Abstract
Tyrosine sulfation is a post-translational modification that facilitates protein-protein interaction. Two sulfated tyrosines (Tys173 and Tys177) were recently identified within the second variable (V2) loop of the major HIV-1 envelope glycoprotein, gp120, and shown to contribute to stabilizing the intramolecular interaction between V2 and the third variable (V3) loop. Here, we report that tyrosine-sulfated peptides derived from V2 act as structural and functional mimics of the CCR5 N-terminus and potently block HIV-1 infection. Nuclear magnetic and surface plasmon resonance analyses indicate that a tyrosine-sulfated V2 peptide (pV2α-Tys) adopts a CCR5-like helical conformation and directly interacts with gp120 in a CD4-dependent fashion, competing with a CCR5 N-terminal peptide. Sulfated V2 mimics, but not their non-sulfated counterparts, inhibit HIV-1 entry and fusion by preventing coreceptor utilization, with the highly conserved C-terminal sulfotyrosine, Tys177, playing a dominant role. Unlike CCR5 N-terminal peptides, V2 mimics inhibit a broad range of HIV-1 strains irrespective of their coreceptor tropism, highlighting the overall structural conservation of the coreceptor-binding site in gp120. These results document the use of receptor mimicry by a retrovirus to occlude a key neutralization target site and provide leads for the design of therapeutic strategies against HIV-1.
Keywords: HIV-1, Envelope, Trimer, Coreceptors, Molecular mimicry, Peptides, Inhibitors, Immune evasion
Highlights
-
•
Tyrosine-sulfated peptides derived from the V2 domain of HIV-1 gp120 mimic the N-terminal domain of the CCR5 coreceptor.
-
•
Tyrosine-sulfated V2 peptides are potent and broad-spectrum inhibitors of HIV-1 infection.
Understanding how HIV-1 protects its outer envelope from the immune system may help devise effective strategies for treatment and vaccine. We derived synthetic peptides from the V2 loop of the external HIV-1 envelope glycoprotein, gp120, which contains sulfate-modified tyrosines that contribute to maintaining the envelope in an antibody-protected configuration. We found that these peptides mimic the structure and function of CCR5, a key cellular coreceptor for HIV-1, interacting with and occluding a major CCR5-binding site in gp120. Tyrosine-sulfated V2 peptides potently block HIV-1 entry and may serve as templates for the design of new antiviral inhibitors.
1. Introduction
Progressive refinement of the molecular anatomy of the native HIV-1 envelope spike, which represents the sole target of neutralizing antibodies, is accruing critical information for the rational design of effective inhibitors and vaccine immunogens (Burton et al., 2012, Mascola and Haynes, 2013, Wyatt and Sodroski, 1998). The envelope spike displayed on the surface of infectious virions is a homotrimer of gp120:gp41 heterodimers (Wyatt and Sodroski, 1998). Due to inherent difficulties in crystallizing the native envelope trimer, most high-resolution structures were initially obtained with monomeric gp120 stabilized in complex with soluble CD4 (sCD4) and/or specific monoclonal antibodies (mAb) (Chen et al., 2009, Huang et al., 2005, Huang et al., 2007, Kwon et al., 2012, Kwong et al., 1998, Kwong et al., 2000, Pancera et al., 2010). Recently, important progress has been made with the design of soluble, cleaved, disulfide bridge-stabilized gp140 trimers (SOSIP.664), which display several features of the pre-fusion envelope spike (Sanders et al., 2013), although the stabilizing mutations were shown to abrogate fusion competence and to induce some alterations of the antigenic profile (Alsahafi et al., 2015). The structure of several SOSIP.664 trimers has been resolved by crystallization or high-resolution cryogenic electron microscopy (cryoEM), providing valuable information on the pre-fusion conformation of the envelope trimer (Bartesaghi et al., 2013, Julien et al., 2013, Kwon et al., 2015, Garces et al., 2015, Lyumkis et al., 2013, Pancera et al., 2014, Stewart-Jones et al., 2016). Additional insights have come with the progressive increase in cryoEM imaging resolution of bona fide native trimers expressed on virion or cellular surface membranes (Hu et al., 2011, Lee et al., 2016, Liu et al., 2008, White et al., 2010, Wu et al., 2010). Despite these remarkable advancements, however, several aspects of the structure-function relationships in the HIV-1 envelope spike remain to be defined, which may be critical for the design of effective inhibitors targeting functional elements of the HIV-1 envelope spike.
We recently reported the identification of two sulfated tyrosines (Tys173 and Tys177) within the second variable (V2) domain of HIV-1 gp120, showing that tyrosine sulfation modulates HIV-1 neutralization sensitivity and, thereby, may facilitate immune evasion (Cimbro et al., 2014), an art mastered by HIV-1 to limit the immune system ability to recognize conserved neutralization epitopes (Chen et al., 2009, Kwong et al., 2002, Liu et al., 2011, Pancera et al., 2010, Pinter et al., 2004). Tyrosine sulfation was documented in gp120 from multiple HIV-1 strains grown in primary human CD4+ T cells, including primary isolates minimally passaged ex vivo, thus corroborating its physiological relevance. Remarkably, the V2 sulfotyrosines are identically spaced as two critical sulfated tyrosines within the N-terminal domain of the CCR5 coreceptor, which were shown to bolster the interaction of CCR5 with the base of the gp120 third variable (V3) domain, facilitating HIV-1 entry (Farzan et al., 1999, Huang et al., 2007). In line with these observations, tyrosine-sulfated peptides derived from the CCR5 N-terminus were shown to specifically inhibit infection by CCR5-tropic HIV-1 isolates (Cormier et al., 2000, Cormier et al., 2001, Farzan et al., 2000). Likewise, a chimeric molecule encompassing a tyrosine-sulfated peptide inserted at the C-terminus of CD4-Ig was recently reported to possess potent antiviral activity against a broad range of HIV-1 isolates (Gardner et al., 2015).
Based on these observations, we hypothesized that the sulfated region of the V2 loop could functionally mimic the sulfated N-terminus of CCR5 and establish an intramolecular interaction with the coreceptor-binding site at the base of the V3 loop. In the unliganded envelope, this interaction could serve as a structural constraint to stabilize the closed pre-fusion conformation of the trimer. Consistent with this hypothesis, recent crystallographic structures obtained with soluble SOSIP trimers show the V2 tyrosines positioned in close proximity to the CCR5-binding site (Julien et al., 2013, Kwon et al., 2015, Pancera et al., 2014, Garces et al., 2015, Stewart-Jones et al., 2016). In the present study, we utilize sulfated V2 mimetic peptides to provide a structural and functional characterization of the sulfotyrosine-mediated intramolecular interaction between V2 and V3. Moreover, we show that tyrosine-sulfated V2 mimetic peptides display potent and broad-spectrum anti-HIV-1 activity, opening new perspectives for the design of effective HIV-1 entry inhibitors.
2. Materials and methods
2.1. Peptides and proteins
Tyrosine-sulfated and control, non-sulfated mimetic peptides were produced using solid-phase synthesis technology by American Peptide Company. Each peptide was purified to > 95% and extensively quality controlled by HPLC and Mass Spectrometry Electro-Spray. The peptide aa. sequences were the following: Lys-Val-Gln-Lys-Glu-Tyr(SO3H)-Ala-Leu-Phe-Tyr(SO3H)-Glu-Leu-Asp-Ile-Val-Pro-Ile-Asp (pV2α-Tys); Lys-Lys-Gln-Lys-Val-His-Ala-Leu-Phe-Tyr(SO3H)-Lys-Leu-Asp-Ile-Val-Pro-Ile-Lys (pV2αΕ-Tys); Met-Asp-Tyr-Gln-Val-Ser-Ser-Pro-Ile-Tyr(SO3H)-Asp-Ile-Asn-Tyr(SO3H)-Tyr-Thr-Ser-Glu-Pro-Ser-Gln-Lys (pCCR5-Tys); Asp-Pro-Val-Leu-Glu-Phe-Tyr(SO3H)-Lys-Ile-Asp-Tyr(SO3H)-Ile-Glu-Leu-Ala-Val-Gln-Lys (pSCR-Tys). The peptides were dissolved in 10 nM NH4OH at 10 mg/mL and stored frozen until use. Recombinant monomeric gp120 (strain BaL) was obtained from the NIH AIDS Reagent Program; the recombinant SOSIP trimer (strain BG505) was produced in 293FS cells and purified by lectin-affinity and size-exclusion chromatography as described (Julien et al., 2013); 2-domain sCD4 was obtained from the NIH AIDS Reagent Program; the CD4-M48U mini-protein (Martin et al., 2003) was a gift of Loic Martin.
2.2. NMR spectroscopy
Peptide pV2α-Tys labeled with 15N was synthesized by American Peptide Company. All NMR samples were prepared by dissolving the tyrosine-sulfated pV2α-Tys in aqueous buffer containing 20 mM sodium phosphate pH 6.8, 50 mM sodium chloride, 0.02% sodium azide and 10% deuterium oxide. Binary complexes of either 2-domain sCD4 or recombinant gp120 (BaL) with pV2α-Tys and the ternary sCD4:gp120:pV2α-Tys peptide complex were prepared by combining all components and incubating on ice for 1 h. The complexes were then exchanged into NMR buffer and concentrated to a final volume of 200 μL. The final concentrations of sCD4, gp120 and pV2α-Tys were 20 μM, 20 μM and 800 μM, respectively. All NMR spectra were collected at 30 °C on a Bruker DRX 600 MHz NMR spectrometer equipped with a 1H/15N/13C cryoprobe.
Peptide assignments were completed using 2D TOCSY and NOESY experiments. NOESY spectra were acquired with τmix of 100, 250 and 400 milliseconds (ms). A τmix of 250 ms was found to give optimal NOE cross peak intensities. To assist in assignment process, we chemically synthesized pV2α-Tys with 15N–labeled Ala, Ile, Leu, Phe, and Val at all 8 occurrences of those amino acids. The 15N–labeled pV2α-Tys peptide, at a concentration of 0.8 mM, was used to collect 2D 1H-15N heteronuclear single quantum coherence (HSQC), 3D 15N–edited TOCSY-HSQC and 3D 15N–edited NOESY-HSQC spectra. trNOESY spectra of pV2α-Tys in the presence of 20 μM sCD4–gp120 were acquired using the same NOESY pulse scheme and parameters.
NMR structures were computed using the torsion angle dynamics program Cyana 3.0. NOE assignments were obtained using an iterative combination of the NOEASSIGN automated peak assignment feature of Cyana and manual editing. A total of 272 unambiguous, non-redundant distance constraints were employed in calculation of the final ensemble of 20 structures, which had an average target function of 0.85 ± 0.11 and average RMSD to the mean of 0.41 ± 0.17 Å (backbone) and 1.24 ± 0.21 Å (heavy atoms).
Competition between pCCR5-Tys and pV2α-Tys for binding to the sCD4:gp120 complex was assessed using 2D NOESY, 2D TOCSY, 2D HSQC and 3D 15N-edited NOESY-HSQC spectra using 15N–labeled V2 peptide. The sample conditions were the same as described above except that the V2 peptide concentration was 0.4 mM. Experimental data sets were collected with 0, 0.8 mM and 1.6 mM unlabeled pCCR5-Tys.
2.3. Circular dichroism spectroscopy
Far-UV CD spectra were measured and recorded on a Jasco-810 Spectropolarimeter (Jasco Inc., Easton, MD) equipped with a temperature controller. Spectra were recorded using a 0.1 cm quartz cuvette, at 0.3 mg/mL (137 μM), with a scan speed of 50 nm/min, response time of 1 s, band width of 4 nm and averaging of 5 scans. Peptide concentrations were determined spectrophotometrically in 30% acetonitrile at 280 nm using a molar extinction coefficient of 2980 M− 1 cm− 1. The raw CD data were analyzed by Dichroweb and using the Contin-LL algorithm (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml), and reference set 3 for secondary structure prediction.
2.4. Surface plasmon resonance
Peptide pV2α-Tys was immobilized on a Biacore CM5 sensor surface. Recombinant gp120 from HIV-1 BaL (at 25-400 nM) or SOSIP trimer (at 12.5–200 nM) were pre-incubated with or without a 2-fold molar excess of CD4-M48U miniprotein (at 200 nM) for 2 min at 25 μL/min in HBS supplemented with 3 mM EDTA and 0.005% Tween-20, followed by a 2 min dissociation phase on a Biacore 3000 instrument (GE Healthcare). Surfaces were regenerated after each cycle by a brief injection of 4.5 M MgCl2.
2.5. Clustal omega alignment
The alignment of V2 domain from 6 different HIV-1 subtypes (A to F) was performed using all the sequences deposited in the Los Alamos HIV Sequence Database and selecting one sequence for each patient isolate. A total of 191 sequences from subtype A, 3306 from subtype B, 2160 from subtype C, 195 from subtype D, 660 from subtype E, and 63 from subtype F were aligned using the Clustal Omega algorithm. Conservative substitutions were defined according to the standard Clustal parameters.
2.6. Secondary structure predictions
Secondary structure predictions were conducted using Agadir (www.agadir.crg.es/links.jsp), GOR V (www.gor.bb.iastate.edu), Jufo 9 (www.meilerlab.org/index.php/servers/), Psi Pred (www.bioinf.cs.ucl.ac.uk/psipred/), and NNPred (www.cmpharm.ucsf.edu/~nomi/nnpredict.html).
2.7. Molecular dynamics simulations
Explicit solvent replica-exchange molecular dynamics (REMD) simulations were performed on peptide pV2α using the NAMD v.2.8 program on the Biowulf Linux cluster at the NIH, Bethesda, MD (http://biowulf.nih.gov/). The peptide was constructed as a random coil, and explicitly solvated with TIP3P water molecules and Na+ and Cl− counterions using the VMD program (http://www.ks.uiuc.edu/Research/vmd/). Periodic boundary conditions were used and electrostatic interactions were calculated using the Particle-Mesh Ewald summation. A non-bonded term cutoff of 12 Å was used. The CHARMM27 forcefield was used with CHARMM atom types and charges. Prior to the start of REMD, the system was minimized using a conjugate gradient method, followed by slow warming to 310 K in 10 K increments. Each increment ran for 5 psec in order to equilibrate the system at a given temperature. Once the 310 K target temperature was reached, the system was equilibrated for an additional 50 psec at the end of which, a total of 40 replicas were run having a temperature range of 300–600 K for a cumulative simulation time of 0.6 μsec. Conformations were obtained at 1 nsec intervals, clustered using an RMS deviation of 5 Å. Representative structures from the clusters were ranked based on energy and the structure with the lowest energy was used for further studies.
The FlexPepDock simulation was performed using 2016 independent trajectories. The pV2a-Tys peptide (residues 168–185) extracted from the lowest-energy REMD model was oriented to the V3 base by all atom superposition of Tys173 and Tys177 to Tys100 and Tys100C of mAb 412d in the co-crystal structure with gp120 (Huang et al., 2007). Trajectories were analyzed using LigPlot by outputting snapshot structures from the trajectories at 1 nsec intervals and computing NACCESS-enhanced LigPlots for each snapshot. The text output was compiled, and the frequencies of observed interactions were used to compare the trajectories. A single view of the trajectory was computed by averaging the snapshots using a custom script in VMD. Following averaging, side chains were reconstructed using the idealize function of Rosetta.
Isothermal-isobaric molecular dynamics simulations of the pV2a-Tys peptide/gp120 complex were done using a forcefield parameters for sulfotyrosine obtained by combining CHARMM27 topologies from related molecules. Isothermal isobaric simulations were conducted as described for the REMD simulation above with the exception that once the 310 K target temperature was reached, the system was not split into replicas. The NAMD default thermostat and barostat was used. Production data was gathered at 1 nsec intervals. Before conducting isothermal-isobaric molecular dynamics simulations of the BG505-SOSIP.664 trimer (PDB ID 4NCO), sulfate groups were added to Tyr173 and Tyr177 using the SYBYL 7.3 program (Tripos). Forcefield parameters for sulfotyrosine were obtained by combining CHARMM27 topologies from related molecules.
2.8. HIV-1 isolates, infection, entry and cell viability assays
Details about the HIV-1 isolates used in this study, the infection assay in primary human CD4+ T cells, the viral entry assay, and the cell viability assay have been reported previously (Auerbach et al., 2012). The HIV-1 entry assay was performed in primary human CD4+ T cells using the CCR5-tropic isolate HIV-1 BaL. The cells (106 per replicate; two replicates per treatment) purified by immunomagnetic beads and previously activated ex vivo with PHA and IL-2 for 5–7 d were preincubated for 15 min at room temperature with the inhibitors in 50 μL of serum-free PBS and then exposed to 500 μL the undiluted viral stocks for 4 h at 37 °C in the continuous presence of the inhibitors. One aliquot of untreated cells was incubated for 4 h at 4 °C and served to determine the background signal level (trypsin-insensitive despite low-temperature conditions preventing virus entry). After incubation, the cells were extensively washed with PBS to remove unbound virus and treated with prewarmed bovine trypsin (Sigma) at 1.25 mg/mL for 10 min at 37 °C, followed by trypsin inactivation by addition of cold RPMI medium containing 10% (vol/vol) FBS. The cells were then washed three times with cold PBS, and the final dry pellets were frozen at − 80 °C overnight. The pellets were lysed using 100 μL of 0.5% (wt/vol) Triton X-100, and the amount of cell-associated p24 protein was quantified. The specific signal was calculated by subtracting from the p24 levels measured in each test sample the background p24 levels measured in cells incubated at 4 °C and treated with trypsin.
2.9. CCR5-binding assay
Cf2Th/syn-CCR5 cells (NIH AIDS Reagent Program), which express high levels of CCR5 on their surface membrane, were used to assess binding of soluble BG505-SOSIP.664 trimers to CCR5. The cells were harvested at ~ 80% confluency with enzyme-free cell dissociation buffer (Gibco). His-tagged BG505-SOSIP.664 trimer and mutants were pre-incubated with or without 2-domain sCD4 for 1 h at 4 °C. After washing with PBS twice, soluble trimers treated with or without sCD4 were incubated with the cells for 1 h at 4 °C, followed by washing with PBS. PE-conjugated mouse anti-His tag antibody (Miltenyi Biotec) was added to the cells for 1-hour at 4 °C. The cells were washed once with PBS, fixed with 2% PFA and analyzed on a BD FACSCanto. Specificity of binding was assessed by abrogation of the signal with an anti-CCR5 mAb (2D6; Becton Dickinson). Data analysis was performed using the FlowJo software.
2.10. Soluble CD4-induced HIV-1 envelope-mediated fusion assay
The HIV-1 envelope-mediated fusion assays were performed as previously described (Salzwedel et al., 2000) with some modification. HeLa cells infected with recombinant vaccinia viruses expressing HIV-1 BaL gp160 were used as effectors and Hos-CCR5 cells (CCR5-positive, CD4-negative) as targets. Soluble CD4 was added at 10 μg/mL prior to mixing effector and target cells.
3. Results
3.1. Direct interaction of a tyrosine-sulfated V2 mimetic peptide with HIV-1 gp120
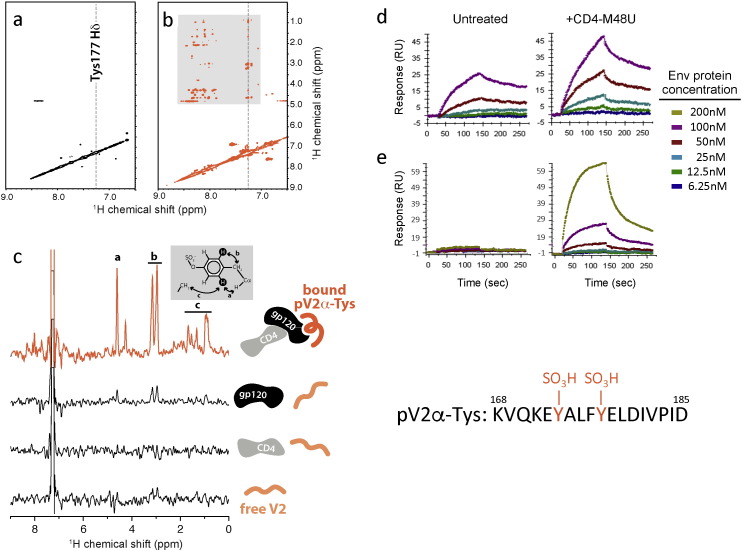
We previously demonstrated that the central region of the V2 loop of HIV-1 gp120 contains two sulfotyrosines (Tys), at position 173 and 177 (Cimbro et al., 2014). Tys177 is highly conserved across all HIV-1 subtypes (> 99%), whereas Tys173 is conserved in subtypes A, B and C, but less so in subtypes D, E and F with an overall cross-subtype conservation of ~ 66% (Fig. S1). To structurally and functionally characterize the sulfated region of V2, a 18-mer peptide (pV2α-Tys) spanning aa. 168–185 from the prototypic CCR5-tropic (R5) isolate BaL, bearing sulfated tyrosines at both positions 173 and 177, was synthesized and tested for its ability to interact with full-length recombinant HIV-1 gp120 by NMR in the presence or absence of sCD4. The 2D nuclear Overhauser effect (NOE) spectrum (NOESY) of 15N-labeled pV2α-Tys was measured and compared with NOESY spectra acquired in the presence of low concentrations of a sCD4:gp120 complex, at a peptide:protein complex molar ratio of 40:1. Since the rate of NOE buildup is proportional to the molecular weight, the relatively featureless NOESY spectrum of a small peptide like pV2α-Tys may, upon binding to a larger protein complex, become populated with NOE crosspeaks reflecting the conformation of the bound peptide. If the dissociation rate is fast enough to permit exchange of free and bound peptide molecules during the NOESY mixing period, intrapeptide NOEs arising while the peptide is bound will be transferred to the larger population of free peptide (trNOE). In the absence of the sCD4:gp120 complex, the NOESY spectrum was devoid of strong NOE signals (Fig. 1a) suggesting that in the unbound state pV2α-Tys has an unstructured random coil conformation. In agreement with this result, circular dichroism (CD) spectral analysis of the unbound peptide yielded no indications of ordered secondary structure (Fig. S2). However, addition of the sCD4:gp120 complex had profound effects on the NOESY spectrum, with the appearance of numerous NOE crosspeaks between pV2α-Tys resonances, consistent with the development of transferred NOEs (trNOE) reporting on the bound conformation (Fig. 1b). To determine whether pV2α-Tys binding depends on CD4-induced conformational changes in gp120, we measured NOESY spectra with either sCD4 or gp120 alone. As expected, no pV2α-Tys trNOEs signals were observed in the presence of sCD4 alone (Fig. S3). A small number of weak trNOE crosspeaks were visible when only gp120 was present, but their number and intensity were dramatically increased when the sCD4:gp120 complex was added. Most of the trNOEs involved the 173–182 segment (Fig. S4), suggesting that residues in this region are primarily responsible for interaction with gp120. Comparisons of the trNOEs detected on the aromatic side chain of Tys177 illustrate the effect of gp120 activation on pV2α-Tys binding and show the prominent role of Tys177 in stabilizing binding to gp120 (Fig. 1c).
Fig. 1.
Nuclear magnetic and surface plasmon resonance analyses of the interaction of a tyrosine-sulfated V2 mimetic peptide with monomeric and trimeric HIV-1 gp120. (a) 2D NOESY spectrum of a tyrosine-sulfated 18-mer V2 mimic, pV2α-Tys, at 0.8 mM. The peptide sequence is indicated at the bottom for reference. (b) 2D trNOESY spectrum of 0.8 mM pV2α-Tys in the presence of 20 μM CD4:gp120 complex. Monomeric gp120 was derived from the CCR5-tropic isolate BaL. (c) 1D slices from the 2D NOESY spectra of 0.8 mM pV2α-Tys corresponding to 7.26 ppm in the ω2 dimension (Tys177 Hδ). Tys177 NOE signals are shown for pV2α-Tys alone (bottom) or in the presence of sCD4, gp120 or the sCD4:gp120 complex as illustrated in the cartoons. (d) Surface plasmon resonance (SPR) analysis of the interaction of the tyrosine-sulfated V2 mimic pV2α-Tys with monomeric HIV-1 gp120 (strain BaL). (e) Surface plasmon resonance analysis of the interaction of the tyrosine-sulfated V2 mimic pV2α-Tys with a soluble cleaved gp140 trimer (BG505 SOSIP.664). The tyrosine-sulfated V2 mimic was immobilized on the surface of the sensor, while gp120 and the SOSIP trimer were used in the flow phase at increasing concentrations either in the unliganded form or after treatment with a CD4 mimic (CD4-M48U).
The interaction of pV2α-Tys with gp120 in both monomeric and trimeric configuration was further investigated by surface plasmon resonance (SPR). The sulfated V2 mimetic peptide was immobilized on the surface of a CM5 sensor, and either recombinant gp120 (strain BaL) or a soluble, stabilized gp140 trimer (BG505-SOSIP.664) was injected in the fluid phase. The tests were performed both in the presence and in the absence of a functional CD4 mimic (CD4-M48U), which induces the same gp120 conformational changes as native CD4 (Martin et al., 2003). Monomeric gp120 bound to the immobilized sulfated peptide with nanomolar affinity even in the absence of CD4-M48U (ka = 2.39 × 104 1/Ms., kd = 1.89 × 10− 3 1/s, KD = 7.91 × 10− 8 M); however, when gp120 was pre-complexed with CD4-M48U, the binding curve showed a greater magnitude and a faster on-rate despite similar affinity (ka = 4.83 × 104 1/Ms., kd = 3.34 × 10− 3 1/s, KD = 6.91 × 10− 8 M) (Fig. 1d), suggesting that CD4-induced (CD4i) conformational changes increase the number of binding sites for pV2α-Tys. No detectable binding of gp120, either in the presence or in the absence of CD4-M48U, was recorded when the non-sulfated homologous peptide (pV2α) was immobilized to the sensor (Fig. S5), confirming the critical role of tyrosine sulfation in bolstering the interaction with gp120. In contrast to monomeric gp120, the soluble gp140 trimer was unable to bind to the sulfated peptide in the absence of the CD4 mimic; however, pre-treatment with CD4-M48U induced binding of the trimer with nanomolar affinity (ka = 3.1 × 104 1/Ms, kd = 5.0 × 10− 3 1/s, KD = 1.61 × 10− 7 M) (Fig. 1e). These observations are consistent with the native-like conformation of the SOSIP trimer, which is not recognized by antibodies directed against CD4i epitopes (Sanders et al., 2013). Altogether, these results corroborate the interaction of the sulfated V2 region with the coreceptor-binding site at the base of the V3 loop, which is partially accessible in the monomeric form but inaccessible in the pre-fusion trimer conformation prior to activation by CD4.
3.2. Competition between tyrosine-sulfated V2 and CCR5 mimetic peptides for binding to gp120
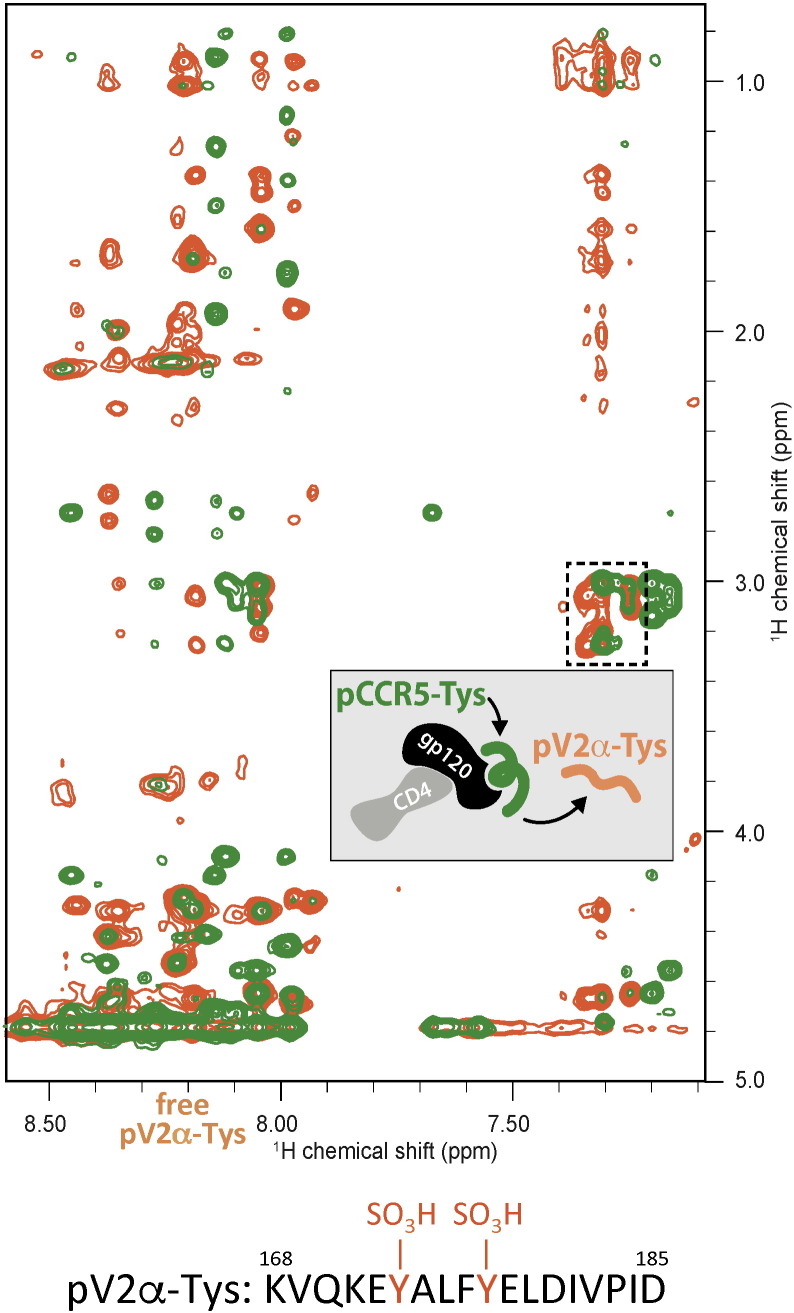
We previously showed that peptide pV2α-Tys competes with mAb 412d (Cimbro et al., 2014), a tyrosine-sulfated antibody that binds to the putative CCR5-binding site at the base of the V3 loop (Huang et al., 2007), suggesting that pV2α-Tys may directly interact with the coreceptor-binding site. However, it was impossible to exclude that pV2α-Tys competed with mAb 412d by binding to a different region of the large molecular footprint of such antibody. To determine whether pV2α-Tys effectively binds to the CCR5-binding site at the base of V3, we used NMR to investigate the direct competition between pV2α-Tys and a tyrosine-sulfated peptide derived from the CCR5 N-terminus (pCCR5-Tys) for gp120 binding. Peptide pCCR5-Tys was added to an NMR sample containing 15N-labeled pV2α-Tys and the sCD4:gp120 complex. Isotope-edited NOESY spectra showed the disappearance of the pV2α-Tys trNOEs upon addition of unlabeled pCCR5-Tys, with no change in the HSQC spectrum (Fig. S6). A similar alteration upon addition of pCCR5-Tys was seen in the 2D NOESY spectrum (Fig. 2), with the pV2α-Tys trNOE signals replaced by a new set of peaks that resemble the previously reported trNOE spectrum of a CCR5 peptide bound to gp120 (Huang et al., 2007). Taken together, these results support the interaction of the tyrosine-sulfated region of V2 with the CCR5 N-terminus-binding site in gp120.
Fig. 2.
Competition of a tyrosine-sulfated V2 mimetic peptide with a sulfated CCR5 N-terminal peptide for gp120 binding. Overlay of 2D trNOESY spectra of 0.4 mM pV2α-Tys in the presence of 20 μM CD4:gp120 complex before (orange) and after (green) displacement by a molar excess of a tyrosine-sulfated CCR5 N-terminal peptide, pCCR5-Tys, at 1.6 mM. The peptide sequence is indicated at the bottom for reference.
3.3. Structural features of a sulfated V2 mimetic peptide interacting with the coreceptor-binding site
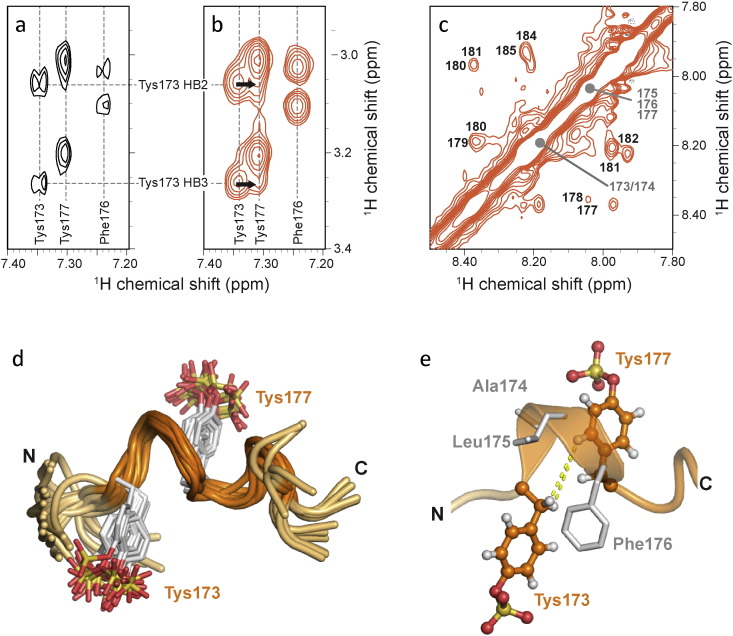
Previous work demonstrated that a CCR5 N-terminal sulfated peptide adopts a helical conformation upon interaction with gp120 (Huang et al., 2007) with distance constraints derived from trNOE signals analogous to those observed in this study for the V2-derived pV2α-Tys in the presence of sCD4:gp120 (Fig. 1d). Although for pV2α-Tys chemical shift degeneracy precluded the unambiguous assignment of medium-range NOEs required for calculation of a high-resolution structure, the NMR signals reporting on the gp120-bound conformation indicated that pV2α-Tys adopts a CCR5-like helical conformation upon interaction with the gp120:CD4 complex. Indeed, NOEs crosspeaks between the Hβ protons of Tys173 and the Hδ protons of Tys177 (Fig. 3a and b) would only arise if the intervening residues adopt a helical turn conformation. Sequential NH to NH NOEs (Fig. 3c) also suggested that gp120-bound pV2α-Tys is in a helical conformation. An ensemble of structural models obtained from torsion angle dynamics calculations using the available distance constraints contained a series of helical turns encompassing the two sulfotyrosines (Fig. 3d), as also illustrated by the NOE distances connecting the side chains of Tys173 and Tys177 (Fig. 3e). In agreement with the NMR data, both secondary structure predictions of the entire V2 region (Fig. S7a) and tertiary structure modeling using a Monte Carlo folding method (ROSETTA) (Fig. S7b) predicted with high confidence the formation of an α-helix encompassing the segment between Tys173 and Tys177. To further evaluate the presence and stability of the α-helix in V2, the conformational space was exhaustively explored starting from a random-coil configuration of peptide pV2α-Tys using replica-exchange molecular dynamics (REMD). An α-helix was frequently detected among the lowest-energy conformations (Fig. S7c). The adoption of a helical structure by the central region of V2 is also supported by the crystal structures of a V2-derived peptide in complex with mAbs CH58 and CH59, two antibodies derived from subjects enrolled in the RV144 vaccine trial (Liao et al., 2013), as well as by molecular modeling of an antibody-bound V2 peptide (Spurrier et al., 2014). Furthermore, a 3/10 helix was recently observed in the distal part of V2 in recent crystal structures of the BG505-SOSIP.664 trimer (Kwon et al., 2015, Pancera et al., 2014, Garces et al., 2015), further supporting the propensity of V2 segments to adopt a helical fold. Altogether, these findings corroborate the concept that the highly flexible V2 loop may interchange between alternative conformations according to its functional state and ligand interaction.
Fig. 3.
NMR characterization of a tyrosine-sulfated V2 mimetic peptide bound to HIV-1 gp120. (a) NOE peaks between the Hb and Hd of Tys173, Phe176 and Tys177 of free pV2a-Tys in a region of the NOESY spectrum that corresponds to the dashed box from Fig. 1D. (b) The same region of the trNOESY spectrum of pV2a-Tys in the presence of 20 μM CD4:gp120 peaks shows stronger NOE signals for Tys173, Phe176 and Tys177 side chains include new NOEs (black arrows) linking Hb of Tys173 and Hd of Tys177. (c) Sequential NH to NH NOEs in the trNOESY spectrum of 0.8 mM pV2a-Tys in the presence of 20 μM CD4:gp120 are consistent with helical conformations. (d) Ensemble of 20 structures of the bound conformation of pV2a-Tys calculated from trNOE distance constraints. Residues 170–182 are shown for clarity. (e) NOEs connecting the side chains of Tys173 and Tys177 (shown in Fig. 1b) define a helical turn.
To further explore the structural features of V2 interaction with the base of V3, we conducted molecular dynamic (MD) simulations of peptide pV2α-Tys docked onto the structure of CD4-bound gp120, as well as of the BG505-SOSIP.664 trimer after sulfate modification of the V2 tyrosines. The lowest-energy structure of peptide pV2α-Tys identified by REMD was docked onto the structure of a CD4-bound gp120 monomer (PDB ID: 2QAD) in the same conformation and orientation as those previously reported for the CCR5 N-terminal peptide (Huang et al., 2007) (Fig. S8a). Flexible docking by ROSETTA documented an energetically favorable interaction of pV2α-Tys with the CCR5-binding site at the base of V3. Analysis by MD confirmed the stability of the complex throughout a 400 nsec simulation. Insertion of Tys177 into a pocket at the base of V3 (Fig. S8b), analogous to that occupied by Tys14 of CCR5 (Fig. S8a), accounted for 25% of the total binding energy, with a preponderance of electrostatic interactions; in contrast, the binding energy of non-sulfated Tyr177 was markedly lower (Fig. S8e). The central role of Tys177 in stabilizing the V2–V3 complex is consistent with very high conservation of this residue across all HIV-1 subtypes (Fig. S1).
To validate the interaction of the tyrosine-sulfated region of V2 with the coreceptor-binding site in the context of the full-length HIV-1 envelope trimer, we performed MD simulations with the BG505-SOSIP.664 trimer (PDB ID: 4NCO) after modification of the V2 tyrosines to sulfotyrosines. All the available high-resolution structures of the BG505-SOSIP.664 trimer (Julien et al., 2013, Kwon et al., 2015, Pancera et al., 2014, Garces et al., 2015) document an extensive interface between V2 and V3, with Tyr177 directly facing the base of V3 in a similar orientation as Tys14 of CCR5 and forming an H-bond with Asn302, a key residue for CCR5 interaction (Fig. S8c). Analysis by MD after addition of sulfate groups to Tyr173 and Tyr177 did not show any significant alteration in both the local and global conformations of the V1V2–V3 loop region, with the side chain of Tys177 fitting well into the sulfotyrosine-binding pocket and establishing additional H-bonds with Arg298 and Asn300 (Fig. S8d). Taken together, these findings support the model of CCR5-like interaction between the tyrosine-sulfated region of V2 and the base of V3 in the intact trimer structure.
3.4. Inhibition of HIV-1 infection by a tyrosine-sulfated V2 mimetic peptide
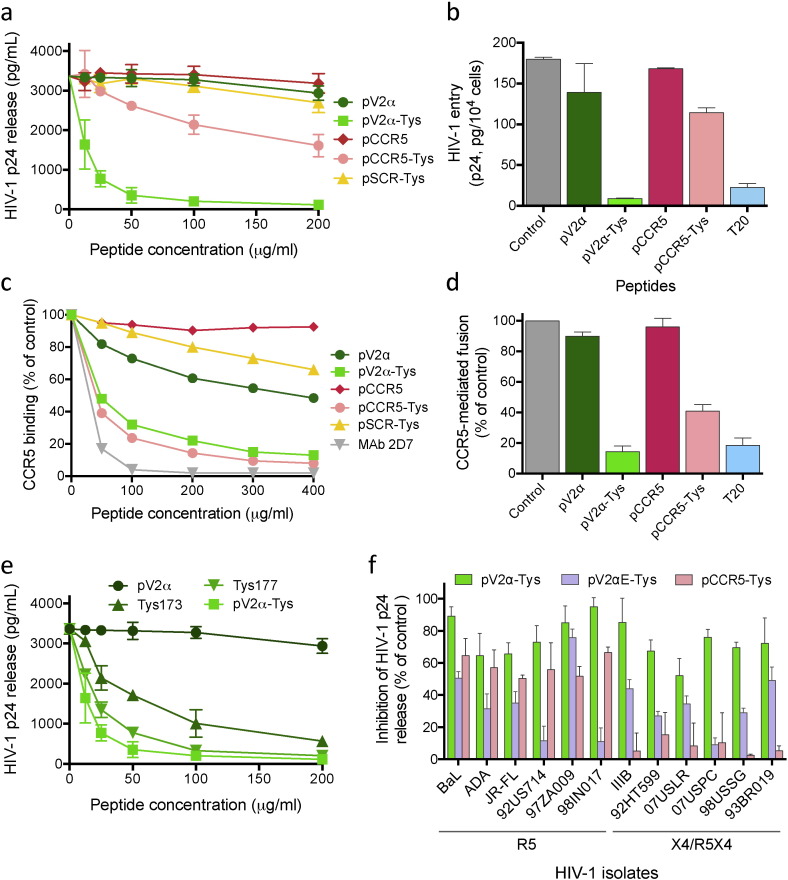
Since the NMR data and the predicted model of V2–V3 interaction documented direct competition between tyrosine-sulfated V2 and CCR5 mimics for binding to gp120, we investigated whether peptide pV2α-Tys can interfere with the coreceptor-gp120 interaction and thereby inhibit HIV-1 infection. In acute infection assays performed in primary human CD4+ T cells, pV2α-Tys potently inhibited infection by a CCR5-tropic HIV-1 isolate, BaL, in a dose-dependent fashion, whereas the non-sulfated homologous peptide, pV2α, was ineffective (Fig. 4a), demonstrating the importance of tyrosine sulfation in bolstering the interaction with gp120. The tyrosine-sulfated CCR5 N-terminal peptide pCCR5-Tys also showed dose-dependent HIV-1 inhibition, while its non-sulfated counterpart was ineffective; a tyrosine-sulfated scrambled peptide with the same aa. content as pV2α-Tys but in random sequence (pSCR-Tys), used as a control, did not show significant inhibitory activity (Fig. 4a). Of note, treatment with either pV2α-Tys or pCCR5-Tys had no detrimental effects on cell viability (Fig. S9).
Fig. 4.
Broad-spectrum inhibition of HIV-1 infection, entry and CCR5 utilization by tyrosine-sulfated V2 mimics. (a) Dose-dependent inhibitory effect of the V2 mimic pV2α-Tys and the CCR5 mimic pCCR5-Tys on cell-free infection of primary human CD4+ T cells by HIV-1, strain BaL; the respective non-sulfated homologous peptides, pV2α and pCCR5, were tested in parallel, as was a control scrambled sulfated peptide (pSCR-Tys). (b) Effect of pV2α-Tys or pCCR5-Tys (both at 50 μM) on HIV-1 BaL entry into activated primary human CD4+ T cells; the gp41-derived peptide T20, a pure HIV-1 entry inhibitor, was used as a control at 50 μg/mL. (c) Dose-dependent inhibitory effect of pV2α-Tys or pCCR5-Tys on binding of a soluble cleaved gp140 trimer (BG505 SOSIP.664) to human CCR5 expressed on the surface of canine thymocytes after pre-activation with sCD4; the scrambled sulfated peptide pSCR-Tys and an anti-CCR5 mAb (2D6) were used as negative and positive controls, respectively. (d) Effect of pV2α-Tys or pCCR5-Tys (both at 50 μM) on soluble CD4-activated fusion between HIV-1 BaL envelope-expressing cells and CCR5+ CD4− cells; the entry inhibitor T20 was used as a control at 50 μg/mL. (e) Dominant role of Tys177 in HIV-1 blockade. Dose-dependent inhibitory effect of peptide pV2α and its modified versions bearing either double (pV2α-Tys) or single (Tys173 and Tys177) sulfated tyrosines on cell-free infection by HIV-1 BaL in primary CD4+ T cells. (f) Broad spectrum of HIV-1 inhibition by tyrosine-sulfated V2 mimetic peptides. Peptides pV2α-Tys (clade B), pV2αΕ-Tys (clade E), or pCCR5-Tys (each at 50 μM) were tested on cell-free infection of purified CD4+ T cells by different strains of HIV-1. Eight primary isolates minimally passaged in vitro (92US714, 92HT599, 07USLR, 07USPC and 07USSG, subtype B; 97ZA009 and 98IN017, subtype C; 93BR019, subtype F) and 4 laboratory-expanded strains (BaL, JR-FL, ADA, IIIB, all subtype B) were tested. The coreceptor-usage phenotype of the various HIV-1 isolates is indicated below the strain designations.
3.5. HIV-1 inhibition by a sulfated V2 mimetic peptide occurs at the level of viral entry and coreceptor utilization
To definitively prove that HIV-1 inhibition by pV2α-Tys occurs via competition with CCR5 coreceptor utilization, we studied the effects of the peptide on viral entry, CCR5 binding and CCR5-mediated fusion. Pretreatment with pV2α-Tys effectively inhibited entry of HIV-1 into primary human CD4+ T cells; the CCR5 N-terminal peptide, pCCR5-Tys, also reduced viral entry, albeit with lower efficiency, while the non-sulfated counterparts of both peptides were ineffective; a potent gp41-derived fusion inhibitor used as a control, T20, exerted a potent inhibitory effect (Fig. 4b). Next, we tested the ability of pV2α-Tys to interfere with binding of the BG505 SOSIP.664 soluble trimer to CCR5 expressed on the surface of a canine thymocytic cell line (CF2). The soluble trimer was pre-activated with sCD4 in order to induce the high-affinity CCR5-binding conformation. Pre-treatment with pV2α-Tys reduced CCR5 binding of the trimer in a dose-dependent fashion with a potency similar to that of the sulfated CCR5 N-terminal peptide or an anti-CCR5 mAb (2D6) used as a control, while the non-sulfated homologous peptide pV2α and a scrambled sulfated peptide (pSCR-Tys) had limited effects (Fig. 4c). To further confirm that HIV-1 inhibition by pV2α-Tys occurs at the level of coreceptor utilization, we used a sCD4-activated fusion assay that selectively evaluates the effects of inhibitors on gp120-CCR5 interaction, downstream of the CD4-binding step. Both pV2α-Tys and pCCR5-Tys efficiently blocked CCR5-mediated fusion, whereas the non-sulfated homologous peptides had no effects and the positive control, peptide T20, was highly effective (Fig. 4d). These results confirmed that the sulfated V2 mimic interferes with the earliest steps of the HIV-1 infectious cycle preventing coreceptor binding and utilization, a critical step for the progression of the viral entry process.
3.6. Dominant role of sulfotyrosine 177 in HIV-1 inhibition by V2 mimetic peptides
To evaluate the relative role of the two V2 sulfotyrosines in the functional interaction with the CCR5-binding site, we tested the HIV-inhibitory activity of peptides bearing sulfate modifications on individual tyrosines (Tys173 and Tys177, respectively). Although both peptides exhibited antiviral activity, Tys177 was more potent than Tys173 (Fig. 4e), suggesting a prominent role of the C-terminal sulfotyrosine, Tys177 in gp120 interaction; the doubly sulfated peptide remained the most potent HIV-1 inhibitor, suggesting that tyrosine sulfation at both positions provides the optimal interactive surface for V2–V3 recognition. These data are consistent with the results of both MD (Fig. S8) and NMR (Fig. 3), highlighting the key role of the highly conserved Tys177 in stabilizing the interaction between the central region of V2 and the base of V3.
3.7. Broad-spectrum HIV-1 inhibition by tyrosine-sulfated V2 mimetic peptides
To assess the breadth of the antiviral activity of pV2α-Tys, we tested its effects on a panel of 12 HIV-1 strains representative of different coreceptor-usage phenotypes (6 CCR5-tropic; 6 CXCR4-tropic or dual-tropic) and genetic subtypes (9 clade B, 2 clade C, 1 clade F), including 8 primary isolates minimally passaged ex vivo; the sulfated CCR5 N-terminal mimic pCCR5-Tys was tested in parallel as a control. Strikingly, all the HIV-1 isolates tested were inhibited by pV2α-Tys regardless of their coreceptor specificity; in contrast, as previously observed (Cormier et al., 2000, Cormier et al., 2001, Farzan et al., 2000), the CCR5 N-terminal peptide showed a restricted activity against CCR5-tropic isolates, but was inactive against CXCR4-using (R5X4 and X4) isolates (Fig. 4f). Moreover, we tested a second tyrosine-sulfated peptide derived from a clade-E HIV-1 envelope (pV2αΕ-Tys), which displays only a single sulfotyrosine (Tys177) in the central region of V2, with Tyr173 substituted by a histidine, as well as a basic aa. (lysine) at position 178. The clade-E sulfated peptide showed a broad antiviral activity similar to that of pV2α-Tys, although 3 HIV-1 strains appeared to be resistant at the peptide concentration utilized; consistent with the presence of a single sulfotyrosine, pV2αΕ-Tys blocked HIV-1 with a generally lower potency compared to pV2α-Tys (Fig. 4f). Altogether, these results suggest that the conserved sulfated region of V2 has the ability to interact with the conserved V3 base of both CCR5- and CXCR4-tropic HIV-1 isolates, bypassing the structural determinants of coreceptor specificity that restrict the activity spectrum of the CCR5 N-terminal peptide.
4. Discussion
The presence of sulfated tyrosines has been documented in different ligands of the external HIV-1 envelope glycoprotein, gp120, including the N-terminal domain of the viral coreceptors (e.g., CCR5) (Farzan et al., 1999) and the CDRH3 region of antibodies specific for the coreceptor-binding site (e.g., 412d) (Choe et al., 2003) and the V1V2 region (e.g., PG16) (Pejchal et al., 2010); in all of these cases, tyrosine sulfation was shown to facilitate binding to gp120. The present study provides another example of tyrosine sulfation relevant to gp120, although the peculiarity of this interaction is that it occurs intramolecularly, locking the central region of V2 onto the coreceptor-binding site at the V3 base in the unliganded trimer. The V1V2 and V3 loops are flexible and movable structures on the gp120 surface, which in the unliganded trimer are physically connected at the apex of the envelope spike, but are rapidly dissociated and dislocated following gp120 activation by CD4 (Wyatt and Sodroski, 1998). Of note, V2 and V3 are believed to play a critical role in stabilizing the pre-fusion envelope structure (Kwon et al., 2012, Sullivan et al., 1993, Xiang et al., 2010), and cooperate in the formation of quaternary epitopes targeted by potent and broadly neutralizing mAbs (Gorny et al., 2005, Moore et al., 2011, Walker et al., 2009, Walker et al., 2011). Moreover, the V2 region was shown to effectively mask neutralization epitopes in V3 (Cao et al., 1997, Liu et al., 2011, Pinter et al., 2004). Our previous work documented the presence of sulfated tyrosines in V2 and demonstrated that enhancement of V2 sulfation by overexpression of the sulfotransferase TPST2 reduced HIV-1 sensitivity to neutralization by sCD4 and antibodies to the V3 tip or other CD4i epitopes, while at the same time increasing the sensitivity to trimer-preferring antibodies such as PG9; conversely, reduction of sulfation by treatment with sodium chlorate had reciprocal effects (Cimbro et al., 2014). These dichotomous effects on neutralization sensitivity imply that the V2 sulfotyrosines play a role in stabilizing the unliganded HIV-1 envelope trimer, shifting the conformational equilibrium toward a more tightly packed structure. Accordingly, recent data from our laboratory demonstrate that the V2 sulfotyrosines provide a mechanism of HIV-1 immune evasion from the bulk of host-produced anti-envelope antibodies (C. Guzzo et al., unpublished data).
The structural and functional features of the tyrosine-sulfated region of V2 described herein suggest that HIV-1 has modeled this region after the N-terminal domain of CCR5, exploiting coreceptor mimicry to optimize the intramolecular interaction between V2 and the CCR5-binding site at the base of the V3 loop. This segment of V2 is inherently flexible and has been reported to adopt two alternative conformations, α-helix (Liao et al., 2013) and β-strand (McLellan et al., 2011, Pancera et al., 2013), when complexed to different antibodies. One possible hypothesis, based on our NMR observations, is that the helical conformation may be preferentially adopted in the CD4-bound trimer, whose high-resolution structure has not yet been resolved, whereas a β-strand may be adopted in the pre-fusion envelope, as observed in the SOSIP trimer structures (Julien et al., 2013, Kwon et al., 2015, Pancera et al., 2014, Garces et al., 2015, Stewart-Jones et al., 2016). Competition studies by NMR demonstrated that V2 and CCR5 N-terminal mimics recognize the same region of gp120, in line with our previous demonstration that a tyrosine-sulfated V2 peptide competes with mAb 412d, a tyrosine-sulfated antibody specific for the coreceptor-binding region (Cimbro et al., 2014). As shown for both CCR5 and mAb 412d, we found that tyrosine sulfation plays an important role in bolstering the interaction between V2 and V3 because mimetic peptides lacking sulfation also lacked biological activity. However, both sulfation and the aromatic ring of the V2 tyrosines appear to play a role in bolstering the V2–V3 interaction. In particular, the aromatic ring of Tys177 appears to establish important hydrophobic contacts within a deep pocket at the base of V3, as previously described for Tys14 of CCR5 (Huang et al., 2007). Since the V2–V3 interaction is a key intramolecular constraint that holds the envelope trimer in its closed, pre-fusion conformation, the efficiency of gp120 sulfation can significantly influence the general conformation and stability of the envelope glycoprotein. This has potential implications both for crystallography studies and in the setting of vaccine manufacturing, because HIV-1 envelope-based immunogens are commonly produced in continuous cell lines that are inefficient, at best, at tyrosine sulfation (Cimbro et al., 2014). Thus, enhancing the levels of tyrosine sulfation might yield more stable envelope glycoproteins more accurately reflecting the antigenic make-up of the native HIV-1 envelope spike.
Our work identifies tyrosine-sulfated V2 mimetic peptides with potent inhibitory activity on HIV-1 entry and fusion, providing potential leads for the design of therapeutic and vaccine strategies against HIV-1. It is remarkable that, unlike CCR5 N-terminus peptides, V2 mimetic peptides are active against a broad range of HIV-1 isolates, irrespective of their coreceptor tropism, highlighting the overall structural conservation of the coreceptor-binding site in gp120. The molecular basis for the coreceptor selectivity of CCR5 N-terminal peptides is unclear at present, but it is likely that, besides the sulfotyrosines, other biochemical or structural features of these peptides may be incompatible with the binding interface with CXCR4-tropic gp120. A possible conflict site is Asp11 in CCR5, which may clash with a negative charge at position 440, as frequently detected in CXCR4-using isolates, while V2 might adapt to both basic and acidic side-chains at this position because it does not contain charged residues between the two sulfotyrosines. Preliminary results from our laboratory support this hypothesis.
In summary, our study documents a strategy of microbial mimicry by a retrovirus with the purpose of occluding a key neutralization target site and, at the same time, of consolidating the metastable pre-fusion envelope conformation. Targeting the interface between the sulfated region of V2 and the base of V3 may represent an effective strategy for the design of effective therapies and vaccines for the control of the HIV-1 pandemic.
Conflict of interest
No conflicts to report.
Contributions
The research was designed by R.C. and P.L. Substantial contributions to the project were made by R.C., C.G., P.Z., Q.L., H.M., D.V.R. and P.L. (peptide binding and inhibition tests and data analysis); F.C.P. and B.F.V. (NMR experiments and data analysis); D.E.H., M.A.D., A.X. and L.D.G. (molecular dynamic simulations and other in silico experiments and analyses).
Acknowledgments
We thank John R. Mascola, James E. Robinson and the IAVI consortium for anti-gp120 mAbs, Peter D. Kwong for anti-gp120 mAbs and 293FS cells, John P. Moore for the BG505 SOSIP.664 plasmid, Loic Martin for CD4-M48U, the NIH AIDS Reagent Program for cell lines and reagents, Andrea Balbo and Davin Jensen for CD spectroscopy analysis, and Yin Lin for technical assistance. This work was supported in part by the Division of Intramural Research of the NIAID, NIH, and by NIH grants AI058072 and AI013225 (to B.F.V.).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.06.037.
Appendix A. Supplementary data
Supplementary figures.
References
- Alsahafi N., Debbeche O., Sodroski J., Finzi A. Effects of the I559P gp41 change on the conformation and function of the human immunodeficiency virus (HIV-1) membrane envelope glycoprotein trimer. PLoS One. 2015;10 doi: 10.1371/journal.pone.0122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach D.J., Lin Y., Miao H., Cimbro R., DiFiore M.J., Gianolini M.E., Furci L., Biswas P., Fauci A.S., Lusso P. Identification of the platelet-derived chemokine CXCL4/PF-4 as a broad-spectrum HIV-1 inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9569–9574. doi: 10.1073/pnas.1207314109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A., Merk A., Borgnia M.J., Milne J.L., Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2013;20:1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Ahmed R., Barouch D.H., Butera S.T., Crotty S., Godzik A., Kaufmann D.E., McElrath M.J., Nussenzweig M.C., Pulendran B., Scanlan C.N., Schief W.R., Silvestri G., Streeck H., Walker B.D., Walker L.M., Ward A.B., Wilson I.A., Wyatt R. A blueprint for HIV vaccine discovery. Cell Host Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Sullivan N., Desjardin E., Parolin C., Robinson J., Wyatt R., Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Kwon Y.D., Zhou T., Wu X., O'Dell S., Cavacini L., Hessell A.J., Pancera M., Tang M., Xu L., Yang Z.Y., Zhang M.Y., Arthos J., Burton D.R., Dimitrov D.S., Nabel G.J., Posner M.R., Sodroski J., Wyatt R., Mascola J.R., Kwong P.D. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H., Li W., Wright P.L., Vasilieva N., Venturi M., Huang C.C., Grundner C., Dorfman T., Zwick M.B., Wang L., Rosenberg E.S., Kwong P.D., Burton D.R., Robinson J.E., Sodroski J.G., Farzan M. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell. 2003;114:161–170. doi: 10.1016/s0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- Cimbro R., Gallant T.R., Dolan M.A., Guzzo C., Zhang P., Lin Y., Miao H., Van Ryk D., Arthos J., Gorshkova I., Brown P.H., Hurt D.E., Lusso P. Tyrosine sulfation in the second variable loop (V2) of HIV-1 gp120 stabilizes V2–V3 interaction and modulates neutralization sensitivity. Proc. Natl. Acad. Sci. U. S. A. 2014;111:3152–3157. doi: 10.1073/pnas.1314718111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier E.G., Persuh M., Thompson D.A., Lin S.W., Sakmar T.P., Olson W.C., Dragic T. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5762–5767. doi: 10.1073/pnas.97.11.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier E.G., Tran D.N., Yukhayeva L., Olson W.C., Dragic T. Mapping the determinants of the CCR5 aminoterminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 2001;75:5541–5549. doi: 10.1128/JVI.75.12.5541-5549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M., Mirzabekov T., Kolchinsky P., Wyatt R., Cayabyab M., Gerard N.P., Gerard C., Sodroski J., Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- Farzan M., Vasilieva N., Schnitzler C.E., Chung S., Robinson J., Gerard N.P., Gerard C., Choe H., Sodroski J. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J. Biol. Chem. 2000;275:33516–33521. doi: 10.1074/jbc.M007228200. [DOI] [PubMed] [Google Scholar]
- Garces F., Lee J.H., de Val N., de la Pena A.T., Kong L., Puchades C., Hua Y., Stanfield R.L., Burton D.R., Moore J.P., Sanders R.W., Ward A.B., Wilson I.A. Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.R., Kattenhorn L.M., Kondur H.R., von Schaewen M., Dorfman T., Chiang J.J., Haworth K.G., Decker J.M., Alpert M.D., Bailey C.C., Neale E.S., Jr., Fellinger C.H., Joshi V.R., Fuchs S.P., Martinez-Navio J.M., Quinlan B.D., Yao A.Y., Mouquet H., Gorman J., Zhang B., Poignard P., Nussenzweig M.C., Burton D.R., Kwong P.D., Piatak M., Jr., Lifson J.D., Gao G., Desrosiers R.C., Evans D.T., Hahn B.H., Ploss A., Cannon P.M., Seaman M.S., Farzan M. AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature. 2015;519:87–91. doi: 10.1038/nature14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny M.K., Stamatatos L., Volsky B., Revesz K., Williams C., Wang X.H., Cohen S., Staudinger R., Zolla-Pazner S. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J. Virol. 2005;79:5232–5237. doi: 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Liu J., Taylor K.A., Roux K.H. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 loop. J. Virol. 2011;85:2741–2750. doi: 10.1128/JVI.01612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.C., Lam S.N., Acharya P., Tang M., Xiang S.H., Hussan S.S., Stanfield R.L., Robinson J., Sodroski J., Wilson I.A., Wyatt R., Bewley C.A., Kwong P.D. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.C., Tang M., Zhang M.Y., Majeed S., Montabana E., Stanfield R.L., Dimitrov D.S., Korber B., Sodroski J., Wilson I.A., Wyatt R., Kwong P.D. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J.P., Cupo A., Sok D., Stanfield R.L., Lyumkis D., Deller M.C., Klasse P.J., Burton D.R., Sanders R.W., Moore J.P., Ward A.B., Wilson I.A. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y.D., Finzi A., Wu X., Dogo-Isonagie C., Lee L.K., Moore L.R., Schmidt S.D., Stuckey J., Yang Y., Zhou T., Zhu J., Vicic D.A., Debnath A.K., Shapiro L., Bewley C.A., Mascola J.R., Sodroski J.G., Kwong P.D. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops0. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y.D., Pancera M., Acharya P., Georgiev I.S., Crooks E.T., Gorman J., Joyce M.G., Guttman M., Ma X., Narpala S., Soto C., Terry D.S., Yang Y., Zhou T., Ahlsen G., Bailer R.T., Chambers M., Chuang G.Y., Doria-Rose N.A., Druz A., Hallen M.A., Harned A., Kirys T., Louder M.K., O'Dell S., Ofek G., Osawa K., Prabhakaran M., Sastry M., Stewart-Jones G.B., Stuckey J., Thomas P.V., Tittley T., Williams C., Zhang B., Zhao H., Zhou Z., Donald B.R., Lee L.K., Zolla-Pazner S., Baxa U., Schön A., Freire E., Shapiro L., Lee K.K., Arthos J., Munro J.B., Blanchard S.C., Mothes W., Binley J.M., McDermott A.B., Mascola J.R., Kwong P.D. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 2015;22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Doyle M.L., Casper D.J., Cicala C., Leavitt S.A., Majeed S., Steenbeke T.D., Venturi M., Chaiken I., Fung M., Katinger H., Parren P.W., Robinson J., Van Ryk D., Wang L., Burton D.R., Freire E., Wyatt R., Sodroski J., Hendrickson W.A., Arthos J. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Kwong P.D., Wyatt R., Majeed S., Robinson J., Sweet R.W., Sodroski J., Hendrickson W.A. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- Kwong P.D., Wyatt R., Robinson J., Sweet R.W., Sodroski J., Hendrickson W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.L., Ozorowski G., Ward A.B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Bonsignori M., Alam S.M., McLellan J.S., Tomaras G.D., Moody M.A., Kozink D.M., Hwang K.K., Chen X., Tsao C.Y., Liu P., Lu X., Parks R.J., Montefiori D.C., Ferrari G., Pollara J., Rao M., Peachman K.K., Santra S., Letvin N.L., Karasavvas N., Yang Z.Y., Dai K., Pancera M., Gorman J., Wiehe K., Nicely N.I., Rerks-Ngarm S., Nitayaphan S., Kaewkungwal J., Pitisuttithum P., Tartaglia J., Sinangil F., Kim J.H., Michael N.L., Kepler T.B., Kwong P.D., Mascola J.R., Nabel G.J., Pinter A., Zolla-Pazner S., Haynes B.F. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Bartesaghi A., Borgnia M.J., Sapiro G., Subramaniam S. Molecular architecture of native HIV-1 gp 120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Cimbro R., Lusso P., Berger E.A. Intraprotomer masking of third variable loop (V3) epitopes by the first and second variable loops (V1V2) within the native HIV-1 envelope glycoprotein trimer. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20148–20153. doi: 10.1073/pnas.1104840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D., Julien J.P., de Val N., Cupo A., Potter C.S., Klasse P.J., Burton D.R., Sanders R.W., Moore J.P., Carragher B., Wilson I.A., Ward A.B. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L., Stricher F., Missé D., Sironi F., Pugniere M., Barthe P., Prado-Gotor R., Freulon I., Magne X., Roumestand C., Menez A., Lusso P., Veas F., Vita C. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat. Biotechnol. 2003;21:71–76. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- Mascola J.R., Haynes B.F. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol. Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Pancera M., Carrico C., Gorman J., Julien J.P., Khayat R., Louder R., Pejchal R., Sastry M., Dai K., O'Dell S., Patel N., Shahzad-ul-Hussan S., Yang Y., Zhang B., Zhou T., Zhu J., Boyington J.C., Chuang G.Y., Diwanji D., Georgiev I., Kwon Y.D., Lee D., Louder M.K., Moquin S., Schmidt S.D., Yang Z.Y., Bonsignori M., Crump J.A., Kapiga S.H., Sam N.E., Haynes B.F., Burton D.R., Koff W.C., Walker L.M., Phogat S., Wyatt R., Orwenyo J., Wang L.X., Arthos J., Bewley C.A., Mascola J.R., Nabel G.J., Schief W.R., Ward A.B., Wilson I.A., Kwong P.D. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.L., Gray E.S., Sheward D., Madiga M., Ranchobe N., Lai Z., Honnen W.J., Nonyane M., Tumba N., Hermanus T., Sibeko S., Mlisana K., Abdool Karim S.S., Williamson C., Pinter A., Morris L., CAPRISA 002 Study Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. J. Virol. 2011;85:3128–3141. doi: 10.1128/JVI.02658-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M., Majeed S., Ban Y.E., Chen L., Huang C.C., Kong L., Kwon Y.D., Stuckey J., Zhou T., Robinson J.E., Schief W.R., Sodroski J., Wyatt R., Kwong P.D. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U S A. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M., Shahzad-Ul-Hussan S., Doria-Rose N.A., McLellan J.S., Bailer R.T., Dai K., Loesgen S., Louder M.K., Staupe R.P., Yang Y., Zhang B., Parks R., Eudailey J., Lloyd K.E., Blinn J., Alam S.M., Haynes B.F., Amin M.N., Wang L.X., Burton D.R., Koff W.C., Nabel G.J., Mascola J.R., Bewley C.A., Kwong P.D. Structural basis for diverse N-glycan recognition by HIV-1–neutralizing V1–V2–directed antibody PG16. Nat. Struct. Mol. Biol. 2013;20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M., Zhou T., Druz A., Georgiev I.S., Soto C., Gorman J., Huang J., Acharya P., Chuang G.Y., Ofek G., Stewart-Jones G.B., Stuckey J., Bailer R.T., Joyce M.G., Louder M.K., Tumba N., Yang Y., Zhang B., Cohen M.S., Haynes B.F., Mascola J.R., Morris L., Munro J.B., Blanchard S.C., Mothes W., Connors M., Kwong P.D. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R., Walker L.M., Stanfield R.L., Phogat S.K., Koff W.C., Poignard P., Burton D.R., Wilson I.A. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A., Honnen W.J., He Y., Gorny M.K., Zolla-Pazner S., Kayman S.C. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 2004;78:5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzwedel K., Smith E., Dey B., Berger E. Sequential CD4/coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J. Virol. 2000;74:326–333. doi: 10.1128/jvi.74.1.326-333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R.W., Derking R., Cupo A., Julien J.P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Peña A.T., Korzun J., Golabek M., de Los R.K., Ketas T.J., van Gils M.J., King C.R., Wilson I.A., Ward A.B., Klasse P.J., Moore J.P. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurrier B., Sampson J., Gorny M.K., Zolla-Pazner S., Kong X.P. Functional implications of the binding mode of a human conformation-dependent V2 monoclonal antibody against HIV. J. Virol. 2014;88:4100–4112. doi: 10.1128/JVI.03153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones G.B., Soto C., Lemmin T., Chuang G.Y., Druz A., Kong R., Thomas P.V., Wagh K., Zhou T., Behrens A.J., Bylund T., Choi C.W., Davison J.R., Georgiev I.S., Joyce M.G., Kwon Y.D., Pancera M., Taft J., Yang Y., Zhang B., Shivatare S.S., Shivatare V.S., Lee C.C., Wu C.Y., Bewley C.A., Burton D.R., Koff W.C., Connors M., Crispin M., Baxa U., Korber B.T., Wong C.H., Mascola J.R., Kwong P.D. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N., Thali M., Furman C., Ho D.D., Sodroski J. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J. Virol. 1993;67:3674–3679. doi: 10.1128/jvi.67.6.3674-3679.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Huber M., Doores K.J., Falkowska E., Pejchal R., Julien J.P., Wang S.K., Ramos A., Chan-Hui P.Y., Moyle M., Mitcham J.L., Hammond P.W., Olsen O.A., Phung P., Fling S., Wong C.H., Phogat S., Wrin T., Simek M.D., Protocol G Principal Investigators, WC K., IA W., DR B., Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Phogat S.K., Chan-Hui P.Y., Wagner D., Phung P., Goss J.L., Wrin T., Simek M.D., Fling S., Mitcham J.L., Lehrman J.K., Priddy F.H., Olsen O.A., Frey S.M., Hammond P.W., Protocol G Principal Investigators, Kaminsky S., Zamb T., Moyle M., WC K., Poignard P., Burton D.R. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.A., Bartesaghi A., Borgnia M.J., Meyerson J.R., de la Cruz M.J., Bess J.W., Nandwani R., Hoxie J.A., Lifson J.D., Milne J.L., Subramaniam S. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.R., Löving R., Lindqvist B., Hebert H., Koeck P.J., Sjöberg M., Garoff H. Single-particle cryoelectron microscopy analysis reveals the HIV-1 spike as a tripod structure. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18844–18849. doi: 10.1073/pnas.1007227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R., Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Xiang S.H., Finzi A., Pacheco B., Alexander K., Yuan W., Rizzuto C., Huang C.C., Kwong P.D., Sodroski J. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J. Virol. 2010;84:3147–3161. doi: 10.1128/JVI.02587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.