MMD1, a reader of histone modifications, is required for progressive compaction of meiotic chromosomes during prophase I in a manner partially dependent on expression of the condensin gene CAP-D3.
Abstract
Chromosome condensation, a process mediated by the condensin complex, is essential for proper chromosome segregation during cell division. Unlike rapid mitotic chromosome condensation, meiotic chromosome condensation occurs over a relatively long prophase I and is unusually complex due to the coordination with chromosome axis formation and homolog interaction. The molecular mechanisms that regulate meiotic chromosome condensation progression from prophase I to metaphase I are unclear. Here, we show that the Arabidopsis thaliana meiotic PHD-finger protein MMD1/DUET is required for progressive compaction of prophase I chromosomes to metaphase I bivalents. The MMD1 PHD domain is required for its function in chromosome condensation and binds to methylated histone tails. Transcriptome analysis and qRT-PCR showed that several condensin genes exhibit significantly reduced expression in mmd1 meiocytes. Furthermore, MMD1 specifically binds to the promoter region of the condensin subunit gene CAP-D3 to enhance its expression. Moreover, cap-d3 mutants exhibit similar chromosome condensation defects, revealing an MMD1-dependent mechanism for regulating meiotic chromosome condensation, which functions in part by promoting condensin gene expression. Together, these discoveries provide strong evidence that the histone reader MMD1/DUET defines an important step for regulating the progression of meiotic prophase I chromosome condensation.
INTRODUCTION
Chromosome condensation is the process of packaging long, thin chromosomes into short, thick entities, ensuring chromosome segregation during both mitosis and meiosis (Wood et al., 2010; Hirano, 2012). Mitotic chromosomes are condensed in an apparently continuous fashion into metaphase chromosomes during a relatively short prophase. Unlike mitosis, meiosis includes an unusually long prophase I, during which chromosome condensation must proceed in coordination with chromosome axis formation and interactions between homologous chromosomes (homologs), including double-strand break (DSB) formation, recombination, pairing, and synapsis (Zickler and Kleckner, 1999; Hamant et al., 2006; Mainiero and Pawlowski, 2014; Mercier et al., 2015). After recombination is completed and the synaptonemal complex (SC) is disassembled, further condensation shortens the chromosomes through late prophase I (diplotene and diakinesis), eventually resulting in metaphase I chromosomes. However, knowledge about factors and mechanisms that regulate the progression of chromosome condensation is rather scarce.
Chromosome condensation requires a large conserved multisubunit complex called condensin (Hirano et al., 1997; Freeman et al., 2000). Most eukaryotes possess two condensin isoforms, condensin I and II (Wood et al., 2010; Mainiero and Pawlowski, 2014), which share a core heterodimer composed of the Structural Maintenance of Chromosome 2 (SMC2) and SMC4 proteins but have different accessory subunits, including CAP-H (Chromosome Associated Protein H), CAP-D2, and CAP-G present in condensin I, and CAP-H2, CAP-D3, and CAP-G2 in condensin II (Hirano, 2005). SMC2 and SMC4 are essential genes, and corresponding mutants in Arabidopsis thaliana, yeast, Drosophila melanogaster, and Caenorhabditis elegans are lethal (Hirano, 2012). In addition, genetic studies using conditional alleles, tissue-specific disruption, and RNAi targeting condensin genes show that they are also important for mitotic and meiotic chromosome condensation and/or segregation in C. elegans (Hagstrom et al., 2002; Chan et al., 2004), D. melanogaster (Hartl et al., 2008), yeast (Yu and Koshland, 2003), and Arabidopsis (Siddiqui et al., 2003; Schubert et al., 2013; Smith et al., 2014).
During mitosis, condensin I and II have distinct contributions to condensation and associate with chromosomes in a sequential manner (Hirota et al., 2004; Ono et al., 2004). Furthermore, the relative ratio of condensin I and II affects metaphase chromosome conformation (Shintomi and Hirano, 2011). Moreover, genes encoding condensin II subunits, such as CAP-D3, have been implicated in meiosis (Lee et al., 2011; Hirano, 2012; Smith et al., 2014). In yeast (Saccharomyces cerevisiae), condensin plays a role in the association of other meiotic chromosome components, to promote SC assembly and facilitate chromosome segregation (Yu and Koshland, 2003; Yu and Koshland, 2005). In the nematode C. elegans, condensin I and II play distinct roles in the regulation of the number and position of DSBs, affecting crossover number and distribution (Mets and Meyer, 2009). Although phosphorylation of condensin I and II can stimulate their activity for mitotic chromosome assembly (Abe et al., 2011), the mechanisms that regulate condensin gene expression are largely unknown.
In Arabidopsis, the MMD1 (MALE MEIOCYTE DEATH1)/DUET PHD-finger protein is required for normal male fertility and meiosis (Reddy et al., 2003; Yang et al., 2003). In particular, the mmd1 mutation was shown to cause abnormal cell death of male meiocytes, as supported by TUNEL signals (indicating the presence of fragmented DNA), which could not be detected in normal meiocytes (Yang et al., 2003). A recent study showed that DUET/MMD1 plays a role in meiotic I-independent cell division, likely by regulating the expression of the male meiosis II-related genes TDM and JAS (Andreuzza et al., 2015), but the mutant chromosome defects in prophase I were not clear, and the underlying molecular mechanism is also unknown. We report here that mmd1 mutant meiocytes in Arabidopsis have partially condensed chromosomes, uncovering a regulatory step of meiotic chromosome condensation and providing an opportunity to study such regulation. We show here that the MMD1 PHD binds to the methylated histone mark H3K4me2/3 in vitro and in vivo, in general agreement with the recent report of the DUET PHD binding to histone peptides (Andreuzza et al., 2015). ChIP-PCR analysis showed that MMD1 is enriched at the CAP-D3 promoter in vivo, supporting a mechanism by which MMD1 promotes the normal CAP-D3 expression in male meiocytes. Furthermore, cap-d3 male meiocytes also exhibit a defect in chromosome condensation, similar to the observation in mmd1, strongly supporting the idea that MMD1 facilitates the progression of meiotic chromosome condensation, at least in part, by promoting the expression of condensin genes. Together, our results uncover an MMD1-dependent regulatory mechanism for progressive meiotic chromosome condensation.
RESULTS
MMD1 Is Required for Normal Progression of Meiotic Chromosome Condensation
MMD1/DUET was found to be required for normal male meiosis and fertility in Arabidopsis (Reddy et al., 2003; Yang et al., 2003; Andreuzza et al., 2015), and a recent study showed that DUET/MMD1 has a role in the delayed meiotic progression and is required for the second meiotic division (Andreuzza et al., 2015), but its role in meiotic prophase I was not studied in depth. To learn more about the mutant meiotic phenotypes, we compared wild-type and mmd1 chromosomes using 4′,6-diamidino-2-phenylindole (DAPI) staining and fluorescent in situ hybridization (FISH) with a centromere probe. At zygotene and pachytene stages in prophase I, DAPI-stained mmd1 meiocytes showed similar chromosome morphologies to those of the wild type (Supplemental Figures 1A and 1B). In addition to the previously reported nonhomologous chromosome association (Yang et al., 2003), mmd1 also had a slight reduction in condensation at diakinesis (Supplemental Figure 1B). At metaphase I, wild-type plants have five highly condensed bivalents (pairs of associated homologs) positioned at the equatorial plate through the force of the spindle (Figure 1A). In contrast, mmd1 metaphase I chromosomes showed varying degrees of reduced condensation, with weak (15/37) (Figure 1B), moderate (10/37) (Figure 1C), and severe defects (12/37) (Figure 1D). Diffuse chromosome signals as well as expanded centromere signals suggest genome-wide alterations in chromosome condensation. We estimated the areas of the centromere fluorescence signals and metaphase I chromosomes from wild-type and mmd1 plants and found both to be significantly larger in mmd1 than those in the wild type (Figures 1E to 1G). Previous studies showed that wild-type and mmd1 chromosomes appeared similar through the pachytene stage, as revealed by DAPI (Siddiqui et al., 2003; Yang et al., 2003), supporting the idea that mmd1 meiotic chromosomes can initiate condensation, but fail to achieve the final highly condensed stage, suggesting a regulatory defect in the progression of condensation.
Figure 1.
Defects in mmd1 in Meiotic Chromosome Condensation.
(A) to (D) Metaphase I chromosome behaviors analyzed by FISH in the wild type (A) and mmd1 ([B] to [D]). Red color represents the centromere FISH signals.
(E) to (G) Estimate of average areas of fluorescent signals in the centromere and metaphase I chromosome area in the wild type and mmd1 determined by Image J.
(E) Determination of the average area of fluorescence in wild-type and mmd1 centromeres. The total number of cells used for analysis of wild-type and mmd1 were 31 and 37, respectively, with weak (15), moderate (10), and severe (12) phenotypes of mutant cells. Images in (F) were used to determine chromosome area. Average metaphase chromosome area for the wild type (n = 53) and mmd1 (n = 67) is shown in (G). Bars = 5 μm. Data are represented as mean ± sd. *P < 0.05 and **P < 0.01, two-tailed Student’s t test.
Chromosome Condensation Defect in mmd1 Is Independent of the Initiation of Meiotic Recombination and Subsequent Synapsis
Because meiotic chromosome condensation occurs in conjunction with pairing and synapsis, it is possible that the defect in mmd1 could be associated with abnormal homolog interaction, which requires DSB formation. In Arabidopsis, DSB formation depends on SPO11-1 (Grelon et al., 2001), a homolog of SPO11 encoding a topoisomerase-like protein conserved in eukaryotes (Keeney et al., 1997), as well as several other proteins: SPO11-2 (Stacey et al., 2006; Hartung et al., 2007), PUTATIVE RECOMBINATION INITIATION DEFECT1 (PRD1), PRD2, and PRD3/Os-PAIR1(Nonomura et al., 2004; De Muyt et al., 2007, 2009), DSB FORMATION (Zhang et al., 2012), and MEIOTIC TOPOISOMERASE VIB-LIKE (Vrielynck et al., 2016). To investigate whether abnormal chromosome condensation in mmd1 depends on meiotic DSB formation, we created an mmd1 spo11-1-3 double mutant, which produced meiocytes with 10 univalents (Figure 2A), similar to the spo11-1-3 single mutant (Figure 2A). In addition, the double mutant chromosomes were also not well condensed at both diakinesis (Figure 2A) and metaphase I (Figure 2B), indicating that the chromosome condensation defect in mmd1 is independent of SPO11-induced meiotic DSBs. The observation of normal appearing pachytene chromosomes suggested that the DSB could still be normal in mmd1. To test this hypothesis, we used an antibody against a DSB marker protein, γH2Ax (Lowndes and Toh, 2005), and found no obvious differences between the wild type (an average of 208 foci, n = 36) and mmd1 (an average of 201 foci, n = 40) at leptotene (P = 0.058) (Figure 2C) and pachytene (wild type average of 59 foci from 28 cells and mmd1 average of 55 foci from 35 cells, P = 0.144) (Figure 2D). The normal formation of DSBs in mmd1 was further supported by additional experiments with antibodies against RAD51 and DMC1, which are recombinases required for single end invasion (Li et al., 2004). At pachytene, both proteins showed similar average numbers of signal foci, with RAD51 showing 53 foci (n = 20) in the wild type and 52 foci (n = 24) in mmd1 (P = 0.63) and DMC1 showing 60 foci (n = 21) in the wild type and 63 foci in mmd1 (n = 32; P = 0.24) (Supplemental Figure 2). Therefore, we conclude that DSB formation does not appear to be affected in mmd1.
Figure 2.
Chromosome Condensation Defect in mmd1 Is Independent of Meiotic Double-Strand Break Formation.
(A) and (B) mmd1 spo11-1-3 double mutant male meiocytes at metaphase I showed 10 univalents, similar to that of spo11-1-3, but with defects in chromosome condensation in diakinesis (A) and metaphase I (B).
(C) and (D) Immunolocalization of γH2Ax showed indistinguishable signals between the wild type and mmd1 in both leptotene (C) and pachytene (D). Bar = 5 μm.
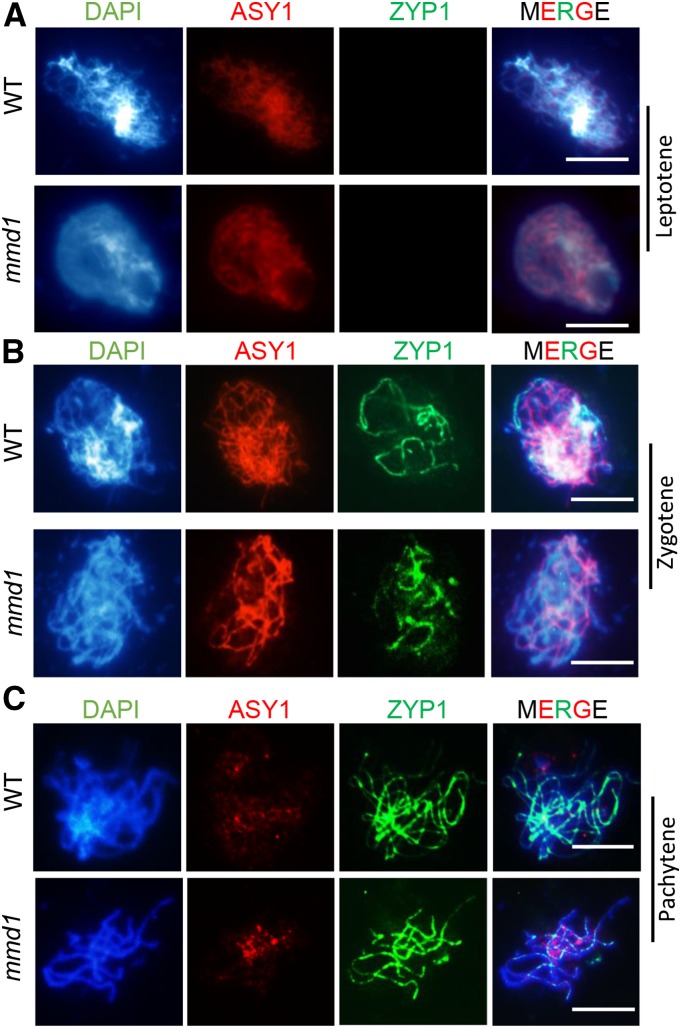
Following DSB-dependent homologous chromosome pairing, the SC, a proteinaceous structure formed between homologs, is required for the stabilization of strand invasion intermediates and facilitates subsequent homolog recombination (Zickler and Kleckner, 1999). We examined the localization patterns of the axial element protein ASY1 (Armstrong et al., 2002) and the central element protein ZYP1 (Higgins et al., 2005) in wild-type and mmd1 meiocytes (Figure 3). At leptotene and zygotene, the labeling patterns of ASY1 with linear signals on chromosomes were similar between the wild type and mmd1 (Figures 3A and 3B). At pachytene, ASY1 signals became diffuse and weak, but no obvious difference was observed in mmd1 compared with the wild type (Figure 3C). Thus, we conclude that mmd1 mutation does not affect ASY1 axis formation. Similar to ASY1 distribution, the initiation of ZYP1 signals at zygotene (Figure 3B) and the linear ZYP1 distribution on chromosomes at pachytene (n = 128) (Figure 3C) appeared normal in mmd1. The normal localization of ZYP1 suggests no obvious defect in SC formation in mmd1.
Figure 3.
Mutation of MMD1 Has No Influence on Axis Element and SC Formation.
The lateral element ASY1 and the central element ZYP1 in the SC appear to be unaffected in mmd1 in leptotene (A), zygotene (B), and pachytene (C). Bar = 5 μm.
The mmd1 Mutation Has a Slight Effect on Meiotic Crossover Formation
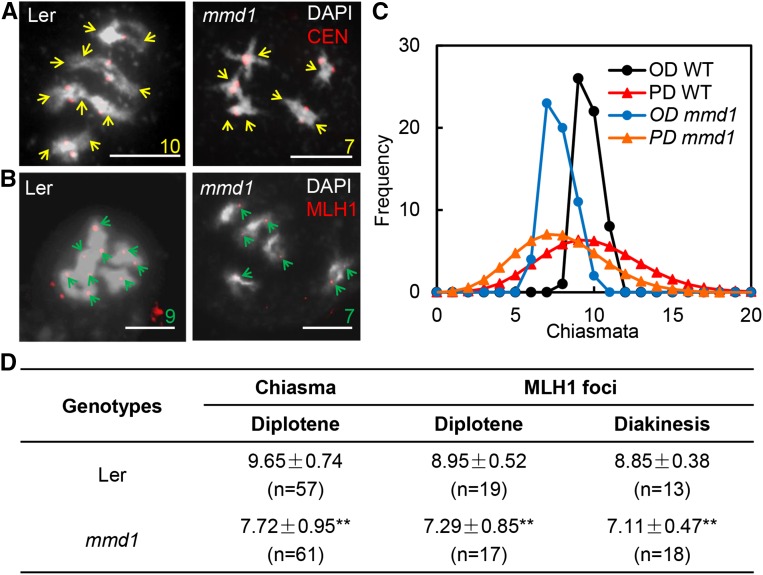
Previous studies in Arabidopsis showed that condensin proteins have a role in the frequency of meiotic recombination (Smith et al., 2014). We thus examined the recombination frequency in the wild type and mmd1 by counting the chiasmata number at diplotene (Figure 4). Wild-type meiocytes showed an average of 9.65 chiasmata (n = 57), while the mmd1 meiocytes exhibited a reduction of ∼2, with an average of 7.72 chiasmata (n = 61) (Figure 4D). Arabidopsis has two types of crossovers (COs): interference-sensitive (Type I) and interference-insensitive (Type II) COs (Copenhaver et al., 2002; Berchowitz et al., 2007). To explore whether the mmd1 mutation has an effect on CO interference, we compared chiasma distributions in the wild type and mmd1. The chiasma distribution in both wild type and mmd1 significantly deviated from Poisson distribution (P < 0.001) (Figure 4C), indicative of interference. To obtain independent evidence for which type of CO was decreased in mmd1, we examined the MLH1 foci, which is a marker for type I CO (Jackson et al., 2006), at both diplotene and diakinesis in the wild type and mmd1 (Figures 4B and 4D). We found that, at both diplotene and diakinesis, mmd1 had reduced MLH1 foci (P < 0.01) compared with the wild type (Figure 4D). Together, these results provide evidence that the mmd1 mutation has a slight, possibly indirect, effect on type I CO formation.
Figure 4.
Meiotic Recombination Frequency Is Slightly Reduced in mmd1.
(A) Images of early diakinesis were used to estimate the positions of chiasmata in Ler and mmd1. Chromosomes were stained by DAPI (white), and red signals represent centromere signals revealed by FISH. The yellow arrows show chiasmata sites on the chromosomes, showing 10 and 7 chiasmata sites in the wild type and mmd1, respectively.
(B) Immunolocalization of MLH1, a type I CO mark, in Ler and mmd1 diakinesis bivalents. Red dots represent the MLH1 signals, and green arrows mark the type I CO positions. Ler has 9 to 10 MLH1 signals, while mmd1 only has 6 to 7 signals.
(C) Observed Poisson distribution of chiasmata in the wild type and mmd1. OD represents observed distribution and PD represents predicted Poisson distribution.
(D) Statistics of chiasmata and number of COs at diplotene and diakinesis in the wild type and mmd1. Data are represented as mean ± sd.
Bar = 10 μm. **P < 0.01, two-tailed Student’s t test.
The MMD1 PHD Finger Binds to the Histone H3K4 Tail and Is Required for Normal Chromosome Condensation
MMD1/DUET encodes a PHD finger protein that is required for normal male fertility and meiosis in Arabidopsis (Reddy et al., 2003; Yang et al., 2003). PHD fingers are known to bind to histone peptides (Patel and Wang, 2013), and a recent study showed that the DUET PHD domain is required for specifically binding to H3K4me2 and rescuing the infertility of the duet mutant (Andreuzza et al., 2015). To further investigate possible binding activities of the MMD1 PHD finger, we first compared the MMD1 PHD finger with different types of known PHD fingers (Patel and Wang, 2013) in an alignment using Molecular Evolutionary Genetics Analysis software (MEGA 6.0) (Tamura et al., 2013) and found that MMD1 PHD contains a Cys4-His-Cys3 conserved motif (Figure 5A) that is predicted to forms an aromatic cage, which can be stabilized by two Zn ions; this conserved motif of known PHD proteins can interact with different modified histone peptides (Patel and Wang, 2013). In addition, the absence of a tyrosine in MMD1 PHD could serve as a multifarious reader of different histone modifications (Li and Li, 2012), suggesting that MMD1 PHD may have binding capacity for multiple modified histone peptides. To test whether the MMD1 PHD finger can bind modified histone peptides, we performed protein gel blot analysis (Figure 5B) and other in vitro peptide binding assays by dot blot (Supplemental Figure 3) and found that a purified recombinant MMD1 PHD finger (601 to 659 amino acids) could bind to H3K4me2/me3, H3K9me2, and H3S10P. These results are similar to the previous finding that other plant PHD fingers could also bind to several histone peptides (Du et al., 2012; Qian et al., 2012), but differ slightly from the specific binding to H3K4me2 observed with a shorter peptide of DUET PHD (606 to 656 amino acids) (Andreuzza et al., 2015). We further quantified these interactions using isothermal titration calorimetry and found that the MMD1 PHD finger had a higher affinity for H3K4me2 (with a Kd of 2.1 μM) than H3K4me3 (with a Kd of 6.5 μM) and the other peptides tested (Figure 5C).
Figure 5.
Histone Binding Assay of the MMD1 PHD Finger, Which Is Required for Condensation.
(A) Alignment of the MMD1 PHD with known PHD fingers from different organisms. The residues marked in green are those in a Cys4-His-Cys3 conserved motif that forms an aromatic cage, which is stabilized by two Zn ions and promotes interaction with different modified histone peptides. The residues marked in yellow are the conserved amino acids for recognition of H3K4me3 through the formation of a pocket, but MMD1 lacks the amino acid Y (Tyr, marked with a red star).
(B) A recombinant GST-PHD (601 to 659 amino acids) fusion protein was bound to biotin-labeled peptides (as indicated above) and precipitated using Streptavidin-tagged beads; the precipitate mixture was separated by SDS-PAGE and the fusion protein was detected with a GST antibody. The biotin-labeled peptides were detected using HRP-Streptavidin, while nonmethylated H3 peptides were used as the control.
(C) The binding affinity was determined by ITC binding curves for complex between PHD and the indicated peptides in terms of Kd values.
(D) Pollen viability in metaphase I determined by Alexander staining and DAPI. MMD1 in mmd1 refers to the expression of full-length MMD1 in the mmd1 mutant background. MMD1ΔPHD in mmd1 refers to the expression of MMD domain (deletion of the PHD domain) in the mmd1 mutant background. The data are based on 21 and 31 transgenic plants harboring full-length MMD1 and MMD1ΔPHD, respectively. Bars = 50 μm in the upper panel and 5 μm in the lower panel.
(E) qRT-PCR analysis of MMD1 expression level in wild-type and transgenic plants. Data are represented as mean ± se from three technical replicates.
To test whether the PHD finger is essential for MMD1 function in vivo, we expressed the full-length MMD1 and MMD1ΔPHD (lacking the PHD domain) transgenes in the mmd1 heterozygous background. Subsequently, we obtained a total of 21 and 31 individual lines expressing the full-length MMD1 and MMD1ΔPHD mRNAs, respectively, in the homozygous mmd1 background. The full-length MMD1 transgene completely rescued the mmd1 mutant defects, including pollen viability and chromatin condensation (Figure 5D), whereas MMD1ΔPHD could not rescue the mmd1 phenotypes (Figure 5D). This failure to rescue was not due to insufficient expression (Figure 5E), indicating that the PHD finger is required for MMD1-dependent fertility and male meiosis.
MMD1 Affects the Expression of Many Genes Involved in Chromosome Organization
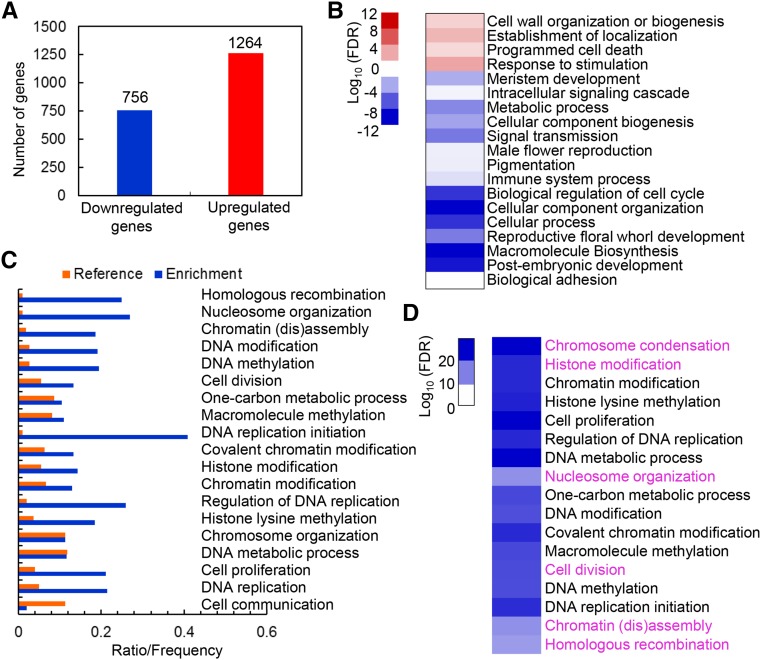
Most characterized PHD finger proteins have been reported to regulate gene transcription (Sanchez and Zhou, 2011). To investigate the potential functions of MMD1 in regulating the expression of genes involved in meiotic chromosome condensation, we implemented high-throughput whole-transcriptome sequencing (RNA-Seq) on the wild type and mmd1 stage 4-7 and 7-12 anthers, with two biological replicates (see Methods). The percentages of mapped reads matching unique genomic positions were on average 81.2 and 80.1% in the wild type and 81.1 and 82.1% in mmd1 for stage 4-7 and 7-12, respectively. Differential gene expression analyses revealed that 756 showed downregulated expression and 1264 had upregulated expression in the mmd1 stage 4-7 anthers relative to the wild type (Generalized fold change, Gfold > 1 or Gfold < −1) (Figure 6A; Supplemental Data Set 1). Similarly, analysis of gene expression in stage 7-12 anthers showed that 1395 and 1243 genes were downregulated and upregulated in mmd1, respectively, compared with the wild type (Gfold > 1 or Gfold < −1) (Supplemental Data Set 2). A comparison of the differentially expressed genes in this study and those reported previously for duet versus the wild-type using microarray data with RNA from floral buds (Andreuzza et al., 2015) indicated that 1.9 and 30% of the genes affected in duet exhibited the same direction of differential expression in stage 4-7 and 7-12 anthers, respectively (Supplemental Figures 4A and 4Band Supplemental Data Set 3). In particular, those genes highlighted as regulated by DUET, such as TDM, JAS, and OSD1 (Andreuzza et al., 2015), were only included in the mmd1 stage 7-12 anther data set (Supplemental Figures 4C and 4D). Moreover, several other genes downregulated in duet (Andreuzza et al., 2015) showed obviously reduced expression in mmd1 stage 7-12 anthers, but not in mmd1 stage 4-7 anthers (Supplemental Figures 4E and 4F), supporting a role for DUET in the second meiotic division subsequent to its function in prophase I chromosome condensation. Thus, we focused on MMD1-regulated genes in stage 4-7 anthers.
Figure 6.
Transcriptome Analysis of wild-type and mmd1 Stage 4-7 Anthers.
(A) The number of differentially expressed genes in mmd1 relative to the wild type (Gfold > 1 or Gfold < −1).
(B) Heat map showing the functional annotation of genes with upregulated and downregulated expression in mmd1 relative to the wild type.
(C) Histogram showing the enrichment of molecular function categories of genes with downregulated expression in mmd1.
(D) Heat map showing the functional annotation in chromosome organization, DNA replication, histone modification, and other enriched categories for genes with downregulated expression in mmd1.
Gene Ontology (GO) was used for the categorization of significantly differentially expressed genes between the wild type and mmd1 to identify distinct biological processes involving genes with increased and reduced expression. Among the genes with upregulated expression, cell wall organization and biogenesis (FDR = 0.0079), programmed cell death (FDR = 0.017), and response to stimulation (FDR < 10−4) were significantly enriched (Figure 6B; Supplemental Table 1). Specifically, the enriched process related to programmed cell death is consistent with the previous characterization of the role of MMD1 in regulating microsporocyte death (Yang et al., 2003). By contrast, among the genes with decreased expression, reproductive process (FDR < 10−6), cellular component organization (FDR < 10−4) and macromolecular biosynthesis (FDR < 10−14) were significantly represented (Figure 6B; Supplemental Table 1). Further analysis of the downregulated genes by AgriGO showed that chromosome organization (FDR < 10−27, 85 genes), chromatin assembly (FDR < 10−11, 23 genes), and nucleosome organization (FDR < 10−11, 18 genes) were significantly enriched categorizes (Figures 6C and 6D; Supplemental Table 2 and Supplemental Data Set 4). Among the chromosome condensation genes, four genes encoding condensin subunits were among the differentially expressed genes (Supplemental Data Set 1) and CAP-D3 (Schubert et al., 2013) showed a more than 2.5-fold difference in expression. Fifty-eight genes related to chromatin or histone modification (FDR < 10−21) were detected, especially for histone lysine methylation. We also found 35 GO-categorized genes involved in DNA replication (FDR < 10−21), especially in the initiation of replication (FDR < 10−20) (Figures 6C and 6D; Supplemental Table 2 and Supplemental Data Set 4). Moreover, the observation of 48 enriched genes associated with cell division (FDR < 10−17) is consistent to the recent discovery of the role of MMD1 in meiotic II cell division (Andreuzza et al., 2015). Finally, we found 17 preferentially expressed genes related to homologous recombination in mmd1 compared with the wild type (Figure 6C; Supplemental Tables 2 and 3 and Supplemental Data Set 4), supporting their role in meiotic recombination. Together, the transcriptome analysis provided evidence that the observed meiotic chromosome condensation defects in mmd1 may be caused by the reduced expression of condensin genes or alteration in chromosome organization.
MMD1 Regulates CAP-D3 Expression Likely by Directly Binding to Its Promoter Region
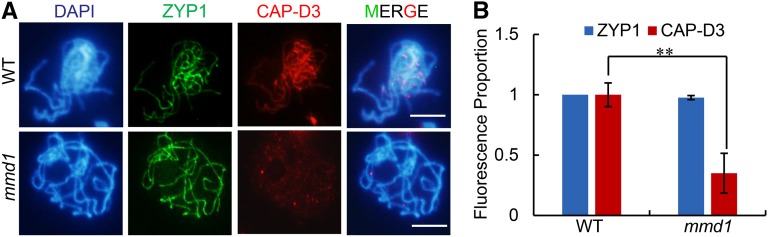
The mmd1 meiotic chromosome condensation phenotype and the mmd1 transcriptome data suggest that MMD1 might directly regulate the expression of genes encoding condensin subunits because these proteins are directly involved in condensation (Figure 7A) (Hirano, 2005). To verify the reduced expression of condensin genes in mmd1, we isolated meiocytes from the wild type and mmd1 and subjected them to qRT-PCR analysis. Consistent with the RNA-seq data, the expression of several condensin genes (CAP-D2, CAP-D3, CAP-H, and CAP-H2) was significantly reduced in mmd1 meiocytes compared with the wild type (Figure 7B), with CAP-D3 showing the greatest reduction (Figure 7B). In previous studies, Arabidopsis CAP-D3 was localized to chromatin in mitotic cells (Schubert et al., 2013; Smith et al., 2014). We therefore performed immunofluorescence experiments in meiocytes and found that the level of CAP-D3 protein was significantly lower in mmd1 chromosomes than in the wild type (Figure 8), providing additional evidence that supports a role for MMD1 in regulating meiotic condensation, in part by affecting CAP-D3 expression levels. This also suggests that the chromosome condensation defect in mmd1 occurs prior to diakinesis, while the mmd1-pachytene chromosomes were similar to those of the wild type. A possible explanation for the normal appearance of pachytene chromosome is that the mmd1 mutant could still express the condensin genes at approximate half of the normal levels, which could be sufficient for the slight condensation of chromosomes at the pachytene stage.
Figure 7.
MMD1 Regulates the Expression of the Condensin Gene CAP-D3 in Meiocytes.
(A) Schematic depiction illustrating the subunit composition of condensin complexes in plants as described in the text.
(B) qRT-PCR analysis of condensin gene expression in wild-type and mmd1 male meiocytes. Data were normalized to ACTIN expression.
(C) CAP-D3 gene structure and the positions of primers.
(D) Validation of MMD1 antibody through analysis of nuclear proteins from core flower buds by protein gel blot analysis.
(E) ChIP-PCR analysis of MMD1 occupancy on CAP-D3 using wild-type inflorescences. Chromatin from leaves was used as a negative control.
(F) ChIP-PCR analysis of MMD1 occupancy on other condensin genes. Data are represented as mean ± se from three (in [B] and [E]) and four (in [F]) independent experiments. *P < 0.05 and **P < 0.01, two-tailed Student’s t test.
Figure 8.
CAP-D3 Levels Are Reduced on mmd1 Pachytene Chromosomes.
Immunolocalization of CAP-D3 and ZYP1 (control) on wild-type and mmd1 pachytene chromosomes.
(A) Wild-type and mmd1 with antibodies against CAP-D3, while ZYP1 was used as a control.
(B) Quantification of fluorescence proportion for ZYP1 and CAP-D3 at pachytene (wild type, n = 35; mmd1, n = 37). Data are represented as mean ± sd. **P < 0.01, two-tailed Student’s t test. Bars = 5 μm.
Thus, we hypothesized that CAP-D3 might be directly regulated by MMD1. To test whether MMD1 binds to the CAP-D3 promoter, we performed ChIP assays using MMD1 antibodies (Figure 7D) and found that MMD1 was enriched at the CAP-D3 promoter, but not at the gene body regions (Figure 7E). We also tested for the binding of MMD1 to other condensin genes but did not observe enrichment at either the promoter or gene body region (Figure 7F). These results strongly support the idea that MMD1 regulates the CAP-D3 expression, probably by binding to H3K4me2 and recruiting other factors. The results also uncover a crucial regulatory step for the progression of partially condensed chromosomes of early meiotic prophase I to the highly condensed chromosomes of late prophase I and metaphase I by promoting the expression and/or accumulation of condensin proteins.
The cap-d3 Mutants Are Defective in Meiotic Chromosome Condensation
The finding that MMD1 binds to the CAP-D3 promoter to regulate its expression raised the possibility that cap-d3 mutations may also cause defects in meiotic chromosome condensation. Arabidopsis CAP-D3 was shown to be required for mitotic chromosome condensation and fertility, and reduced CAP-D3 function caused abnormal interactions between meiotic chromosomes (Smith et al., 2014). We obtained two T-DNA insertion lines (Figure 9A). Plants homozygous for cap-d3-1, a strong allele, exhibited a severe dwarf phenotype and fertility defects (Figures 9C and 9D), consistent with the pervious findings (Smith et al., 2014). In contrast, cap-d3-2, a potentially weaker allele, had partial transcripts prior to the T-DNA insertion site (Figure 9B) and produced a truncated protein detectable by anti-CAP-D3 antibodies (Figure 9G). Consistently, cap-d3-2 plants appeared normal during vegetative growth, but had dramatically reduced fertility (Figure 9D).
Figure 9.
Molecular Characterization and Phenotypic Analysis of cap-d3 Mutants.
(A) An illustration of the gene structure of (and T-DNA insertions in) CAP-D3.
(B) qRT-PCR analysis of CAP-D3 expression in male meiocytes of the wild type, cap-d3-1, and cap-d3-2. P1-5 primer pairs were located as indicated in (A).
(C) Pollen viability for the wild type, cap-d3-1, and cap-d3-2, as revealed by Alexander staining. The lack of red staining indicates dead pollen.
(D) cap-d3-1 showed severe vegetative growth and fertility defects, while cap-d3-2 showed normal growth but defects in fertility.
(E) to (G) Detection of CAP-D3 protein on chromosomes in the wild type (E), cap-d3-1 (F), and cap-d3-2 (G).
Data are represented as mean ± se from three independent experiments. Bar = 50 μm in (C) and 5 μm in (E) to (G).
Further analysis of chromosome morphology by DAPI showed that both mutants had no obvious abnormality before or at diakinesis (Supplemental Figure 1). At metaphase I, the chromosome condensation defect in cap-d3-2 resembled that of mmd1 (Figure 10D); in addition, cap-d3-1 displayed defects in centromere orientation (Figure 10C). Quantitative analyses of the areas of florescent centromere signals and chromosomes at metaphase I showed that these areas were significantly larger in both the cap-d3-1 and cap-d3-2 mutants than in the wild type (Figures 10G to 10I), but no significant difference was observed compared with mmd1 (Figures 10G to 10I). Although there was no significant difference between cap-d3-1 and cap-d3-2, the latter had a slightly larger centromere area than the former (Figures 10C, 10D, and 10G to 10I). A possible explanation is that cap-d3-1 displayed slender chromosome features, while the cap-d3-2 chromosome features were broad (Figures 10C and 10D). This result suggests that the condensin level has an important role in shaping chromosome features. Thus, the observed meiotic condensation defect in cap-d3 is in agreement with the reduced CAP-D3 expression in mmd1. Considering that the different T-DNA insertional alleles had different degrees of phenotypic defects, these results also suggest that the N-terminal SCOP domain of CAP-D3 is important in mitotic cell division, while the C-terminal Cnt1 domain may only be required for meiosis; the Cnt1 domain was absent in both cap-d3-1 and cap-d3-2, while the SCOP domain could be present in cap-d3-2 due to the detected transcript prior to the T-DNA insertion (Figure 9B). It is also possible that disruption of the Cnt1 domain causes a reduction in CAP-D3 stability, similar to the reduced level of CAP-D3 in mmd1.
Figure 10.
The cap-d3 Mutants Are Defective in Meiotic Chromosome Condensation, Similar to mmd1.
(A) to (F) FISH of metaphase I chromosome in the wild type (A), mmd1 (B), cap-d3-1 (C), capd-3-2 (D), mmd1 cap-d3-1 (E), and mmd1 cap-d3-2 (F). Red color represents centromeres signals.
(G) to (I) Estimate of average areas of fluorescence in centromeres and metaphase I chromosome area in the wild type, mmd1, cap-d3 single mutants, and mmd1 cap-d3 double mutants determined by Image J. Determination of average areas of fluorescence in centromeres of the wild type, mmd1, cap-d3 single mutants, and mmd1 cap-d3 double mutants in (H). The total number of cells used for analysis for the wild type mmd1, cap-d3-1, cap-d3-2, mmd1 cap-d3-1, and mmd1 cap-d3-2 was 22, 19, 23, 27, 34, and 26, respectively. Images in (G) were used to determine chromosome area. Average metaphase chromosome area (circled by blue lines) is shown in (I), and the total number of the wild type, mmd1, cap-d3-1, cap-d3-2, mmd1 cap-d3-1, and mmd1 cap-d3-2 cells was 13, 40, 36, 45, 42, and 32, respectively. Bar = 5 μm. Data are represented as mean ± sd. **P < 0.01, two-tailed Student’s t test.
To examine the genetic interaction of MMD1 with CAP-D3, we generated double mutants of mmd1 with two different cap-d3 alleles, a putative null allele cap-d3-1 and a less severe allele cap-d3-2, and compared meiotic chromosome condensation defects between the wild type, each single mutant, and the double mutants. The chromosome defects of the mmd1 cap-d3-1 double mutant were more severe than those of the single mutants, but those of the mmd1 cap-d3-2 double mutant was similar to those of the single mutants (Figures 10E to 10G and 10I), suggesting that MMD1 and CAP-D3 both play a role in regulating condensation. Because chromosome condensation defects in the mmd1 cap-d3-1 double mutant were more severe than those of single mutants (Figures 10G to 10I), the relationship between these genes is more complex than single linear upstream/downstream regulation. For example, expression analysis of the mmd1 mutant indicated that other condensin genes besides CAP-D3 were also affected. It is also likely that factors other than MMD1 contribute to the expression of CAP-D3 because CAP-D3 expression is reduced only partially in the mmd1 mutant.
Therefore, our genetic analyses provide additional evidence that supports the control of meiotic chromosome condensation progression by MMD1 via the partial regulation of CAP-D3 expression. To further test this hypothesis, we expressed CAP-D3 under the control of the exogenous CaMV 35S promoter in the mmd1 mutant background and found that the meiotic chromosomes in the transgenic lines exhibited a greater extent of condensation than those in the mmd1 mutant (Supplemental Figure 5), indicating that MMD1-independent CAP-D3 expression could partially rescue the chromosome condensation defects in the mmd1 mutant and providing additional evidence to support the role of MMD1 in meiotic chromosome condensation progression, at least in part in a CAP-D3-dependent fashion.
DISCUSSION
MMD1 Defines an Important Regulatory Step in Meiotic Chromosome Condensation
Both mitotic and meiotic chromosome condensation require the condensin complex (Hirano, 2012). Unlike the rapid, continuous mitotic chromosome condensation, meiotic chromosome condensation proceeds in a stepwise fashion to accommodate the complex homolog interactions that occur during the unusually long prophase I. Early condensation starting from the onset of prophase I results in thin, thread-like chromosomes. Each chromatid is organized into a linear array of loops, forming a proteinaceous structural axis called the axial element. Homologous axes are linked along their entire lengths via transverse filaments, forming the SC, which is crucial for stabilizing homolog interactions during prophase I. Premature formation of the highly condensed metaphase chromosome would severely impede pairing, synapsis, and recombination. The completion of homologous recombination and dissolution of the SC allow further condensation during late prophase I (diplotene and diakinesis), ultimately forming highly condensed metaphase I chromosomes, which can then be efficiently segregated by the spindle. However, little has been reported on the regulation of this stepwise condensation of meiotic chromosomes.
Although MMD1 (also called DUET) was first identified as a gene required for normal meiosis and fertility in Arabidopsis (Siddiqui et al., 2003; Yang et al., 2003), a recent study showed that DUET/MMD1 functions in meiosis II cell division through regulating the expression of TDM1 and JAS (Andreuzza et al., 2015). However, the role of MMD1 in meiosis prophase I chromosome condensation was unclear. The detailed analysis of mmd1 presented here indicates that, while MMD1 is not required for the formation of axial elements (Figure 3), it is crucial for shaping metaphase I bivalents during late condensation. This role is further supported by the reduction in CAP-D3 protein levels and the abnormally low expression levels of the corresponding genes in mmd1 meiotic cells compared with the wild type (Figures 7B and 8). The observation that the chromosome behaviors were not obviously abnormal in mmd1 from early to mid prophase I suggests that the relatively low level of CAP-D3 expression in mmd1 meiocytes might still be sufficient for early condensation (Figure 8). It is also likely that the other condensin genes are still functional. It was reported that the relative amounts of condensin I and II can affect the ratio of chromosome length to width (Green et al., 2012), with condensin II favoring relatively short, thick chromosomes. A recent study also showed that Arabidopsis condensin complexes have an important role in maintaining chromosome morphogenesis, such as condensin subunit I SMC4 and CAP-D2, as well as subunit II CAP-D3 (Smith et al., 2014). Mouse condensin II is required for normal morphology and the rigidity of meiotic bivalents in oocytes (Houlard et al., 2015). Our finding that expression of the condensin II-specific gene CAP-D3 is dramatically reduced in mmd1 meiocytes is consistent with the idea that formation of short, thick late meiotic chromosomes is facilitated by condensin II activity. Therefore, MMD1 is required for the progression of male meiotic chromosome condensation from partially condensed early prophase I chromosomes to the densely packed metaphase I state, thereby revealing a key regulatory step for meiotic chromosome condensation.
A Working Model for the Role of MMD1 in Meiotic Chromosome Condensation
PHD fingers bind to (read) histones in a sequence-dependent manner (Sanchez and Zhou, 2011). PHD domain-containing proteins have been shown to participate in epigenetic regulation of multiple biological processes (Del Rizzo and Trievel, 2014), but how they regulate any meiotic process in any organism was unknown until the recent discovery that DUET/MMD1 regulates meiosis II cell division (Andreuzza et al., 2015). The defects in the progression of meiotic chromosome condensation in mmd1 demonstrate the importance of MMD1. The only known domain in MMD1 is a PHD finger, which is required to rescue the mmd1 mutant phenotypes in chromosome condensation. Therefore, it is possible that MMD1 acts as a histone reader via its PHD finger to regulate the expression of genes required for meiotic progression in male meiocytes.
Consistent with the previous finding (Andreuzza et al., 2015), our study also demonstrated that MMD1 PHD is a histone reader, mainly on H3K4me2 (Figure 4). We showed here that MMD1 binds to the CAP-D3 promoter to regulate its expression, and our results support a model for how MMD1 functions to regulate the progression of meiotic chromosome condensation (Figure 11). In this model, during chromosome condensation in early meiotic prophase I, the MMD1 PHD finger binds to H3K4me2 marks at the CAP-D3 promoter and recruits other factors to the same site, thereby activating CAP-D3 expression. Normal CAP-D3 expression is necessary for the progression of meiotic chromosome condensation, especially for the highly condensed bivalents from pachytene to metaphase I (Figure 11). In the absence of MMD1, the lower than normal levels of CAP-D3 can still ensure partial meiotic chromosome condensation during prophase I, but they are unable to promote the formation of highly condensed bivalents at metaphase I (Figure 11). Consistently, cap-d3 mutants exhibit meiotic chromosome condensation defects similar to those of mmd1. However, the mmd1 cap-d3-1 double mutant showed more severe chromosome condensation defects than each single mutant, suggesting that other factors besides MMD1 and CAP-D3 might also function in chromosome condensation. Therefore, it is plausible that MMD1 serves as a histone reader to positively regulate the expression of CAP-D3 and other genes, ultimately ensuring chromosome condensation, while CAP-D3 mediates much, if not all, of the role of MMD1 in promoting condensation from late prophase I through metaphase I.
Figure 11.
Illustration Showing MMD1-Dependent Meiotic Chromosome Condensation.
At leptotene, the replicated sister chromatids held by cohesion are compacted into thin thread-like chromosomes by condensin complexes including CAP-D3 (purple dots). In the wild type, the maintenance of normal CAP-D3 levels during meiotic prophase I is necessary for the formation of the highly condensed bivalents at metaphase I and depends on MMD1, which regulates its expression. The absence of MMD1 due to insufficient levels of CAP-D3 or other condensin subunits results in the partial compaction of chromosomes or bivalents at metaphase I.
Divergent Roles of Condensin Subunits in Meiotic Recombination
Homologous chromosome interaction is a crucial event during meiotic prophase I comprising paring, synapsis, and recombination (Zickler and Kleckner, 1999; Hamant et al., 2006; Mercier et al., 2015). Several studies have shown that pairing, synapsis, and recombination are coordinately regulated. Normal pairing and synapsis have a positive role in promoting recombination, while recombination also occurs independently of paring and synapsis (Hamant et al., 2006; Mercier et al., 2015). However, whether the compaction of chromosomes from a long, thread-like structure to the highly condensed bivalent has a role in meiotic recombination remains unclear. Chromosome condensation requires the condensin complex, which is highly conserved among human, Drosophila, C. elegans, yeast, and plants (Legagneux et al., 2004; Hirano, 2012). However, the current information about the effect of condensin on meiotic recombination is quite limited. Studies in C. elegans showed that condensin I or II plays an important role in the distribution of DSB formation and increases the number of COs and CO interference (Mets and Meyer, 2009). The underlying mechanism likely involves the role of condensing in chromosome axis formation (Tsai et al., 2008). In budding yeast, homologs of CAP-D2 and CAP-G were implicated in the resolution of linkages between homologs and their mutations, resulting in increased meiotic recombination (Yu and Koshland, 2003). Similarly, the failed resolution of homologous conjunction was also observed in the Drosophila cap-h2 mutant (Hartl et al., 2008), suggesting that the resolution of homolog conjunction by condensins could be conserved. In human mitotic cells, depletion of condensin II CAP-D3 caused defects in chromosome condensation, accompanied by am ∼3-fold reduction in recombination efficiency (Wood et al., 2008). The Arabidopsis condensin I SMC4 was reported to be required for maintaining chromosome morphology and crossover number (Smith et al., 2014). Mutation in SMC4 caused a slight reduction in meiotic COs (Smith et al., 2014). Although we show here the condensin II subunit CAP-D3 has a role in mitotic and meiotic chromosome condensation, which is consistent with previous results (Schubert et al., 2013; Smith et al., 2014), its role in CO formation is unknown. We showed here that there is a slight reduction in CO formation in mmd1, suggesting that normal chromosome condensation ensures meiotic recombination, similar to the previous finding of the role of SMC4 in CO formation (Smith et al., 2014). However, both a previous study (Smith et al., 2014) and this study provided evidence that mutation of CAP-D3 alone seems to have no influence on meiotic crossover formation, suggesting that the role of MMD1 in CO formation is likely not mediated by CAP-D3. Therefore, we conclude that MMD1 affects meiotic CO formation, likely by coordinately regulating condensin gene expression, including condensin I and II, as observed in the transcriptome data. This result is also in agreement with the previous finding that the ratio of condensin I and II is critical for shaping metaphase I bivalents (Lee et al., 2011). However, understanding the mechanisms by which MMD1 regulates the expression of condensin I genes and CO formation will require future study.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana mutants used here were as follows: mmd1 (Yang et al., 2003), Spo11-1-3 (SALK_045787), cap-d3-1 (SAIL_826_B06), and cap-d3-2 (SALK_094776). Double mutants were identified in the F2 generation by PCR using primers as described in Supplemental Data Set 5. Plants were grown in a greenhouse at 22°C under a 16-h white and yellow light (bulb type: 36W/54-765 ISL/25; Philips)/8-h dark cycle.
FISH and Immunofluorescence
FISH and immunofluorescence were conducted as described previously (Wang et al., 2014). The primary antibodies used for the immunofluorescence experiment were diluted as follows: 1:100 (γH2Ax) (Miao et al., 2013), 1:100 (ZYP1), 1:300 (ASY1), 1:200 (RAD51), and 1:200 (DMC1) (Li et al., 2012), 1:100 (MLH1) (Chelysheva et al., 2010), and 1:200 (CAP-D3) (Schubert et al., 2013). The MMD1 antibody was used with 1:400 dilution and its specificity was validated by protein gel blot analysis using wild-type and mmd1 early inflorescences with male meiocytes (Figure 7D). Images were obtained under a Zeiss Axio Imager A2 microscope and organized using Photoshop CS3 (Adobe Systems). The fluorescence intensity of chromosomes was quantified by Image J (Collins, 2007) and normalized to DAPI signals. The areas of centromere fluorescence were scanned by Image J, and the ratio was calculated for the value of each mmd1 cell to the average value of wild-type cells. For convenience, we defined 1.2 < ratio < 2 as weak, 2 < ratio < 2.8 as moderate, and ratio > 2.8 as severe. The outlines of chromosome areas were drawn by hand, and the areas were screened by Image J.
For normalization and calculation of fluorescence proportions, we estimated fluorescent signals using circle drawing tools in Image J software and defined “set measurements” (including areas, integrated density, and mean gray value) and then selected “Measure” from the analyze menu. At the same time, we measured the dual-immunostaining ZYP1 signals at the same cell as the control, which was set to “1.”
For chromatin area values in transgenic plants harboring CAP-D3 in the mmd1 background, we first quantified the chromatin area in the wild type and mmd1 as 0.23 ± 0.05 cm2 and 0.41 ± 0.07 cm2. For convenience, we defined the area < 0.28 cm2 as good, 0.28 cm2 < the area < 0.34 cm2 as moderate and the 0.34 cm2 < the area < 0.48 cm2 as weak, with abnormal condensation similar to that of mmd1.
Histone Peptide Binding Assay
The MMD1 cDNA region encoding the PHD finger (residues 601 to 659) was cloned into vector pGEX6P-1 (pGEX Vectors 28-9546-48; GE Healthcare) by EcoRI/SalI to produce a GST fusion protein, which was expressed in Escherichia coli and purified using a glutathione Sepharose TM 4B column (GE Healthcare). For binding with individual peptides, 0.5 μg of biotin-labeled peptide was incubated overnight at 4°C with 4 μg of the fusion protein comprising MMD1 PHD-finger with GST in binding buffer (Li et al., 2012). Streptavidin beads (Invitrogen) were then added, followed by further incubation for 1.5 h. After washing, the binding mixture was analyzed by SDS-PAGE and the PHD-GST fusion protein was detected using protein gel blot analysis with anti-GST antibodies. A fusion protein of the MMD1 Pre-PHD domain with GST was used as a negative control. For isothermal titration calorimetry (ITC), GST-tagged PHD protein (601 to 680 amino acids) was digested by PreScission Protease overnight in 4°C, purified by gel filtration, and used for the subsequent peptide binding assay by ITC as described previously (Horton et al., 2010). The histone peptides used are listed in Supplemental Table 3.
For protein dot blot analysis, 5 μg GST-PHD (601 to 659 amino acids) was incubated with 2 μg biotin-conjugated peptide in 1.2 mL pull-down buffer (Zhang et al., 2006) overnight at 4°C. One hundred microliters settled glutathione-Sepharose beads were then added and the mixture was incubated on a rotation wheel for more than 2 h at 4°C. The beads were washed four times with pull-down buffer, and protein was eluted by elution buffer (pull-down buffer supplemented with 1 M NaCl). The GST-Pre-PHD protein was used as a negative control. The eluted sample was transferred onto PVDF membrane (Millipore), bound with HRP-labeled streptavidin (Cat#A0303; Beyotime), and detected by electro-chemical luminescence.
Plasmid Construction and Plant Transformation
For complementation plasmid construction, the promoter of MMD1 (by SacI/NcoI) and the full-length cDNA of MMD1 (by NcoI/SpeI) or MMD1ΔPHD (by NcoI/SpeI) were cloned into pCAMBIA1304 (Hajdukiewicz et al., 1994), and the 35CaMV promoter driving the full-length cDNA of CAP-D3 was cloned into pCAMBIA 1306 (provided by Ge’ Lab at Fudan University) by KpnI/SmaI. The constructs were individually introduced into Agrobacterium tumefaciens GV3101 (Cat. # CH5002A; 2nd Lab) to produce MMD1/+ heterozygous Arabidopsis transformants using the floral dip method (Clough and Bent, 1998). Positive T1 plants were screened on 0.5± MS medium containing 25 mg/L hygromycin and genotyped using MMD1 genotyping primers (Supplemental Data Set 5).
RNA-Seq and GO Enrichment Analysis
For each of two biological replicates (from different batches of plants), more than 600 anthers at stage 4-7 including male meiocytes and more than 650 anthers at stage 7-12 in the wild type (Landsberg erecta) and mmd1 (also in Ler) were used for transcriptome analysis. Total RNA was isolated using a ZR Plant RNA Miniprep Kit (Zymo Research) and examined using an Agilent 2100 bioanalyzer. cDNA libraries were constructed following the manufacturer’s instructions (Illumina) using approximately the same amounts of mRNA purified from wild-type and mmd1 samples, and the cDNAs were fragmented into short fragments. The first-strand and second-strand cDNA were synthesized using random hexamer-primer and added to the adapter. Finally, cDNA was amplified by PCR and sequenced with the Illumina HiSeq 2000 system. The RNA-seq data were detected and mapped to TAIR10 as descried previously (Trapnell et al., 2012), and RPKM (reads per kilobases per million reads) values were then calculated. We used Gfold (Feng et al., 2012) for ranking the differentially expressed genes between the wild type and mmd1 and defined genes with Gfold > 1 as upregulated genes and those with Gfold < −1 as downregulated genes. GO terms defined by the TAIR 10 GO slim database were used as the reference for GO analysis. GO enrichment analysis was conducted using the online AgriGO database with Fisher’s test, and statistics were compiled by FDR correction (http://bioinfo.cau.edu.cn/agriGO/). The heat maps of expression patterns were constructed by Cluster 3.0 and Java Treeview (http://sourceforge.net/projects/jtreeview/). The histograms were produced using Microsoft Excel 2013.
Quantitative RT-PCR
Arabidopsis male meiocytes were isolated as described previously (Wang et al., 2014). Total RNA extraction was conducted using a ZR Plant RNA MiniPrep Kit (The Epigenetics Company) with male meiocytes and inflorescences. Reverse transcription was conducted using a FastQuant RT Kit (with gDNase) (Cat. # KR106-02; Tiangen), and RT-PCR was performed as described previously (Yang et al., 2011) with SuperReal PreMix Plus (SYBR Green) (Cat#FP205-02; Tiangen) on a StepOnePlus real-time PCR system (Applied Biosystems). The statistical significance (P value) of the difference in expression levels for a gene was calculated using a two-sample t test. The primers used are listed in Supplemental Data Set 5.
MMD1 Antibody Production
Full-length MMD1 cDNA was cloned into pET22b (Novagen) and transformed into BL21(DE3) pLysS cells for protein expression. Protein purification was performed according to Harlow and Lane (1988). Briefly, upon induction, the overexpressed histidine-tagged MMD1 protein accumulated in the insoluble fraction. Inclusion bodies were collected from overexpressing cells, washed, and solubilized in PBS containing 8 M urea. The protein was purified using nickel chromatography, followed by SDS-PAGE. The purified MMD1 protein was used to inject New Zealand White rabbits using standard procedures (Harlow and Lane, 1988).
Protein Extraction and Protein Gel Blot Analysis
Protein extraction and protein gel blot analysis were performed as described previously (Wang et al., 2012). Total protein and nuclear protein were extracted in protein extraction buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM PMSF, and 1× protease inhibitor cocktail [Roche]) from flower buds. Nuclear protein samples were collected after filtering through Miracloth (Millipore) and centrifuged at 3500 rpm at 4°C for 30 min. Antibodies were the same as those used for immunofluorescence. Goat anti-rabbit (Cat# M21002) or goat anti-mouse (Cat#M21001) HRP-conjugated secondary antibodies (Abmart) were used against the primary antibodies. Signals were visualized with a CLiNX Science Instruments imager. The protein band intensity was calculated using Chemi Analysis software.
Chromatin Immunoprecipitation
ChIP was conducted as described previously (Saleh et al., 2008). The antibody used in this study was anti-MMD1. ChIP products were dissolved in 100 μL of water, and 0.5 μL of the solution was used for each PCR. The primers use for the normalization gene UBQ and other primers for detecting enrichment or binding efficiency are listed in Supplemental Data Set 5.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database and the GenBank/EMBL libraries under the following accession numbers: MMD1/DUET (AT1G66170), CAP-D3 (AT4G15890), and RNA-seq data (SRR3584322).
Supplemental Data
Supplemental Figure 1. Chromosome Behaviors of mmd1, cap-d3, and Their Double Mutants in Early Prophase I Revealed by DAPI Staining.
Supplemental Figure 2. Detection of RAD51 and DMC1 Foci in Wild-Type and mmd1 Pachytene Chromosomes.
Supplemental Figure 3. Histone Binding Assay of the MMD1 PHD Finger by Protein Dot Blot Analysis.
Supplemental Figure 4. Comparison of MMD1-Affected Genes in Anthers with DUET-Regulated Genes in Buds.
Supplemental Figure 5. Expression of CAP-D3 in mmd1 Rescues Meiotic Chromosome Condensation Defects.
Supplemental Table 1. Primary GO Categorization of Genes with Preferentially Upregulated and Downregulated Expression in mmd1.
Supplemental Table 2. GO Annotation of Genes with Downregulated Expression in mmd1.
Supplemental Table 3. Peptides Used in This Study.
Supplemental Data Set 1. Genes with Downregulated and Upregulated Expression in mmd1 Stage 4-7 Anthers.
Supplemental Data Set 2. Genes with Downregulated and Upregulated Expression in mmd1 Stage 7-12 Anthers.
Supplemental Data Set 3. Coregulated Genes between duet and mmd1 Stage 4-7 Anthers, as well as duet and mmd1 Stage 7-12 Anthers.
Supplemental Data Set 4. GO Category Analysis of Genes with Downregulated Expression in mmd1 Stage 4-7 Anthers.
Supplemental Data Set 5. Primers Used in This Study.
Supplementary Material
Acknowledgments
We greatly appreciate the kind gifts of MLH1 antibody from Raphaël Mercier at INRA, Centre de Versailles-Grignon (France) and SMC3 and CAP-D3 antibodies from Veit Schubert at the Leibniz Institute of Plant Genetics and Crop Plant Research (Gatersleben, Germany). We thank Gregory Copenhaver from University of North Carolina at Chapel Hill for valuable comments and editing on the manuscript. We also thank the ABRC at Ohio State University for the Arabidopsis mutant seeds. This work was supported by grants from the Ministry of Science and Technology of China (2012CB910503), the National Natural Science Foundation of China (31370347 and 31570314), and by funds from State Key Laboratory of Genetic Engineering, Fudan University, and Rijk Zwaan.
AUTHOR CONTRIBUTIONS
Y.W., J.W., and H.M. designed the research. J.W. performed most of experiments. B.N. and Y.W. performed mutant phenotypic analysis. H.W. collected samples for RNA-seq and plant growth. J.H. analyzed high-throughput sequencing datasets. C.M. provided MMD1 antibody. A.D. provided technical assistance. J.W., C.M., Y.W., and H.M. analyzed data and wrote the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Abe S., Nagasaka K., Hirayama Y., Kozuka-Hata H., Oyama M., Aoyagi Y., Obuse C., Hirota T. (2011). The initial phase of chromosome condensation requires Cdk1-mediated phosphorylation of the CAP-D3 subunit of condensin II. Genes Dev. 25: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreuzza S., Nishal B., Singh A., Siddiqi I. (2015). The chromatin protein DUET/MMD1 controls expression of the meiotic gene TDM1 during male meiosis in Arabidopsis. PLoS Genet. 11: e1005396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong S.J., Caryl A.P., Jones G.H., Franklin F.C. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645–3655. [DOI] [PubMed] [Google Scholar]
- Berchowitz L.E., Francis K.E., Bey A.L., Copenhaver G.P. (2007). The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R.C., Severson A.F., Meyer B.J. (2004). Condensin restructures chromosomes in preparation for meiotic divisions. J. Cell Biol. 167: 613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L., Grandont L., Vrielynck N., le Guin S., Mercier R., Grelon M. (2010). An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. 129: 143–153. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Collins T.J. (2007). ImageJ for microscopy. Biotechniques 43 (suppl.): 25–30. [DOI] [PubMed] [Google Scholar]
- Copenhaver G.P., Housworth E.A., Stahl F.W. (2002). Crossover interference in Arabidopsis. Genetics 160: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A., Vezon D., Gendrot G., Gallois J.L., Stevens R., Grelon M. (2007). AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. EMBO J. 26: 4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A., Pereira L., Vezon D., Chelysheva L., Gendrot G., Chambon A., Lainé-Choinard S., Pelletier G., Mercier R., Nogué F., Grelon M. (2009). A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet. 5: e1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rizzo P.A., Trievel R.C. (2014). Molecular basis for substrate recognition by lysine methyltransferases and demethylases. Biochim. Biophys. Acta 1839: 1404–1415. [DOI] [PubMed] [Google Scholar]
- Du J., et al. (2012). Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151: 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Meyer C.A., Wang Q., Liu J.S., Shirley Liu X., Zhang Y. (2012). GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 28: 2782–2788. [DOI] [PubMed] [Google Scholar]
- Freeman L., Aragon-Alcaide L., Strunnikov A. (2000). The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 149: 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L.C., et al. (2012). Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J. Cell Sci. 125: 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M., Vezon D., Gendrot G., Pelletier G. (2001). AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K.A., Holmes V.F., Cozzarelli N.R., Meyer B.J. (2002). C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16: 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994. [DOI] [PubMed] [Google Scholar]
- Hamant O., Ma H., Cande W.Z. (2006). Genetics of meiotic prophase I in plants. Annu. Rev. Plant Biol. 57: 267–302. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Laboratory Press; ). [Google Scholar]
- Hartl T.A., Sweeney S.J., Knepler P.J., Bosco G. (2008). Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet. 4: e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Wurz-Wildersinn R., Fuchs J., Schubert I., Suer S., Puchta H. (2007). The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19: 3090–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Sanchez-Moran E., Armstrong S.J., Jones G.H., Franklin F.C.H. (2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19: 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. (2005). Condensins: organizing and segregating the genome. Curr. Biol. 15: R265–R275. [DOI] [PubMed] [Google Scholar]
- Hirano T. (2012). Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 26: 1659–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Kobayashi R., Hirano M. (1997). Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89: 511–521. [DOI] [PubMed] [Google Scholar]
- Hirota T., Gerlich D., Koch B., Ellenberg J., Peters J.M. (2004). Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 117: 6435–6445. [DOI] [PubMed] [Google Scholar]
- Horton J.R., Upadhyay A.K., Qi H.H., Zhang X., Shi Y., Cheng X. (2010). Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 17: 38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlard M., Godwin J., Metson J., Lee J., Hirano T., Nasmyth K. (2015). Condensin confers the longitudinal rigidity of chromosomes. Nat. Cell Biol. 17: 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson N., Sanchez-Moran E., Buckling E., Armstrong S.J., Jones G.H., Franklin F.C. (2006). Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J. 25: 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Giroux C.N., Kleckner N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Lee J., Ogushi S., Saitou M., Hirano T. (2011). Condensins I and II are essential for construction of bivalent chromosomes in mouse oocytes. Mol. Biol. Cell 22: 3465–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legagneux V., Cubizolles F., Watrin E. (2004). Multiple roles of Condensins: a complex story. Biol. Cell 96: 201–213. [DOI] [PubMed] [Google Scholar]
- Li W., Chen C., Markmann-Mulisch U., Timofejeva L., Schmelzer E., Ma H., Reiss B. (2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. USA 101: 10596–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Qian W., Zhao Y., Wang C., Shen J., Zhu J.K., Gong Z. (2012). Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 11425–11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li H. (2012). Many keys to push: diversifying the ‘readership’ of plant homeodomain fingers. Acta Biochim. Biophys. Sin. (Shanghai) 44: 28–39. [DOI] [PubMed] [Google Scholar]
- Lowndes N.F., Toh G.W. (2005). DNA repair: the importance of phosphorylating histone H2AX. Curr. Biol. 15: R99–R102. [DOI] [PubMed] [Google Scholar]
- Mainiero S., Pawlowski W.P. (2014). Meiotic chromosome structure and function in plants. Cytogenet. Genome Res. 143: 6–17. [DOI] [PubMed] [Google Scholar]
- Mercier R., Mézard C., Jenczewski E., Macaisne N., Grelon M. (2015). The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66: 297–327. [DOI] [PubMed] [Google Scholar]
- Mets D.G., Meyer B.J. (2009). Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139: 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C., Tang D., Zhang H., Wang M., Li Y., Tang S., Yu H., Gu M., Cheng Z. (2013). Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25: 2998–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K., Nakano M., Fukuda T., Eiguchi M., Miyao A., Hirochika H., Kurata N. (2004). The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16: 1008–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Fang Y., Spector D.L., Hirano T. (2004). Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell 15: 3296–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D.J., Wang Z. (2013). Readout of epigenetic modifications. Annu. Rev. Biochem. 82: 81–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W., et al. (2012). A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336: 1445–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy T.V., Kaur J., Agashe B., Sundaresan V., Siddiqi I. (2003). The DUET gene is necessary for chromosome organization and progression during male meiosis in Arabidopsis and encodes a PHD finger protein. Development 130: 5975–5987. [DOI] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Sanchez R., Zhou M.M. (2011). The PHD finger: a versatile epigenome reader. Trends Biochem. Sci. 36: 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V., Lermontova I., Schubert I. (2013). The Arabidopsis CAP-D proteins are required for correct chromatin organisation, growth and fertility. Chromosoma 122: 517–533. [DOI] [PubMed] [Google Scholar]
- Shintomi K., Hirano T. (2011). The relative ratio of condensin I to II determines chromosome shapes. Genes Dev. 25: 1464–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui N.U., Stronghill P.E., Dengler R.E., Hasenkampf C.A., Riggs C.D. (2003). Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development 130: 3283–3295. [DOI] [PubMed] [Google Scholar]
- Smith S.J., Osman K., Franklin F.C. (2014). The condensin complexes play distinct roles to ensure normal chromosome morphogenesis during meiotic division in Arabidopsis. Plant J. 80: 255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey N.J., Kuromori T., Azumi Y., Roberts G., Breuer C., Wada T., Maxwell A., Roberts K., Sugimoto-Shirasu K. (2006). Arabidopsis SPO11–2 functions with SPO11-1 in meiotic recombination. Plant J. 48: 206–216. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.J., et al. (2008). Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 22: 194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrielynck N., Chambon A., Vezon D., Pereira L., Chelysheva L., De Muyt A., Mézard C., Mayer C., Grelon M. (2016). A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 351: 939–943. [DOI] [PubMed] [Google Scholar]
- Wang Y., Cheng Z., Lu P., Timofejeva L., Ma H. (2014). Molecular cell biology of male meiotic chromosomes and isolation of male meiocytes in Arabidopsis thaliana. Methods Mol. Biol. 1110: 217–230. [DOI] [PubMed] [Google Scholar]
- Wang Y., Cheng Z., Huang J., Shi Q., Hong Y., Copenhaver G.P., Gong Z., Ma H. (2012). The DNA replication factor RFC1 is required for interference-sensitive meiotic crossovers in Arabidopsis thaliana. PLoS Genet. 8: e1003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A.J., Severson A.F., Meyer B.J. (2010). Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 11: 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.L., Liang Y., Li K., Chen J. (2008). Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J. Biol. Chem. 283: 29586–29592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Lu P., Wang Y., Ma H. (2011). The transcriptome landscape of Arabidopsis male meiocytes from high-throughput sequencing: the complexity and evolution of the meiotic process. Plant J. 65: 503–516. [DOI] [PubMed] [Google Scholar]
- Yang X., Makaroff C.A., Ma H. (2003). The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15: 1281–1295. [PMC free article] [PubMed] [Google Scholar]
- Yu H.G., Koshland D.E. (2003). Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages. J. Cell Biol. 163: 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.G., Koshland D. (2005). Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis. Cell 123: 397–407. [DOI] [PubMed] [Google Scholar]
- Zhang C., Song Y., Cheng Z.H., Wang Y.X., Zhu J., Ma H., Xu L., Yang Z.N. (2012). The Arabidopsis thaliana DSB formation (AtDFO) gene is required for meiotic double-strand break formation. Plant J. 72: 271–281. [DOI] [PubMed] [Google Scholar]
- Zhang P., Du J., Sun B., Dong X., Xu G., Zhou J., Huang Q., Liu Q., Hao Q., Ding J. (2006). Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 34: 6621–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. (1999). Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33: 603–754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.