Abstract
Anti-VEGF antibody bevacizumab has prolonged progression-free survival in several cancer types, however acquired resistance is common. Adaption has been observed pre-clinically, but no human study has shown timing and genes involved, enabling formulation of new clinical paradigms.
In a window-of-opportunity study in 35 ductal breast cancer patients for 2 weeks prior to neoadjuvant chemotherapy, we monitored bevacizumab response by Dynamic Contrast-Enhanced Magnetic Resonance [DCE-MRI], transcriptomic and pathology.
Initial treatment response showed significant overall decrease in DCE-MRI median Ktrans, angiogenic factors such ESM1 and FLT1, and proliferation. However, it also revealed great heterogeneity, spanning from downregulation of blood vessel density and central necrosis to continued growth with new vasculature. Crucially, significantly upregulated pathways leading to resistance included glycolysis and pH adaptation, PI3K-Akt and immune checkpoint signaling, for which inhibitors exist, making a strong case to investigate such combinations.
These findings support that anti-angiogenesis trials should incorporate initial enrichment of patients with high Ktrans, and a range of targeted therapeutic options to meet potential early resistance pathways. Multi-arm adaptive trials are ongoing using molecular markers for targeted agents, but our results suggest this needs to be further modified by much earlier adaptation when using drugs affecting the tumor microenvironment.
Keywords: Anti-angiogenic treatment, Breast cancer, Resistance, DCE-MRI, Radiogenomics
Highlights
-
•
DCE-MRI two weeks post-treatment of primary breast cancer shows heterogeneity of initial response to bevacizumab.
-
•
Gene expression shows heterogeneous changes but an overall decrease in angiogenic factors ESM1 and FLT1, and proliferation.
-
•
Increased PI3K-Akt and immune checkpoint signaling suggests combination of Bevacizumab with inhibitors of these pathways.
-
•
Increased PI3K-Akt and immune checkpoint signaling suggests combination of Bevacizumab with inhibitors of these pathways.
Available evidence is that the anti-angiogenic treatment Bevacizumab currently has a limited range of indications in breast cancer compared to other cancers. Here, we show that multimodality dynamic monitoring of treatment response allow classification of patient's response as early as two weeks after treatment, and reveals new mechanisms associated with resistance. The implications, are that the role of bevacizumab in neoadjuvant breast cancer needs to be re-evaluated using early classification of resistant patients, but more generally that trials need to be designed prospectively, taking into account dynamic monitoring to be able to rapidly adapt available new agents for combination.
1. Introduction
Angiogenesis is a key process in cancer providing oxygen and nutrients to the growing tumor mass. Key player is the vascular endothelial growth factor [VEGF], targeted by antibodies or small molecule kinase inhibitors. First such agent was bevacizumab [Avastin], monoclonal antibody against VEGF. Clear activity in clinical trials has led to implementation of such therapies as standard of care. However, effects are usually on progression–free rather than overall survival, and often measured in weeks (Carmeliet and Jain, 2000). Whilst benefit has been shown in some cancers, in breast cancer there has been controversy on the role of antiangiogenic therapy (Rossari et al., 2012). Early studies showed improvement of response in metastatic disease with Taxol (Jain, 2014), whilst neoadjuvant studies showed contrasting results (Jain, 2014, Bear et al., 2012, Earl et al., 2015, Sikov et al., 2015, von Minckwitz et al., 2012).
Lack of consensus between clinical trials has questioned the effectiveness of bevacizumab in breast cancer, and the drug is no longer in use in several countries. However, there is large evidence of heterogeneity of response. Therefore we undertook a study in the situation where bevacizumab was approved to understand mechanisms behind this heterogeneity, and clinical implications. Circulating levels of short VEGFA isoforms, neuropilin-1 and VEGF receptor 1 expression in tumours or plasma, and VEGFA genetic variants have been reported as potential biomarkers of bevacizumab response (Lambrechts et al., 2013, Hegde et al., 2013). Changes in DCE-MRI parameters with therapy have been shown, such as volume transfer constant (Ktrans), complex function of vessel permeability, surface area and tumor blood flow; however, how to use MRI to impact on drug therapy modulation is controversial (O'Connor and Jayson, 2012); reflecting limited insight into molecular correlates. A small window study of 21 patients combining DCE-MRI monitoring with limited number of molecular markers, showed reduction of perfusion parameters with increase in tumor apoptosis (Wedam et al., 2006). However, studies to date have been hindered by either combination with chemotherapy, or focus on advance cancers where multiple resistance mechanisms are likely to have developed, or else limited pharmacodynamics assessments probing only some aspects of tumor response.
The present single-agent bevacizumab study, in previously untreated primary breast cancer patients, asks whether DCE-MRI complemented with molecular profiling of response could provide criteria for patient stratification and insight into early actionable pathways of resistance.
2. Methods
2.1. Study Design and Participants
A phase II, non-randomized, open-label study sponsored by Oxford Radcliffe Hospitals NHS Trust (ORH/PID/5575) was given National Research Ethics Services, UK, approval (Oxford REC no. 08/H0604/69). Patient characteristics are shown in Suppl. Table-1 and Consort diagram in Fig. S1. Previously untreated breast cancer patients with either histologically proven locally advanced breast cancer or tumor > 3 cm in diameter were included. Bevacizumab was administered as single infusion (15 mg/kg) 2 weeks prior to neoadjuvant chemotherapy. Multiparametric DCE-MRI scans, core biopsies and blood samples were obtained immediately before and 2 weeks after bevacizumab. 47 previously untreated locally advanced primary breast cancer patients were enrolled between July 2008 and November 2010 by the Oxford Radcliffe Hospital NHS Trust (n = 30) and Mount Vernon Hospital (n = 17). The present study focuses on ductal cancers (n = 36); analysis of other subtypes will be presented elsewhere. DCE-MRI data was analysable for 35 patients only.
2.2. Outcomes: DCE-MRI
Treatment response was measured by DCE-MRI. High temporal resolution T1-weighted acquisition was used to images 8–12 central slices of the tumor region. DCE-MRI setup, imaging acquisition details and all image processing and derivation of PK parameters are reported in detail in the Suppl. Methods. Furthermore, all scripts and codes used for the analysis have been submitted to the GitHub public directory: https://github.com/nph/MRI.Side effects reporting followed CTCAEv3.0 (http://ctep.cancer.gov).
2.3. Molecular Profiling
An experienced radiologist obtained the ultrasound guided core biopsies using 14 gauge core biopsy needles with 22 mm throw from the margin of tumours. Affymetrix Human Exon 1.0 ST arrays were used to measure gene expression. To confirm gene expression results, quantitative real-time PCR (qRT-PCR) was performed using same cDNA as used for gene expression array work. Another ultrasound guided core biopsy sample was collected directly on biopsy cassettes immediately immersed in 50mLs of 10% Formalin in standard biopsy pot for immunohistochemistry (IHC) analysis. All samples were processed within 7 days of collection to render formalin-fixed paraffin-embedded (FFPE) blocks (stored at room temperature). Further details on samples handling and storage, and analysis methods are in Suppl. Methods.
2.4. Statistical Analysis
Statistical analysis for DCE-MRI, gene expression profiling and PCR are reported in Suppl. Methods. Changes of immunohistochemistry parameter scores after bevacizumab were analyzed using paired non-parametric method (Wilcoxon Signed Rank Sum, WSRS). Correlation analyses between gene expression scores, MRI parameters, clinical variables, IHC parameters scores were also performed using non-parametric methods (Spearman for continuous, Kruskal-Wallis for categorical variables). GraphPad PRISM v4.0c, Matlab and R v2.13.0 were used.
2.5. Funding
Breast Cancer Research Foundation, Breast Cancer Research Trust, NIHR Oxford Biomedical Research Centre, and Oxford Cancer Research UK Imaging Centre. Roche provided bevacizumab free of charge. EU FP7 p-medicine and Eureca projects supported FMB and RVS.
3. Results
3.1. Highly Heterogeneous Early Response to Bevacizumab
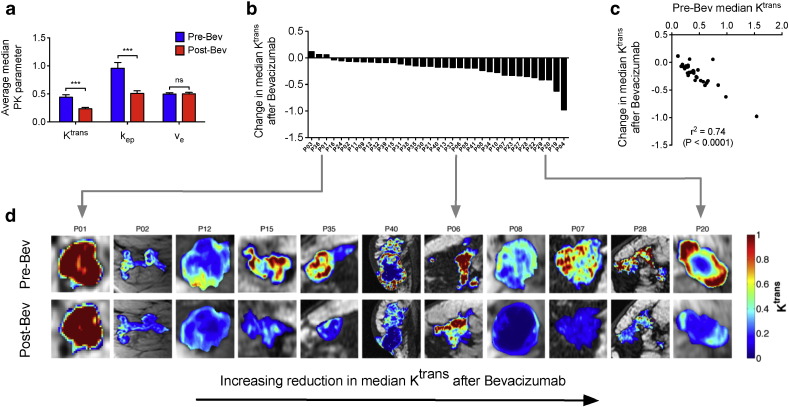
Single-dose bevacizumab administration (15 mg/kg) was well tolerated with no grade 3 toxicity in 36 patients (Table S1). Of six patients presenting erythematous breast mass, four showed reduced erythema (Fig. S2). No changes after treatment could be detected by standard clinical evaluation (Fig. S2); however DCE-MRI analysis revealed significant decrease in median Ktrans and kep (WSRS, P < 0.001) (Fig. 1a), and Minkowski–Bouligand fractal dimension (WSRS P = 0.012, Fig. S3). This suggests general improvement in hierarchical architecture, hence tendency to vasculature normalisation.
Fig. 1.
a) Graph show average median pharmacokinetic parameters. Blue - pre-bevacizumab; red = post-bevacizumab; Bev = bevacizumab; *** = significant; ns = not significant. b) Waterfall plot showing absolute change in Median Ktrans after bevacizumab across whole study population. c) Graph show significant negative correlation of absolute change in median Ktrans with pre-bevacizumab median Ktrans. d) Ktrans maps of eleven representative patients showing change in Ktrans after bevacizumab. Arranged in increasing reduction in Median Ktrans after bevacizumab. Pre-Bev = pre-bevacizumab; Post-Bev = post-bevacizumab; “P” = patient.
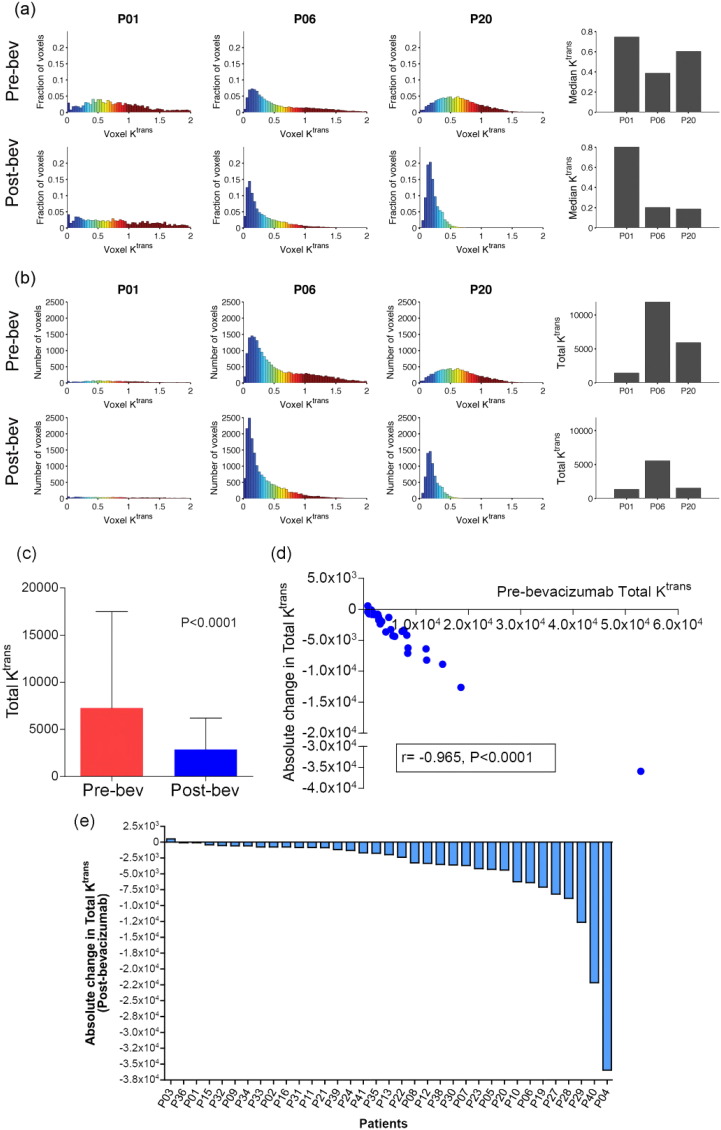
DCE-MRI response was heterogeneous (Fig. 1b, S3), with high baseline median Ktrans tumours showing greatest reduction (Spearman ρ = − 0.92, P = 1e − 08, Fig. 1c). Variability in median Ktrans was also observed within individual tumours (Fig. 1d). Visual assessment of parametric maps confirmed this heterogeneity, revealing patterns from not-significant change to strong median Ktrans reduction across complete tumor mass (Fig. 1d). As different parameter distributions could result in similar median Ktrans values, we investigated cumulative Ktrans over the central tumor region. This new parameter, defined here as total Ktrans, provides an estimate of total target mass presented for modification by bevacizumab, and might help defining heterogeneity (Fig. 2). Total Ktrans decreased post-bevacizumab (WSRS, P < 0.0001) (Fig. 2c), changes were correlated with pre-bevacizumab Total Ktrans (Fig. 2d), and highly variable (Fig. 2e).
Fig. 2.
a) Change in Median Ktrans after bevacizumab in three representative patients. b) Change in Total Ktrans after bevacizumab in same patients; c) Graph showing significant reduction in Total Ktrans after bevacizumab. d) Graph show significant negative correlation of absolute change in Total Ktrans with pre-bevacizumab Total Ktrans. e) Waterfall plot showing absolute change in Total Ktrans after bevacizumab across whole study population.
3.2. Angiogenesis is Downregulated Whilst VEGFA Transcript is Induced
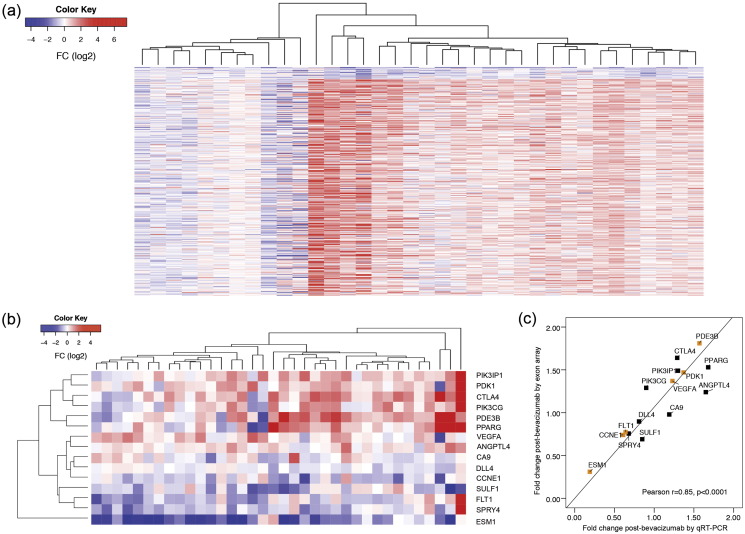
Gene-wise paired differential expression analysis revealed 23 significantly (FDR < 0.05) downregulated genes post-bevacizumab (Fig. 3a) (Table S2). High correlation between gene expression arrays and qRT-PCR confirmation was observed (Fig. 3b–c, Figs. S4, S5). Notably, several (6/23) were also downregulated post-bevacizumab in our xenograft models (Masiero et al., 2013) including GPR56, inhibitor of VEGF production; EXO1, exonuclease promoting cleavage of transcribed immunoglobulin switch regions; ILF2, coding for nuclear factor of activated T-cells (NFAT) component.
Fig. 3.
a) Heatmap of differentially expressed genes after bevacizumab. Each row represents a differentially expressed gene arranged by ascending order of fold change from top to bottom and each column represents fold change for single patients, red = up-regulation and blue = down-regulation, samples were visually clustered using hierarchical clustering. (b) Heatmap showing high variation in gene expression (as analyzed by exon array) of genes validated by qRT-PCR. Each row represents a gene and each column represents single patient's fold change for each gene post-bevacizumab. Red = up-regulation, Blue = down-regulation. (c) Significant correlation between fold changes in genes of interest as observed by exon array and qRT-PCR. Yellow boxes represents genes significantly changing by exon array as well as qRT-PCR analysis.
Top downregulated transcript was endothelial cell specific molecule-1 (ESM1), regulated by VEGFA, mediating angiogenesis and invasion. Interestingly, we observed significantly greater ESM1 downregulation in high grade tumours (Fig. S5). Other downregulated genes confirmed by qRT-PCR were fms-related tyrosine kinase 1 (FLT1), encoding a VEGF receptor (VEGFR) family member, and delta like ligand 4 (DLL4), vascular-specific Notch ligand (Fig. S4). Significant reduction in microvascular area was also observed. Namely, plasmalemma vascular associated protein (PLVAP) showed an average 20% percentage decrease post-treatment (WSRS P = 0.014) (Figs. 4, S6).
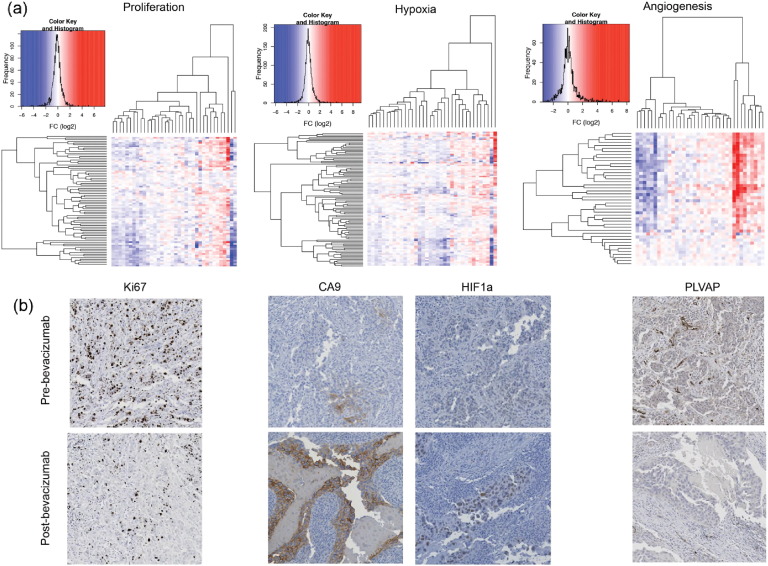
Fig. 4.
a) Heatmap showing high variability in genes involved in Proliferation, Hypoxia and Angiogenesis signatures. Each row represents a gene and each column represents single patient. Colors reflect the fold change for each gene post-bevacizumab. With whole cohort of study patients (n = 36), significant global down-regulation of the proliferation signature (paired t-test: t = − 2.09, P-value = 0.04) but no significant change in the specific breast cancer hypoxia signature (paired t-test: t = − 0.20, P-value = 0.84), angiogenesis signature (paired t test: t = 0.81, P-value = 0.42). b) IHC sections showing significant downregulation of proliferation marker (Ki67) (P = 0.01), upregulation of carbonic anhydrase-9 (P = 0.04), upregulation of Hypoxia inducible factor 1 alpha (HIF1a) (P = 0.001) and down-regulation of plasmalemma vascular associated protein (PLVAP) (P = 0.02) in response to bevacizumab.
A 393 significantly upregulated genes were also identified (Fig. 3a, Table S3), including VEGFA itself (confirmed by qRT-PCR, Fig. S7) suggesting a negative feedback loop. Interestingly, chemokine receptor CXCR4, upregulated by VEGFA, and its ligand chemokine stromal cell-derived factor-1 (SDF-1)/CXCL12 were also upregulated (Table S3). Transcription factor (TF) over-representation analysis (Table S4) showed activation of ETS2 post-treatment, required for endothelial cell survival during embryonic angiogenesis, and whose expression in fibroblasts modulates angiogenesis in breast cancer (Wallace et al., 2013). Similarly for, Lymphoid Enhancer-Binding Factor, LEF1, binding to T-cell receptor-alpha enhancer and involved in Wnt signaling; and for Nuclear factor of activated T-cells, NFAT, regulator of signaling between cancer and stroma cells [as reviewed in (Mancini and Toker, 2009)].
Notably, 59 of these 393 genes were also upregulated post-bevacizumab in our xenograft models (Table S5); including interleukins (ILs) such as IL5, IL receptors IL1R1 and IL7R. Interestingly, IPA upstream regulator analysis (Table S5) found IL12 (P = 1.63E − 06), IL4 (P = 7.18E − 06), IL13 (P = 1.07E − 05), interferon alpha (P = 1.61E − 05) and IL7 (P = 1.18E − 05) as most enriched upstream regulators. The first four are inhibitors of angiogenesis; the latter controls proliferation by influencing the tumor microenvironment, is over-expressed in triple negative breast cancers (Lehmann et al., 2011) and has been found to induce VEGFD and increase lymphangiogenic in preclinical systems (Al-Rawi et al., 2005) which could highlight potential escape mechanism.
3.3. Decrease in Tumor Proliferation After Bevacizumab
Cyclin E coding gene CCNE1, essential for cell cycle control at G1/S transition, was also consistently downregulated (Figs. 1, S4). Cyclin E is associated with chromosome instability, is prognostic in breast cancer (Scaltriti et al., 2011), and low molecular weight isoforms increased angiogenesis in human melanoma cells in-vivo (Bales et al., 2005). Of note, TF over-representation analysis (Table S4) predicted increased activity of FOXO4, negative cell cycle regulator and inhibitor of proliferation. Concordantly, a human breast cancer proliferation signature (Desmedt et al., 2008) (paired t-test P = 0.04, Figs. 4a1 and S8), and percentage of Ki-67 positive cells decreased (WSR test P = 0.006) (Figs. 4b1 and S6). Interestingly, reduction was greater in grade 3 than grade 2 cases (Fig. S9).
3.4. Immune Checkpoint Signaling and Metabolism Upregulation After Bevacizumab
Transcriptional upregulation after bevacizumab was more variable than downregulation (Fig. 1), suggesting after initial general response to bevacizumab, distinct adaptation mechanisms are rapidly induced in patient subgroups. Key immune response pathways were induced (Tables S3–S5), including CD28 and its activating binding ligand CD86. Importantly, expression levels of immune checkpoint receptor CTLA4, inhibitory binding partner of CD86, and negative regulator of T-cell activation and function, were also increased after treatment (Table S3), as was IL10 receptor alpha, IL10RA, suppressor of anti-tumor T-cell response (Table S3).
Top upregulated metabolism genes, confirmed by qRT-PCR, included pyruvate dehydrogenase kinase-1 (PDK1) and phosphodiesterase 3B cGMP-inhibited (PDE3B) (Fig. S7). We analyzed whether these metabolism changes could result from increased tumor hypoxia due to decreased blood vessel supply. Several HIF1A target genes were significantly upregulated post-treatment (Fig. S10), similarly to xenograft models (Table S3). These included PHD3 (EGLN3), regulator of HIF1 degradation; HIF1A regulated ENO2, HK2, PDK1 and VEGFA; and SPP1 and DDIT4, involved in PI3K-Akt signaling. A human cancer hypoxia signature (Buffa et al., 2010) was increased in a patient subgroup (Figs. 4a2, S8). Furthermore, percentage of CA9 and HIF1A positive cells were increased (WSRS P = 0.04) (Figs. 4b2, S11). Notably, in 7/36 patients (20%) both CA9 and HIF1A were upregulated, whilst no consistent downregulation was observed (Fig. S11).
3.5. DCE-MRI Response Highlights Differential Regulation of VEGF-blockade
Firstly, we divide patients into responders and non-responders using a DCE-MRI binary classification. Specifically, only patients showing 30% or greater reduction in median Ktrans were categorised as responders (Ah-See et al., 2008). This classification might seem conservative, however it considers the variability in DCE-MRI parameter estimation which has been discussed extensively in previous work (Othman et al., 2016, Beuzit et al., 2016, Wang et al., 2015). In particular, it defines as significant changes, hence response to treatment, only those changes in median Ktrans which are above the average level of noise observed in our (Ah-See et al., 2008) and other group's (Li et al., 2014) previous breast cancer studies.
To assess this classification, we exploited recently published human cancer angiogenesis (Masiero et al., 2013) and VEGF-blockade (Bais et al., 2011) signatures. These identified coherent response groups (Fig. S12), and a group of 12 patients with consistent downregulation of DCE-MRI and these signatures. Conversely, a coherent resistance profile was identified in 5 patients (Fig. S12). Differential pathway activation analysis was undertaken (Table S6, Fig. S12) which considers linked expression of genes within the same pathway, recovering some statistical power. This revealed decreased activation of the Hippo pathway in responders; namely, downregulation of STK3 and linked upregulation of LATS2, an inhibitor angiogenesis (Dai et al., 2013). Furthermore, activation of axon guidance, including VEGFA-regulated RND1 and PLXNA1, decreased in responders whilst increased in non-responders; and mTOR signaling and carbohydrate metabolism, including PGM1, increased in non-responders whilst decreased in responders. Although agreement between DCE-MRI and molecular profiling is encouraging, it is clear that when using such DCE-MRI binary classification, the non-responder group is small (n = 8), hindering statistical power.
Next, we considered Ktrans as continuous response variable and discovered 471 genes whose expression changes post-bevacizumab were positively correlated with total Ktrans changes (FDR < 0.05), showing greater downregulation in responder cases (Table S7). Pathways analysis (Table S8) indicated downregulation of Wnt signaling (FDR = 0.002), extracellular matrix (ECM) receptor interaction (FDR < 0.0001), cell adhesion molecules (FDR = 0.0009), MAPK (FDR = 0.0105), and VEGF (FDR = 0.0123) signaling. This was confirmed by gene set analysis (GSA) where changes in MAPK, ECM and NFAT gene sets were positively correlated with change in total Ktrans (Fig. 5). Furthermore, human cancer angiogenesis and VEGF-blockade signatures were both downregulated in cases with greatest decrease in total Ktrans (Spearman rho = 0.40, P = 0.02; and rho = 0.39, P = 0.02 respectively). This confirms greater DCE-MRI response is characterised by downregulation of angiogenesis and proliferation.
Fig. 5.

Heatmap of gene set analysis for expression changes post-bevacizumab associated with DCE-MRI parameters. The DCE-MRI parameters considered were: baseline median and total Ktrans; and changes (Δ) in median and total Ktrans. Gene Set Analysis (GSA) algorithm was used (see Methods); the GSA score is shown and colour-bar is provided with absolute values. This represents the strength of the positive and/or negative association between the specific gene set (y-axis) and the specific DCE-MRI parameter (x-axis). Gene sets that were significant (FDR < 0.05) in at least one of the four analyses are shown. The sets included were the full GSEA Biocarta gene set V3 (http://www.broadinstitute.org/gsea/msigdb/collections.jsp), a previously derived hypoxia signature (Buffa et al., 2010) (not shown in the heatmap as never significant), ER and ERBB2 signaling signatures (not shown as never significant), proliferation and Wnt signatures (“Wnt signaling”) (Desmedt et al., 2008), a recently derived angiogenesis signature (“Angiogenesis”) (Masiero et al., 2013) and VEGF-blockade signature (“Anti-VEGF”) (Bais et al., 2011).
Finally, strong increase in hypoxia and HIF1A regulated genes was seen in non-responder cases (Fig. S12), and increase in hypoxia marker CA-9 by IHC was correlated with increased VEGFA protein levels by ELISA (Spearman r = 0.51, P = 0.002) (Fig. S13). This suggests that reduced angiogenesis in these tumours might cause disrupted rather than normalized blood vessel network, with consequent increase in hypoxia.
3.6. Baseline DCE-MRI Predicts Downregulation of Angiogenesis
Initial DCE-MRI Ktrans status was the strongest predictor of response, namely tumours with high median Ktrans at baseline demonstrated the greatest decrease in Ktrans post-bevacizumab (Fig. 1c). To exclude regression to mean artefacts, this was confirmed using transcriptional profiles, which showed that tumours with high baseline median Ktrans respond to bevacizumab with downregulation of angiogenesis, whilst patients with low initial values of median Ktrans show no or little variation in angiogenesis. Specifically, 110 genes showed changes after bevacizumab that were significantly negatively correlated with baseline median Ktrans, namely downregulated post-bevacizumab in tumours with higher baseline median Ktrans (Table S9). Pathways included angiogenesis, cell adhesion and NFAT (Table S10).
Similar analysis using total Ktrans identified 1080 genes whose changes were negatively correlated with baseline total Ktrans, namely downregulated post-bevacizumab in patients with high baseline total Ktrans (Table S11). All but one (TMEM129, an uncharacterized transmembrane protein) of 110 negatively correlated genes in median Ktrans analysis were confirmed in total Ktrans analysis, indicating high agreement between these two parameters but possibly greater power when using total Ktrans. Similarly, GSA highlighted 11 gene sets whose post-bevacizumab changes were negatively correlated with baseline median Ktrans, and 29 with baseline total Ktrans (Fig. 5). Of note, angiogenesis signatures were top-downregulated sets, followed by wide spectra of angiogenesis-related pathways.
4. Discussion
Monitoring and early prediction of patient response to therapy is increasingly important, particularly because of heterogeneity of response to expensive or toxic agents, and missed opportunity of effective therapy by early switching. Many drugs, although targeted to highly specific processes, are not targeted to genetic lesion, so it is difficult to select patients on baseline genetic criteria. For anti-angiogenic treatment, this is even more important as effectiveness is small overall but likely to have substantial benefit in minority of patients. It is notable that median progression-free interval in many trials of bevacizumab is often only 4–6 weeks, and 1–2 weeks overall survival benefit, suggesting this early pattern is what sets agenda for final benefit. Our aim was to link macroscopic heterogeneity of response to molecular pathways that are targetable.
We combined monitoring by DCE-MRI and molecular profiling with short-term first-line treatment in primary breast cancer to measure early response to bevacizumab. Clearly when analysing whole tumor by imaging we can never sample to the same extent, but only conduct small number of biopsies. Furthermore, previous work from our (Ah-See et al., 2008) and other groups (Othman et al., 2016, Beuzit et al., 2016, Wang et al., 2015) has demonstrated DCE-MRI parameter estimation can depend on a variety of factors including normal physiological variability, MRI set-up and acquisition protocols, and different image processing methods. Strikingly, in spite of this, we observed strong correlations of functional imaging with molecular changes, regardless of the DCE-MRI method used to categorize the patients (binary response, continuous median Ktrans and total Ktrans), reflecting heterogeneity of response and providing added molecular information.
DCE-MRI showed initial response to bevacizumab in most patients. Specifically, 91% (32/35) patients showed decrease in median Ktrans, and 77% (27/35) had a > 30% decrease. This indicates a general initial vascular normalisation, and downregulation of angiogenic factors ESM1 and FLT1. Interestingly, another family member, FLT4, was predictive for bevacizumab response in ovarian cancer (Collinson et al., 2013).
Importantly, strong downregulation of proliferation post-bevacizumab suggests a critical role of even aberrant leaky vessels in providing enough nutrients and oxygen for growth. This has implication for current routine practice, and supports combination of anti-angiogenic treatment with chemotherapy to further inhibit growth as is usually the case, but with little previous rationale.
Whilst this work was submitted, a study was published (Tolaney et al., 2015) with similar window design, but a lower dose of bevacizumab. Decreased microvessel density [MVD] at 2 weeks was observed. We, however, reveal the complex response to anti-bevacizumab therapy at an early timepoint, when this response can be evaluated alone.
Notwithstanding initial response, our study revealed heterogeneity ranging from tumours developing central necrosis to continuous growth. In fact, it is clear that a subgroup of patients experienced vessels normalisation, with decreased proliferation. Equally, there is another group of patients where HIF1α and regulated genes (e.g. CA9), necrosis and angiogenesis increase. So we showed that vascular normalisation is likely to be a mechanism of response, but there is a subgroup of patients where changes are induced in response to hypoxia which can actually induce further vessel production and resistance, such as induction of CXCR4 and metabolism. This suggests a feedback loop in these patients where an initial response potentially shuts down, rather than normalize, blood supply with consequent induction of hypoxia. These resistance mechanisms offer great opportunities for combination therapy as we have already demonstrated in preclinical models (McIntyre et al., 2012, Favaro et al., 2012, Li et al., 2007).
Importantly, immune checkpoint genes were induced on treatment, including CTLA4, and its ligand CD86, which could antagonize CD28 T-cell activation. Although CTLA4 has greater CD86 affinity than CD28 (Linsley et al., 1991); co-presence of activating/inhibiting signaling suggests co-existence of recruited versus non-recruited T-cell pockets in these tumours. Recently, Programmed Death Ligand 1, PD-L1, not showing significant changes in this study but member of the same CD28/CTLA4 family of receptors, has been shown to be direct target of HIF1A, and when blocked under hypoxia it enhanced myeloid-derived suppressor cells-mediated T-cell activation (Noman et al., 2014). We cannot ascertain at this stage whether this is to specific antibody effects or interaction with hypoxia; however, these findings support benefit from combination of bevacizumab with novel immune checkpoint inhibitors to restore and boost T-cell immune response.
Finally, we found that macroscopic analysis of whole tumours could predict response, and baseline Ktrans was the strongest predictor, which suggests VEGF is main determinant of vascular leakiness, though not necessarily angiogenesis. Although baseline gene expression did not strongly correlate with MRI variation, once an environmental stress was induced there was strong concordance between imaging and mRNA changes, enabling patient classification by gene response linked to imaging changes with therapy implications. Control theory indicates difficulty of relating response to baselines if rules for connection are unknown, but our results show how quickly tumours adapt and then allow the characteristics to be defined.
We conclude that bevacizumab has been prematurely discontinued, rather than focusing on finding subgroups of patients who most benefit using monitoring during 2 week window before continuing therapy. This would be cost-effective and help stratify patients for combination or other targeted therapies.
Finally, we suggest new paradigms for clinical research. Firstly, trials should incorporate appropriate initial enrichment of patients with high Ktrans, and a range of therapeutic options to meet potential early resistance pathways induced. Then, early imaging will be needed to stratify patients into categories likely to have different mechanism of adaptation, and biopsies to select patients for appropriate combinations. Repeatability of these assays makes this widely feasible. Multi-arm adaptive trials are ongoing using molecular markers for targeted agents, but we suggest this needs to be further modified by much earlier adaptation when using drugs affecting the tumor microenvironment.
Author Contributions
SM, FMB, NPH, ALH, AP, AM designed the study. AM, AP and ALH co-supervised the clinical implementation of the study. SM, SLi and SL collected the clinical data; SM and AJ performed experiments. FMB performed the transcriptomic data analysis, with contributions from RvS and LK. NPH analyzed imaging data with contribution from RA. FMB supervised the analysis and integration of molecular, clinical and imaging data. FMB wrote the manuscript with contribution from all authors. All authors approved the final version of the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.07.017.
Contributor Information
Francesca M. Buffa, Email: francesca.buffa@oncology.ox.ac.uk.
Adrian L. Harris, Email: adrian.harris@oncology.ox.ac.uk.
Appendix A. Supplementary data
Supplementary figures.
Supplementary tables.
Supplementary material.
References
- Ah-See M.L., Makris A., Taylor N.J. Early changes in functional dynamic magnetic resonance imaging predict for pathologic response to neoadjuvant chemotherapy in primary breast cancer. Clin. Cancer Res. 2008;14(20):6580–6589. doi: 10.1158/1078-0432.CCR-07-4310. [DOI] [PubMed] [Google Scholar]
- Al-Rawi M.A., Watkins G., Mansel R.E., Jiang W.G. Interleukin 7 upregulates vascular endothelial growth factor D in breast cancer cells and induces lymphangiogenesis in vivo. Br J Surg. 2005;92(3):305–310. doi: 10.1002/bjs.4832. [DOI] [PubMed] [Google Scholar]
- Bais C., Singh M., Kaminker J., Brauer M. 2011. Biological Markers for Monitoring Patient Response to VEGF Antagonists. Google Patents. [Google Scholar]
- Bales E., Mills L., Milam N. The low molecular weight cyclin E isoforms augment angiogenesis and metastasis of human melanoma cells in vivo. Cancer Res. 2005;65(3):692–697. [PubMed] [Google Scholar]
- Bear H.D., Tang G., Rastogi P. Bevacizumab added to neoadjuvant chemotherapy for breast cancer. N. Engl. J. Med. 2012;366(4):310–320. doi: 10.1056/NEJMoa1111097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuzit L., Eliat P.A., Brun V. Dynamic contrast-enhanced MRI: study of inter-software accuracy and reproducibility using simulated and clinical data. Journal of magnetic resonance imaging : JMRI. 2016;43(6):1288–1300. doi: 10.1002/jmri.25101. [DOI] [PubMed] [Google Scholar]
- Buffa F.M., Harris A.L., West C.M., Miller C.J. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br. J. Cancer. 2010;102(2):428–435. doi: 10.1038/sj.bjc.6605450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P., Jain R.K. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- Collinson F., Hutchinson M., Craven R.A. Predicting response to bevacizumab in ovarian cancer: a panel of potential biomarkers informing treatment selection. Clin. Cancer Res. 2013;19(18):5227–5239. doi: 10.1158/1078-0432.CCR-13-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., She P., Chi F. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J. Biol. Chem. 2013;288(47):34041–34051. doi: 10.1074/jbc.M113.518019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmedt C., Haibe-Kains B., Wirapati P. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin. Cancer Res. 2008;14(16):5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- Earl H.M., Hiller L., Dunn J.A. Efficacy of neoadjuvant bevacizumab added to docetaxel followed by fluorouracil, epirubicin, and cyclophosphamide, for women with HER2-negative early breast cancer (ARTemis): an open-label, randomised, phase 3 trial. Lancet Oncol. 2015;16(6):656–666. doi: 10.1016/S1470-2045(15)70137-3. [DOI] [PubMed] [Google Scholar]
- Favaro E., Bensaad K., Chong M.G. Glucose utilization via glycogen phosphorylase sustains proliferation and prevents premature senescence in cancer cells. Cell Metab. 2012;16(6):751–764. doi: 10.1016/j.cmet.2012.10.017. [DOI] [PubMed] [Google Scholar]
- Hegde P.S., Jubb A.M., Chen D. Predictive impact of circulating vascular endothelial growth factor in four phase III trials evaluating bevacizumab. Clin. Cancer Res. 2013;19(4):929–937. doi: 10.1158/1078-0432.CCR-12-2535. [DOI] [PubMed] [Google Scholar]
- Jain R.K. Antiangiogenesis strategies revisited: from starving tumors to alleviating hypoxia. Cancer Cell. 2014;26(5):605–622. doi: 10.1016/j.ccell.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts D., Lenz H.J., de Haas S., Carmeliet P., Scherer S.J. Markers of response for the antiangiogenic agent bevacizumab. J. Clin. Oncol. 2013;31(9):1219–1230. doi: 10.1200/JCO.2012.46.2762. [DOI] [PubMed] [Google Scholar]
- Lehmann B.D., Bauer J.A., Chen X. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.L., Sainson R.C., Shi W. Delta-like 4 Notch ligand regulates tumor angiogenesis, improves tumor vascular function, and promotes tumor growth in vivo. Cancer Res. 2007;67(23):11244–11253. doi: 10.1158/0008-5472.CAN-07-0969. [DOI] [PubMed] [Google Scholar]
- Li X., Arlinghaus L.R., Ayers G.D. DCE-MRI analysis methods for predicting the response of breast cancer to neoadjuvant chemotherapy: pilot study findings. Magn. Reson. Med. 2014;71(4):1592–1602. doi: 10.1002/mrm.24782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley P.S., Brady W., Urnes M., Grosmaire L.S., Damle N.K., Ledbetter J.A. CTLA-4 is a second receptor for the B cell activation antigen B7. J. Exp. Med. 1991;174(3):561–569. doi: 10.1084/jem.174.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini M., Toker A. NFAT proteins: emerging roles in cancer progression. Nat. Rev. Cancer. 2009;9(11):810–820. doi: 10.1038/nrc2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiero M., Simoes F.C., Han H.D. A core human primary tumor angiogenesis signature identifies the endothelial orphan receptor ELTD1 as a key regulator of angiogenesis. Cancer Cell. 2013;24(2):229–241. doi: 10.1016/j.ccr.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre A., Patiar S., Wigfield S. Carbonic anhydrase IX promotes tumor growth and necrosis in vivo and inhibition enhances anti-VEGF therapy. Clin. Cancer Res. 2012;18(11):3100–3111. doi: 10.1158/1078-0432.CCR-11-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noman M.Z., Desantis G., Janji B. PD-L1 is a novel direct target of HIF-1alpha, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J. Exp. Med. 2014;211(5):781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor J.P., Jayson G.C. Do imaging biomarkers relate to outcome in patients treated with VEGF inhibitors? Clin. Cancer Res. 2012;18(24):6588–6598. doi: 10.1158/1078-0432.CCR-12-1501. [DOI] [PubMed] [Google Scholar]
- Othman A.E., Falkner F., Kessler D.E. Comparison of different population-averaged arterial-input-functions in dynamic contrast-enhanced MRI of the prostate: effects on pharmacokinetic parameters and their diagnostic performance. Magn. Reson. Imaging. 2016;34(4):496–501. doi: 10.1016/j.mri.2015.12.009. [DOI] [PubMed] [Google Scholar]
- Rossari J.R., Metzger-Filho O., Paesmans M. Bevacizumab and breast cancer: a meta-analysis of first-line phase III studies and a critical reappraisal of available evidence. J Oncol. 2012;2012:417673. doi: 10.1155/2012/417673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaltriti M., Eichhorn P.J., Cortes J. Cyclin E amplification/overexpression is a mechanism of trastuzumab resistance in HER2 + breast cancer patients. Proc. Natl. Acad. Sci. U. S. A. 2011;108(9):3761–3766. doi: 10.1073/pnas.1014835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikov W.M., Berry D.A., Perou C.M. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance) J. Clin. Oncol. 2015;33(1):13–21. doi: 10.1200/JCO.2014.57.0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolaney S.M., Boucher Y., Duda D.G. Role of vascular density and normalization in response to neoadjuvant bevacizumab and chemotherapy in breast cancer patients. Proc. Natl. Acad. Sci. U. S. A. 2015;112(46):14325–14330. doi: 10.1073/pnas.1518808112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Minckwitz G., Eidtmann H., Rezai M. Neoadjuvant chemotherapy and bevacizumab for HER2-negative breast cancer. N. Engl. J. Med. 2012;366(4):299–309. doi: 10.1056/NEJMoa1111065. [DOI] [PubMed] [Google Scholar]
- Wallace J.A., Li F., Balakrishnan S. Ets2 in tumor fibroblasts promotes angiogenesis in breast cancer. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0071533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Su Z., Ye H. Reproducibility of dynamic contrast-enhanced MRI in renal cell carcinoma: a prospective analysis on intra- and Interobserver and scan-rescan performance of pharmacokinetic parameters. Medicine. 2015;94(37) doi: 10.1097/MD.0000000000001529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedam S.B., Low J.A., Yang S.X. Antiangiogenic and antitumor effects of bevacizumab in patients with inflammatory and locally advanced breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2006;24(5):769–777. doi: 10.1200/JCO.2005.03.4645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.
Supplementary tables.
Supplementary material.