Myosin motor mechanism. Annu Rev Biophys. 2010;39:539–57. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 12.Schuster M, Treitschke S, Kilaru S, Molloy J, Harmer NJ, Steinberg G. Myosin-5, kinesin-1 and myosin-17 cooperate in secretion of fungal chitin synthase. EMBO J. 2011;31(1):214–27. doi: 10.1038/emboj.2011.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn RD, Bateman A, Clements J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40(Database issue):D302–5. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchler-Bauer A, Lu S, Anderson JB, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database issue):D225–9. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22(3):356–8. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 17.Yusuf NA, Green JL, Wall RJ, et al. The Plasmodium class XIV Myosin, MyoB, has a distinct subcellular location in invasive and motile stages of the malaria parasite and an unusual light chain. J Biol Chem. 2015;290(19):12147–64. doi: 10.1074/jbc.M115.637694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syamaladevi DP, Sowdhamini R. Evolutionary traces decode molecular mechanism behind fast pace of myosin XI. BMC Struct Biol. 2011;11(1):35. doi: 10.1186/1472-6807-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keeling PJ, Slamovits CH. Simplicity and complexity of microsporidian genomes. Eukaryot Cell. 2004;3(6):1363–9. doi: 10.1128/EC.3.6.1363-1369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang G, Cowled C, Shi Z, et al. Comparative analysis of bat genomes. Science. 2013 Jan;339:456–60. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanner F, Sorensen CM, Holstein-Rathlou NH, Peti-Peterdi J. Connexins and the kidney. Am J Physiol Regul Integr Comp Physiol. 2010;298(5):R1143–55. doi: 10.1152/ajpregu.00808.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu KC, Jacobs DT, Dunn BD, Fanning AS, Cheney RE. Myosin-X functions in polarized epithelial cells. Mol Biol Cell. 2012;23:1675–87. doi: 10.1091/mbc.E11-04-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J Cell Sci. 2000;113(pt 19):3439–51. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- 24.Spudich JA, Sivaramakrishnan S. Myosin VI: an innovative motor that challenged the swinging lever arm hypothesis. Nat Rev Mol Cell Biol. 2010 Feb;11:128–37. doi: 10.1038/nrm2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Syamaladevi DP, Kalaimathy S, Pasha N, Subramonian S, Sowdhamini R. A three-step validation following genome-wide data mining for myosin family members improves search efficiency; 2011 IEEE 11th International Conference on Data Mining Workshops; New York. 2011. pp. 1071–4. [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 29.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conesa A, Götz S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;2008:1–12. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(10):4465–82. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. ML tree of myosin head domains. The tree is collapsed at key nodes and rooted using the midpoint-rooted tree option. Myosin classes are indicated. Nodal support was obtained using RAxML with 1,000 bootstrap replicates. Different colors have been used to indicate different myosin classes. The arrows indicate the direction in which the phylogenetic tree has to be read.
Supplementary Figure 2. Alignments of the ATP-binding conserved regions in the different myosin classes. The residues that are completely conserved are shaded in black. The green box in the alignment indicates the motif region. The consensus sequence is based on MultAlin31: uppercase is identity, lowercase is consensus level >0.5, ! is any one of the amino acids I or V, $ is anyone of L/M, % is anyone of F/Y, and # is anyone of NDQEBZ. Lowercase is consensus level > SimilarityGlobalScore if S, M, or E are used as Similarity Type. Please see Supplementary Fig. 3 for alignment of all the novel myosin classes.
Supplementary Figure 3. Alignments of the ATP- binding conserved regions within the novel myosin classes. The residues that are completely conserved are shaded in black. The consensus sequence is based on MultAlin: uppercase is identity, lowercase is consensus level >0.5, !is anyone of IV, $ is anyone of LM, % is anyone of FY, and # is anyone of NDQEBZ. Lowercase is consensus level >SimilarityGlobalScore if S, M, or E are used as Similarity Type
Supplementary Figure 4. Diversity in domain architectures observed among each myosin classes. Pfam, CDD, and SMART searches were used to identify putative gene architectures. The gene identifier and four-letter code are elaborated at the end.
Supplementary Figure 5. Diagram of protein domain composition of myosins in different eukaryotic groups. The lineage-specific Pfam domains are illustrated. Darker circles denote a frequency of occurrence in more than five sequences and lighter circles indicate a lesser frequency. Detailed information is provided in Supplementary Figure 6.
Supplementary Figure 6. Detailed diagram of protein domain composition of myosins in different eukaryotic groups. The lineage-specific Pfam domains are illustrated. Darker circles denote a frequency of occurrence in more than five sequences and lighter circles indicate a lesser frequency.
Supplementary Figure 7. The domain-tree view of diverse domain architectures observed in different fungal myosin classes. Schematic representation of domain architecture for each myosin is mapped onto the tree. The tree is based on the distance matrix computed by Jaccard similarity coefficient using the DoMosaic tool. Values marked above the branches refer to Jaccard index.
Supplementary Figure 8. Domain Architecture of the orphan myosins identified in our study. The four-letter code refers to the following organisms: Nvec (Nematostella vectensis), Mbre (Monosiga brevicollis MX1), Esil (Ectocarpus siliculosus), Csin (Clonorchis sinensis), Cowc (Capsaspora owczarzaki ATCC 30864), Acas (Acanthamoeba castellanii str. Neff), Ehux (Emiliania huxleyi CCMP1516), Sros (Salpingoeca rosetta), and Sdic (Saprolegnia diclina). The gene identifiers are also indicated in the figure.
Supplementary File 1. The fasta file of myosin head domains used to compute the phylogenetic tree shown in Supplementary Figure 1.
Supplementary Table 1. The gene identifier, accession Id, organism name, taxa Id, and P-loop motif for all the novel myosin sequences are provided. The sequences having differences in the GESGAGKT motif are highlighted.
Supplementary Table 2. Mapping of Gene Ontology terms (cellular component, molecular function, and biological process) for all the novel myosin domains.